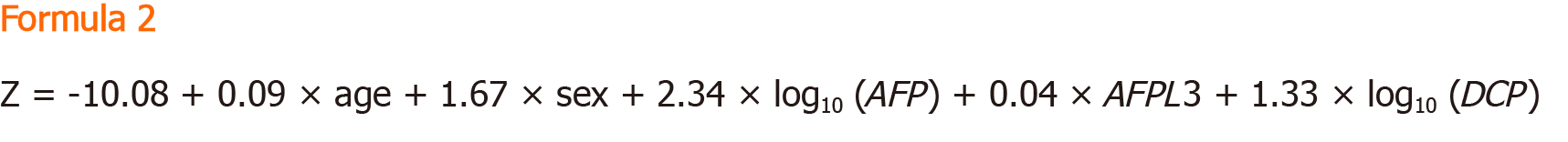Published online Sep 27, 2020. doi: 10.4254/wjh.v12.i9.558
Peer-review started: May 27, 2020
First decision: June 15, 2020
Revised: July 6, 2020
Accepted: August 24, 2020
Article in press: August 24, 2020
Published online: September 27, 2020
Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related deaths worldwide. If diagnosed early, curative treatment options such as surgical resection, loco-regional therapies, and liver transplantation are available to patients, increasing their chances of survival and improving their quality of life. Unfortunately, most patients are diagnosed with late stage HCC where only palliative treatment is available. Therefore, biomarkers which could detect HCC early with a high degree of sensitivity and specificity, may play a crucial role in the diagnosis and management of the disease. This review will aim to provide an overview of the different biomarkers of HCC comprising those used in the diagnosis of HCC in at risk populations, as well as others with potential for prognosis, risk predisposition and prediction of response to therapeutic intervention.
Core Tip: The use of ultrasound with/without alpha-fetoprotein in the context of screening patients with chronic liver disease for the development of early stage hepatocellular carcinoma that is treatable, remains problematic. Consequently there has been considerable work done to examine biomarkers either individually, or in combination to address this deficiency. Whilst there are several promising targets (discussed in this manuscript) for this indication it appears that Gender, Age: Lens culinaris agglutinin-reactive of alpha-fetoprotein, Alpha-fetoprotein, and Des-γ-carboxy prothrombin, which has been established in Europe and Japan, and remains to be so in North America, may be clinically the best performer available.
- Citation: Singh G, Yoshida EM, Rathi S, Marquez V, Kim P, Erb SR, Salh BS. Biomarkers for hepatocellular cancer. World J Hepatol 2020; 12(9): 558-573
- URL: https://www.wjgnet.com/1948-5182/full/v12/i9/558.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i9.558
Liver cancer is the sixth most common type of cancer globally and is ranked third for the most cancer-related deaths[1]. Liver cancer is more prominent in men being the second leading cause for cancer-related deaths and sixth in women[1]. Hepatocellular carcinoma (HCC) accounts for 85%-90% of primary liver cancer cases[2]. The prevalence of HCC is disproportionately high in areas with a high incidence of hepatitis B virus (HBV). These areas mainly include sub-Saharan Africa and Eastern Asia[3]. It is estimated that almost 80% of all HCCs are viral in etiology induced by both HBV and hepatitis C virus (HCV)[4]. Other risk factors include chronic alcohol consumption, non-alcoholic steatohepatitis (NASH) and cirrhosis arising from a variety of other causes.
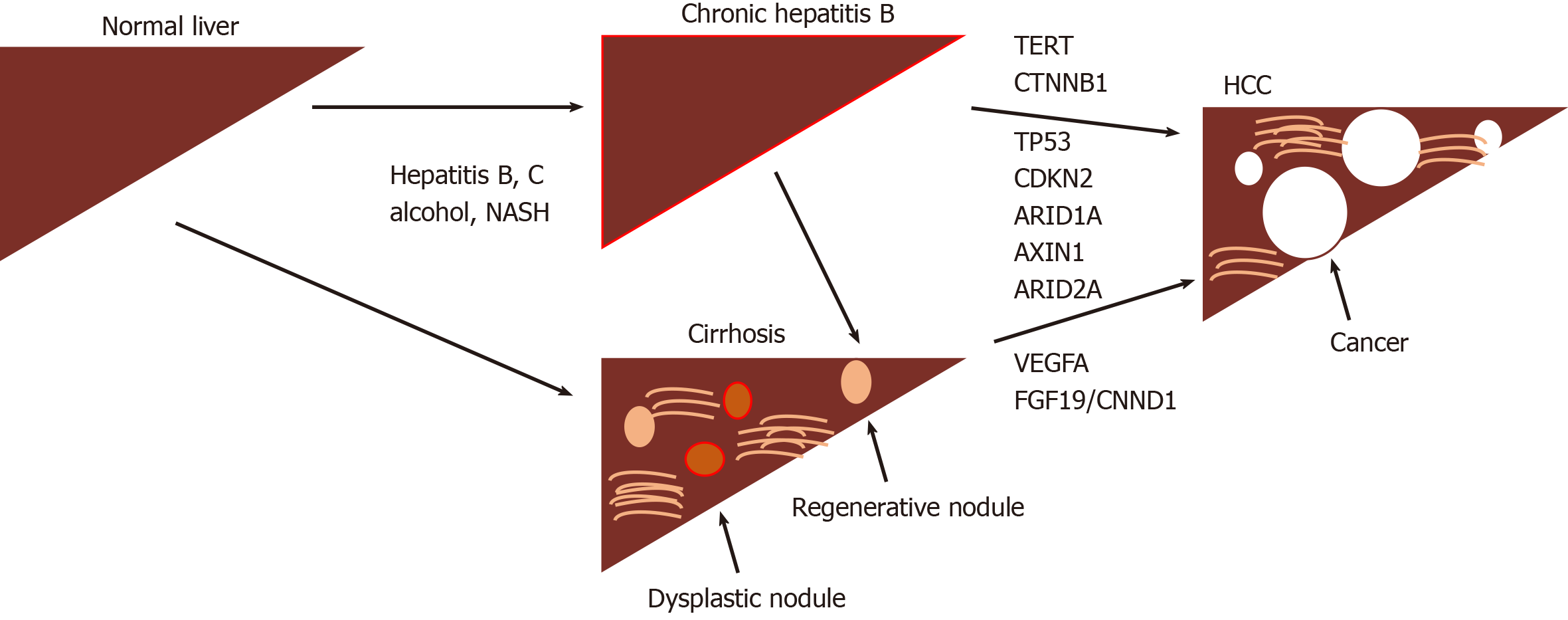
The development of HCC is recognized to be a multistep progress with dysplastic macronodules transforming into early and then more aggressive tumors. A number of driver mutations are associated with this process (Figure 1) including TERT (most frequent, 60%), CTNNB1, TP53, CDKN2, ARID1A, AXIN1 as well as DNA gene amplifications involving VEGFA (6p21) and FGF19/CNND1 (11q13), with continuing in-depth exome sequencing turning up novel mutational signatures and risk associations[5,6]. Unfortunately, none of these have any demonstrated value as biomarkers for early disease.
The most widely used classification system, used for treatment decision making is the Barcelona Clinic Liver Cancer (BCLC) algorithm[7]. Ideally, screening would enable pick-up of lesions at early stages (O, A). Disappointingly, HCC is often diagnosed during the advanced stages of the disease as the tumour is often asymptomatic until it progresses and becomes large and infiltrating (stages B, C and D). Curative treatment options such as liver resection, transplantation, and radio-frequency ablation are then precluded due to the poor prognosis of the disease. Advanced-stage HCC non-operative patients are often prescribed a vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor β (PDGFR-β) tyrosine kinase inhibitor, of which sorafenib is the prototypical drug. Sorafenib inhibits tumour angiogenesis and increases median survival on average three months longer than placebo, as reported in a phase 3, double-blind study[8].
A meta-analysis study revealed a median survival of less than one year for patients with HCC diagnosed at advanced stage. However, if diagnosed early, the five-year survival rate of HCC is estimated to be over 70%[9]. There is also a need for screening patients who undergo successful treatment/resection, as HCC disease recurrence and death occur in upto 70% and 50% respectively, after 5 years. Currently, transabdominal ultrasound is the recommended modality for surveillance of patients with cirrhosis[10], however this is of limited sensitivity for smaller lesions, where repeated US scanning (USS) and/or further imaging in the form of either CT or MRI may be required. On-going surveillance of high-risk individuals such as those who have cirrhosis or chronic viral hepatitis can help increase the survival rates of those diagnosed with HCC[7,11].
A biomarker, as defined by the World Health Organisation is: “Any substance, structure, or process that can be measured in the body or its products and influence or predict the incidence of outcome or disease[12]”. Biomarkers can be broadly categorized into four types, each of which have received attention in HCC: Diagnostic, prognostic, predisposing/risk and predictive[13]. Diagnostic biomarkers aim to detect, at an earlier stage, the presence of a disease or condition. Prognostic biomarkers are used to identify the likely outcome of disease progression or recurrence. Predisposition biomarkers are used to indicate the likelihood of developing a certain condition or disease, most commonly through identification of genetic mutations or subtypes. Lastly, predictive biomarkers aim to evaluate the likelihood of a specific medical intervention to have a favourable or unfavourable effect.
