Published online May 27, 2021. doi: 10.4254/wjh.v13.i5.557
Peer-review started: January 16, 2021
First decision: February 24, 2021
Revised: February 28, 2021
Accepted: April 22, 2021
Article in press: April 22, 2021
Published online: May 27, 2021
Gut dysbiosis is common in cirrhosis.
To study the influence of gut dysbiosis on prognosis in cirrhosis.
The case-control study included 48 in-patients with cirrhosis and 21 healthy controls. Stool microbiome was assessed using 16S ribosomal ribonucleic acid gene sequencing. We used modified dysbiosis ratio (MDR): [Bacilli (%) + Proteobacteria (%)]/[Clostridia (%) + Bacteroidetes (%)]. Patients with MDR more the median made up the group with severe dysbiosis, others did the group with non-severe dysbiosis. The follow-up period was 4 years.
The mortality rate of patients with severe dysbiosis was significantly higher than that of patients with non-severe dysbiosis (54.2% vs 12.5%; P = 0.001). The presence of severe dysbiosis was independent risk factors for death [hazard ratio = 8.6 × (1.9-38.0); P = 0.005]. The abundance of Enterobacteriaceae (P = 0.002), Proteobacteria (P = 0.002), and Lactobacillaceae (P = 0.025) was increased and the abundance of Firmicutes (P = 0.025) and Clostridia (P = 0.045) was decreased in the deceased patients compared with the survivors. The deceased patients had a higher MDR value than the survivors [0.131 × (0.069-0.234) vs 0.034 × (0.009-0.096); P = 0.004]. If we applied an MDR value of 0.14 as the cutoff point, then it predicted patient death within the next year with a sensitivity of 71.4% and a specificity of 82.9% [area under the curve = 0.767 × (0.559-0.974)]. MDR was higher in patients with cirrhosis than in health controls [0.064 × (0.017-0.131) vs 0.005 × (0.002-0.007); P < 0.001], and in patients with decompensated cirrhosis than in patients with compensated cirrhosis [0.106 × (0.023-0.211) vs 0.033 × (0.012-0.074); P = 0.031]. MDR correlated negatively with prothrombin (r = -0.295; P = 0.042), cholinesterase (r = -0.466; P = 0.014) and serum albumin (r = -0.449; P = 0.001) level and positively with Child–Turcotte–Pugh scale value (r = 0.360; P = 0.012).
Gut dysbiosis is associated with a poorer long-term prognosis in cirrhosis.
Core Tip: The mortality rate of patients with severe dysbiosis was significantly higher than that of patients with non-severe dysbiosis. The abundance of Enterobacteriaceae, Proteobacteria, and Lactobacillaceae was increased and the abundance of Firmicutes and Сlostridia was decreased in the deceased patients compared with survivors. The abundance of Bacilli, Enterococcaceae and Lactobacillaceae was higher and the abundance of Clostridia was lower in those who died during the first year of follow-up compared with those who survived this year. The abundance of Enterobacteriaceae and Proteobacteria was higher in those who died in 2nd-4th years of follow-up compared with survivors.
- Citation: Maslennikov R, Ivashkin V, Efremova I, Alieva A, Kashuh E, Tsvetaeva E, Poluektova E, Shirokova E, Ivashkin K. Gut dysbiosis is associated with poorer long-term prognosis in cirrhosis. World J Hepatol 2021; 13(5): 557-570
- URL: https://www.wjgnet.com/1948-5182/full/v13/i5/557.htm
- DOI: https://dx.doi.org/10.4254/wjh.v13.i5.557
Microbiota are stable ecological communities of microorganisms in certain habitats[1]. Recently, the human microbiota has attracted the attention of researchers. Previous studies have shown that its composition varies in different diseases, and it has been hypothesized that the pathology of the human microbiota (dysbiosis) can participate in the pathogenesis of these diseases[2].
As the gut microbiota is the richest human microbiota, most research has been devoted to it. The gut microbiota plays an important role in human life; it digests non-digestible carbohydrates as well as generates vitamins and short-chain fatty acids (butyrate is particularly prominent), which are used as a source of energy by colonocytes. This function is performed by strict anaerobes of the main taxa of normal microbiota, which belong to the Clostridia class and Bacteroidetes phylum. Nevertheless, the gut microbiota can also play a pathogenic role because it has potential pathogenic bacteria, which belong to the Bacilli class (Streptococcaceae, Enterococcaceae) and Proteobacteria phylum (Enterobacteriaceae). In addition, facultative anaerobes of the Bacilli class and Proteobacteria phylum can enter the gut wall, mesenteric lymph nodes, portal, and systemic blood flow. This phenomenon is called bacterial translocation. The gut microbiota is also the main source of endotoxin (lipopolysaccharide), a toxic substance of gram-negative bacteria, primarily Proteobacteria[3].
To date, several articles[4-6] have been published that describe alterations of the gut microbiome in cirrhosis. Researchers have shown that the abundance of harmful Proteobacteria increases, whereas the abundances of useful Ruminococcaceae and Lachnospiraceae belonging to the Clostridia class decrease in the gut microbiome in cirrhosis.
Analysis of the relationship between gut dysbiosis and the course of cirrhosis is complicated by several problems. The first is the fact that the only reliable method for analyzing the gut microbiota is sequencing, which is very expensive and requires a rare bioinformatics specialist. Therefore, the study of dysbiosis has not yet transcended the walls of scientific laboratories and entered clinical medicine.
The second problem is the interpretation of obtained data. The researcher acquires a huge amount of redundant data after sequencing. A generalizing indicator should be used to simplify the analysis. Several such indicators have been proposed, including the richness and diversity of microbiota, the Firmicutes/Bacteroidetes ratio[7], and the cirrhosis dysbiosis ratio (CDR)[5]. However, these indicators have various disadvantages; first of all, many of them have a weak theoretical basis. Thus, the proliferation of harmful bacteria can lead to an increase in the richness and diversity of microbiota. However, the proliferation of beneficial bacteria can lead to similar changes; therefore, an increase or decrease in these indicators cannot be correctly interpreted. Firmicutes is too heterogeneous and represented by useful members of the Сlostridia class as well as potentially pathogenic members of the Bacilli class. In addition, the Firmicutes/Bacteroidetes ratio does not take into account Proteobacteria that are the main potentially pathogenic bacteria. Bacteroidetes has a multifaceted effect on the macroorganism and cannot be considered as only harmful bacteria. Therefore, the Firmicutes/Bacteroidetes ratio may be useful for comparing the gut microbiota between countries, between individuals on different diets, or for assessing changes in the microbiota with age, but it cannot show how much better or worse the composition of the microbiota has become.
The CDR proposed by Bajaj et al[5] is based on the ratio of “good” to “bad” bacteria. However, it also has some disadvantages. Its values decrease with an increase in the severity of dysbiosis, which can lead to misinterpretation. Bacteroidaceae were among the “bad” bacteria, but they play a rather neutral role in the gut microbiome and are widely represented in the microbiomes of healthy individuals, especially in studies from Asian countries[4]. In addition, Bacteroidaceae, being strict anaerobes, cannot be subjects of bacterial translocation[8]. At the same time, the list of “bad” bacteria did not include Bacilli, which together with Enterobacteriaceae are responsible for bacterial translocation[8] and secondary infections[3,9,10] in cirrhosis.
Thus, the development and testing of a pathogenetically-based dysbiosis ratio remains an important task. With this ratio, it will be possible to replace expensive and inaccessible sequencing with polymerase chain reaction (PCR) for selected taxa via automatic ratio calculation, which will allow for the introduction of gut dysbiosis tests into clinical practice.
The second important task of studying gut dysbiosis in cirrhosis is to clarify whether its presence affects the prognosis of patients.
Identifying a solution to these two problems is the aim of the present research.
We used the CDR as a basis but flipped the equation such that the “bad” bacteria were in the numerator and the “good” bacteria were in the denominator. Therefore, our modified dysbiosis ratio (MDR) increased with aggravation of dysbiosis, which was less confusing in its interpretation. We considered Proteobacteria and Bacilli as “bad” bacteria since they are responsible for bacterial translocation as well as the development of secondary infections[3,8-10] and their contents increase in cirrhosis[4,5]. We used the dominant taxa in healthy individuals, Clostridia and Bacteroidetes, as “good” bacteria. These taxa are strict anaerobes; therefore, they do not undergo bacterial translocation and do not cause extraintestinal secondary infections in cirrhosis[3,8-10]. Clostridia predominate in the American population, where the Western diet is widespread, whereas Bacteroidetes are more common in the Asian population, where the Eastern diet is widespread[4,5]. Thus, the total accounting of these taxa is also intended to reduce the effect of diet on the value of the MDR. The abundance of Clostridia has been found to decrease with the development of cirrhosis[4,5]. The changes in Bacteroidetes abundance in cirrhosis appear to vary across different studies[4-6].
Thus, the pathogenesis- and evidence-based MDR was calculated as follows: [Bacilli (%) + Proteobacteria (%)]/[Clostridia (%) + Bacteroidetes (%)].
In this case-control prospective study, 113 consecutive patients with cirrhosis were admitted to the Department of Hepatology of Clinic for Internal Diseases, Gastroenterology and Hepatology at Sechenov University (Moscow, Russia) and screened for inclusion. The study procedures were explained to potential participants, and written informed consent was obtained before enrollment. The present study was approved by the Ethics Committee of Sechenov University in accordance with the Declaration of Helsinki (№03-16).
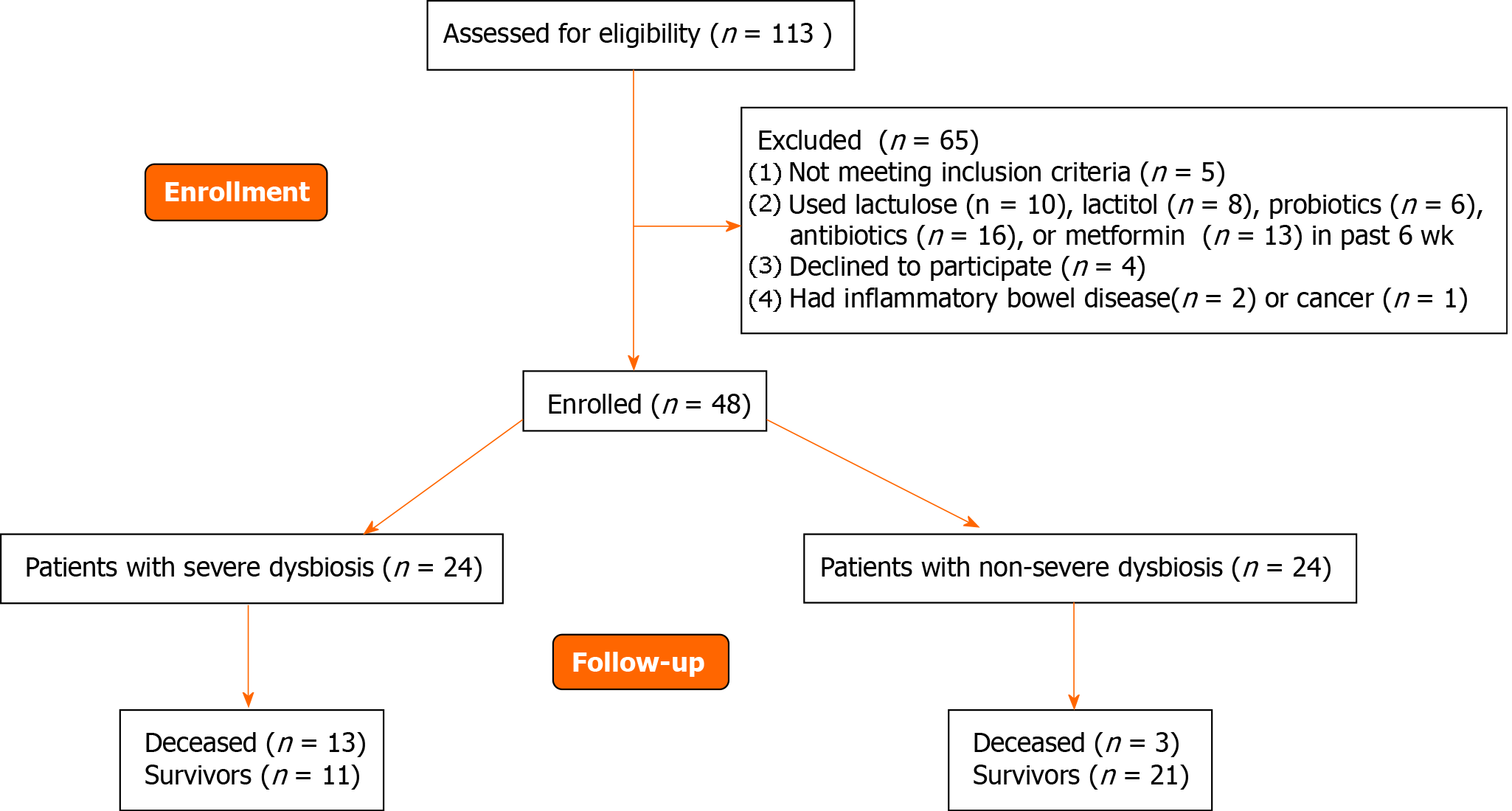
The inclusion criteria were as follows: diagnosis of cirrhosis verified by histology or clinical, biochemical, and ultrasound findings; and age between 18 and 70 years. The exclusion criteria were as follows: use of lactulose, lactitol, or other prebiotics, probiotics, antibiotics, or metformin in the past 6 wk; alcohol consumption in the past 6 wk; or inflammatory bowel disease, cancer, or any other serious disease. Of the original 113 patients screened for inclusion, 48 met the criteria and were enrolled in the study while 65 were excluded (Figure 1).
A study control group consisted of 21 healthy individuals who visited the clinic for routine health examinations during the same period.
The severity of liver disease was determined using the Child–Turcotte–Pugh (CTP) scoring system[11], in which class A was defined as compensated cirrhosis and classes B and C were defined as decompensated cirrhosis.
The morning after admission, a stool sample was taken into a sterile disposable container and immediately frozen at -80 °C[12].
Deoxyribonucleic acid from the stool was isolated using the MagNa Pure Compact Nucleic Acid Isolation Kit I (Roche, Basel, Switzerland) according to the manufacturer’s instructions. Libraries for sequencing were prepared by two rounds of PCR amplification. In the first round, specific primers for the v3-v4 region of the 16S ribosomal ribonucleic acid (RNA) gene were used: 16S-F: TCGTCGGCA-GCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG and 16S-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC.
After amplification, the PCR product was purified using AMPure XP magnetic beads (Beckman Coulter, Brea, CA, United States). Then, a second round of PCR was performed to attach specific adapters and enable multiplexing of the samples. To begin, 5 μL of the first PCR product was added to the reaction after ball cleaning with primers containing Illumina indices (Nextera XT Index v2 Primers; San Diego, CA, United States) and adapter sequences as well as 2 × KAPA HiFi HotStart ReadyMix. The amplification products were also purified using AMPure XP beads (Beckman Coulter). The concentrations of the prepared libraries were then measured using a Qubit 2.0 fluorimeter (London, United Kingdom) and quantitative PCR. The quality of the libraries was assessed using the Agilent 2100 Bioanalyzer (Santa Clara, CA, United States). The libraries were mixed in equal proportions and diluted to the required concentration to be run on a MiSeq (Illumina) device. Pair-end readings of 300 + 300 nucleotides were obtained. Reads were trimmed from the 3’-tail with Trimmomatic (Illumina) and then merged into a single amplicon with the MeFiT tool[13,14]. We did not perform operational taxonomic unit picking; instead, we classified amplicon sequences with the Ribosomal Database Project (RDP) classifier and RDP database[15].
The patients were contacted by phone every 3 mo to confirm that they were alive. If there was no answer, we contacted the patient’s relatives by phone to find out if the patient was alive or dead. If it was not possible to contact them, we studied patient electronic medical records in the United Medical Information and Analytical System of Moscow, in which death registration data are entered. The follow-up period was 4 years.
Statistical analysis was performed with STATISTICA 10 (StatSoft Inc., Tulsa, OK, United States) and SPSS Statistics (IBM SPSS, Armonk, NY, United States) software. The data are presented as medians (interquartile ranges). Differences between continuous variables were assessed with the Mann-Whitney test because many variables were not distributed normally. Fisher’s exact test was used to assess the differences between categorical variables. Survival was assessed using the Kaplan-Meier estimator and Cox regression test. A Cox regression model was constructed to assess the influence of various factors on patient survival and hazard ratios (HRs). Correlations between variables were computed using Spearman’s rank correlation. P values ≤ 0.05 were considered as statistically significant.
Participants with cirrhosis and healthy controls were comparable in age [51 (40-59) vs 46 (42-54) years; Р = 0.489], body mass index [24.6 × (22.7-27.7) vs 26.3 × (25.1-29.0) kg/m2; Р = 0.110], and sex distribution (male/female: 23/25 vs 8/13; Р = 0.313).
Seventeen participants with cirrhosis had compensated cirrhosis (CTP class А), and the remaining 31 had decompensated cirrhosis (19 class В and 12 class С). Participants with compensated cirrhosis and decompensated cirrhosis were also comparable in age [49 (38-55) years vs 52 (40-59) years, P = 0.316], body mass index [24.8 × (21.8-27.8) kg/m2 vs 24.4 × (22.8-27.7) kg/m2; P = 0.771], and sex distribution (6/11 vs 17/14; P = 0.160).
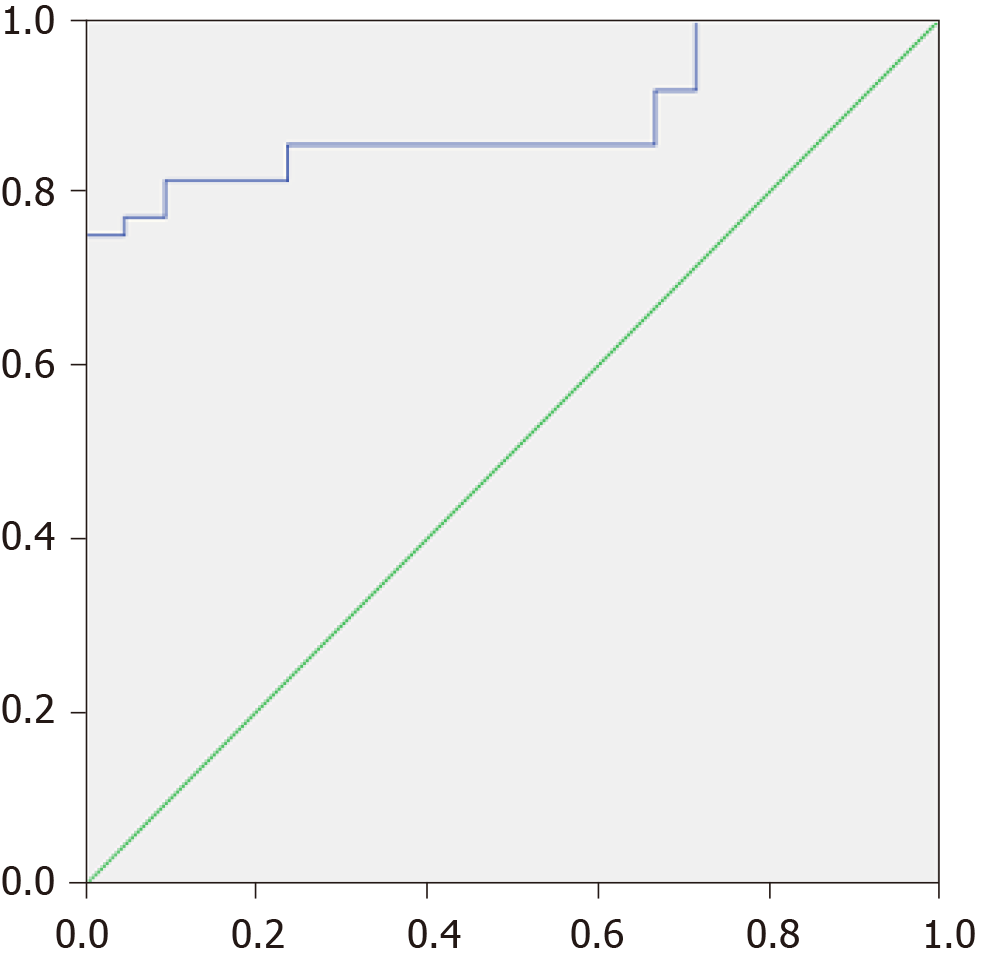
The MDR was higher in patients with cirrhosis than in healthy controls [0.064 × (0.017-0.131) vs 0.005 × (0.002-0.007); P < 0.001] and in patients with decompensated cirrhosis than in those with compensated cirrhosis [0.106 × (0.023-0.211) vs 0.033 × (0.012-0.074); P = 0.031]. When taken as the cutoff point, an MDR value of 0.01 made it possible to distinguish patients with cirrhosis from healthy individuals with a sensitivity of 81.3% and a specificity of 90.5% [AUC = 0.884 × (0.806-0.962)] (Figure 2). The specificity approached nearly 100% with a cutoff value of 0.02.
If we used the median MDR (0.064) as a cutoff point, then the group of patients with cirrhosis could be divided into patients with severe (n = 24) and non-severe (n = 24) dysbiosis (Figure 1).
The abundance of useful Clostridia was reduced and that of harmful Bacilli was increased, whereas the abundance of harmful Enterobacteriaceae was not significantly changed in patients with non-severe dysbiosis compared to healthy controls. The abundance of Clostridia further decreased, the abundance of Bacilli further increased, and the abundance of Enterobacteriaceae also increased in patients with severe dysbiosis. Interestingly, an increase in the abundance of Bifidobacteriaceae considered beneficial to the gut microbiome was also observed in cirrhosis without significant differences between groups with different degrees of dysbiosis. The abundance of Bacteroidetes did not differ significantly between patients with cirrhosis and healthy individuals (Table 1).
| Taxa | Heath controls (n = 21) | Cirrhosis with non-severe dysbiosis (n = 24) | Cirrhosis with severe dysbiosis (n = 24) | P value, Non-severe dysbiosis vs controls | P value, Severe dysbiosis vs controls | P value, Severe dysbiosis vs non-severe one |
| Firmicutes | 91.8 (89.3-96.4) | 89.7 (73.0-93.6) | 80.1 (62.7-88.1) | 0.074 | < 0.001 | 0.028 |
| Clostridia | 88.0 (86.6-91.7) | 83.5 (69.8-88.7) | 69.8 (57.4-77.2) | 0.008 | < 0.001 | 0.001 |
| Ruminococcaceae | 33.9 (28.1-41.6) | 27.6 (19.2-36.5) | 18.8 (7.9-31.7) | 0.086 | 0.002 | 0.081 |
| Lachnospiraceae | 43.8 (37.2-54.6) | 37.6 (27.2-60.5) | 31.0 (22.1-46.0) | 0.488 | 0.030 | 0.190 |
| Bacilli | 0.1 (0.0-0.2) | 0.5 (0.2-1.9) | 7.1 (1.3-14.8) | < 0.001 | < 0.001 | < 0.001 |
| Streptococcaceae | 0.03 (0.02-0.10) | 0.29 (0.12-0.52) | 3.20 (0.38-10.4) | < 0.001 | < 0.001 | 0.002 |
| Lactobacillaceae | 0.00 (0.00-0.01) | 0.02 (0.01-0.22) | 0.47 (0.12-1.50) | 0.002 | < 0.001 | < 0.001 |
| Enterococcaceae | 0.00 (0.00-0.00) | 0.00 (0.00-0.03) | 0.03 (0.01-0.08) | 0.067 | 0.001 | < 0.001 |
| Bacteroidetes | 5.6 (2.8-8.1) | 5.7 (1.8-12.9) | 6.1 (3.2-8.2) | 0.954 | 0.829 | 0.959 |
| Bacteroidaceae | 2.5 (0.8-3.4) | 1.3 (0.6-4.3) | 1.4 (0.2-3.8) | 0.991 | 0.432 | 0.261 |
| Actinobacteria | 0.2 (0.1-0.3) | 0.8 (0.3-2.8) | 0.7 (0.4-2.9) | < 0.001 | < 0.001 | 0.687 |
| Bifidobacteriaceae | 0.1 (0.0-0.2) | 0.6 (0.1-2.6) | 0.5 (0.2-2.3) | 0.002 | 0.001 | 0.687 |
| Proteobacteria | 0.39 (0.14-0.51) | 0.15 (0.10-0.81) | 3.57 (1.77-6.65) | 0.869 | < 0.001 | < 0.001 |
| Enterobacteriaceae | 0.03 (0.01-0.05) | 0.04 (0.02-0.61) | 2.70 (1.58-6.24) | 0.104 | < 0.001 | < 0.001 |
There were no significant differences in age, body mass index, sex distribution, and etiology of cirrhosis between patients with severe and non-severe dysbiosis. Patients with severe dysbiosis had lower serum albumin and cholinesterase levels, higher CTP scale values, and higher C-reactive protein levels. Although the incidences of ascites, esophageal varices, and hepatic encephalopathy were higher in patients with severe dysbiosis than in those with non-severe dysbiosis, these differences did not reach the significance level. There were no differences between the groups of patients in red blood cell, white blood cell, and platelet counts; creatinine, sodium, potassium, and glucose levels; and aminotransferase, alkaline phosphatase, and gamma-glutamate transferase activities (Table 2).
| Severe dysbiosis (n = 24) | Non-severe dysbiosis (n = 24) | P value | |
| Age, yr | 51.5 (42.0-59.0) | 50.0 (35.0-57.5) | 0.392 |
| Body mass index, kg/m2 | 24.6 (22.8-27.7) | 24.2 (22.7-27.7) | 0.837 |
| Male/female | 12/12 | 11/13 | 0.500 |
| Etiology of cirrhosis: Alcohol | 9 | 9 | 0.617 |
| Autoimmune | 2 | 7 | 0.068 |
| HBV | 7 | 2 | 0.068 |
| HCV | 5 | 3 | 0.350 |
| Cryptogenic | 1 | 3 | 0.304 |
| Child–Turcotte–Pugh score | 9 (8-10) | 7 (6-9) | 0.047 |
| Death | 13 | 3 | 0.001 |
| Death within the first year of follow-up | 5 | 2 | 0.092 |
| Death during the following years of follow-up | 8 | 1 | 0.002 |
| Esophageal varices (present/absent) | 20/4 | 18/6 | 0.477 |
| Hepatic encephalopathy (overt/minimal/absent) | 11/9/4 | 6/11/7 | 0.288 |
| Number connection test, seconds | 87 (65-118) | 79 (59-92) | 0.248 |
| Ascites (present/absent) | 16/8 | 11/13 | 0.122 |
| Spontaneous bacterial peritonitis (present/absent) | 0/24 | 0/24 | 1.000 |
| Red blood cells, 1012 cell/L | 3.8 (3.4-4.0) | 3.9 (3.6-4.5) | 0.370 |
| White blood cells, 109 cell/L | 3.8 (2.7-5.3) | 3.8 (3.1-5.2) | 0.628 |
| Platelets, 109 cell/L | 87 (55-120) | 76 (60-108) | 0.860 |
| Serum total protein, g/L | 70 (61-76) | 73 (64-78) | 0.599 |
| Serum albumin, g/L | 31 (28-37) | 38 (34-41) | 0.009 |
| Serum total bilirubin, μmol/L | 47 (31-62) | 31 (24-63) | 0.375 |
| Prothrombin index (Quick test), % | 58 (48-67) | 64 (54-71) | 0.239 |
| Creatinine, mg/dL | 0.69 (0.53-0.87) | 0.73 (0.66-0.90) | 0.187 |
| Serum sodium, mmol/L | 141 (139-144) | 141 (138-143) | 0.795 |
| Serum potassium, mmol/L | 4.3 (4.0-4.7) | 4.3 (4.1-4.7) | 0.844 |
| Serum glucose, mmol/L | 5.1 (4.7-5.6) | 5.3 (4.7-6.0) | 0.260 |
| Alanine aminotransferase, U/L | 36 (25-72) | 37 (23-60) | 0.804 |
| Aspartate aminotransferase, U/L | 54 (41-98) | 40 (26-67) | 0.219 |
| Gamma glutamyl transferase, U/L | 77 (40-148) | 76 (36-131) | 0.621 |
| Alkaline phosphatase, U/L | 221 (188-340) | 222 (166-298) | 0.542 |
| Cholinesterase, kU/L | 2.7 (1.9-3.7) | 4.0 (3.6-4.5) | 0.031 |
| C-reactive protein, mg/L | 10.1 (2.1-16.1) | 2.1 (0.3-8.9) | 0.032 |
| Splenic length, cm | 15.4 (14.0-17.6) | 16.1 (13.3-19.2) | 0.841 |
The MDR correlated negatively with prothrombin (r = -0.295; P = 0.042), cholinesterase (r = -0.466; P = 0.014) and serum albumin (r = -0.449; P = 0.001) levels and positively with CTP scale values (r = 0.360; P = 0.012).
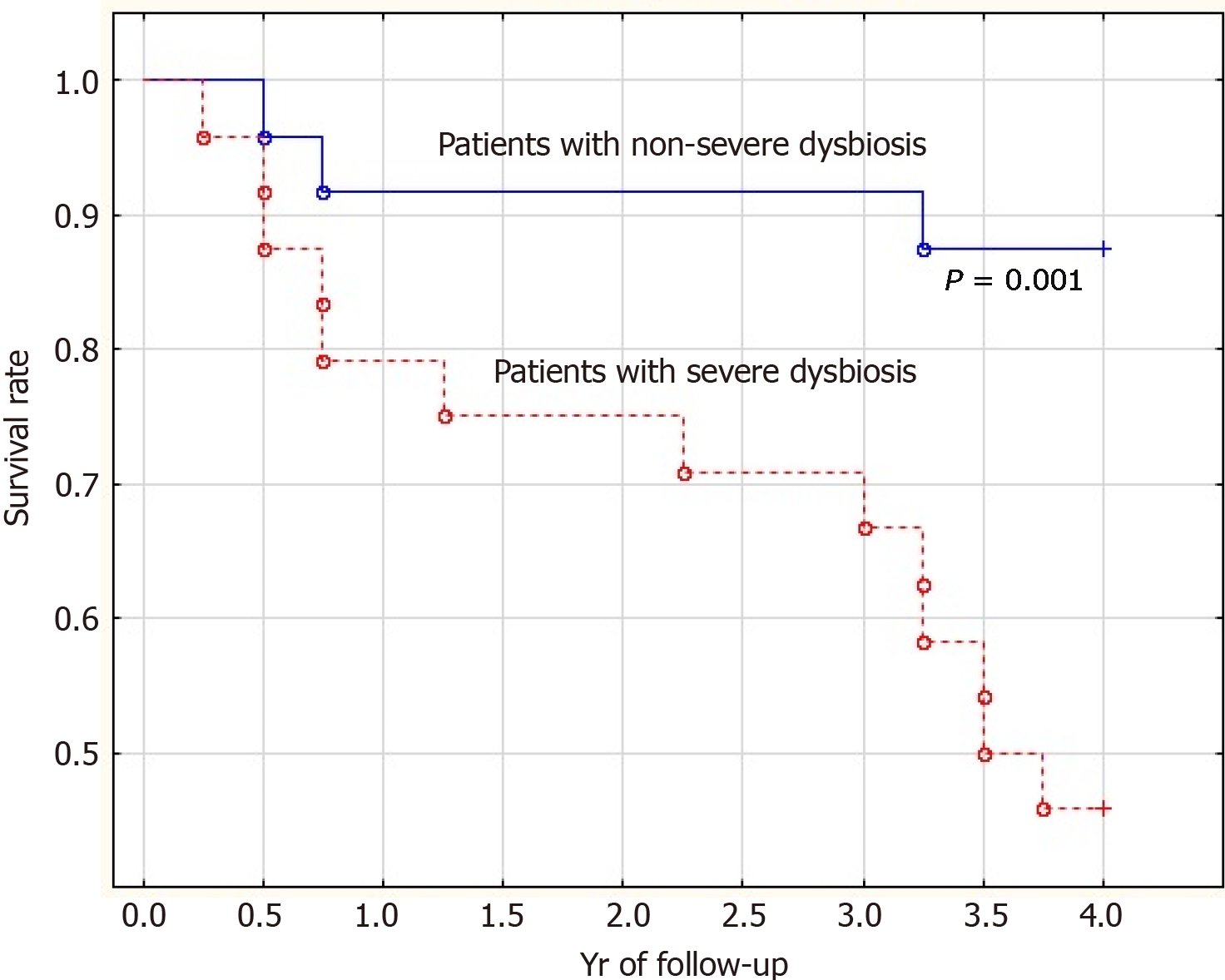
The mortality rate of patients with severe dysbiosis was significantly higher than that of patients with non-severe dysbiosis (54.2% vs 12.5%; P = 0.001). Moreover, the difference in mortality was insignificant in the first year of follow-up (20.8% vs 8.3%; P = 0.092) and significant in subsequent years of follow-up (33.4% vs 4.2%; P = 0.002) (Figure 3).
Deceased patients had a higher MDR value than the survivors [0.131 × (0.069-0.234) vs 0.034 × (0.009-0.096); P = 0.004]. Moreover, this was observed in the deceased in the first year of follow-up [0.191 × (0.035-1.126) vs 0.046 × (0.012-0.115); P = 0.022] as well as in subsequent years [0.115 × (0.074-0.144) vs 0.034 × (0.009-0.096); P = 0.044].
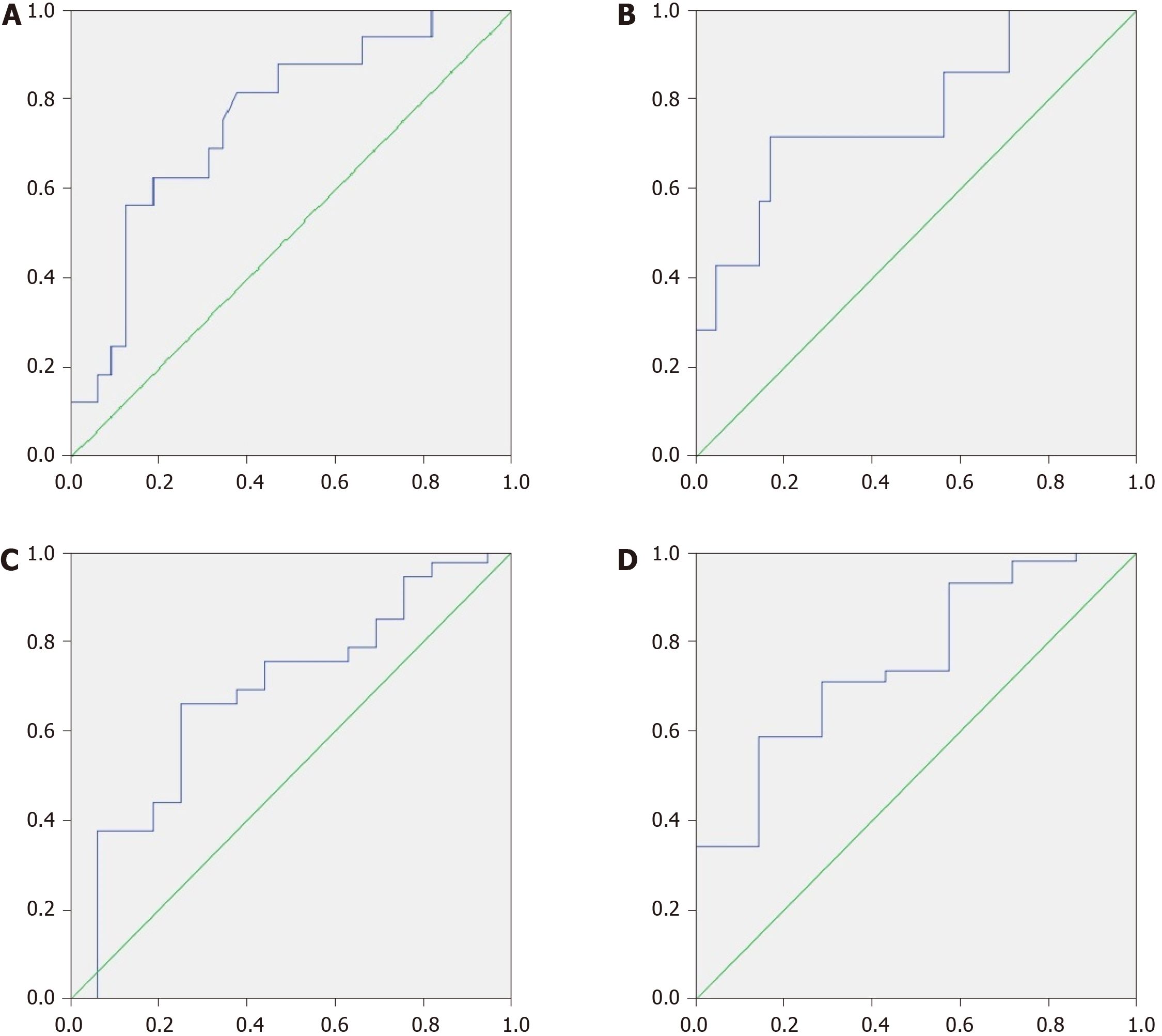
If we took an MDR value of 0.05 as the cutoff point, it predicted patient death within the next 4 years with a sensitivity of 65.2% and a specificity of 81.3%. If we used 0.11 for this, then the sensitivity was 81.3% and the specificity was 62.5% [AUC = 0.755 × (0.611-0.899); Figure 4A].
If we applied an MDR value of 0.14 as the cutoff point, then it predicted patient death within the next year with a sensitivity of 71.4% and a specificity of 82.9% [AUC = 0.767 × (0.559-0.974); Figure 4B].
The presence of severe dysbiosis [HR = 8.6 × (1.9-38.0); P = 0.005] and total serum bilirubin level [HR = 1.005 × (1.001-1.010); P = 0.015] were independent risk factors for death, unlike serum albumin (P = 0.870) and prothrombin (P = 0.167) levels, degrees of ascites (P = 0.752), and esophageal varices (P = 0.230).
In addition, death in the first year of follow-up was significantly determined by serum albumin level [HR = 0.83 × (0.71-0.97); P = 0.020], unlike degrees of ascites (P = 0.619), dysbiosis (P = 0.241), total serum bilirubin (P = 0.742) and prothrombin levels (P = 0.386), and esophageal varices (P = 0.125). However, mortality in subsequent years of follow-up was determined significantly by the degree of dysbiosis only [HR = 24.8 × (2.3-269.6); P = 0.008].
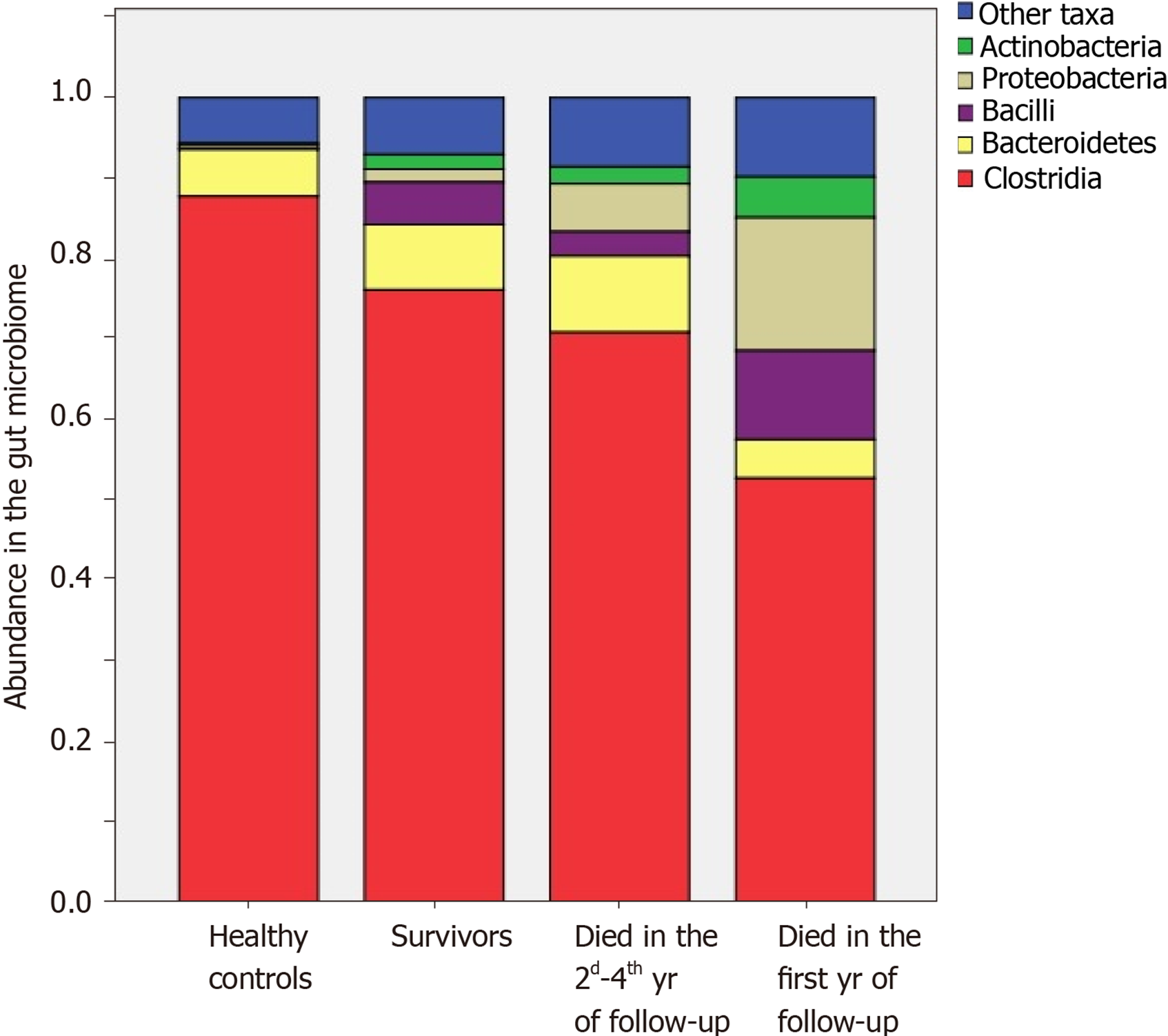
The abundances of Enterobacteriaceae [2.4 × (1.6-7.6) vs 0.4 × (0.0-1.7)%; P = 0.002], Proteobacteria [3.4 × (1.9-8.2) vs 0.6 × (0.1-2.0)%; P = 0.002], and Lactobacillaceae [0.35 × (0.12-0.81) vs 0.06 × (0.01-0.31)%; P = 0.025] were increased, and the abundances of Firmicutes [78.8 × (62.7-85.6) vs 87.1 × (71.7-93.6)%; P = 0.025] and Clostridia [73.0 × (51.9-78.2) vs 80.1 × (68.5-87.2)%; P = 0.045] were decreased in the gut microbiome of deceased patients compared to the survivors.
The abundances of Bacilli [14.0 × (1.4-18.4) vs 1.1 × (0.3-4.6)%; P = 0.017], Enterococcaceae [0.09 × (0.04-0.38) vs 0.01 × (0.00-0.04)%; P = 0.005], and Lactobacillaceae [0.45 × (0.24-1.52) vs 0.09 × (0.01-0.38)%; P = 0.021] were higher, and the abundance of Clostridia [67.1 × (31.2-78.2) vs 77.5 × (68.5-86.8)%; P = 0.047] was lower in those who died during the first year of follow-up compared to those who survived the first year. The abundances of Enterobacteriaceae [2.2 × (1.8-6.5) vs 0.4 (0.0-1.7)%; P = 0.009] and Proteobacteria [3.8 × (2.5-7.0) vs 0.6 × (0.1-2.0)%; P = 0.010] were higher in those who died in the second through fourth years of follow-up compared to the survivors. The deceased during the first year of follow-up had higher abundances of Bacilli [14.0 × (1.4-18.4) vs 0.5 × (0.4-4.2)%; P = 0.026] and Enterococcaceae [0.09 × (0.04-0.38) vs 0.00 × (0.00-0.05)%; P = 0.002] than those who died in the next 3 years of follow-up (Figures 5 and 6).
There was no significant difference in the Firmicutes/Bacteroidetes ratio between patients with cirrhosis and healthy individuals [13.3 × (7.8-40.9) vs 15.8 × (11.2-33.1); P = 0.469], the survivors and deceased patients [14.0 × (6.1-51.7) vs 12.7 × (8.0-26.4); P = 0.938], and patients with compensated and decompensated cirrhosis [16.0 × (7.8-68.7) vs 13.1 × (7.9-35.6); P = 0.846].
The CDR was significantly lower in patients with cirrhosis than in healthy individuals [16.4 × (7.2-39.0) vs 34.9 × (23.0-101.1); P = 0.002], in deceased patients than in the survivors [10.5 × (4.5-18.9) vs 19.7 × (10.7-57.6); P = 0.041], and in decompensated cirrhosis than in compensated cirrhosis [13.1 × (5.0-27.4) vs 22.5 × (14.1-65.4); P = 0.039]. Using the cutoff value of this ratio equal to 22, we could distinguish between patients with cirrhosis and healthy individuals with a sensitivity of 64.6% and a specificity of 85.7% [AUC = 0.735 × (0.620-0.850)]. The CDR was lower in patients who died in the first year of follow-up compared to those who survived the first year [9.4 × (1.7-15.4) vs 17.7 × (9.0-54.8); P = 0.035] but did not differ significantly between those who died in the following years and those who survived [13.6 × (7.3-22.5) vs 19.7 × (10.7-57.6); P = 0.321].
If we used a CDR value of 15 as the cutoff point, then it predicted patient death within the next 4 years with a sensitivity of 68.8% and a specificity of 62.5% [AUC = 0.684 × (0.522-0.845); Figure 4C] as well as within the first year with a sensitivity of 85.7% and a specificity of 58.5% [AUC = 0.753 × (0.569-0.936); Figure 4D].
Translating scientific developments into clinical practice is a rather difficult task. The study of the gut microbiome in various diseases is becoming mainstream in modern science, but thus far, it has no applications in clinical practice. It is hindered by the high cost of sequencing the fecal microbiome and the shortage of bioinformatics specialists.
Therefore, an important step in introducing the study of gut dysbiosis into clinical practice is to replace this expensive method with a simpler and more affordable one. PCR is an ideal candidate to determine the content of selected taxa in feces, followed by a comprehensive assessment of the state of the gut microbiome.
The idea to conduct a comprehensive assessment of the state of the gut microbiome in cirrhosis originated with Bajaj and colleagues[5]. However, their CDR can be improved, which was one of the aims of our study.
Here, we modified the CDR to improve its analytical performance and show that it can be used to predict the death of patients.
First, we inverted the CDR equation, placing the abundance of “bad” bacteria in the numerator and the abundance of “good” bacteria in the denominator. Thus, the value of our MDR increases with the aggravation of dysbiosis, which is more logical. The original CDR decreases with the aggravation of dysbiosis, which can be confusing to interpret.
Our MDR is based on the data regarding the role of various taxa in the pathogenesis of cirrhosis complications and changes in their abundance in cirrhosis. We excluded Bacteroidaceae from the list of “bad” bacteria since their role in the pathogenesis of cirrhosis is not clear, and the change in their abundance in the gut microbiome in cirrhosis varies according to different researchers. According to our data, it does not change significantly, according to Chen et al[4], it decreases, and according to Bajaj et al[5], it increases in compensated cirrhosis and decreases in decompensated cirrhosis, becoming almost the same as that in healthy individuals. On the contrary, in a study by Kakiyama et al[6], the abundance of Bacteroidaceae decreased with compensated cirrhosis and increased with decompensated cirrhosis. Instead, we added Bacilli to the list of “bad” bacteria, which, like Proteobacteria/Enterobacteriaceae, are responsible for bacterial translocation and the development of extraintestinal infections in cirrhosis[8-10]. The abundances of both of these taxa increased with cirrhosis according to all studies[4-6], including ours.
As “good” bacteria, we used the higher-level taxon Clostridia, which includes all taxa accounted as “good” bacteria in the CDR. The main problem is that the abundance of these taxa is highly dependent on diet[16,17]. Among healthy individuals, it was 90% in the Russian population (our data), approximately 45% in the American population[5], and approximately 30% in the Chinese population[4]. However, if you add to them to the abundance of Bacteroidetes, which changes in the opposite direction relative to Clostridia and Firmicutes[16,17], then the differences were not so large: 95%, 80%, and 90%, respectively. This dependence of the Bacteroidetes and Clostridia abundances on diet led to the fact that the value of the CDR in our population was more than an order of magnitude higher than in the original study. Thus, the addition of Bacteroidetes to the group of “good” bacteria can neutralize the effect of diet on MDR and allow it to be used in different populations.
In our study, we were able to show that despite the change in the order of values, the CDR retained its main characteristics: it was higher in healthy individuals, lower in patients with compensated cirrhosis, and minimal in patients with decompensated cirrhosis.
Both the CDR and MDR were useful in assessing the prognosis of patients with cirrhosis, but the analytical characteristics of our modification were higher. In particular, the MDR, unlike the CDR, made it possible to assess the long-term (more 1 year) prognosis of patients.
Interestingly, the different taxa included in the MDR had different effects on prognosis. Clostridia and Bacilli mainly determined the medium-term prognosis (death within a year), and Proteobacteria and Enterobacteriaceae determined the long-term prognosis (death over the subsequent 3 years). This finding may be due to the fact that Bacilli provide a more powerful translocation of living bacteria, which leads to faster death, whereas Enterobacteriaceae act mainly by translocating their endotoxin, which leads to a more delayed death.
Thus, we were able to show that the gut microbiome in cirrhosis can be comprehensively and reliably evaluated using targeted analysis of the most significant taxa, which will allow for replacing expensive and poorly available sequencing with cheaper and more affordable PCR for four indicators (Proteobacteria, Bacilli, Clostridia, and Bacteroidetes) that does not require interpretation by rare bioinformatics specialists.
This will be a big step forward in introducing the achievements of fundamental hepatology into clinical practice, as it will give doctors an instrument for assessing the state of the gut microbiome in their patients as well as determining how it is affected by drugs that are prescribed for the correction of dysbiosis. This reality reinforces the strength of our study.
In addition, our study is the first to describe the effect of gut dysbiosis on the prognosis of patients with cirrhosis, thereby confirming existing hypotheses about the important role of the gut-liver axis in the course of cirrhosis[3,18-21]. This is its second strong point.
The limitation of our study is its small sample size, which did not prevent us from obtaining significant results. It should also be noted that patients with severe hepatic encephalopathy (grades 2-4) are typically not admitted to our clinic, so these patients were not included in our study. The question of whether our results can be transferred to this cohort of patients remains open. Since patients with infections received antibiotics before admission, which could change the composition of the gut microbiota, we excluded them from the study. None of the included patients developed infectious complications of cirrhosis during hospitalization. Thus, patients with infectious complications of cirrhosis were not included in our study, and it is not clear whether the results can be generalized to them. A larger study involving non-included patient populations should be provided to confirm the findings.
New studies are needed to evaluate how various methods (e.g., probiotics, prebiotics, antibiotics, and fecal transplantation) can correct dysbiosis by analyzing the MDR and how this correction can improve the prognosis of patients with cirrhosis.
In conclusion, we were able to improve the CDR as well as show that gut dysbiosis is associated with poor prognosis in cirrhosis. Thus, we have developed a methodological apparatus and scientific basis for the correction of gut dysbiosis in such patients.
Gut dysbiosis is common in cirrhosis.
The aim is to study the influence of gut dysbiosis on prognosis in cirrhosis.
The objectives include the development and test of a modified dysbiosis ratio (MDR) to distinguish between patients with cirrhosis and healthy controls, patients with compensated and decompensated cirrhosis, deceased and surviving patients.
The case-control study included 48 in-patients with cirrhosis and 21 healthy controls. Stool microbiome was assessed using 16S ribosomal ribonucleic acid gene sequencing. We used MDR: [Bacilli (%) + Proteobacteria (%)]/[Clostridia (%) + bacteroidetes (%)]. Patients with MDR more its median made up the group with severe dysbiosis, others did the group with non-severe dysbiosis. The follow-up period was 4 years.
The mortality rate of patients with severe dysbiosis was significantly higher than that of patients with non-severe dysbiosis. The presence of severe dysbiosis was independent risk factors for death. The deceased patients had a higher MDR value than the survivors. MDR was higher in patients with cirrhosis than in health controls and in patients with decompensated cirrhosis than in patients with compensated cirrhosis.
Gut dysbiosis is associated with a poorer long-term prognosis in cirrhosis.
A larger study involving non-included patient populations should be provided to confirm the findings. New studies are needed to evaluate how various methods (e.g., probiotics, prebiotics, antibiotics, and fecal transplantation) can correct dysbiosis by analyzing the MDR and how this correction can improve the prognosis of patients with cirrhosis.
We are grateful to the staff of the Post-Genomic Research Laboratory of the Institute of Molecular Biology V.A.Engelgardt, headed by Anna Kudryavtseva, and bioinformatics Georgy Krasnov, without whose qualified work this study would be impossible.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Dai YC, Elshaarawy O, Li CP, Vegetti A S-Editor: Zhang L L-Editor: A P-Editor: Wang LL
| 1. | Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499-504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 557] [Cited by in F6Publishing: 669] [Article Influence: 133.8] [Reference Citation Analysis (0)] |
| 2. | Kåhrström CT, Pariente N, Weiss U. Intestinal microbiota in health and disease. Nature. 2016;535:47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 120] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Giannelli V, Di Gregorio V, Iebba V, Giusto M, Schippa S, Merli M, Thalheimer U. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20:16795-16810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 156] [Cited by in F6Publishing: 149] [Article Influence: 14.9] [Reference Citation Analysis (1)] |
| 4. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 674] [Cited by in F6Publishing: 701] [Article Influence: 53.9] [Reference Citation Analysis (1)] |
| 5. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 659] [Cited by in F6Publishing: 715] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 6. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 503] [Cited by in F6Publishing: 549] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 7. | Mariat D, Firmesse O, Levenez F, Guimarăes V, Sokol H, Doré J, Corthier G, Furet JP. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 997] [Cited by in F6Publishing: 1097] [Article Influence: 73.1] [Reference Citation Analysis (0)] |
| 8. | Berg RD, Garlington AW. Translocation of certain indigenous bacteria from the gastrointestinal tract to the mesenteric lymph nodes and other organs in a gnotobiotic mouse model. Infect Immun. 1979;23:403-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 528] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 9. | Jalan R, Fernandez J, Wiest R, Schnabl B, Moreau R, Angeli P, Stadlbauer V, Gustot T, Bernardi M, Canton R, Albillos A, Lammert F, Wilmer A, Mookerjee R, Vila J, Garcia-Martinez R, Wendon J, Such J, Cordoba J, Sanyal A, Garcia-Tsao G, Arroyo V, Burroughs A, Ginès P. Bacterial infections in cirrhosis: a position statement based on the EASL Special Conference 2013. J Hepatol. 2014;60:1310-1324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 568] [Cited by in F6Publishing: 580] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 10. | Bhattacharya C, Das-Mondal M, Gupta D, Sarkar AK, Kar-Purkayastha S, Konar A. Infection in cirrhosis: A prospective study. Ann Hepatol. 2019;18:862-868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Pugh RN, Murray-Lyon IM, Dawson JL, Pietroni MC, Williams R. Transection of the oesophagus for bleeding oesophageal varices. Br J Surg. 1973;60:646-649. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5490] [Cited by in F6Publishing: 5499] [Article Influence: 107.8] [Reference Citation Analysis (1)] |
| 12. | Fouhy F, Deane J, Rea MC, O'Sullivan Ó, Ross RP, O'Callaghan G, Plant BJ, Stanton C. The effects of freezing on faecal microbiota as determined using MiSeq sequencing and culture-based investigations. PLoS One. 2015;10:e0119355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 13. | Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114-2120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30322] [Cited by in F6Publishing: 33472] [Article Influence: 3347.2] [Reference Citation Analysis (0)] |
| 14. | Parikh HI, Koparde VN, Bradley SP, Buck GA, Sheth NU. MeFiT: merging and filtering tool for illumina paired-end reads for 16S rRNA amplicon sequencing. BMC Bioinformatics. 2016;17:491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 54] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261-5267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12842] [Cited by in F6Publishing: 12166] [Article Influence: 715.6] [Reference Citation Analysis (0)] |
| 16. | De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691-14696. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3584] [Cited by in F6Publishing: 3703] [Article Influence: 264.5] [Reference Citation Analysis (0)] |
| 17. | Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2004] [Cited by in F6Publishing: 2039] [Article Influence: 145.6] [Reference Citation Analysis (0)] |
| 18. | Fukui H. Gut-liver axis in liver cirrhosis: How to manage leaky gut and endotoxemia. World J Hepatol. 2015;7:425-442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 116] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 19. | Maslennikov R, Pavlov C, Ivashkin V. Small intestinal bacterial overgrowth in cirrhosis: systematic review and meta-analysis. Hepatol Int. 2018;12:567-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Maslennikov R, Pavlov C, Ivashkin V. Is small intestinal bacterial overgrowth a cause of hyperdynamic circulation in cirrhosis? Turk J Gastroenterol. 2019;30:964-975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 21. | Simbrunner B, Mandorfer M, Trauner M, Reiberger T. Gut-liver axis signaling in portal hypertension. World J Gastroenterol. 2019;25:5897-5917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 55] [Cited by in F6Publishing: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |