Published online Aug 8, 2015. doi: 10.4254/wjh.v7.i16.2020
Peer-review started: April 21, 2015
First decision: July 1, 2015
Revised: July 6, 2015
Accepted: July 23, 2015
Article in press: July 27, 2015
Published online: August 8, 2015
Similar to other cancers, a multistep process of carcinogenesis is observed in hepatocellular carcinoma (HCC). Although the mechanisms underlying the development of HCC have been investigated in terms of oncology, virology, and stem cell biology, the whole picture of hepatocarcinogenesis remains to be elucidated. Recent progress in molecular biology has provided clues to the underlying cause of various diseases. In particular, sequencing technologies, such as whole genome and exome sequencing analyses, have made an impact on genomic research on a variety of cancers including HCC. Comprehensive genomic analyses have detected numerous abnormal genetic alterations, such as mutations and copy number alterations. Based on these findings, signaling pathways and cancer-related genes involved in hepatocarcinogenesis could be analyzed in detail. Simultaneously, a number of novel biomarkers, both from tissue and blood samples, have been recently reported. These biomarkers have been successfully applied to early diagnosis and prognostic prediction of patients with HCC. In this review, we focus on the recent developments in molecular cancer research on HCC and explain the biological features and novel biomarkers.
Core tip: Recent progress in molecular biology enabled understanding of the mechanisms underlying hepatocarcinogenesis and identification of useful biomarkers. According to these findings, further efforts would be needed to improve understanding of these molecular mechanisms and to establish novel therapeutic approaches.
- Citation: Chiba T, Suzuki E, Saito T, Ogasawara S, Ooka Y, Tawada A, Iwama A, Yokosuka O. Biological features and biomarkers in hepatocellular carcinoma. World J Hepatol 2015; 7(16): 2020-2028
- URL: https://www.wjgnet.com/1948-5182/full/v7/i16/2020.htm
- DOI: https://dx.doi.org/10.4254/wjh.v7.i16.2020
Hepatocellular carcinoma (HCC) is a major cause of cancer-related deaths, accounting for approximately 600000 deaths annually worldwide and more than 30000 deaths annually in Japan[1,2]. It is well-known that hepatitis B virus (HBV), hepatitis C virus (HCV), alcohol abuse, and nonalcoholic fatty liver disease are the major risk factors for hepatocarcinogenesis[3]. Majority of patients with HCC are HBV or HCV carriers[4,5]. In HBV X protein (HBx) transgenic mice, HCC developed within one year after birth[6]. Similarly, HCV core transgenic mice exhibited hepatic steatosis several months after birth and eventually developed HCC[7]. These findings implicate that chronic infection with HBV and HCV have a direct action on hepatocarcinogenesis. Moreover, the incidence of HCC in patients with metabolic syndrome or nonalcoholic steatohepatitis has been increasing[8].
It is now widely considered that accumulation of genetic and/or epigenetic alterations transforms normal cells into cancer cells through a neoplastic state. This clinically well recognized process is called “stepwise carcinogenesis”[9]. The transformed cells usually acquire unique properties, such as sustained proliferative signaling, evasion growth suppressors, resistance to cell death, ability for replicative immortality, induction of angiogenesis, and activation of invasion and metastasis[10,11]. Recent progress in molecular biology and translational science enabled characterization of cancer cells and establishment of therapeutic approaches in a wide range of cancers. Sorafenib, an oral multikinase inhibitor, has been recognized as a new molecular-targeted therapy for HCC. The agent suppresses tumor growth and angiogenesis by inhibiting the RAS/RAF/MAPK signaling pathway and tyrosine kinase receptors including vascular endothelial growth factor receptor (VEGFR)[12]. However, the prognosis of patients with HCC treated with sorafenib has not been essentially satisfactory[13,14]. Therefore, further understanding of the molecular mechanisms underlying hepatocarcinogenesis and establishment of novel therapeutic approaches remain the most important challenges.
In this review, we will summarize the recent progress in molecular cancer research on HCC and explain the molecular mechanisms underlying hepatocarcinogenesis. We will also highlight the serological and pathological biomarkers of HCC for diagnosis and prognostication.
It has been documented that dysregulation of several signaling pathways including p53/RB, Wnt/β-catenin, PI3K/PTEN/Akt/mTOR pathways plays an important role in the development and progression of HCC[15,16]. It is considered that the aberrant activation or inactivation of these pathways is attributable to somatic alterations, such as mutations, changes of copy numbers, and chromosomal rearrangements[17]. These results could be applied to the classification and prognostication of HCC[18]. Among them, mutation of p53 and β-catenin has long been recognized as a common genetic alteration in HCC[19,20], and it is observed in approximately 30% and 20% of HCC samples, respectively.
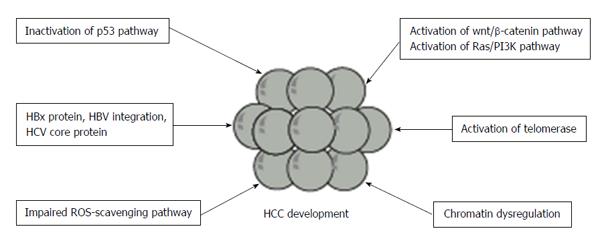
Recent whole genome and exome sequencing analyses enabled the surveillance of the signature of genomic alteration and identification of somatically mutated genes (Figure 1). In a study on exome sequencing of 24 HCC samples, Guichard et al[21] demonstrated that major pathways including Wnt/β-catenin signaling, p53/cell cycle signaling, PI3K/RAS signaling, chromatin regulation, and oxidative and endoplasmic reticulum stress signaling were commonly altered by somatic mutations or homozygous gene deletions. They found recurrent alterations in four genes (ARID1A, RPS6KA3, NFE2L2 and IRF2), which were not previously reported in HCC. Particularly, ARID1A, a chromatin remodeling gene, was shown to be frequently mutated in alcohol-related HCC. Whole-genome sequencing (WGS) of HCC samples has also revealed recurrent somatic mutations in several genes associated with chromatin regulation, such as ARID1A, ARID1B, ARID2, MLL, MLL3, BAZ2B, BRD8, BPTF, BRE and HIST1H4B[22]. Mutations in at least one of these chromatin regulator genes were detected in more than 50% of HCC tissues. Taken together, dysregulated chromatin remodeling plays a critical role in HCC development.
Genetic and functional heterogeneity in tumor-constituent cells has been observed in a wide range of cancers[23]. To explain this, a hierarchical model or cancer stem cell (CSC) model has been proposed and debated on[24]. This model postulates that a small population generates a hierarchical structure containing descendant tumor cells. However, clonal evolution model suggests that a series of clonal expansions accompanied by accumulated genetic alterations contribute to intratumor heterogeneity[25]. Recent sequencing technologies have successfully demonstrated that most tumors exhibit extensive intratumoral heterogeneity characterized by individual tumor cells showing different somatic mutation pattern[26]. Gerlinger et al[27] conducted exome sequencing analysis of resected renal cell carcinoma samples and demonstrated not only intratumoral heterogenity but also genetic alternations between primary tumor and metastatic lesions.
Multicentric tumor development is one of the most vital aspects of hepatocarcinogeneis[28]. Fujimoto et al[22] performed WGS on two pairs of multicentric HCV-associated HCCs and unexpectedly found out that neither common somatic mutations in coding region nor common structural alterations were observed in these tumors. These results may indicate that multicentric HCCs were derived from cells with different genetic alterations. On the other hand, Huang et al[29] employed exome sequencing of nine pairs of matched primary HBV-associated HCCs and portal vein tumor thrombus (PVTT) and demonstrated that more than 90% of nonsynonymous somatic mutations were shared between primary HCC and PVTT. Furthermore, 65 genes with mutations either in primary HCC or PVTT were identified. Among them, mutations in KDM6A, CUL9, FGD6, AKAP3 and RNF139 were detected in PVTT, but not in primary tumors.
It is well known that HBx, a multifunctional protein encoded by the HBV genome, has the ability of the transcriptional transactivation[30]. Moreover, HBx could activate the JAK/STAT signaling pathway but impair the p53 function[31,32]. Thus, HBx is deeply involved in hepatocarcinogenesis.
The integration of HBV DNA into the host genome is one of the important factors in hepatocarcinongenesis in patients with chronic HBV infection[33]. This appears to contribute to oncogene activation and/or tumor-suppressor gene inactivation. HBV integration at the Cyclin A and retinoic acid receptor β gene has been reported approximately 20 years ago[34,35]. Recently, a novel sequencing technology was successfully applied to the analyses of HBV genome integration. HBV genome integration in the telomerase reverse transcriptase (TERT) locus was observed in 4 of 11 HBV-related HCC samples examined[22]. Because activation of telomerase, encoded by the TERT, is associated with cellular immortalization, the dysregulation of TERT expression may play a crucial role in hepatocarcinogenesis. Sung et al[36] conducted WGS of 88 Chinese patients with HCC and noted that nearly 40% of HBV breakpoints were located around the X and core gene. Further, they revealed that in some HCC samples, five genes (TERT, MLL4, CCNE1, SENP5, and ROCK1) were recurrently affected by HBV integration. They reported that the number of HBV integration sites per tumor significantly correlates with serum levels of HBsAg and alfa-fetoprotein (AFP). In addition, patients with HCC with several HBV integration sites exhibited shorter survival time than those with few integration sites.
Genome-wide association studies are microarray-oriented technologies that have been utilized to identify single nucleotide polymorphisms (SNPs) associated with many traits and diseases. Intronic SNPs in KIF1B and STAT4 have been shown to be highly associated with HCC occurrence in Chinese chronic HBV carriers[37,38]. Similarly, it has been recently reported that SNPs in MICA and DEPDC5 are associated with HCC development in Japanese patients with chronic HCV infection[39,40]. To confirm the utility of these SNPs as risk markers for HCC, further analyses of different populations would be needed.
It has been reported that signaling pathways and molecular mechanisms operating in stem cells are similar to those in cancer[41]. For example, BMI1, a polycomb gene product, is a general regulator in normal stem cell systems[42]. On the other hand, high expression levels of BMI1 were observed in various cancers. We have previously reported that fetal hepatic stem/progenitor cells transduced with BMI1 acquired enhanced self-renewal capability and tumorigenicity to generate combined HCC in a mouse transplant model[43]. Glinsky et al[44] analyzed gene expression profiles in wild type and BMI1-/- neurospheres. By comparing those to the data obtained from primary and metastatic prostate cancer, they successfully selected 11 BMI1-associated genes. This 11-gene death-from-cancer signature was validated in both epithelial and hematological malignancies. The gene set has been shown to predict unfavorable outcomes in patients with these malignancies, which indicates that BMI1-driven pathways are closely associated with an aggressive cancer phenotype.
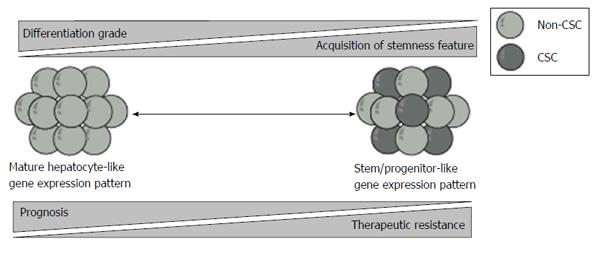
In a microarray-based gene expression analyses, HCC with similar expression patterns as that of hepatic stem/progenitor cells was associated with poor prognosis[45]. c-MET is a tyrosine kinase receptor for hepatocyte growth factor (HGF). HGF/c-MET signaling plays a crucial role in the development and regeneration of the liver[46]. Kaposi-Novak et al[47] conducted global gene expression profiling of wild type and Met-deficient mouse hepatocytes and showed that Met-regulated gene expression signature is associated with aggressive phenotypes in HCC, such as vascular invasion. Similarly, it has been reported that epithelial cell adhesion molecule (EpCAM) served as a surface marker in both hepatic stem cell and CSCs[48]. Furthermore, HCC is subclassified into four groups on the basis of the expression of EpCAM and AFP. These subtypes displayed distinct gene expression patterns with features resembling certain stages of hepatic lineages. EpCAM+AFP+ HCCs exhibit hepatocytic progenitor-like expression patterns and have poor prognoses. In contrast, EpCAM-AFP- HCCs exhibit mature hepatocyte-like expression patterns and have favorable prognoses.
Taken together, prognostic stratification based on the expression of surface markers and molecules in hepatic stem/progenitor cells revealed that stemness features are closely associated with unfavorable prognosis in patients with HCC (Figure 2).
AFP, a plasma protein produced by the yolk sac and fetal liver cells[49], has been the most widely used biomarker for the detection of HCC[50]. However, AFP is not a necessarily specific marker for HCC, considering that its levels of may also be observed in patients with chronic hepatitis and cirrhosis. In contrast, lens culinaris agglutinin-reactive fraction of AFP (AFP-L3) is specific for HCC and has been available in clinical settings[51]. In addition, AFP-L3+ HCC frequently exhibits biologically malignant characteristics, such as portal vein invasion and undifferentiated pathology[52]. Protein induced by vitamin K absence or antagonist-II (PIVKA-II), also called des-gamma-carboxy prothrombin, is an abnormal prothrombin induced by vitamin K shortage. Hepatocytes with malignant transformation impair the vitamin K-dependent γ-glutamyl carboxylation and produce PIVKA-II[53]. The serum levels of PIVKA-II as well as AFP in patients with HCC were significantly higher than those in patients with chronic hepatitis and cirrhosis. A marked increase in serum PIVKA-II level is also observed in patients receiving anticoagulation therapy with warfarin. Serum PIVKA-II level is shown to be closely associated with large tumor diameters and vascular invasion compared with that of AFP and AFP-L3[54]. However, these markers have been shown to be insufficient for the detection of small HCC. Simultaneous measurement of these markers, such as AFP and PIVKA-II[55] and AFP-L3 and PIVKA-II[56], contributes to the improved diagnostic value for HCC detection.
Recently, dickkopf-1 (DKK1) has been shown to be a promising serum marker for the detection of HCC[57]. DKK1 is a secreted protein with two cysteine-rich regions and functions as a negative modulator of the Wnt/β-catenin pathway by interacting with the co-receptor[58]. It has been shown that serum DKK1 levels were significantly higher in patients with HBV-related HCC than those in the controls[59]. In addition, simultaneous measurement of DKK1 and AFP was shown to improve diagnostic accuracy. Further analyses would be necessary to determine whether DKK1 contributes to the diagnosis of HCV-related HCC.
Polycomb group gene products: Polycomb group (PcG) complexes regulate epigenetic cellular memory and establish and maintain cellular identities during embryogenesis, development, and tumorigenesis[60,61]. PcG complexes can be functionally divided into at least two distinct complexes: a maintenance complex, polycomb repressive complex (PRC) 1 and an initiation complex, PRC2. BMI1, one of the components of PRC1, is essential for maintaining the self-renewal capability of somatic stem cells including hepatic stem cells[62,63]. We have previously shown that BMI1 regulates CSCs in HCC cell lines[64]. These findings suggest that BMI1 regulates self-renewal of both normal stem cells and CSCs by repressing the transcription of negative regulator genes for stem cell maintenance, such as Ink4a and Arf[65]. Furthermore, BMI1 expression levels in HCC tumor tissues are well correlated with the progression and prognosis of the disease[66].
Ezh2, one of the components of PRC2, shows catalytic activity specific for the trimethylation of histone H3 at lysine 27. We have previously reported that Ezh2 tightly regulates the self-renewal and differentiation of murine hepatic stem/progenitor cells[67]. Similar to BMI1, EZH2 is also overexpressed in tumor-initiating HCC cells and HCC tumor tissues[66]. In addition, EZH2-knockdown using short-hairpin RNA and the pharmacological inhibition of EZH2 by an S-adenosylhomocysteine hydrolase inhibitor, 3-deazaneplanocin A (DZNep) markedly impaired the growth and tumorigenic ability of HCC cells[68].
Taken together, PcG proteins such as BMI1 and EZH2 may be encouraging therapeutic targets for HCC. Considering that highly selective PcG protein inhibitors have been developed, clinical trials would be of importance[69,70].
Glypican-3: Glypican-3 (GPC3), a member of the family of glypican heparan sulfate proteoglycans, is an oncofetal protein expressed in fetal liver and HCC[71].
Abnormal expression of GPC3 was observed in approximately 70% of HCC tumor samples and in approximately 50% of serum samples of patients with HCC[72]. Additionally, increased GPC3 expression detected by immunohistochemical analyses correlated with poor prognosis among patients with HC[73]. Considering that a clinical trial using a GPC3 peptide vaccine in patients with advanced HCC has also been carried out[74], this appears to serve not only as a tumor marker but also as a therapeutic target.
Heat shock protein 70: Heat shock proteins (HSPs) are highly conserved protein serve as multifunctional molecular chaperones[75]. Their expression is usually upregulated in response to stressful stimuli such as heat stress. Increased expression of HSP70 has been reported in a wide range of cancers including HCC[76]. Chuma et al[77] conducted oligonucleotide array analyses to compare expression profiles among seven pairs of early components and progressed components of nodule- in-nodule type HCCs. They successfully demonstrated that HSP70 expression was upregulated according to the differentiation grade. It is possible that HSP70 could be a sensitive marker for the differential diagnosis of early HCC from precancerous lesions. In addition, combination of markers, such as HSP70, GPC3, and glutamine synthetase[78] and HSP70, GPC3, and EZH2[79] may contribute to the accurate diagnosis of HCC by immunohistochemical analyses.
Sal-like protein 4: Sal-like protein 4 (Sall4), a member of the zinc finger transcription factor family, is highly expressed in murine hepatic stem/progenitor cells and functions as a regulator of cell fate decisions. Overexpression of Sall4 in these cells significantly inhibits hepatocyte-lineage maturation and adversely accelerates cholangiocyte-lineage terminal differentiation[80]. Overexpression of Sall4 in HCC cell lines suppresses hepatocytic differentiation and leads to the acquisition of stem cell-like phenotypes, such as chemo-resistance[81]. Together, Sall4 is closely associated with the properties of both normal stem cells and CSCs in liver. Recently, Yong et al[82] performed clinicopathological and gene-expression microarray analyses in terms of Sall4 expression and revealed that increased expression of Sall4 was closely associated with aggressive phenotypes of HCC and unfavorable survival. Gene set enrichment analysis showed that Sall4-high HCC were significantly enriched with genes involved in embryonic stem cell signature, metastasis, hepatoblastoma, and progressive HCC compared with Sall4-low HCC. In addition, Sall4 peptide, consisting of 12 amino acids, has been shown to block the oncogenic role of Sall4 in part by modulating PTEN/PI3K/AKT signaling in HCC cells.
Sorafenib, an oral multi-kinase inhibitor, is available as a new molecular-targeted therapy against HCC. Global guidelines currently recommend sorafenib as first-line therapy for Child-Pugh A patients with advanced HCC[83-86]. Sorafenib demonstrates its anti-HCC effect by suppressing tumor growth factors and angiogenesis through the inhibition of the RAF/MEK/ERK signaling pathway and tyrosine kinase receptors, such as VEGFR[87]. The safety and efficacy of sorafenib on patients with advanced HCC has been demonstrated in phase III studies[88,89].
Some investigators have tried to determine the predictive markers for the response to sorafenib, which also serve as predictive indicators of prognosis in patients with HCC. Physical findings, such as hand-foot-skin reaction (HFSR) and hypertension have shown to predict favorable outcomes in patients treated with sorafenib[90,91]. Alternatively, the presence of lung metastasis could predict poor response to sorafenib[92]. It has been also reported that early decrease in AFP level and increase in PIVKA-II level determine the therapeutic efficacy of sorafenib[93,94]. Concordant with these findings, our analyses demonstrated that increase in AST or AFP levels, existence of MVI, and lack of HFSR serves as independent predictors of poor prognosis[95]. Recently, it has been reported that genetic amplification of FGF3/4 and VEGF-A was frequently observed in responders to sorafenib in HCC[96,97]. Rudalska et al[98] conducted in vivo RNAi screening to identify sorafenib-response genes and reported that both shRNA-mediated and pharmacological silencing of MAPK14 (p38α) sensitize HCC cells to sorafenib therapy. They also highlighted the importance of inhibiting MAPK14-dependent activation of MEK/ERK and ATF2 signaling to overcome sorafenib-resistance.
Taken together, these findings may be useful as prognostic biomarkers and are breakthroughs for understanding of the molecular mechanisms underlying the development and progression of HCC.
Progress in molecular biology, such as next-generation sequencing, unveils the biological features of HCC. These analyses were conducted on tissue and blood samples. Recently, the use of “liquid biopsy” to analyze circulating tumor DNA in peripheral blood has been documented[99,100]. This approach is minimally invasive and enables the detection of the sequence and mutations of target genes. Combined use of novel approaches and biomarkers contributes to early diagnosis and the selection of the appropriate treatment for patients with HCC. Further efforts would be needed to improve the prognosis of patients with advanced HCC.
We thank the members of HCC group of the Gastroenterology Section of Chiba University Hospital for their valuable discussion and helpful support.
P- Reviewer: Sazci A S- Editor: Ji FF L- Editor: A E- Editor: Liu SQ
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893-2917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11128] [Cited by in F6Publishing: 11614] [Article Influence: 893.4] [Reference Citation Analysis (4)] |
| 2. | Omata M, Tateishi R, Yoshida H, Shiina S. Treatment of hepatocellular carcinoma by percutaneous tumor ablation methods: Ethanol injection therapy and radiofrequency ablation. Gastroenterology. 2004;127:S159-S166. [PubMed] [Cited in This Article: ] |
| 3. | El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118-1127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2881] [Cited by in F6Publishing: 2977] [Article Influence: 229.0] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2183] [Cited by in F6Publishing: 2337] [Article Influence: 194.8] [Reference Citation Analysis (0)] |
| 5. | Webster DP, Klenerman P, Dusheiko GM. Hepatitis C. Lancet. 2015;385:1124-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 382] [Cited by in F6Publishing: 306] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 6. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 880] [Cited by in F6Publishing: 855] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 7. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 931] [Cited by in F6Publishing: 893] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 8. | Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820-1832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 968] [Cited by in F6Publishing: 964] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 9. | Land H, Parada LF, Weinberg RA. Cellular oncogenes and multistep carcinogenesis. Science. 1983;222:771-778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 814] [Cited by in F6Publishing: 929] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 10. | Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19834] [Cited by in F6Publishing: 18704] [Article Influence: 779.3] [Reference Citation Analysis (0)] |
| 11. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42985] [Article Influence: 3306.5] [Reference Citation Analysis (4)] |
| 12. | Wilhelm SM, Carter C, Tang L, Wilkie D, McNabola A, Rong H, Chen C, Zhang X, Vincent P, McHugh M. BAY 43-9006 exhibits broad spectrum oral antitumor activity and targets the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in tumor progression and angiogenesis. Cancer Res. 2004;64:7099-7109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2966] [Cited by in F6Publishing: 3035] [Article Influence: 151.8] [Reference Citation Analysis (0)] |
| 13. | Ogasawara S, Kanai F, Obi S, Sato S, Yamaguchi T, Azemoto R, Mizumoto H, Koushima Y, Morimoto N, Hirata N. Safety and tolerance of sorafenib in Japanese patients with advanced hepatocellular carcinoma. Hepatol Int. 2011;5:850-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Ogasawara S, Chiba T, Ooka Y, Kanogawa N, Motoyama T, Suzuki E, Tawada A, Kanai F, Yoshikawa M, Yokosuka O. Efficacy of sorafenib in intermediate-stage hepatocellular carcinoma patients refractory to transarterial chemoembolization. Oncology. 2014;87:330-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 140] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 15. | Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312-1327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 801] [Cited by in F6Publishing: 840] [Article Influence: 52.5] [Reference Citation Analysis (0)] |
| 16. | Kudo M. Signaling pathway/molecular targets and new targeted agents under development in hepatocellular carcinoma. World J Gastroenterol. 2012;18:6005-6017. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 37] [Cited by in F6Publishing: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Shibata T, Aburatani H. Exploration of liver cancer genomes. Nat Rev Gastroenterol Hepatol. 2014;11:340-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 18. | Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385-7392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 753] [Cited by in F6Publishing: 851] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 19. | Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166-2176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 411] [Cited by in F6Publishing: 543] [Article Influence: 31.9] [Reference Citation Analysis (1)] |
| 20. | Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245-250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 703] [Cited by in F6Publishing: 697] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 21. | Guichard C, Amaddeo G, Imbeaud S, Ladeiro Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M, Degos F. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694-698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 962] [Cited by in F6Publishing: 1059] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 22. | Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760-764. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 631] [Cited by in F6Publishing: 681] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 23. | Heppner GH. Tumor heterogeneity. Cancer Res. 1984;44:2259-2265. [PubMed] [Cited in This Article: ] |
| 24. | Chiba T, Kamiya A, Yokosuka O, Iwama A. Cancer stem cells in hepatocellular carcinoma: Recent progress and perspective. Cancer Lett. 2009;286:145-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 25. | Neu N, Ploier B, Ofner C. Cardiac myosin-induced myocarditis. Heart autoantibodies are not involved in the induction of the disease. J Immunol. 1990;145:4094-4100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2054] [Cited by in F6Publishing: 2016] [Article Influence: 168.0] [Reference Citation Analysis (0)] |
| 26. | Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos CR. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1008] [Cited by in F6Publishing: 933] [Article Influence: 93.3] [Reference Citation Analysis (0)] |
| 27. | Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883-892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6057] [Cited by in F6Publishing: 5637] [Article Influence: 469.8] [Reference Citation Analysis (0)] |
| 28. | Matsumoto Y, Fujii H, Matsuda M, Kono H. Multicentric occurrence of hepatocellular carcinoma: diagnosis and clinical significance. J Hepatobiliary Pancreat Surg. 2001;8:435-440. [PubMed] [Cited in This Article: ] |
| 29. | Huang J, Deng Q, Wang Q, Li KY, Dai JH, Li N, Zhu ZD, Zhou B, Liu XY, Liu RF. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117-1121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 293] [Cited by in F6Publishing: 319] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 30. | Ng SA, Lee C. Hepatitis B virus X gene and hepatocarcinogenesis. J Gastroenterol. 2011;46:974-990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 31. | Lee YH, Yun Y. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J Biol Chem. 1998;273:25510-25515. [PubMed] [Cited in This Article: ] |
| 32. | Elmore LW, Hancock AR, Chang SF, Wang XW, Chang S, Callahan CP, Geller DA, Will H, Harris CC. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc Natl Acad Sci USA. 1997;94:14707-14712. [PubMed] [Cited in This Article: ] |
| 33. | Brechot C, Pourcel C, Louise A, Rain B, Tiollais P. Presence of integrated hepatitis B virus DNA sequences in cellular DNA of human hepatocellular carcinoma. Nature. 1980;286:533-535. [PubMed] [Cited in This Article: ] |
| 34. | Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Bréchot C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene. 1992;7:1653-1656. [PubMed] [Cited in This Article: ] |
| 35. | Dejean A, Bougueleret L, Grzeschik KH, Tiollais P. Hepatitis B virus DNA integration in a sequence homologous to v-erb-A and steroid receptor genes in a hepatocellular carcinoma. Nature. 1986;322:70-72. [PubMed] [Cited in This Article: ] |
| 36. | Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765-769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 619] [Cited by in F6Publishing: 660] [Article Influence: 55.0] [Reference Citation Analysis (0)] |
| 37. | Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia W, Ma F, Huang W, Yu L, Yue W. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755-758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 280] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 38. | Jiang DK, Sun J, Cao G, Liu Y, Lin D, Gao YZ, Ren WH, Long XD, Zhang H, Ma XP. Genetic variants in STAT4 and HLA-DQ genes confer risk of hepatitis B virus-related hepatocellular carcinoma. Nat Genet. 2013;45:72-75. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 226] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 39. | Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 267] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 40. | Miki D, Ochi H, Hayes CN, Abe H, Yoshima T, Aikata H, Ikeda K, Kumada H, Toyota J, Morizono T. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers. Nat Genet. 2011;43:797-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 41. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6844] [Cited by in F6Publishing: 6706] [Article Influence: 291.6] [Reference Citation Analysis (0)] |
| 42. | Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 204] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 43. | Chiba T, Zheng YW, Kita K, Yokosuka O, Saisho H, Onodera M, Miyoshi H, Nakano M, Zen Y, Nakanuma Y. Enhanced self-renewal capability in hepatic stem/progenitor cells drives cancer initiation. Gastroenterology. 2007;133:937-950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 180] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 44. | Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503-1521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 683] [Cited by in F6Publishing: 698] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 45. | Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ, Sun Z. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410-416. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 729] [Cited by in F6Publishing: 719] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 46. | Michalopoulos GK, DeFrances MC. Liver regeneration. Science. 1997;276:60-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2495] [Cited by in F6Publishing: 2398] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 47. | Kaposi-Novak P, Lee JS, Gòmez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582-1595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 284] [Cited by in F6Publishing: 288] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 48. | Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, Budhu A, Zanetti KA, Chen Y, Qin LX. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 545] [Cited by in F6Publishing: 561] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 49. | Mackiewicz A, Breborowicz J. The in vitro production of alpha--fetoprotein variants by human fetal organs. Oncodev Biol Med. 1980;1:251-261. [PubMed] [Cited in This Article: ] |
| 50. | Johnson PJ. Role of alpha-fetoprotein in the diagnosis and management of hepatocellular carcinoma. J Gastroenterol Hepatol. 1999;14 Suppl:S32-S36. [PubMed] [Cited in This Article: ] |
| 51. | Yamashita F, Tanaka M, Satomura S, Tanikawa K. Prognostic significance of Lens culinaris agglutinin A-reactive alpha-fetoprotein in small hepatocellular carcinomas. Gastroenterology. 1996;111:996-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 121] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 52. | Oka H, Saito A, Ito K, Kumada T, Satomura S, Kasugai H, Osaki Y, Seki T, Kudo M, Tanaka M. Multicenter prospective analysis of newly diagnosed hepatocellular carcinoma with respect to the percentage of Lens culinaris agglutinin-reactive alpha-fetoprotein. J Gastroenterol Hepatol. 2001;16:1378-1383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 114] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 53. | Liebman HA. Isolation and characterization of a hepatoma-associated abnormal (des-gamma-carboxy)prothrombin. Cancer Res. 1989;49:6493-6497. [PubMed] [Cited in This Article: ] |
| 54. | Yamamoto K, Imamura H, Matsuyama Y, Kume Y, Ikeda H, Norman GL, Shums Z, Aoki T, Hasegawa K, Beck Y. AFP, AFP-L3, DCP, and GP73 as markers for monitoring treatment response and recurrence and as surrogate markers of clinicopathological variables of HCC. J Gastroenterol. 2010;45:1272-1282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 120] [Cited by in F6Publishing: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 55. | Ishii M, Gama H, Chida N, Ueno Y, Shinzawa H, Takagi T, Toyota T, Takahashi T, Kasukawa R. Simultaneous measurements of serum alpha-fetoprotein and protein induced by vitamin K absence for detecting hepatocellular carcinoma. South Tohoku District Study Group. Am J Gastroenterol. 2000;95:1036-1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 55] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 56. | Choi JY, Jung SW, Kim HY, Kim M, Kim Y, Kim DG, Oh EJ. Diagnostic value of AFP-L3 and PIVKA-II in hepatocellular carcinoma according to total-AFP. World J Gastroenterol. 2013;19:339-346. [PubMed] [Cited in This Article: ] |
| 57. | Tung EK, Mak CK, Fatima S, Lo RC, Zhao H, Zhang C, Dai H, Poon RT, Yuen MF, Lai CL. Clinicopathological and prognostic significance of serum and tissue Dickkopf-1 levels in human hepatocellular carcinoma. Liver Int. 2011;31:1494-1504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 58. | Sato N, Yamabuki T, Takano A, Koinuma J, Aragaki M, Masuda K, Ishikawa N, Kohno N, Ito H, Miyamoto M. Wnt inhibitor Dickkopf-1 as a target for passive cancer immunotherapy. Cancer Res. 2010;70:5326-5336. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 124] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 59. | Shen Q, Fan J, Yang XR, Tan Y, Zhao W, Xu Y, Wang N, Niu Y, Wu Z, Zhou J. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13:817-826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 289] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 60. | Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409-418. [PubMed] [Cited in This Article: ] |
| 61. | Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 503] [Cited by in F6Publishing: 526] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 62. | Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843-851. [PubMed] [Cited in This Article: ] |
| 63. | Chiba T, Seki A, Aoki R, Ichikawa H, Negishi M, Miyagi S, Oguro H, Saraya A, Kamiya A, Nakauchi H. Bmi1 promotes hepatic stem cell expansion and tumorigenicity in both Ink4a/Arf-dependent and -independent manners in mice. Hepatology. 2010;52:1111-1123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 64. | Chiba T, Miyagi S, Saraya A, Aoki R, Seki A, Morita Y, Yonemitsu Y, Yokosuka O, Taniguchi H, Nakauchi H. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008;68:7742-7749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 65. | Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12:328-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 247] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 66. | Yonemitsu Y, Imazeki F, Chiba T, Fukai K, Nagai Y, Miyagi S, Arai M, Aoki R, Miyazaki M, Nakatani Y. Distinct expression of polycomb group proteins EZH2 and BMI1 in hepatocellular carcinoma. Hum Pathol. 2009;40:1304-1311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 39] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 67. | Aoki R, Chiba T, Miyagi S, Negishi M, Konuma T, Taniguchi H, Ogawa M, Yokosuka O, Iwama A. The polycomb group gene product Ezh2 regulates proliferation and differentiation of murine hepatic stem/progenitor cells. J Hepatol. 2010;52:854-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 68. | Chiba T, Suzuki E, Negishi M, Saraya A, Miyagi S, Konuma T, Tanaka S, Tada M, Kanai F, Imazeki F. 3-Deazaneplanocin A is a promising therapeutic agent for the eradication of tumor-initiating hepatocellular carcinoma cells. Int J Cancer. 2012;130:2557-2567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 69. | McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1320] [Cited by in F6Publishing: 1342] [Article Influence: 111.8] [Reference Citation Analysis (0)] |
| 70. | Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG, Allison DF, Cai L, Rockowitz S, Liu S, Liu Y, Li F, Vedadi M, Frye SV, Garcia BA, Zheng D, Jin J, Wang GG. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood. 2015;125:346-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 163] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 71. | Grozdanov PN, Yovchev MI, Dabeva MD. The oncofetal protein glypican-3 is a novel marker of hepatic progenitor/oval cells. Lab Invest. 2006;86:1272-1284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 81] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 72. | Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 629] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 73. | Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, Nakagohri T, Konishi M, Kobayashi N, Kinoshita T. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2009;100:1403-1407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 74. | Sawada Y, Yoshikawa T, Nobuoka D, Shirakawa H, Kuronuma T, Motomura Y, Mizuno S, Ishii H, Nakachi K, Konishi M. Phase I trial of a glypican-3-derived peptide vaccine for advanced hepatocellular carcinoma: immunologic evidence and potential for improving overall survival. Clin Cancer Res. 2012;18:3686-3696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 208] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 75. | Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631-677. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3762] [Cited by in F6Publishing: 3571] [Article Influence: 99.2] [Reference Citation Analysis (0)] |
| 76. | Jego G, Hazoumé A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2013;332:275-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 287] [Cited by in F6Publishing: 314] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 77. | Chuma M, Sakamoto M, Yamazaki K, Ohta T, Ohki M, Asaka M, Hirohashi S. Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology. 2003;37:198-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 236] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 78. | Di Tommaso L, Franchi G, Park YN, Fiamengo B, Destro A, Morenghi E, Montorsi M, Torzilli G, Tommasini M, Terracciano L. Diagnostic value of HSP70, glypican 3, and glutamine synthetase in hepatocellular nodules in cirrhosis. Hepatology. 2007;45:725-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 273] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 79. | Cai MY, Tong ZT, Zheng F, Liao YJ, Wang Y, Rao HL, Chen YC, Wu QL, Liu YH, Guan XY. EZH2 protein: a promising immunomarker for the detection of hepatocellular carcinomas in liver needle biopsies. Gut. 2011;60:967-976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 80. | Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000-1011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469-1483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 82. | Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266-2276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 83. | Omata M, Lesmana LA, Tateishi R, Chen PJ, Lin SM, Yoshida H, Kudo M, Lee JM, Choi BI, Poon RT. Asian Pacific Association for the Study of the Liver consensus recommendations on hepatocellular carcinoma. Hepatol Int. 2010;4:439-474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 732] [Cited by in F6Publishing: 813] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 84. | Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020-1022. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5972] [Cited by in F6Publishing: 6338] [Article Influence: 487.5] [Reference Citation Analysis (1)] |
| 85. | Kudo M, Izumi N, Kokudo N, Matsui O, Sakamoto M, Nakashima O, Kojiro M, Makuuchi M. Management of hepatocellular carcinoma in Japan: Consensus-Based Clinical Practice Guidelines proposed by the Japan Society of Hepatology (JSH) 2010 updated version. Dig Dis. 2011;29:339-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 549] [Cited by in F6Publishing: 630] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 86. | European Association For The Study Of The Liver; European Organisation For Research And Treatment Of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908-943. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4059] [Cited by in F6Publishing: 4345] [Article Influence: 362.1] [Reference Citation Analysis (2)] |
| 87. | Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol. 2006;33:392-406. [PubMed] [Cited in This Article: ] |
| 88. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9016] [Cited by in F6Publishing: 9507] [Article Influence: 594.2] [Reference Citation Analysis (1)] |
| 89. | Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3854] [Cited by in F6Publishing: 4336] [Article Influence: 271.0] [Reference Citation Analysis (0)] |
| 90. | Reig M, Torres F, Rodriguez-Lope C, Forner A, LLarch N, Rimola J, Darnell A, Ríos J, Ayuso C, Bruix J. Early dermatologic adverse events predict better outcome in HCC patients treated with sorafenib. J Hepatol. 2014;61:318-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 186] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 91. | Estfan B, Byrne M, Kim R. Sorafenib in advanced hepatocellular carcinoma: hypertension as a potential surrogate marker for efficacy. Am J Clin Oncol. 2013;36:319-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 92. | Yau T, Chan P, Ng KK, Chok SH, Cheung TT, Fan ST, Poon RT. Phase 2 open-label study of single-agent sorafenib in treating advanced hepatocellular carcinoma in a hepatitis B-endemic Asian population: presence of lung metastasis predicts poor response. Cancer. 2009;115:428-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 93. | Kuzuya T, Asahina Y, Tsuchiya K, Tanaka K, Suzuki Y, Hoshioka T, Tamaki S, Kato T, Yasui Y, Hosokawa T. Early decrease in α-fetoprotein, but not des-γ-carboxy prothrombin, predicts sorafenib efficacy in patients with advanced hepatocellular carcinoma. Oncology. 2011;81:251-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 94. | Ueshima K, Kudo M, Takita M, Nagai T, Tatsumi C, Ueda T, Kitai S, Ishikawa E, Yada N, Inoue T. Des-γ-carboxyprothrombin may be a promising biomarker to determine the therapeutic efficacy of sorafenib for hepatocellular carcinoma. Dig Dis. 2011;29:321-325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 95. | Ogasawara S, Chiba T, Ooka Y, Kanogawa N, Saito T, Motoyama T, Suzuki E, Tawada A, Kanai F, Yokosuka O. Sorafenib treatment in Child-Pugh A and B patients with advanced hepatocellular carcinoma: safety, efficacy and prognostic factors. Invest New Drugs. 2015;33:729-739. [PubMed] [Cited in This Article: ] |
| 96. | Arao T, Ueshima K, Matsumoto K, Nagai T, Kimura H, Hagiwara S, Sakurai T, Haji S, Kanazawa A, Hidaka H. FGF3/FGF4 amplification and multiple lung metastases in responders to sorafenib in hepatocellular carcinoma. Hepatology. 2013;57:1407-1415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 97. | Horwitz E, Stein I, Andreozzi M, Nemeth J, Shoham A, Pappo O, Schweitzer N, Tornillo L, Kanarek N, Quagliata L. Human and mouse VEGFA-amplified hepatocellular carcinomas are highly sensitive to sorafenib treatment. Cancer Discov. 2014;4:730-743. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 142] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 98. | Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang TW, Hohmeyer A, Pesic M, Leibold J, von Thun A. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med. 2014;20:1138-1146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 205] [Cited by in F6Publishing: 218] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 99. | Leary RJ, Sausen M, Kinde I, Papadopoulos N, Carpten JD, Craig D, O’Shaughnessy J, Kinzler KW, Parmigiani G, Vogelstein B. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 460] [Cited by in F6Publishing: 477] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 100. | Chan KC, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, Siu SS, Chan WC, Chan SL, Chan AT. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 369] [Cited by in F6Publishing: 395] [Article Influence: 35.9] [Reference Citation Analysis (0)] |