Alpha-fetoprotein: Alpha-fetoprotein (AFP), a glycoprotein and an oncofetal antigen has been the most widely used biomarker to aid in the diagnosis of HCC[14]. Aberrant production of AFP is observed in almost 50% of all HCCs[15]. However, this biomarker is not specific for HCC, and can also be upregulated during chronic liver disease, pregnancy, other malignancies of the gastrointestinal tract, as well as of the gonadal region[16]. A meta-analysis report found that AFP assays for HCC diagnosis had a pooled sensitivity of 51.9% and a specificity of 94%, yielding an area under the curve (AUC) value of 0.81[17], (Table 1). Unfortunately, it lacks specificity at low levels (20 ng/dL), and may fail to detect small HCCs. AFP levels of 400 ng/dL or greater in plasma are thought to be diagnostic of HCC and may indicate medical complications such as greater tumor size, portal vein thrombosis, and an overall lower median survival rate[18]. Due to these limitations, as well as being impacted by liver inflammation thereby hindering the test’s ability to predict a true positive or a true negative, active research to find a better alternative has yielded several potential alternatives.
| Marker | Comparison groups | Sensitivity | Specificity | AUC | Ref. |
| AFP | Meta-analysis | 51.9 | 94 | 0.81 | Xu et al[17] |
| AFP-L3 | Meta-analysis | 48.3 | 92.9 | 0.76 | Yi et al[21] |
| DCP | Meta-analysis | 71 | 84 | 0.89 | Zhu et al[25] |
| Glypican-3 | Meta-analysis | 55 | 58 | 0.78 | Xu et al[34] |
| AKR1B10 | HCC vs controls | 72.7 | 95.7 | 0.9 | Ye et al[36] |
| HMGB3 | HCC vs controls | 75.6 | 85.6 | 0.79 | Zheng et al[41] |
| GALAD | HCC vs controls | 91.6 | 89.7 | 0.97 | Berhane et al[49] |
AFP-L3: Aberrant glycosylation of proteins is a known hallmark of cancer[19]. Lens culinaris agglutinin-reactive AFP (AFP-L3) is the glycosylated isoform of AFP, which has been found to be more sensitive than the widely-used AFP biomarker, when AFP is elevated. The AFP-L3 value is reported as a percentage of the total AFP with a cut-off value of more than 10% commonly used[20]. Twelve articles included in a meta-analysis reported a sensitivity and a specificity of 48.3% and 92.9%, respectively and a summary receiver operating characteristic (SROC) of 0.7564[21].
AFP-L3 has also been found to be a prognostic biomarker after hepatic resection to determine the likelihood of recurrent HCC. Individuals with a low AFP-L3 level 15% after the second hepatic resection were found to have a 5-year survival rate of 91.7%. In contrast, individuals with an AFP-L3 level 15% had a 5-year survival rate of 23.8%[22].
Des-gamma carboxy-prothrombin: Des-gamma carboxy-prothrombin (DCP), also known as “protein induced by vitamin K absence or antagonist-II” (PIVKA-II), is an abnormal prothrombin protein which lack γ-carboxy residues[23]. A cross-sectional study was used to compare the commonly used clinical marker, AFP to DCP. Sensitivity and specificity values of 89% and 95%, respectively, have been reported with a DCP cut-off value of 125 mAU/mL[24]. A meta-analysis based on 20 publications reported sensitivity and specificity of 71% and 84%, respectively, with a SROC of 0.8930[25]. Elevated DCP levels have been associated with a high prevalence of portal vein invasion[26].
DCP has been found to be a predictive marker of response to treatment with the pharmaceutical tyrosine kinase inhibitor, gefitinib, which targets epidermal growth factor receptor (EGFR) and other proteins such as c-Met and hepatocyte growth factor (HGF)[22]. Gefitinib was found to induce apoptosis in HCC cells, however when treated in the presence of elevated DCP levels, an antagonizing effect was observed reducing the gefitinib-induced apoptosis of the tumorous hepatocytes[27], which occurred through its ability to upregulate EGFR, c-Met and HGF. High DCP has also been shown to be associated with tumor recurrence, metastases, and overall large tumour burden[28,29].
Golgi protein 73: Golgi protein 73 (GP73) is a type II Golgi transmembrane protein has received attention as a diagnostic marker for HCC. A study with 68 patients diagnosed with HCC were evaluated for GP73 levels 2 d prior to transcatheter arterial chemoembolization (TACE) and 7 and 30 d following the procedure. Using ELISA, the protein expression was observed to be markedly higher on average in patients with HCC compared to the controls, 152.5 µg/L vs 49.3 µg/L, respectively. Two days following TACE the levels decreased to 99.2 µg/L. After 30 days, the levels were 115.2 µg/L in those with a good response (CT evidence of good lipiodol retention and no active lesions) vs 183.2 µg/L where there was a poor response. In regards to the Barcelona clinic liver cancer stages, increasing GP73 concentrations were observed for progressively more advanced stages of the disease[30], BCLC stage A, 92.1 µg/L, stage B, 122.9 µg/L, and stage C, 162.6 µg/L.
Glypican-3: Glypican-3 (GPC3), a cell surface protein, with a 70 kDa core protein mass has been shown to display diagnostic and therapeutic utility for HCC[29,31]. GPC3 has been found to be highly expressed in HCC’s – a study which analyzed the GPC3 mRNA transcript, found 74.8% of HCC samples vs 3.2% non-tumor liver control samples expressed this[32]. A meta-analysis analyzing the prognostic abilities of GPC3 found overexpression to be an indicator of poor overall survival with a hazard-ratio (HR) of 2.18 (95%CI: 1.47-3.24), poor disease-free survival (HR = 2.05, 95%CI: 1.43-2.93), tumor vascular invasion with an odds-ratio (OR) of 2.74 (95%CI: 1.15-6.52), and hepatic cirrhosis (OR = 2.10, 95%CI: 1.31-3.36)[33].
Although GPC3 has proven to be a good prognostic biomarker, its diagnostic abilities on the other hand are below par. A meta-analysis study comparing the performance of AFP and GPC3 found the latter marker to be inferior than the commonly used marker, AFP. The pooled sensitivity and specificity of GPC3 was found to be 0.55 (95%CI: 0.52-0.58) and 0.58 (95%CI: 0.54-0.61), whereas AFP’s sensitivity and specificity was 0.54 (95%CI: 0.51-0.57) and 0.83 (95%CI: 0.80-0.85), respectively. The combination of GCP3 and AFP increased the tests’ sensitivity and specificity to 0.85 (95%CI: 0.81-0.89) and 0.79 (95%CI: 0.73-0.84), respectively[34].
Aldo-Keto Reductase family 1 member 10: The aldo-keto reductase family 1 member 10 (AKR1B10) has been linked as a potential biomarker indicating diagnostic and prognostic value[35]. AKR1B10, is a part of a family of NAD(P)H linked oxidoreductases; found on chromosome 7 (7p33), AKR1B10 is involved in the reduction of aldehyde to alcohol, converting retinal to retinol. A large multicenter study from three independent hospitals in China recruited a total of 1224 participants to validate the role of AKR1B10 in the diagnosis of HCC[36]. Serum levels of AKR1B10 were assessed in the cohort and found an AUC of 0.896 (95%CI: 0.867-0.921) a sensitivity of 72.7%, and a specificity of 95.7% with a diagnostic cutoff value of AKR1B10 at 267.9 pg/mL. Interestingly, although knockdown of AKRB1B10 has been found to decrease cell proliferation, invasiveness, and tumour growth, a high expression of AKR1B10 unexpectedly indicated better overall and disease-free survival[37]. Furthermore, it is worth noting that AKR1B10 expression is increased in the early-stage HCC[38].
High mobility group box 3: The high mobility group box 3 (HMGB3) is a part of the high mobility group (HMG) family of chromosomal proteins involved in chromatin replication, recombination, transcription, DNA repair and stability[39]. Downregulation of microRNA-200b, which is a direct target of HMGB3, occurs in HCC and increases the proliferation and migration of cells in HCC[40]. In a study of 225 patients, the serum HMGB3 levels were assessed at a cutoff value of > 2.0 ng/mL. The AUC for HMGB3 was found to be 0.791 (95%CI: 0.730–0.853) with a sensitivity of 75.6%, and a specificity of 81.6%. This was found to be slightly better than the clinical marker commonly used with an AUC of 0.743 (95%CI: 0.679-0.808), a sensitivity of 56.7%, and a specificity of 76.5% at a cut-off value of 20 ng/mL[41]. High HMGB3 expression was also correlated with poor overall-survival and disease-free survival.
Dickkopf 1: Serum levels of Dickkopf 1 (DKK1), which is a secretory antagonist of the Wnt pathway, have been investigated in HCC, cirrhosis, chronic hepatitis B and healthy controls. Good sensitivities and specificities were reported in both the test and validation cohorts, with DKK1 being reported as positive in early stage disease (< 2 cm tumors) as well as in AFP negative patients. Unfortunately, despite a correlation between DKK1 and tumor size there was none seen with BCLC stage[42].
SALL4: A promising biomarker SALL4, which like AFP is an oncofetal protein, has been correlated with outcomes in HCC, in separate cohorts of patients from Hong Kong and Singapore[43]. This marker appears to be associated with a progenitor, more aggressive form of HCC, and the findings have been confirmed independently[44]. Of possible therapeutic importance is SALL4’s property of recruiting the nucleosomal remodeling complex ((NuRD) thereby repressing tumor suppressors such as PTEN. This interaction has been exploited and an inhibitory peptide found with a target affinity of 23nM which has been demonstrated to have significant antitumor effects in xenograft mouse models (85% growth reduction)[45].
Phe-Trp and GCA: Using a modification of LC-MS, Luo et al[46] have reported that a biomarker panel comprising phenylalanyl-tryptophan (Phe-Trp) and glycocholate (GCA) performed well in distinguishing HCC from cirrhosis and healthy controls. In particular, the panel could detect AFP negative HCC as well as small HCC (S-HCC), defined as a solitary HCC nodule, or at most 2 nodules less than 3cm in diameter.
BALAD: The BALAD model was first introduced in 2006 by Toyoda et al[47] to aid in the staging of HCC using five serum markers: Bilirubin, Albumin, Lens culinaris agglutinin-reactive of alpha-fetoprotein, Alpha-fetoprotein, and Des-γ-carboxy prothrombin (BALAD). A multicenter study recruited 2600 HCC patients while excluding those on warfarin or Vitamin K, as these may alter the serum DCP levels[47]. Cutoff values of 400 ng/dL for AFP, 15% for AFP-L3, and 100 milli-arbitrary unit/mL for DCP were found to optimally predict patient survival. Although the system seems promising, further studies need to be conducted to validate this model.
The BALAD-2 model is refined from the previous BALAD model, which combines raw data with the previous Japanese cohort along with a newly added United Kingdom cohort[48]. The major difference between the BALAD and BALAD-2 model is the statistical analysis; the BALAD-2 model assumes the variables to be continuous rather than assuming a linear relationship. In addition, the original BALAD model divided cohorts into six classes (0-5), whereas the BALAD-2 model divided the cohort into four classes (1-4). A study by Berhane et al[49] aimed to validate the BALAD-2 model by collecting patient outcomes of 2,430 individuals diagnosed with HCC, 4404 individuals diagnosed with chronic liver disease, 229 individuals diagnosed with hepatobiliary tract cancer, and 92 healthy individuals. The Formula 1 is used to group individuals into four prognostic groups.
AFP and DCP were modelled as per 1000 units, measured in ng/mL where AFP was capped at 50000 units. Bilirubin and albumin were measured in mmol/L and g/LS, respectively. The 4 prognostic groups were based on score > 0.24 (risk 4, high), between 0.24 and > -0.91 (risk 3), from -0.91 to -1.74 (risk 2) and < -1.74 (risk 1, low).
GALAD: The GALAD model was established to aid in the diagnosis of HCC, first developed by Johnson et al[50] through a United Kingdom cohort. Similar to the BALAD model, the GALAD model uses three tumour serological markers: Lens culinaris agglutinin-reactive of alpha-fetoprotein, Alpha-fetoprotein, and Des-γ-carboxy prothrombin. However, the GALAD model replaces the two liver function tests with Gender and Age[50]. The GALAD score is calculated using the Formula 2.
Where gender is assigned an arbitrary score of 0 for females and 1 for males.
The aforementioned study by Berhane et al[49] found a GALAD score cut-off of -0.63 to yield an AUC of 0.97 (95%CI: 0.96–0.98) with a sensitivity and specificity of 91.6% and 89.7%, respectively in the United Kingdom cohort. The Japanese cohort was found to demonstrate optimal performance at a GALAD score cut-off of -1.95 with an AUC of 0.93 (95%CI: 0.92–0.94) and sensitivity and specificity of 81.4% and 89.1%, respectively[49].
This score has been used to investigate early HCC developing in patients with NASH in centres in Germany and Japan. In a case control study involving 125 patients with HCC and 231 patients with NASH from 8 centres in Germany, as well as 389 patients under surveillance in Japan, of whom 26 patients developed HCC, it was found that GALAD identified HCC patients with a significantly greater AUC than any of AFP, AFP-L3 or DCP[51].
Death receptor 4: Tumour necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis by binding to the TRAIL receptor 1 (TRAILR1) also known as the death receptor 4 (DR4)[52]. Genetic alteration of DR4 suggest a higher susceptibility to a number of cancers such as bladder, ovarian, and HCC[52-54]. A study conducted by Körner et al[52] examined SNPs at C626G (Thr209Arg, rs20575) and A638C (Glu228Ala, rs20576) in individuals affected with HBV and HCC (n = 56); HCV, cirrhosis, and HCC (n = 159); HCV, cirrhosis, without HCC (n = 75); HCV without cirrhosis and HCC (n = 159); HCV (n = 234); and healthy controls (n = 359). Overall the study found an increased risk of HCC in individuals that carried the 626C allele and the homozygous 638AA genotype mutant who were infected with HCV genotype 1 with an odds ratio of 1.975 (95%CI: 1.205-3.236) (P = 0.007). Another study, conducted in an Egyptian population examined the A1322G SNP of the DR4 gene and found an odds ratio of 2.34 (95%CI: 1.56-3.51) and 3.51 (95%CI: 2.33-5.28) for the AG genotype and GG genotype, respectively with an increased risk of individuals affect with HCV-related HCCs[55] (Table 2).
| Marker | Comparison groups | SNP | Genotype | OR | P value | Ref. |
| DR4 | HCC/HBV, HCC/HCV, HCV, controls | C626G/A638C | 626C + 638AA | 1.98 (1.20-3.24) | 0.007 | Körner et al[52] |
| DR4 | HCC/HCV, HCV, Controls | A1322G | AG | 2.34 (1.56-3.51) | Zayed et al[55] | |
| GG | 3.51 (2.33-5.28) | 0.001 | ||||
| TLL1 | HCV-treated | rs17047200 | 2.37 (1.74-3.23) | 2.66 × 10-8 | Matsuura et al[64] | |
| MHC class 1 | HCC/HCV, Controls | Rs2596542G>A | 1.39 (1.27-1.52) | 4.21 × 10-13 | Kumar et al[66] | |
| (MICA) | (Japanese cohort) | |||||
| MHC class II | HCC-HCV, HCV | DQB1*03:01 (HCV-Geno 1) | 0.65 (0.48-0.89) | 0.007 | Lee et al[70] | |
| DQB1*06:02 | 3.03 (1.18-7.74) | 0.02 | ||||
| (HCV-non-Geno1) | ||||||
Kinesin family member 1B: Kinesin Family member 1B (KIF1B), part of the kinesin superfamily, is involved in axon myelination, growth, and transport of organelles, proteins, and RNAs to specific locations in the cell[56]. With two alternative splice isoforms, KIF1Bα and KIF1Bβ, KIF1B is found on chromosome 1 (1p36.22)[57]. KIF1Bβ has been found to function as a haplo-insufficient tumour-suppressor gene inducing apoptosis, independent from p53[58]. The downregulation of KIF1B mRNA has been shown to correlate with poor prognosis of HCC in different clinicopathologic situations such as vascular invasion, recurrence, and overall-survival[59]. A genome-wide association (GWAS) study by Zhang et al[57] identified an intronic SNP – rs17401966 – in the KIF1B gene. Samples were collected from 1962 individuals with 1430 HBV-related HCC cases and 159 family trios of Chinese ancestry. The study identified this polymorphism has a protective effect on HCC, decreasing the likelihood of developing HCC with an odds-ratio of 0.61 (95%CI: 0.55–0.67). However, conflicting studies examining the KLF1B polymorphism in individuals derived from Saudi Arabian, Japanese, and Thai populations found no significant associations[60-62]. A meta-analysis study of the KLF1B polymorphism determined that the polymorphism decreases the risk of HCC for Chinese populations[57].
Although HCV may be eradicated after sustained virologic response (SVR) is achieved, a likelihood of developing HCC exists in cases with more advanced fibrosis. A study by Nagata et al[63] examined the risk of developing HCC after SVR using interferon-therapy and interferon-free therapy and found the probability of novel HCC development to be 2.5% and 1.1%, respectively.
Tolloid-like 1: A GWAS identified a SNP variant in the gene tolloid-like 1 (TLL1) found on chromosome 4 within the intronic region (rs17047200)[64]. TLL1 was originally found to play a role in the formation of the interventricular septum of the heart and is now a marker of interest in HCC[65]. The study recruited 457 patients in Japan who underwent SVR through interferon-based treatments and found an odds-ratio of 2.37 (95%CI: 1.74-3.23; P = 2.66 × 10−8)[64].
Major histocompatibility complex class 1: A study by Kumar et al[66] in 2011 identified a susceptibility locus for individuals who developed HCV-related HCC through a GWAS conducted in a Japanese cohort. DNA was genotyped in 721 individuals with HCV-related HCC and 2890 HCV-negative controls which identified eight SNPs (P < 1 × 10−5). In the replication stage, 673 cases of HCC and 2596 HCV-negative controls were genotyped at the eight SNPs and identified a polymorphism found on the 5’ flanking region of the major histocompatibility complex (MHC) class I polypeptide-related sequence A gene (MICA) on chromosome 6 (6p21.33). The polymorphism, rs2596542G>A, with an OR of 1.39 (95%CI: 1.27–1.52) (P = 4.21 × 10−13), was found to be associated with a susceptibility to develop HCC from chronic hepatitis-C. It is worth noting that although the polymorphism is associated with developing HCC from chronic hepatitis C, it is not associated with a susceptibility to develop chronic hepatitis C. The polymorphism was also associated with a decrease in expression of the MICA protein in the HCV-related HCC cohort. Conversely, a study by Lange et al[67] conducted in Switzerland found the rs2596542G>A to have a protective effect on HCC in patients with HCV throughout the Caucasian population (OR = 0.52, 95%CI: 0.26-1.08). A meta-analysis study analyzing a total of 11 articles with 4528 HCC cases and 16625 controls, found the A/A allele to increase the risk among Asian and African population and an increased risk of HCC in the G/G allele among the Caucasian populations[68]. Increased risk of HCC with individuals who carry the polymorphism was not found in HBV-related HCC’s.
MHC class II: The human leukocyte antigen (HLA) is the human MHC, which is found on chromosome 6 (6p21) and is divided into three classes, class I, class, II, and class III. Class II contains the DQ gene family made up of the α and β chains, DQA1 and DQB1, respectively[69]. A GWAS genotyped 502 HCC patients and 749 controls identifying a SNP present in the HLA gene, specifically the HLA-DQB1 gene. An additional 994 HCV seropositive participants were genotyped, specifically in the HLA-DQB1 gene and found that DQB1*03:01 had protective effects for individuals with HCV genotype 1 with an odds-ratio of 0.43 (95%CI: 0.23-0.81) (P=0.0095). However, a DQB1*06:02 indicated a risk of developing HCC for non-genotype 1 HCV patients with an odds-ratio of 3.03 (95%CI: 1.18-7.74) (P = 0.0208)[70].
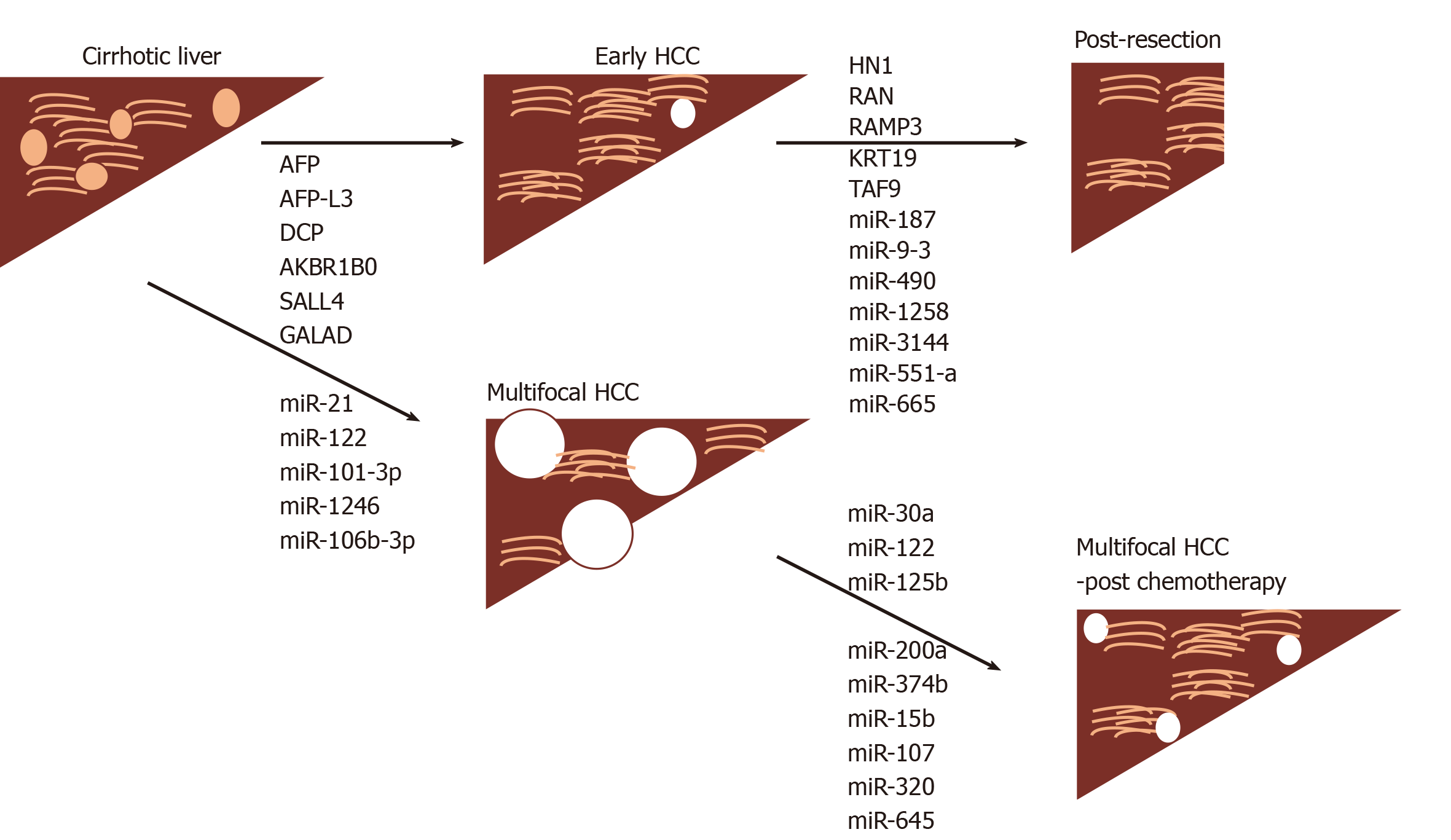
For postoperative HCC, Nault et al[71] have described a 5-gene signature consisting of HN1, RAN, RAMP3, KRT19, and TAF9 (of which 4 were upregulated in tumor) that had prognostic ability for postoperative tumor recurrence and survival. The effect was apparent whether a Western cohort of hepatitis C- or an Eastern cohort of hepatitis B-related HCC were analyzed.
The human genome encodes for many more RNA molecules than proteins, which are known as non-coding RNAs (ncRNA). These comprise both short molecules between 20 to 30 nucleotides long, known as mi-RNA, si-RNA and pi-RNA, together with long non-coding RNA (lncRNA) which are greater than 200 nucleotides. Both types have been investigated in HCC and are discussed below. This section will also briefly describe circRNAs that are formed through the back-splicing of the 3’ and 5’ ends to form a loop that can sequester miRNAs and proteins to affect gene expression.
Mi-RNAs in diagnosis: Micro-RNAs are usually 21-23 nucleotides in length and function via the RNA-induced silencing complex (RISC) to regulate gene expression through mRNA degradation, or alternatively by translational repression. They are recognized to be highly useful tools in the diagnosis and prognosis, as well as serving as therapeutic targets for diseases[72]. Aberrant production or alterations of mi-RNAs have been associated with a number of pathologies including cancer, diabetes and cardiovascular diseases[73].
Initial studies utilized different methodologies to investigate miRNA expression in hepatitis B, and HCC, with findings in one study[74] where miR-25, miR-375 and let7f could significantly separate HCC from controls (AUC = 0.997), were not reproducible in another[75]. The seven miRNAs discovered in the latter study, miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a and miR-801 were shown to distinguish between HCC and all of healthy controls, hepatitis B and cirrhosis. Moreover, the changes persisted across the range of BCLC stages O, A, B and C. Of interest, 4 of the identified miRNAs (26a, 223, 21 and 122) had also been previously reported as capable of achieving this. Notably, 2 of these, miR-21 and miR-122, have been underscored as useful biomarkers in a subsequent meta-analysis involving 50 studies that included 3423 cases of HCC, 2403 cases of chronic hepatitis and 1887 healthy controls[76]. The pooled analyses indicated that they were slightly better at distinguishing HCC from controls than those with chronic hepatitis. Of these two molecules miR-122 appeared to be particularly compelling since mice with a genetic deletion of this in the liver were found to be prone to the development of NASH, fibrosis and HCC with expression of oncofetal molecules such as AFP and IGF2[77,78].
Moshiri et al[79] have used RNA sequencing of plasma to approach the problem differently. After identification of 38 differentially expressed miRNAs with at least a 3-fold change between HCC and cirrhosis and/or controls, 9 were chosen for further validation steps by droplet digital PCR (ddPCR) technology. Of these, miR-101-3p, miR-1246, miR-106b-3p, miR411-5p were evaluated in independent cohorts. Whether they analyzed plasma or serum, diagnostic accuracies of well over AUC > 0.90 were achieved for miR-101-3p, miR-1246 and miR-106b-3p, individually or in combination.
Role in treatment of HCC: Previous work with sorafenib did not identify any biomarkers that were predictive of treatment response in patients with HCC, however, baseline levels of angiopoietin 2 and VEGF were independent predictors of survival[80]. A more recent study has shown an inverse correlation between levels of miR-221 and sorafenib resistance in animal models and a small cohort of patients[81]. Bruix et al[82] and Teufel et al[83] investigated tissue and baseline plasma samples in patients involved in a trial to investigate the response to regorafenib, which is another multikinase inhibitor. Levels of miR-30a, miR-122, mir-125b, miR-200a, miR-374b, miR-15b, miR-107, miR-320 and miR-645 were all associated with survival time with regorafenib treatment. The study also found survival time associations with reduced baseline levels of Ang1, cystatin B, LAP-TGFb1, Lox-1, MIP1a, after treatment.
The objective of using screening in a cirrhotic population is to detect HCC early, however, in an analogous situation, what happens to patients that undergo HCC resection and is there a way of then predicting recurrence? Mi-RNAs may be useful in this scenario as shown by Fu et al[84]. By analyzing data on 318 patients from The Cancer Genome Atlas (TGCA) they uncovered a 7-miRNA signature correlated with 5year survival which comprised miR-187, miR-9-3, miR-490, miR-1258, miR-3144, miR-551-a and miR-665. These findings will require replication in larger prospective cohorts to validate them further.
LncRNAs in HCC: In a search for novel genes in HCC, Panzitt et al[85] discovered the lncRNA HULC (highly upregulated in liver cancer). This has been investigated in HCC and found to be associated with clinical stage and intrahepatic metastasis[86]. Other lncRNAs found to be overexpressed in HCC and associated with a poor prognosis include ZEB1-AS1 and DANCR which function to repress cadherin expression and CTNNB1 degradation respectively[87,88]. Conversely, other lncRNAs exhibit reduced expression in HCC and affect tumor progression by EMT[89].
CircRNAs in HCC: With respect to CircRNAs, several have been shown to affect key aspects of tumor biology. As an example, CircMAT2B has been implicated in altering tumor metabolism under hypoxia; it does so by sequestering miR-338-3p which leads to increased PKM which is involved in glycolysis[90]. Another is CircASAP1 which affects miR-326 and miR-532-5p thereby enhancing MAPK (mitogen-activated protein kinase) signaling and TAM infiltration. This has been shown to be associated with a poor prognosis[91].
A ‘liquid biopsy’, performed through a blood collection, may be the simplest means whereby physicians can collect information from patients in a minimally invasive manner[92]. It may become an alternative to the time-consuming surgical biopsies which place patients at risk of developing complications. The risk of needle-track tumour seeding is another disadvantage of surgical biopsies, which precludes their routine use to evaluate suspected liver cancer. Moreover, serial collections would be feasible, allowing evaluation of tumour progression in real time[93]. A liquid biopsy analyzes a range of molecular data such as circulating tumor cells (CTCs), cell-free circulating tumor DNA (ctDNA), and exosomes released from necrotic tumor cells, thus providing insight into tumour behaviour.
Circulating tumor cells in HCC: Circulating tumor cells are extremely rare, estimated to be as low as 10 cells in 10 mL of blood, making them difficult to detect[94]. However, CTCs can provide a wealth of information on multiple DNA abnormalities, gene fusion transcripts, and RNA expression of the cancerous cells when isolated. Flow cytometry is commonly used to search for CTCs through fluorescently labelled cellular tags, many which target stem cell markers such as: Epithelial cell adhesion molecule (EpCAMs), CD133, CD90, CD44, CD13, and cytokeratin 19[95,96]. A study by Sun et al[97] demonstrated the clinical significance of CTCs in 123 HCC patients by analyzing EpCAM, which showed a high probability of tumour recurrence in individuals with ≥ 2 CTCs in 7.5 mL of blood. Recurrence was found in 26 of 51 patients with ≥ 2 CTCs, whereas only 15 of 72 patients with < 2 CTCs showed recurrence after curative resections. The mean follow-up time was reported to be 15.1 ± 2.3 mo[97]. Perioperative analysis of CTCs for individuals with HCC may provide insight on prognosis and can tailor clinical treatment decisions.
Circulating tumor DNA: Cell-free circulating tumor DNA (ctDNA) is typically 180-200 basepairs (bp) in length (approximately the size of mononucleosomal unit), released when tumour cells are phagocytosed or undergo apoptosis. The difficulty of analyzing ctDNA is due to its low concentration in the blood. Moreover, cell-free DNA (cfDNA) is released by normal cells further decreasing ctDNA concentrations. Healthy subjects are found to have a peripheral cfDNA concentration of 10 ng/mL to around 100 ng/mL, with a half-life between 16 min and 2.5 h[95]. Quantitative analysis from several studies have revealed the mean concentration of cfDNA to be 3-4 times elevated in HCC patients as compared with chronic hepatitis patients, and almost 20 times higher compared to healthy controls[98]. Clinico-pathological parameters such as tumor grade, size of tumor, shorter overall survival, and metastatic ability have been found to correlate with elevated cfDNA levels[98]. Aberrant epigenetic alterations, through DNA methylation, have been found to be one of the universal hallmarks of cancer which is being investigated in ctDNAs[99]. A study by Chan et al[100] analyzed the hypermethylation of the RASSF1A [Ras association (RalGDS/AF-6) domain family member 1A] gene, observed in 93% of HCC patents vs 58% of HBV patients, and 8% of the healthy controls. Interestingly, with a cut off value of 1 × 106 copies/L of the hypermethylated RASSF1A, 50% of AFP-negative HCCs are identified. This may indicate a role for its use in promising combinatorial techniques to help in the diagnosis of HCC. Other features of elevated RASSF1A concentrations may predict poor disease-free survival.
The basic aims of HCC biomarker research are to find novel molecules, as well as optimizing use of existing ones, to be able to diagnose the disease earlier in at risk populations, and furthermore, to be able to predict disease outcome in response to treatment, as well as provide prognostic information (Figure 2). At the same time the process needs to safeguard patients against unnecessary testing and follow-up where there are abnormal biomarker findings but no defined algorithms for further management, given that only a fraction of the at-risk population will eventually develop the disease[101]. A systematic review has indicated that USS-based screening is indeed capable of improving mortality associated with HCC[102]. Conversely, recent work has questioned the validity of any screening for patients with cirrhosis, in a matched case control study of the VA health care system, using either USS, AFP or both, where no difference was found for HCC-related mortality[103]. The finding of significant heterogeneity in approaches to HCC management in 18031 patients from 14 countries, together with distinct demographics and outcomes, indicates a need for earlier diagnosis[104].
Serum biomarkers such as AFP allow for a minimally-invasive and rapid evaluation of at risk patients. However, there are no recommendations for its regular use outside Japan, where in conjunction with USS, and AFP-L3 and DCP, it is used every 6 months. A recent meta-analysis utilizing 32 studies and 13367 patients showed that US, with AFP vs without, exhibited greater sensitivity at detecting early stage HCC (63%, 95%CI: 48-75% vs 45%, 95%CI: 30%-62%, P = 0.002). However, US alone was more specific (RR = 1.08; 95%CI: 1.05-1.09) and detected any stage HCC with a sensitivity of 84%[105].
Some markers of more aggressive HCC may evolve into developing management algorithms and SALL4 may be one such example. This may also offer some insight into disease management, as demonstrated with the aid of molecular reconstruction and subsequent investigation of interfering with its interaction with NuRD[45]. Similarly, in evaluating markers indicative of advanced HCC it was uncovered that Ang2 and Vegf independently marked cases with more rapidly progressive disease but had no bearing on sorafenib response[80]. In comparable work conducted with regorafenib, survival changes correlated with levels of several miRNAs including miR-30a, miR-122 and miR-200a.
As discussed in this article, there are a number of other serological markers such as GP73, GPC3, AKR1B10, which appear promising but all require further validation. It may be that there is no advantage for any of these over AFP alone, as demonstrated in a meta-analysis for another interesting biomarker, osteopontin[106]. Alternatively, they may require to be combined with other markers such as albumin, to improve their performance.
Micro-RNAs can be used as diagnostic or prognostic tools and may also serve as therapeutic targets for HCC. Unfortunately, despite the wealth of data generated in this area, this approach has failed to show consistency for the molecules assessed. Micro-RNAs appear to exhibit variability according to whether measured in plasma or serum hence in part explaining the discrepancies observed in numerous earlier studies. It appears that mi-RNAs are found in higher concentrations in plasma vs serum, with platelet mediated degradation during the clotting process speculated to be a possible explanation for this.
Of those that were initially found to be promising, specifically miR-21 and miR-122, these were not reported to be differentially expressed in an analysis utilizing RNA sequencing. The work by Moshiri et al[79] has shown that some additional mi-RNAs that may have potential greater accuracy. However, given the lack of overall reproducibility of findings so far in this field, these observations remain preliminary and will require follow-up. The same conclusion may apply to the analysis of CTCs in HCC. In one study 95% of 195 HCC patients demonstrated a correlation between hybrid and mesenchymal CTCs (EpCAM/Twist/Snail) with BCLC stage, AFP, recurrence and metastasis[107]. However, a retrospective study that analyzed 113 HCC patients before curative treatment and 143 HCC patients after curative treatment, found no correlation of total CTCs or the EMT phenotype with AFP, BCLC stage, tumor size or vascular invasion[108].
From a practical perspective, perhaps the composite scoring systems GALAD and BALAD-2 currently exhibit the most favorable diagnostic accuracy over conventional methods. The findings have been replicated in cohorts of patients in both Europe and Japan, where the etiology of HCC differs. Further prospective analysis in North America may help to establish this approach as shown by the recent work for GALAD in NASH related HCC[51]. A retrospective analysis has also indicated its superiority over USS in detecting HCC including patients with negative AFP, and its performance remained excellent for early stage HCC[109].
In conclusion, despite the plethora of studies so far, and the promise of different classes of biomarkers, there appears to be no specific one that currently fulfils the need to pick up early HCC (BCLC stage O/A) with any advance over USS with or without AFP. Similarly, patients undergoing curative resection or chemo-/immuno-therapy may benefit from comparable analyses. Future prospective multicentre trials are required in defined at risk populations for HCC to assess the various classes of agents discussed.
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Gastroenterological Association; American College of Gastroenterology; Canadian Association of Gastroenterology; and Royal College of Physicians United Kingdom.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Cheng J, Wang YG, Yang L S-Editor: Ma YJ L-Editor: A P-Editor: Li JH
| 1. | Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R, Wolfe C, Hamadeh RR, Moore A, Werdecker A, Gessner BD, Te Ao B, McMahon B, Karimkhani C, Yu C, Cooke GS, Schwebel DC, Carpenter DO, Pereira DM, Nash D, Kazi DS, De Leo D, Plass D, Ukwaja KN, Thurston GD, Yun Jin K, Simard EP, Mills E, Park EK, Catalá-López F, deVeber G, Gotay C, Khan G, Hosgood HD 3rd, Santos IS, Leasher JL, Singh J, Leigh J, Jonas JB, Sanabria J, Beardsley J, Jacobsen KH, Takahashi K, Franklin RC, Ronfani L, Montico M, Naldi L, Tonelli M, Geleijnse J, Petzold M, Shrime MG, Younis M, Yonemoto N, Breitborde N, Yip P, Pourmalek F, Lotufo PA, Esteghamati A, Hankey GJ, Ali R, Lunevicius R, Malekzadeh R, Dellavalle R, Weintraub R, Lucas R, Hay R, Rojas-Rueda D, Westerman R, Sepanlou SG, Nolte S, Patten S, Weichenthal S, Abera SF, Fereshtehnejad SM, Shiue I, Driscoll T, Vasankari T, Alsharif U, Rahimi-Movaghar V, Vlassov VV, Marcenes WS, Mekonnen W, Melaku YA, Yano Y, Artaman A, Campos I, MacLachlan J, Mueller U, Kim D, Trillini M, Eshrati B, Williams HC, Shibuya K, Dandona R, Murthy K, Cowie B, Amare AT, Antonio CA, Castañeda-Orjuela C, van Gool CH, Violante F, Oh IH, Deribe K, Soreide K, Knibbs L, Kereselidze M, Green M, Cardenas R, Roy N, Tillmann T, Li Y, Krueger H, Monasta L, Dey S, Sheikhbahaei S, Hafezi-Nejad N, Kumar GA, Sreeramareddy CT, Dandona L, Wang H, Vollset SE, Mokdad A, Salomon JA, Lozano R, Vos T, Forouzanfar M, Lopez A, Murray C, Naghavi M. The Global Burden of Cancer 2013. JAMA Oncol. 2015;1:505-527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1729] [Cited by in F6Publishing: 1954] [Article Influence: 244.3] [Reference Citation Analysis (0)] |
| 2. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3846] [Cited by in F6Publishing: 4102] [Article Influence: 241.3] [Reference Citation Analysis (2)] |
| 3. | Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2-S6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 770] [Cited by in F6Publishing: 832] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 4. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2183] [Cited by in F6Publishing: 2337] [Article Influence: 194.8] [Reference Citation Analysis (0)] |
| 5. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1006] [Cited by in F6Publishing: 1175] [Article Influence: 130.6] [Reference Citation Analysis (0)] |
| 6. | Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450-1462. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2066] [Cited by in F6Publishing: 2565] [Article Influence: 513.0] [Reference Citation Analysis (0)] |
| 7. | Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301-1314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2800] [Cited by in F6Publishing: 3379] [Article Influence: 563.2] [Reference Citation Analysis (3)] |
| 8. | Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Häussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J; SHARP Investigators Study Group. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378-390. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9016] [Cited by in F6Publishing: 9507] [Article Influence: 594.2] [Reference Citation Analysis (1)] |
| 9. | Llovet JM, Fuster J, Bruix J. Intention-to-treat analysis of surgical treatment for early hepatocellular carcinoma: resection versus transplantation. Hepatology. 1999;30:1434-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1331] [Cited by in F6Publishing: 1393] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 10. | Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723-750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2121] [Cited by in F6Publishing: 2675] [Article Influence: 445.8] [Reference Citation Analysis (1)] |
| 11. | Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol. 2004;130:417-422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 751] [Cited by in F6Publishing: 848] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 12. | Strimbu K, Tavel JA. What are biomarkers? Curr Opin HIV AIDS. 2010;5:463-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1112] [Cited by in F6Publishing: 1037] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 13. | Carlomagno N, Incollingo P, Tammaro V, Peluso G, Rupealta N, Chiacchio G, Sandoval Sotelo ML, Minieri G, Pisani A, Riccio E, Sabbatini M, Bracale UM, Calogero A, Dodaro CA, Santangelo M. Diagnostic, Predictive, Prognostic, and Therapeutic Molecular Biomarkers in Third Millennium: A Breakthrough in Gastric Cancer. Biomed Res Int. 2017;2017:7869802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 14. | Attwa MH, El-Etreby SA. Guide for diagnosis and treatment of hepatocellular carcinoma. World J Hepatol. 2015;7:1632-1651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 154] [Article Influence: 17.1] [Reference Citation Analysis (2)] |
| 15. | Bai DS, Zhang C, Chen P, Jin SJ, Jiang GQ. The prognostic correlation of AFP level at diagnosis with pathological grade, progression, and survival of patients with hepatocellular carcinoma. Sci Rep. 2017;7:12870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 130] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 16. | Meany DL, Sokoll LJ, Chan DW. Early Detection of Cancer: Immunoassays for Plasma Tumor Markers. Expert Opin Med Diagn. 2009;3:597-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 17. | Xu C, Yan Z, Zhou L, Wang Y. A comparison of glypican-3 with alpha-fetoprotein as a serum marker for hepatocellular carcinoma: a meta-analysis. J Cancer Res Clin Oncol. 2013;139:1417-1424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol. 2006;12:1175-1181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 222] [Cited by in F6Publishing: 225] [Article Influence: 12.5] [Reference Citation Analysis (2)] |
| 19. | Kirwan A, Utratna M, O'Dwyer ME, Joshi L, Kilcoyne M. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. Biomed Res Int. 2015;2015:490531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 137] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 20. | Huang TS, Shyu YC, Turner R, Chen HY, Chen PJ. Diagnostic performance of alpha-fetoprotein, lens culinaris agglutinin-reactive alpha-fetoprotein, des-gamma carboxyprothrombin, and glypican-3 for the detection of hepatocellular carcinoma: a systematic review and meta-analysis protocol. Syst Rev. 2013;2:37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Yi X, Yu S, Bao Y. Alpha-fetoprotein-L3 in hepatocellular carcinoma: a meta-analysis. Clin Chim Acta. 2013;425:212-220. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Matsuda M, Asakawa M, Amemiya H, Fujii H. Lens culinaris agglutinin-reactive fraction of AFP is a useful prognostic biomarker for survival after repeat hepatic resection for HCC. J Gastroenterol Hepatol. 2011;26:731-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 23. | Sumi A, Akiba J, Ogasawara S, Nakayama M, Nomura Y, Yasumoto M, Sanada S, Nakashima O, Abe T, Yano H. Des-γ-carboxyprothrombin (DCP) and NX-DCP expressions and their relationship with clinicopathological features in hepatocellular carcinoma. PLoS One. 2015;10:e0118452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 24. | Marrero JA, Su GL, Wei W, Emick D, Conjeevaram HS, Fontana RJ, Lok AS. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology. 2003;37:1114-1121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 304] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 25. | Zhu R, Yang J, Xu L, Dai W, Wang F, Shen M, Zhang Y, Zhang H, Chen K, Cheng P, Wang C, Zheng Y, Li J, Lu J, Zhou Y, Wu D, Guo C. Diagnostic Performance of Des-γ-carboxy Prothrombin for Hepatocellular Carcinoma: A Meta-Analysis. Gastroenterol Res Pract. 2014;2014:529314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Koike Y, Shiratori Y, Sato S, Obi S, Teratani T, Imamura M, Yoshida H, Shiina S, Omata M. Des-gamma-carboxy prothrombin as a useful predisposing factor for the development of portal venous invasion in patients with hepatocellular carcinoma: a prospective analysis of 227 patients. Cancer. 2001;91:561-569. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 6] [Reference Citation Analysis (0)] |
| 27. | Cui SX, Zhang YS, Chu JH, Song ZY, Qu XJ. Des-gamma-carboxy prothrombin (DCP) antagonizes the effects of gefitinib on human hepatocellular carcinoma cells. Cell Physiol Biochem. 2015;35:201-212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Shimada M, Yamashita Y, Hamatsu T, Hasegawa H, Utsunomiya T, Aishima S, Sugimachi K. The role of des-gamma-carboxy prothrombin levels in hepatocellular carcinoma and liver tissues. Cancer Lett. 2000;159:87-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 29. | Song P, Gao J, Inagaki Y, Kokudo N, Hasegawa K, Sugawara Y, Tang W. Biomarkers: evaluation of screening for and early diagnosis of hepatocellular carcinoma in Japan and china. Liver Cancer. 2013;2:31-39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 78] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 30. | Ai N, Liu W, Li ZG, Ji H, Li B, Yang G. High expression of GP73 in primary hepatocellular carcinoma and its function in the assessment of transcatheter arterial chemoembolization. Oncol Lett. 2017;14:3953-3958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 31. | Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125:89-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 644] [Cited by in F6Publishing: 629] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 32. | Hsu HC, Cheng W, Lai PL. Cloning and expression of a developmentally regulated transcript MXR7 in hepatocellular carcinoma: biological significance and temporospatial distribution. Cancer Res. 1997;57:5179-5184. [PubMed] [Cited in This Article: ] |
| 33. | Li J, Gao JZ, Du JL, Wei LX. Prognostic and clinicopathological significance of glypican-3 overexpression in hepatocellular carcinoma: a meta-analysis. World J Gastroenterol. 2014;20:6336-6344. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 25] [Cited by in F6Publishing: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 34. | Xu D, Su C, Sun L, Gao Y, Li Y. Performance of Serum Glypican 3 in Diagnosis of Hepatocellular Carcinoma: A meta-analysis. Ann Hepatol. 2019;18:58-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | DiStefano JK, Davis B. Diagnostic and Prognostic Potential of AKR1B10 in Human Hepatocellular Carcinoma. Cancers (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 36. | Ye X, Li C, Zu X, Lin M, Liu Q, Liu J, Xu G, Chen Z, Xu Y, Liu L, Luo D, Cao Z, Shi G, Feng Z, Deng H, Liao Q, Cai C, Liao DF, Wang J, Jin J, Cao D. A Large-Scale Multicenter Study Validates Aldo-Keto Reductase Family 1 Member B10 as a Prevalent Serum Marker for Detection of Hepatocellular Carcinoma. Hepatology. 2019;69:2489-2501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 37. | Liu TA, Jan YJ, Ko BS, Wu YJ, Lu YJ, Liang SM, Liu CC, Chen SC, Wang J, Shyue SK, Liou JY. Regulation of aldo-keto-reductase family 1 B10 by 14-3-3ε and their prognostic impact of hepatocellular carcinoma. Oncotarget. 2015;6:38967-38982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Han C, Gao L, Zhao L, Sheng Q, Zhang C, An Z, Xia T, Ding Y, Wang J, Bai H, Dou X. Immunohistochemistry Detects Increased Expression of Aldo-Keto Reductase Family 1 Member B10 (AKR1B10) in Early-Stage Hepatocellular Carcinoma. Med Sci Monit. 2018;24:7414-7423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 39. | Stros M. HMGB proteins: interactions with DNA and chromatin. Biochim Biophys Acta. 2010;1799:101-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 421] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 40. | Wang LK, Xie XN, Song XH, Su T, Chang XL, Xu M, Liang B, Huang DY. Upregulation of miR-200b Inhibits Hepatocellular Carcinoma Cell Proliferation and Migration by Targeting HMGB3 Protein. Technol Cancer Res Treat. 2018;17:1533033818806475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 41. | Zheng W, Yang J, Dong Z, Wang L, Fang M, Wu W, Yao D, Yao M. High mobility group box 3 as an emerging biomarker in diagnosis and prognosis of hepatocellular carcinoma. Cancer Manag Res. 2018;10:5979-5989. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Shen Q, Fan J, Yang XR, Tan Y, Zhao W, Xu Y, Wang N, Niu Y, Wu Z, Zhou J, Qiu SJ, Shi YH, Yu B, Tang N, Chu W, Wang M, Wu J, Zhang Z, Yang S, Gu J, Wang H, Qin W. Serum DKK1 as a protein biomarker for the diagnosis of hepatocellular carcinoma: a large-scale, multicentre study. Lancet Oncol. 2012;13:817-826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 289] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 43. | Yong KJ, Gao C, Lim JS, Yan B, Yang H, Dimitrov T, Kawasaki A, Ong CW, Wong KF, Lee S, Ravikumar S, Srivastava S, Tian X, Poon RT, Fan ST, Luk JM, Dan YY, Salto-Tellez M, Chai L, Tenen DG. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266-2276. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 183] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 44. | Yin F, Han X, Yao SK, Wang XL, Yang HC. Importance of SALL4 in the development and prognosis of hepatocellular carcinoma. World J Gastroenterol. 2016;22:2837-2843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 21] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 45. | Liu BH, Jobichen C, Chia CSB, Chan THM, Tang JP, Chung TXY, Li J, Poulsen A, Hung AW, Koh-Stenta X, Tan YS, Verma CS, Tan HK, Wu CS, Li F, Hill J, Joy J, Yang H, Chai L, Sivaraman J, Tenen DG. Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide. Proc Natl Acad Sci USA. 2018;115:E7119-E7128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 39] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 46. | Luo P, Yin P, Hua R, Tan Y, Li Z, Qiu G, Yin Z, Xie X, Wang X, Chen W, Zhou L, Wang X, Li Y, Chen H, Gao L, Lu X, Wu T, Wang H, Niu J, Xu G. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 233] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 47. | Toyoda H, Kumada T, Osaki Y, Oka H, Urano F, Kudo M, Matsunaga T. Staging hepatocellular carcinoma by a novel scoring system (BALAD score) based on serum markers. Clin Gastroenterol Hepatol. 2006;4:1528-1536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 48. | Johnson PJ. The BALAD-2 and GALAD biomarker models for hepatocellular carcinoma. Clin Adv Hematol Oncol. 2017;15:452-454. [PubMed] [Cited in This Article: ] |
| 49. | Berhane S, Toyoda H, Tada T, Kumada T, Kagebayashi C, Satomura S, Schweitzer N, Vogel A, Manns MP, Benckert J, Berg T, Ebker M, Best J, Dechêne A, Gerken G, Schlaak JF, Weinmann A, Wörns MA, Galle P, Yeo W, Mo F, Chan SL, Reeves H, Cox T, Johnson P. Role of the GALAD and BALAD-2 Serologic Models in Diagnosis of Hepatocellular Carcinoma and Prediction of Survival in Patients. Clin Gastroenterol Hepatol. 2016;14:875-886.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 170] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 50. | Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, Morse J, Hull D, Patman G, Kagebayashi C, Hussain S, Graham J, Reeves H, Satomura S. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomarkers Prev. 2014;23:144-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 171] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 51. | Best J, Bechmann LP, Sowa JP, Sydor S, Dechêne A, Pflanz K, Bedreli S, Schotten C, Geier A, Berg T, Fischer J, Vogel A, Bantel H, Weinmann A, Schattenberg JM, Huber Y, Wege H, von Felden J, Schulze K, Bettinger D, Thimme R, Sinner F, Schütte K, Weiss KH, Toyoda H, Yasuda S, Kumada T, Berhane S, Wichert M, Heider D, Gerken G, Johnson P, Canbay A. GALAD Score Detects Early Hepatocellular Carcinoma in an International Cohort of Patients With Nonalcoholic Steatohepatitis. Clin Gastroenterol Hepatol. 2020;18:728-735.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 137] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 52. | Körner C, Riesner K, Krämer B, Eisenhardt M, Glässner A, Wolter F, Berg T, Müller T, Sauerbruch T, Nattermann J, Spengler U, Nischalke HD. TRAIL receptor I (DR4) polymorphisms C626G and A683C are associated with an increased risk for hepatocellular carcinoma (HCC) in HCV-infected patients. BMC Cancer. 2012;12:85. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Hazra A, Chamberlain RM, Grossman HB, Zhu Y, Spitz MR, Wu X. Death receptor 4 and bladder cancer risk. Cancer Res. 2003;63:1157-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 54. | Dick MG, Versmold B, Engel C, Meindl A, Arnold N, Varon-Mateeva R, Sutter C, Niederacher D, Deissler H, Preisler-Adams S, Kast K, Schäfer D, Gadzicki D, Heinritz W, Wappenschmidt B, Schmutzler RK. Association of death receptor 4 variant (683A > C) with ovarian cancer risk in BRCA1 mutation carriers. Int J Cancer. 2012;130:1314-1318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 55. | Zayed SA, Zahran NM, Khorshied MM, Abdel-Aziz AO, Mahmoud O, Morsy SA, Shousha HI, Elbaz TM, Nabeel MM, Harb ARK. Genetic variations in death receptor domain 4 gene and the susceptibility to hepatitis C related hepatocellular carcinoma. J Med Virol. 2019;91:1537-1544. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 56. | Wang ZC, Gao Q, Shi JY, Yang LX, Zhou J, Wang XY, Shi YH, Ke AW, Shi GM, Ding ZB, Dai Z, Qiu SJ, Fan J. Genetic polymorphism of the kinesin-like protein KIF1B gene and the risk of hepatocellular carcinoma. PLoS One. 2013;8:e62571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 57. | Zhang Z. Association between KIF1B rs17401966 polymorphism and hepatocellular carcinoma risk: a meta-analysis involving 17,210 subjects. Tumour Biol. 2014;35:9405-9410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 58. | Munirajan AK, Ando K, Mukai A, Takahashi M, Suenaga Y, Ohira M, Koda T, Hirota T, Ozaki T, Nakagawara A. KIF1Bbeta functions as a haploinsufficient tumor suppressor gene mapped to chromosome 1p36.2 by inducing apoptotic cell death. J Biol Chem. 2008;283:24426-24434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 80] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 59. | Yang SZ, Wang JT, Yu WW, Liu Q, Wu YF, Chen SG. Downregulation of KIF1B mRNA in hepatocellular carcinoma tissues correlates with poor prognosis. World J Gastroenterol. 2015;21:8418-8424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 60. | Al-Qahtani A, Al-Anazi M, Viswan NA, Khalaf N, Abdo AA, Sanai FM, Al-Ashgar H, Al-Ahdal M. Role of single nucleotide polymorphisms of KIF1B gene in HBV-associated viral hepatitis. PLoS One. 2012;7:e45128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 61. | Sawai H, Nishida N, Mbarek H, Matsuda K, Mawatari Y, Yamaoka M, Hige S, Kang JH, Abe K, Mochida S, Watanabe M, Kurosaki M, Asahina Y, Izumi N, Honda M, Kaneko S, Tanaka E, Matsuura K, Itoh Y, Mita E, Korenaga M, Hino K, Murawaki Y, Hiasa Y, Ide T, Ito K, Sugiyama M, Ahn SH, Han KH, Park JY, Yuen MF, Nakamura Y, Tanaka Y, Mizokami M, Tokunaga K. No association for Chinese HBV-related hepatocellular carcinoma susceptibility SNP in other East Asian populations. BMC Med Genet. 2012;13:47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 62. | Sopipong W, Tangkijvanich P, Payungporn S, Posuwan N, Poovorawan Y. The KIF1B (rs17401966) single nucleotide polymorphism is not associated with the development of HBV-related hepatocellular carcinoma in Thai patients. Asian Pac J Cancer Prev. 2013;14:2865-2869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 63. | Nagata H, Nakagawa M, Asahina Y, Sato A, Asano Y, Tsunoda T, Miyoshi M, Kaneko S, Otani S, Kawai-Kitahata F, Murakawa M, Nitta S, Itsui Y, Azuma S, Kakinuma S, Nouchi T, Sakai H, Tomita M, Watanabe M; Ochanomizu Liver Conference Study Group. Effect of interferon-based and -free therapy on early occurrence and recurrence of hepatocellular carcinoma in chronic hepatitis C. J Hepatol. 2017;67:933-939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 154] [Article Influence: 22.0] [Reference Citation Analysis (1)] |
| 64. | Matsuura K, Sawai H, Ikeo K, Ogawa S, Iio E, Isogawa M, Shimada N, Komori A, Toyoda H, Kumada T, Namisaki T, Yoshiji H, Sakamoto N, Nakagawa M, Asahina Y, Kurosaki M, Izumi N, Enomoto N, Kusakabe A, Kajiwara E, Itoh Y, Ide T, Tamori A, Matsubara M, Kawada N, Shirabe K, Tomita E, Honda M, Kaneko S, Nishina S, Suetsugu A, Hiasa Y, Watanabe H, Genda T, Sakaida I, Nishiguchi S, Takaguchi K, Tanaka E, Sugihara J, Shimada M, Kondo Y, Kawai Y, Kojima K, Nagasaki M, Tokunaga K, Tanaka Y; Japanese Genome-Wide Association Study Group for Viral Hepatitis. Genome-Wide Association Study Identifies TLL1 Variant Associated With Development of Hepatocellular Carcinoma After Eradication of Hepatitis C Virus Infection. Gastroenterology. 2017;152:1383-1394. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 85] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 65. | Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977-2986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 123] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 66. | Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, Otsuka M, Tateishi R, Omata M, Nakagawa H, Koike K, Kamatani N, Kubo M, Nakamura Y, Matsuda K. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet. 2011;43:455-458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 267] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 67. | Lange CM, Bibert S, Dufour JF, Cellerai C, Cerny A, Heim MH, Kaiser L, Malinverni R, Müllhaupt B, Negro F, Semela D, Moradpour D, Kutalik Z, Bochud PY; Swiss Hepatitis C Cohort Study Group. Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma. J Hepatol. 2013;59:504-509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 68. | Wang H, Cao H, Xu Z, Wang D, Zeng Y. SNP rs2596542G>A in MICA is associated with risk of hepatocellular carcinoma: a meta-analysis. Biosci Rep. 2019;39. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 69. | Choo SY. The HLA system: genetics, immunology, clinical testing, and clinical implications. Yonsei Med J. 2007;48:11-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 265] [Cited by in F6Publishing: 201] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 70. | Lee MH, Huang YH, Chen HY, Khor SS, Chang YH, Lin YJ, Jen CL, Lu SN, Yang HI, Nishida N, Sugiyama M, Mizokami M, Yuan Y, L'Italien G, Tokunaga K, Chen CJ; REVEAL-HCV Cohort Study Group. Human leukocyte antigen variants and risk of hepatocellular carcinoma modified by hepatitis C virus genotypes: A genome-wide association study. Hepatology. 2018;67:651-661. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 71. | Nault JC, De Reyniès A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, Decaens T, Franco D, Imbeaud S, Rousseau F, Azoulay D, Saric J, Blanc JF, Balabaud C, Bioulac-Sage P, Laurent A, Laurent-Puig P, Llovet JM, Zucman-Rossi J. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology. 2013;145:176-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 247] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 72. | Huang W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. Methods Mol Biol. 2017;1617:57-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 138] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 73. | Pogribny IP. MicroRNAs as biomarkers for clinical studies. Exp Biol Med (Maywood). 2018;243:283-290. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 74. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 365] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 75. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X, Wang Z, Qiu S, Wang X, Yang G, Sun H, Tang Z, Wu Y, Zhu H, Fan J. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 469] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 76. | Huang JT, Liu SM, Ma H, Yang Y, Zhang X, Sun H, Zhang X, Xu J, Wang J. Systematic Review and Meta-Analysis: Circulating miRNAs for Diagnosis of Hepatocellular Carcinoma. J Cell Physiol. 2016;231:328-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 77. | Hsu SH, Wang B, Kota J, Yu J, Costinean S, Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, Mao H, Wei M, Clark KR, Mendell JR, Caligiuri MA, Jacob ST, Mendell JT, Ghoshal K. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871-2883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 583] [Cited by in F6Publishing: 591] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 78. | Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ, Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, Hsu MT, Wu JC, Huang HD, Shiao MS, Hsiao M, Tsou AP. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884-2897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 612] [Cited by in F6Publishing: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 79. | Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, De Petro G, Bassi C, Lupini L, Sattari A, Cheung D, Veneziano D, Nigita G, Shankaraiah RC, Portolani N, Carcoforo P, Fornari F, Bolondi L, Frassoldati A, Sabbioni S, Colombo M, Croce CM, Negrini M. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018;9:15350-15364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 65] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 80. | Llovet JM, Peña CE, Lathia CD, Shan M, Meinhardt G, Bruix J; SHARP Investigators Study Group. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res. 2012;18:2290-2300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 406] [Cited by in F6Publishing: 436] [Article Influence: 36.3] [Reference Citation Analysis (2)] |
| 81. | Fornari F, Pollutri D, Patrizi C, La Bella T, Marinelli S, Casadei Gardini A, Marisi G, Baron Toaldo M, Baglioni M, Salvatore V, Callegari E, Baldassarre M, Galassi M, Giovannini C, Cescon M, Ravaioli M, Negrini M, Bolondi L, Gramantieri L. In Hepatocellular Carcinoma miR-221 Modulates Sorafenib Resistance through Inhibition of Caspase-3-Mediated Apoptosis. Clin Cancer Res. 2017;23:3953-3965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 113] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 82. | Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, Gerolami R, Masi G, Ross PJ, Song T, Bronowicki JP, Ollivier-Hourmand I, Kudo M, Cheng AL, Llovet JM, Finn RS, LeBerre MA, Baumhauer A, Meinhardt G, Han G; RESORCE Investigators. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2160] [Cited by in F6Publishing: 2366] [Article Influence: 338.0] [Reference Citation Analysis (0)] |
| 83. | Teufel M, Seidel H, Köchert K, Meinhardt G, Finn RS, Llovet JM, Bruix J. Biomarkers Associated With Response to Regorafenib in Patients With Hepatocellular Carcinoma. Gastroenterology. 2019;156:1731-1741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 84. | Fu Q, Yang F, Xiang T, Huai G, Yang X, Wei L, Yang H, Deng S. A novel microRNA signature predicts survival in liver hepatocellular carcinoma after hepatectomy. Sci Rep. 2018;8:7933. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 85. | Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 587] [Cited by in F6Publishing: 613] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 86. | Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, Yao Z, Shen ZY. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431-42446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 208] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 87. | Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575-1584. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 232] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 88. | Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499-511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 298] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 89. | Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT, Liang KH, Shieh TM, Chen CY, Hsueh C. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1α activity and inhibiting epithelial-mesenchymal transition. Oncotarget. 2016;7:43588-43603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 90. | Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B Promotes Glycolysis and Malignancy of Hepatocellular Carcinoma Through the miR-338-3p/PKM2 Axis Under Hypoxic Stress. Hepatology. 2019;70:1298-1316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 185] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 91. | Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, Mao L, Luo CB, Yu SY, Huang XW, Cao Y, Jia F, Zhou J. Circular RNA Sequencing Identifies CircASAP1 as a Key Regulator in Hepatocellular Carcinoma Metastasis. Hepatology. 2019;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 147] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 92. | Palmirotta R, Lovero D, Cafforio P, Felici C, Mannavola F, Pellè E, Quaresmini D, Tucci M, Silvestris F. Liquid biopsy of cancer: a multimodal diagnostic tool in clinical oncology. Ther Adv Med Oncol. 2018;10:1758835918794630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 262] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 93. | Lianidou E, Pantel K. Liquid biopsies. Genes Chromosomes Cancer. 2019;58:219-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 92] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 94. | Micalizzi DS, Maheswaran S, Haber DA. A conduit to metastasis: circulating tumor cell biology. Genes Dev. 2017;31:1827-1840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 272] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 95. | Okajima W, Komatsu S, Ichikawa D, Miyamae M, Ohashi T, Imamura T, Kiuchi J, Nishibeppu K, Arita T, Konishi H, Shiozaki A, Morimura R, Ikoma H, Okamoto K, Otsuji E. Liquid biopsy in patients with hepatocellular carcinoma: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol. 2017;23:5650-5668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 61] [Cited by in F6Publishing: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 96. | Chiappini F. Circulating tumor cells measurements in hepatocellular carcinoma. Int J Hepatol. 2012;2012:684802. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 97. | Sun YF, Xu Y, Yang XR, Guo W, Zhang X, Qiu SJ, Shi RY, Hu B, Zhou J, Fan J. Circulating stem cell-like epithelial cell adhesion molecule-positive tumor cells indicate poor prognosis of hepatocellular carcinoma after curative resection. Hepatology. 2013;57:1458-1468. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 282] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 98. | Ng CKY, Di Costanzo GG, Terracciano LM, Piscuoglio S. Circulating Cell-Free DNA in Hepatocellular Carcinoma: Current Insights and Outlook. Front Med (Lausanne). 2018;5:78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 99. | Schnekenburger M, Florean C, Dicato M, Diederich M. Epigenetic alterations as a universal feature of cancer hallmarks and a promising target for personalized treatments. Curr Top Med Chem. 2016;16:745-776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 100. | Chan KC, Lai PB, Mok TS, Chan HL, Ding C, Yeung SW, Lo YM. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin Chem. 2008;54:1528-1536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 122] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 101. | Sherman M. How to improve HCC surveillance outcomes. JHEP Rep. 2019;1:460-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 102. | Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med. 2014;11:e1001624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 516] [Cited by in F6Publishing: 538] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 103. | Moon AM, Weiss NS, Beste LA, Su F, Ho SB, Jin GY, Lowy E, Berry K, Ioannou GN. No Association Between Screening for Hepatocellular Carcinoma and Reduced Cancer-Related Mortality in Patients With Cirrhosis. Gastroenterology. 2018;155:1128-1139.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 104. | Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, Sherman M. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35:2155-2166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 569] [Cited by in F6Publishing: 724] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 105. | Tzartzeva K, Singal AG. Testing for AFP in combination with ultrasound improves early liver cancer detection. Expert Rev Gastroenterol Hepatol. 2018;12:947-949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 106. | Wan HG, Xu H, Gu YM, Wang H, Xu W, Zu MH. Comparison osteopontin vs AFP for the diagnosis of HCC: a meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38:706-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 107. | Chen J, Cao SW, Cai Z, Zheng L, Wang Q. Epithelial-mesenchymal transition phenotypes of circulating tumor cells correlate with the clinical stages and cancer metastasis in hepatocellular carcinoma patients. Cancer Biomark. 2017;20:487-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 108. | Chen Y, Li S, Li W, Yang R, Zhang X, Ye Y, Yu J, Ye L, Tang W. Circulating tumor cells undergoing EMT are poorly correlated with clinical stages or predictive of recurrence in hepatocellular carcinoma. Sci Rep. 2019;9:7084. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 109. | Yang JD, Addissie BD, Mara KC, Harmsen WS, Dai J, Zhang N, Wongjarupong N, Ali HM, Ali HA, Hassan FA, Lavu S, Cvinar JL, Giama NH, Moser CD, Miyabe K, Allotey LK, Algeciras-Schimnich A, Theobald JP, Ward MM, Nguyen MH, Befeler AS, Reddy KR, Schwartz M, Harnois DM, Yamada H, Srivastava S, Rinaudo JA, Gores GJ, Feng Z, Marrero JA, Roberts LR. GALAD Score for Hepatocellular Carcinoma Detection in Comparison with Liver Ultrasound and Proposal of GALADUS Score. Cancer Epidemiol Biomarkers Prev. 2019;28:531-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |