Published online Oct 27, 2020. doi: 10.4254/wjh.v12.i10.775
Peer-review started: April 27, 2020
First decision: August 9, 2020
Revised: August 18, 2020
Accepted: September 14, 2020
Article in press: September 14, 2020
Published online: October 27, 2020
The recent rise in the incidence of hepatitis B virus (HBV) infections in a densely populated city of eastern India (“mixing vessel” of people of varied socio-economic and immune status) prompted this study. Applying saliva on fingers for enumerating bank notes is a common practice in the Indian subcontinent. Paper notes may be a potential source of “horizontal” transmission of this virus, especially if there are cuts/bruises on the oral mucous membrane or skin.
To investigate whether paper currencies could be a plausible mode of horizontal transmission of HBV infection.
Polymerase chain reactions (PCR) followed by nucleotide sequencing was done for the detection of HBV. Hepatitis B virus surface antigen enzyme-linked immunosorbent assay(HBsAg ELISA) was performed on all HBV deoxyribonucleic acid-positive samples to check the detectability of the virus. Atomic force microscopy (AFM) was carried out for visual confirmation of HBV particles in ultracentrifuged/immunoprecipitated samples from currency paper washings.
HBV-specific PCRs on pellets obtained after ultracentrifugation/ immunoprecipitation of the currency paper washings detected potentially intact/viable HBV (genotype D2) in 7.14% of samples (n = 70). AFM gave the visual confirmation of HBV particles in ultracentrifuged/immunoprecipitated samples from currency paper washings. However, HBV isolates from the currency notes could not be detected by HBsAg ELISA.
It is a common practice in the Indian subcontinent to count paper currencies by applying saliva on fingertips. Paper notes may be a potential source of “horizontal” transmission of this virus, especially if there are cuts/bruises on the oral mucous membrane or skin, but it was practically not possible to demonstrate experimentally such transmission. Detection of potentially intact/viable and “occult” HBV from currency poses potential risk of silent transmission of this virus among the general population.
Core Tip: The recent upsurge in hepatitis B virus (HBV) infections in eastern India prompted the search for this virus in low denomination paper notes in this region. Applying saliva on finger tips for enumerating currency notes is a common practice. Thus, paper currencies may be a potential source of “horizontal” HBV transmission, especially if there are cuts/bruises on the oral mucous membrane or skin. We discovered that intact HBV particles are present in about 7.14% of the currencies. Molecular analysis and immunoassays suggested that the circulating HBV are “occult” in nature, hence capable of “silent transmission” in the general population.
- Citation: Das P, Supekar R, Chatterjee R, Roy S, Ghosh A, Biswas S. Hepatitis B virus detected in paper currencies in a densely populated city of India: A plausible source of horizontal transmission? World J Hepatol 2020; 12(10): 775-791
- URL: https://www.wjgnet.com/1948-5182/full/v12/i10/775.htm
- DOI: https://dx.doi.org/10.4254/wjh.v12.i10.775
Transaction of paper currency occurs rampantly in exchange of goods and other services. Chances of microbial contamination in the currencies of lower denominations are higher as they are more widespread and exchanged frequently among people irrespective of socio-economic status within a population[1]. While enumerating paper money, people often apply saliva on fingers, a potential means of contaminating the currency notes as well as exposure to microbes already present on the currency notes.
In developing and densely populated countries like India, where people are of diverse immune and hygiene status, handling of currency may lead to transfer of different kinds of microorganisms. Paper currency may be contaminated with droplets from sneezing or coughing and even by touching with contaminated hands[2]. Microbes may also be introduced into the paper notes due to unhygienic habits like keeping currency notes in socks, shoes and pockets, putting them under the carpet or rugs and squeezing them in the hand[3]. Storing these notes in polythene, cotton or leather bags in humid and dark conditions favor the growth of microorganisms on them[4].
Cases of currency contamination with pathogenic microbes have been reported from many countries like Bangladesh, Ghana, Nigeria, United States, Myanmar, Egypt, Nepal and Pakistan[5].
Rhinovirus, rotavirus and influenza virus have been detected on currency notes and coins[5], but till date there had been no reports on detection of hepatitis B virus (HBV) in currency sample. Hepatitis viruses may cause acute or chronic liver disease. The latter condition often leads to cirrhosis and may eventually culminate in liver cancer. Hepatitis viruses may be transmitted via the fecal-oral route (hepatitis A and E) or via blood (hepatitis B, C and D).
The recent unusual rise in HBV infection in India[6] prompted the search for HBV in currency notes as they are in circulation among people of varied immune status and may serve as a potential medium of HBV transmission.
Paper currency samples, seventy in number, of a single denomination (INR 10) were used for the present study. They were collected during 2016-2017 from hospitals, grocery stores, fish-meat markets and public transport. Five of each such places were chosen; currency samples were collected in duplicate or more, stored in sterile zip lock packs and quickly transported to the laboratory for further processing (Table 1). Two currency samples fresh from the mint, which were not yet into circulation, and two used samples that have been autoclaved were also included in the present study as negative controls. Surfaces of each of the samples were thoroughly washed with 5 mL 1 × phosphate buffered saline (PBS) buffer and each washed solution was stored at 4 °C until used. For the screening of HBV, washed out solutions were ultra-centrifuged at 64000 g for 90 min at 4 °C. The resultant pellets were supposed to contain all intact microbes (bacteria, viruses, etc.) present in the currency washings, and all extraneous and lighter materials like free nucleic acids should be in the supernatant. The precipitate for each sample, collected at the bottom of the ultracentrifuge tube, was washed thoroughly, resuspended in 1 × PBS buffer and collected in a fresh tube.
| Types of locations of sample collection | Detailed description of the actual sites of collection | Co-ordinates | Distance from central Kolkata1 (in Km) | Number of currency samples collected (n = 70) | Number of samples that are hepatitis B virus-positive |
| Hospitals | Nil Ratan Sircar Medical College and Hospital | 22.5638° N, 88.3690° E | 2.6 | 2 | 1 HBV-positive (S5) |
| SSKM Hospital | 22.5396° N, 88.3439° E | 4.8 | 2 | - | |
| Chittaranjan National Cancer Institute (CNCI) | 22.5254° N, 88.3465° E | 3.7 | 2 | - | |
| Calcutta National Medical College and Hospital (CNMC) | 22.5467° N, 88.3704° E | 4.5 | 3 | 1HBV-positive (S8) | |
| KPC Medical College and Hospital | 22.4956° N, 88.3706° E | 9.3 | 4 | - | |
| Public transport routes (bus): First stop and last stop | 2302(Kamarhati to Alipore Zoo) | 22.6847° N, 88.3706° E-22.5248° N, 88.3312° E | 13.4 - 5.1 | 4 | - |
| 47B (Lake town to Super Market, Prince Anwar Shah Road) | 22.6070° N, 88.4028° E-22.5015° N, 88.3617° E | 19.4-7.5 | 5 | - | |
| 3C/1 (Anandapur to Nagerbazar) | 22.5148° N, 88.4098° E-22.6218° N, 88.4180° E | 11.5 -19.7 | 5 | - | |
| 45 (Dumdum to Baishnabghata) | 22.6471° N, 88.4317° E-22.4729° N, 88.3764° E | 22.8-11.4 | 5 | - | |
| S9 (Jadavpur to Karunamoyee) | 22.4956° N, 88.3706° E-22.5851° N, 88.4222° E | 8.8 -13.5 | 5 | 1 HBV-positive (S6) | |
| Grocery shop | Shyam Bazar | 22.5982° N, 88.3687° E | 6.2 | 3 | - |
| Big Bazaar, Ganguli Bagan | 22.4800° N, 88.3757° E | 18.2 | 3 | - | |
| South City Mall | 22.5015° N, 88.3617° E | 7.5 | 4 | - | |
| Dunlop | 22.6519° N, 88.3786° E | 12.4 | 4 | - | |
| Gariahat | 22.5170° N, 88.3658° E | 6.7 | 4 | - | |
| Fish-meat market | Howrah | 22.5958° N, 88.2636° E | 6.1 | 2 | 1 HBV-positive (S7) |
| Baguihati | 22.6107° N, 88.4271° E | 18.3 | 4 | - | |
| Hazra | 22.5228° N, 88.3500° E | 4.3 | 4 | - | |
| Jadavpur | 22.4956° N, 88.3706° E | 8.8 | 2 | 1 HBV-positive (S9) | |
| Gariahat | 22.5170° N, 88.3658° E | 6.7 | 3 | - |
None of the treatments to which the paper currency samples were subjected amounted to purposeful act of destroying, willful defacing, disfiguring or mutilation or any other kind of violation of currency handling procedures laid down by the Government of India. Washing of the samples with 1 × PBS buffer did not destroy the currency samples and were still suitable for re-use after washing and subsequent drying in air.
Total nucleic acid was extracted from the 1 × PBS buffer wash of the currency samples using High Pure Viral Nucleic acid kit (Roche, Mannheim, Germany). Extracted DNA was quantified by means of Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA, United States).
Two sets of primers specific to HBV S gene were used for HBV detection[7]. To amplify HBV S gene, the first round polymerase chain reaction (PCR) was performed with SPL-3 and SPL-2 primers while the nested PCR was performed using the second set of internal primers SPL-4 and SPL-5 (Table 2). The cycling conditions for the first round PCR comprised initial denaturation at 95 °C (5 min) for one cycle followed by 30 cycles of heating at 95 °C (40 s), primer annealing at 55 °C (1.5 min) and extension for 2 min at 72 °C. The final extension was done at 72 °C (10 min) for one cycle followed by hold at 4 °C. The first round PCR product was diluted ten-fold and subjected to the second round nested PCR with SPL-4 & SPL-5 primers. This PCR was programmed as follows: Initial denaturation at 95 °C (10 min) for one cycle followed by 30 cycles of heating at 94 °C (1 min), primer annealing at 55 °C (1 min) and extension at 72 °C (1.5 min). The final extension was carried out at 72 °C for one cycle (10 min).
| Name | Sequence 5’-3’ | Amplicon size (bp) | Reference |
| SPL-3-F | GCGCGCGCTAGCACCATGGGGARCAYCRYATCRGGA | 1652 | [7] |
| SPL-2-R | GCCTTTGCAAGCTTCASACCAATTTATGCCTAC | ||
| SPL-4-F | ACCACAGAGTCTAGACTYGTGGT | 1277 | |
| SPL-5-R | GGTCGGAACRRCAGRCGRAGAAG | ||
| HBVRT-F | GTGTCTGCGGCGTTTTATCA | 98 | This study |
| HBVRT-R | GACAAACGGGCAACATACCTT |
PCR products were resolved by 1% agarose gel electrophoresis. Bands were observed under ultraviolet light (Carestream Gel Logic 212 Pro, Rochester, NY, United States), and the gel images were recorded. PCR bands of correct size were gel-purified (Qiagen Gel Extraction Kit, Hilden, Germany), eluted in nuclease-free water and subjected to bi-directional DNA sequencing using the same primers used for PCR amplification. All PCRs contained GoTaq® Green Master Mix (M7122, Promega, Madison, WI, United States), 0.2-0.4 µM forward and reverse primers and 100–300 ng DNA.
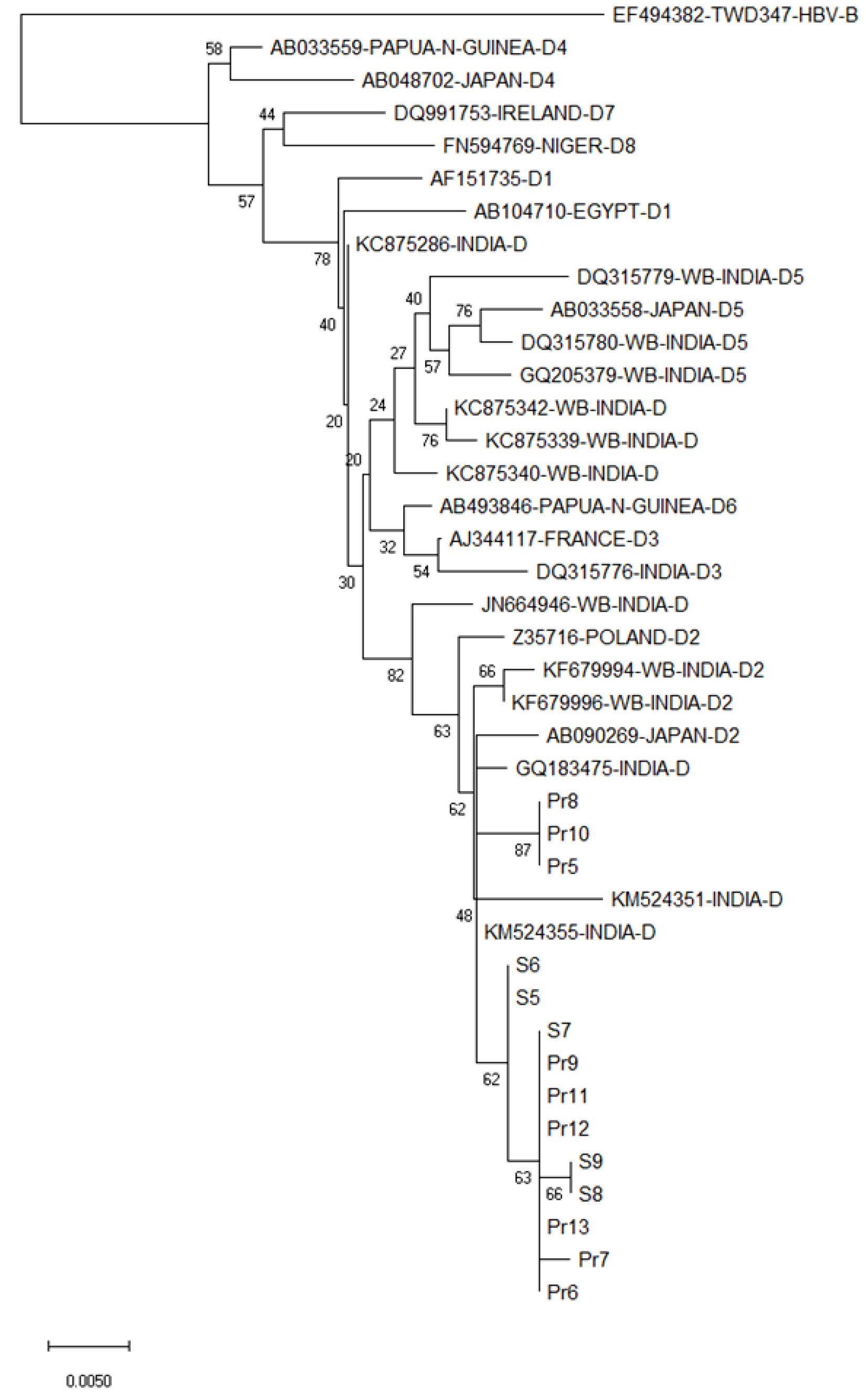
Nucleotide (nt) sequences, confirmed by bi-directional sequencing of the PCR products were subjected to National Center for Biotechnology Information-Basic Local Alignment Search Tool (BLAST) for determining genetic matches with sequences available in the database. They were then aligned using MEGAX with other closely related sequences identified from the BLAST search. A phylogenetic tree was generated by the Neighbor-Joining method with Bootstrap test (2000 replicates) using MEGAX on 510 nt of S gene (pertaining to nt positions 326-835 as in HBV34 genotype D2 isolate, accession number KC875340, West Bengal, India) in order to characterize the HBV isolates from the currency samples and study their genetic distances with other closely related HBV strains[8].
Quantitative real-time PCR was performed on the HBV-positive DNA samples to determine the HBV DNA copy number in the currency samples. A pcDNA3.1 (+) plasmid construct (Invitrogen, Carlsbad, CA, United States) containing the HBVRT-F and HBVRT-R (Table 2) product of HBV genotype D “S” gene cloned into it, was used as the standard for the real-time PCR. Serial dilutions of this recombinant plasmid were used as known standards to calculate the HBV DNA copy number of the currency samples tested. A 98 bp fragment of the “S” gene (pertaining to nt position 379 to 476 as in HBV isolate KC875340) was amplified using the primers HBVRT-F (nt position 379 to 398) and HBVRT-R (nt position 456 to 476). The DNA copy number of each PCR product was determined from its DNA concentration. Four replicates were run for each standard and sample. The real-time PCR mix for each sample contained 10 µL of Luna Universal qPCR Master Mix (New England BioLabs, Ipswich, MA, United States), 1 µL of a mix of primers (10 µM each), 1 μL of sample (< 100 ng DNA) or 2.5 μL of standard and nuclease-free water to a final volume of 20 µL. The thermal cycling conditions in the Quant Studio 5 (Applied Biosystem, Foster City, CA, United States) consisted of initial hold at 95 °C (1 min), followed by 40 cycles of 15 s at 95 °C and 30 s at 58 °C. Fluorescence was monitored during the 55 °C annealing phase. Formation of bands of expected size was further confirmed by agarose gel electrophoresis of the pooled replicates per sample after completion of real-time PCR run.
In order to rule out that the laboratory reagents or equipment were not contaminated with HBV, control nested PCRs were performed with two sets of primers: SPL-3 and SPL-2 as first round primers and SPL-4 and SPL-5 as second round primers, as described previously. Ultracentrifuged pellet suspensions from 25 representative currency samples, including all HBV DNA-positive and remaining HBV DNA-negative samples were tested for the presence of the antigen marker of HBV [i.e., hepatitis B virus surface antigen (HBsAg) through enzyme-linked immunosorbent assay (ELISA)]. The assay was performed using the Monolisa HBsAg Ultra kit (Hercules, CA, Bio-Rad) following manufacturer’s instructions. The samples were tested in duplicates, diluted in the ratio of 1:5 in the supplied sample diluent.
HBV DNA-positive samples that had higher HBV DNA copy number were selected and subjected to atomic force microscopy (AFM) to gather visual evidence for the presence of approximately 42 nm HBV particles (Dane particles)[9].
For this, two representative HBV DNA-positive ultracentrifuged samples and one HBV DNA-negative ultracentrifuged sample (as negative control) were selected. All three samples were put through an identical procedure for AFM sample preparation. Furthermore, immunoprecipitation was performed on the above mentioned three samples. Immunoprecipitation technique was adopted to trap selectively the HBV particles from the ultracentrifuged pellet and to enrich the particles in the available volume for more authentic imaging. For this, Immunoprecipitation Kit-DynabeadsTM Protein G (Invitrogen) was used and the assay was performed following the standard protocol suggested by the manufacturer of the kit. Monoclonal anti-HBV antigen HBsAg (Cat No. SAB4700767, Sigma-Aldrich, St Louis, MO, United States) was used as the trapping antibody for immunoprecipitating the target antigen (i.e. HBsAg on HBV virion surface).
The final eluate (antigen-antibody complexes) obtained was treated with 0.2% Triton X-100 in the ratio 1:1 for 30 min followed by pulse sonication to disrupt the virus-antibody complexes. The solution was then centrifuged at 13000 rpm for 5 min at 4 °C. The supernatant was carefully transferred to a fresh centrifuge tube. Both, immunoprecipitated and non-immunoprecipitated samples were diluted in nuclease free water as required, and 5 µL of each diluted sample was applied on freshly cleaved muscovite Ruby mica sheet (ASTM V1 grade) and allowed to air dry. Once the sample was fixed, the mica sheet was put through AFM.
Acoustic-AC mode AFM was performed by means of a Pico plus 5500 AFM (Agilent Technologies, Santa Clara, CA, United States) with a piezo scanner with maximum range of 9 µm. Images were processed by flattening, using Pico view 1.4 version software (Agilent Technologies). Image manipulation was done through Pico Image Advanced version software (Agilent Technologies).
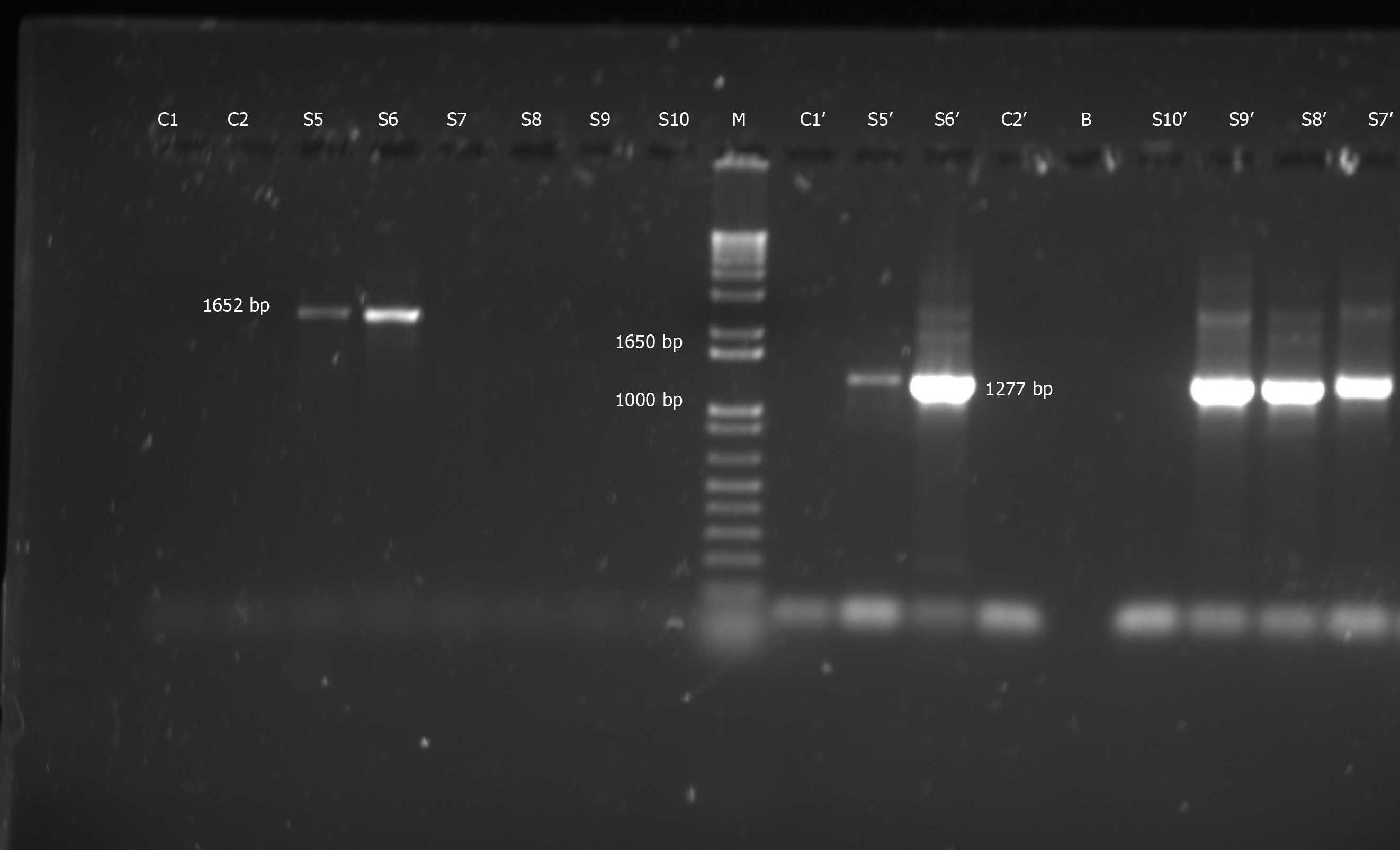
HBV “S” gene-specific PCRs on these DNA samples revealed the presence of HBV in five out of 70 samples screened (7.14%). PCR products of expected size (i.e. 1652 bp for SPL-3-2 and/or 1277 bp for SPL-4-5) were observed in all HBV-positive samples (Figure 1).
The second round 1277 bp bands, obtained in optimum concentration and purity, upon bi-directional sequencing (using SPL-4 and SPL-5 primers) confirmed that the observed PCR products were indeed part of the “S” gene of HBV (Figure 2).
All the HBV isolates from the currency samples were of genotype D2, as evident from their close clustering with other genotype D2 strains in the phylogenetic tree (Figure 3). Only partial S and Pol protein open reading frames were retrievable from the nucleotide sequences obtained for the five HBV-positive samples (GenBank accession numbers: S5: MN158164; S6: MN158165; S7: MN158166; S8: MN158167; S9: MN158168).
It was estimated from quantitative real-time PCR results that the number of HBV copies ranged from (6-10) × 108 for sample S5; (2-3) × 109 for S6 and S7; (3-4) × 105 for S8 and (3-6) × 103 for S9. The above copy numbers were estimates of total number of HBV copies present in the entire 5 mL washing from each paper currency sample.
Laboratory reagents (e.g., water for PCR, PCR mix and primers) or equipment (e.g., PCR machines and centrifuges) showed no evidence of HBV-contamination. This was proved by the absence of any visible band in case of control PCRs (water control; primers control) with SPL-3-2 and SPL-4-5 nested PCRs (Figure 1).
All the HBV positive and negative samples tested were found to be negative for HBsAg by ELISA, suggesting that the genotype D2 HBV strains present in the ultracentrifuged pellets were not detectable by Monolisa ELISA. The ELISA performed, passed the qualitative test criteria as per the manufacturer’s guidelines. The assay tested each sample in duplicate, and the mean of sample-to-cut off ratio (S/CO) was considered to arrive at the results (data not shown).
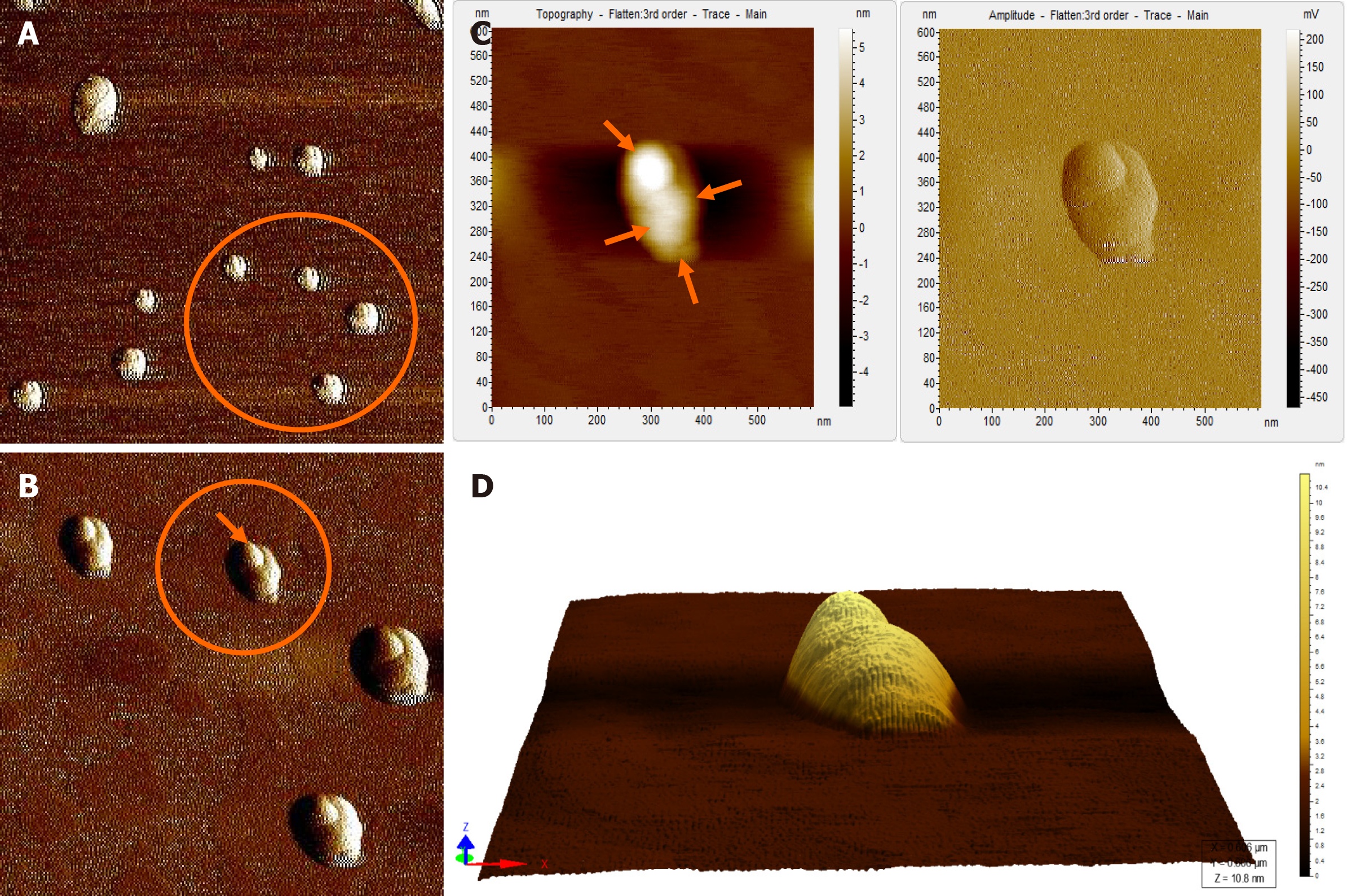
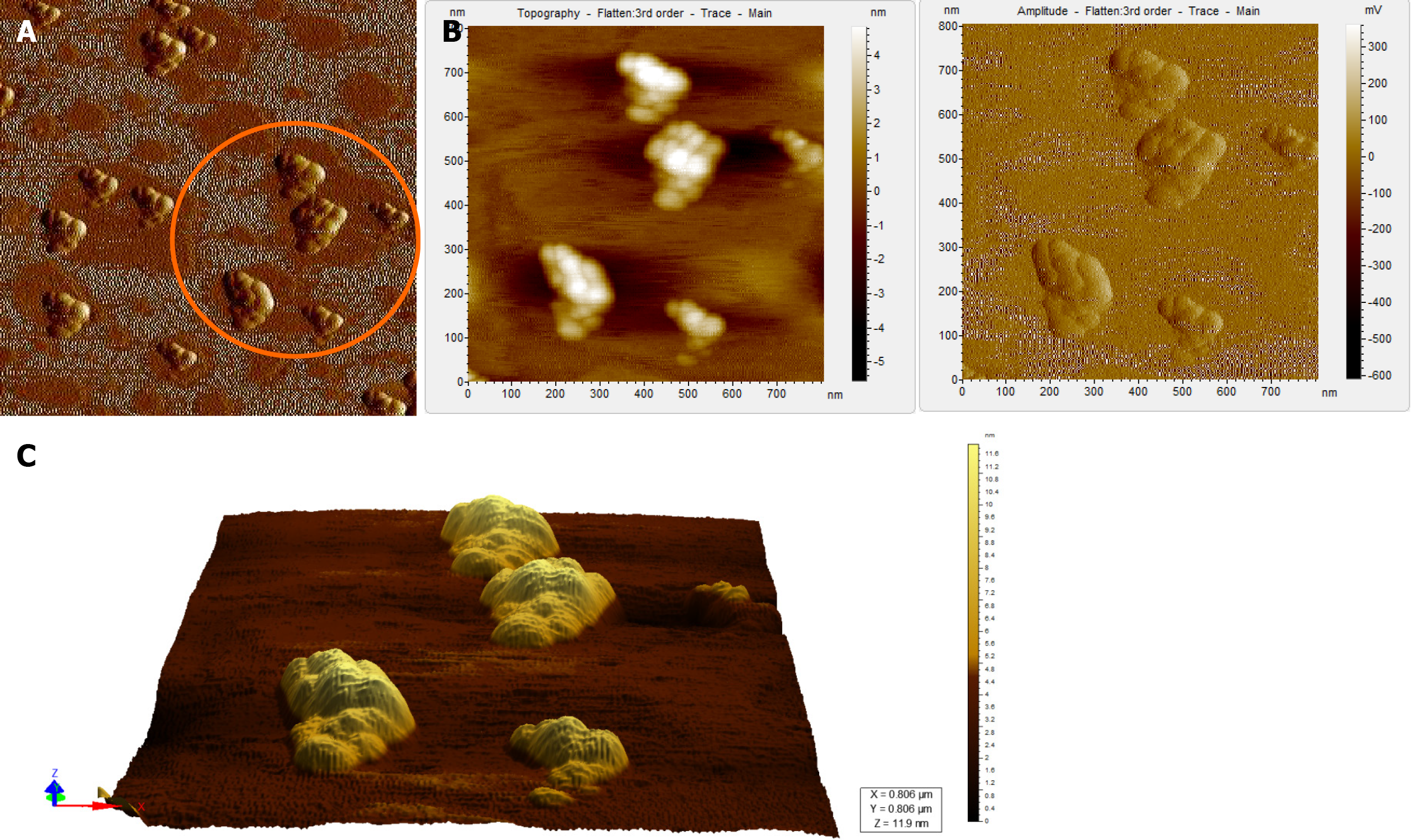
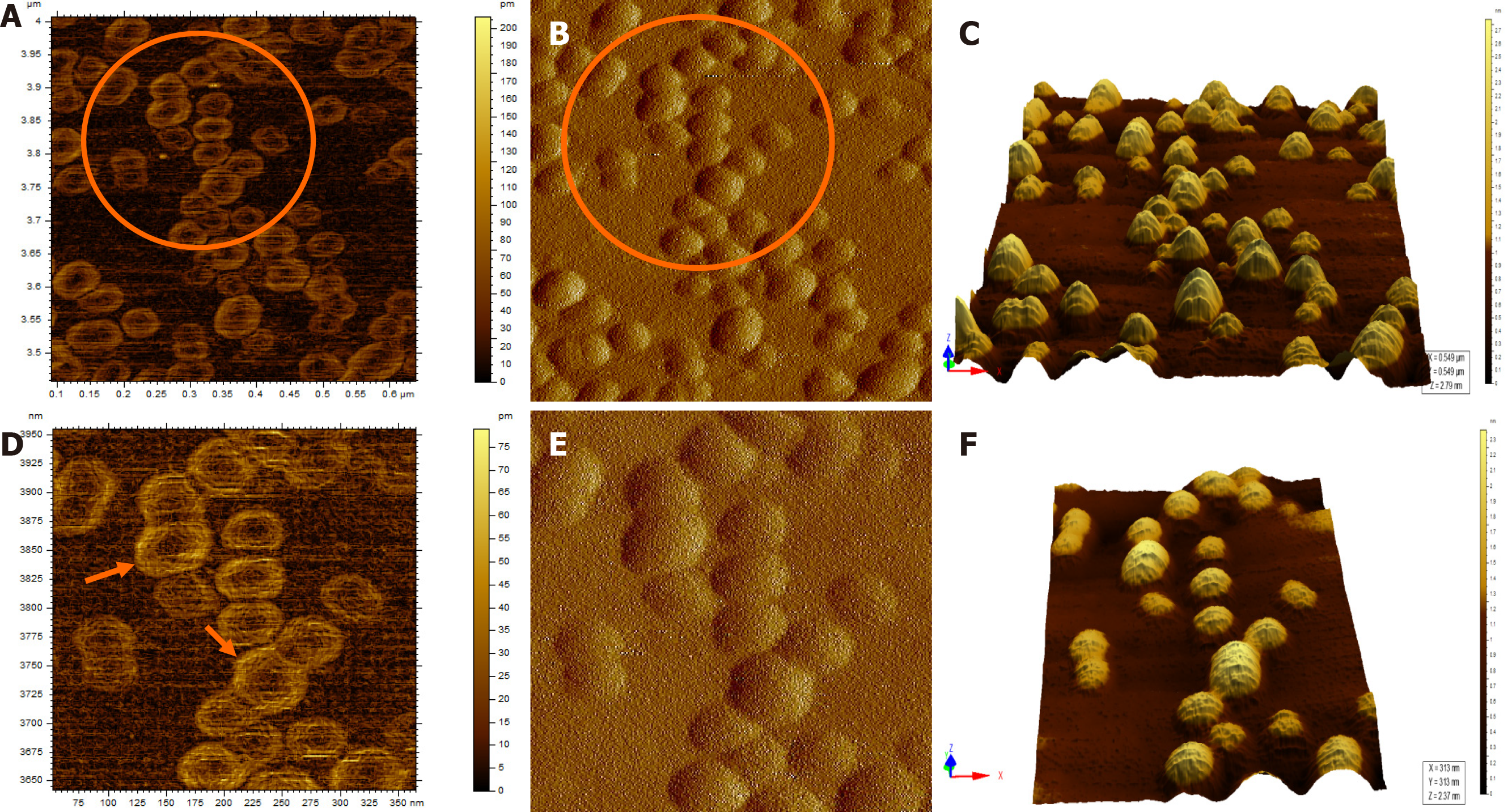
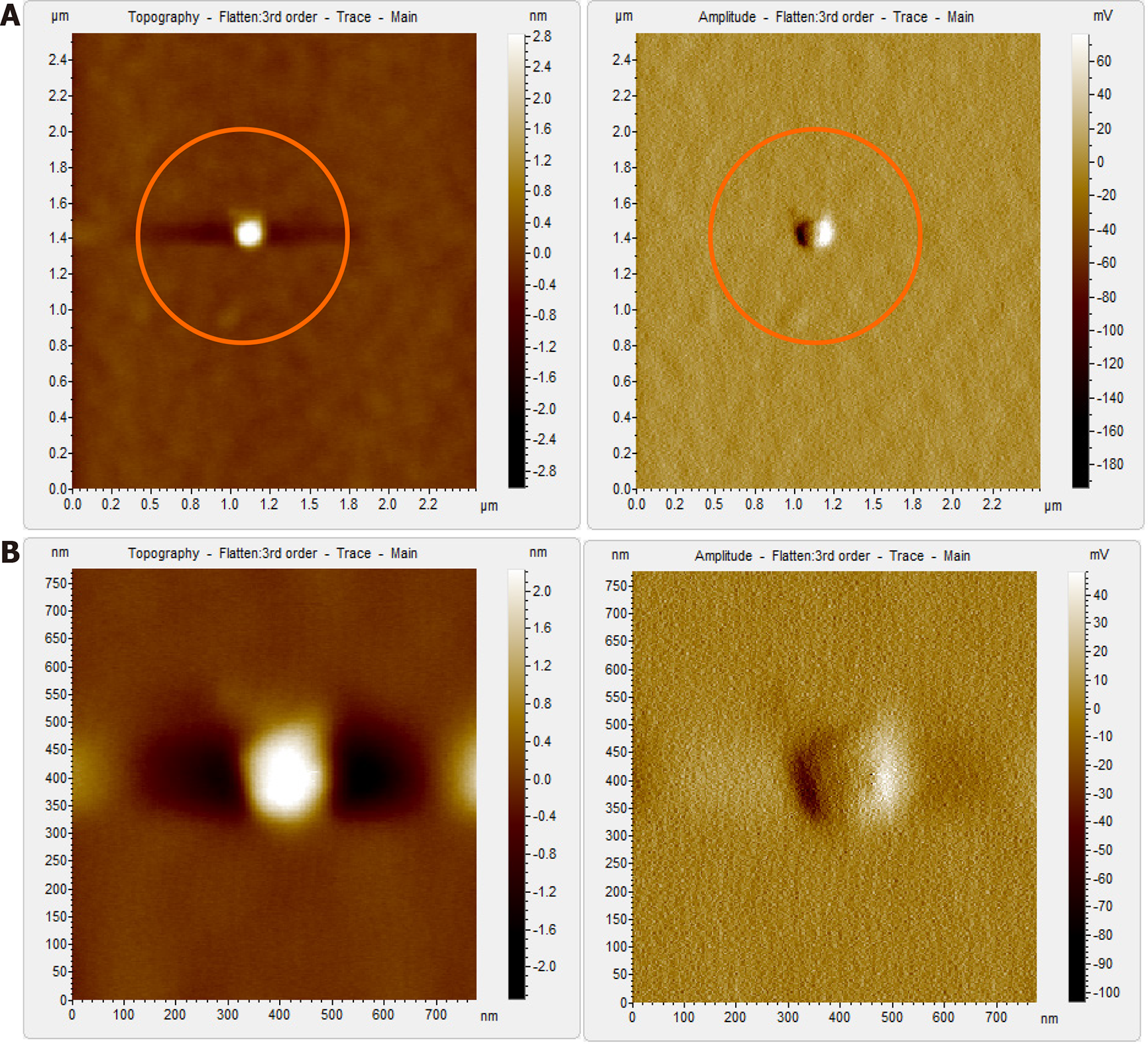
When the diluted (1:100) non-immunoprecipitated ultracentrifuged HBV DNA- positive samples were imaged through AFM, clusters of HBV virion-like particles were visible (Figure 4). The diameter of the virus-like particles ranged from 40 to 60 nm (Figure 4).
The HBV DNA- negative sample displayed globular clumps, comprising of globule-like structures much smaller than HBV particles (Figure 5). The topographical demarcation for both the HBV DNA-positive (Figure 4D) and negative samples (Figure 5C) is clearly evident from the images.
AFM imaging of the Triton X-100-treated immunoprecipitated HBV DNA positive-sample validated the aforementioned results (Figure 6). Triton X-100 treatment followed by sonication led to the disruption of the virion clusters into individual particles. Complete/partial disintegration of virus envelope attached to the immunoprecipitating antibody exposed the icosahedral core particle of HBV (Figure 6A, D). The size of the individual virion particle observed was approximately 42 nm in accordance to the diameter of HBV and the number of particles observed corroborated with the high copy number of HBV DNA found in the samples.
In contrast, the HBV DNA- negative sample, which showed globular structures previously (Figure 5), after being subjected to immunoprecipitation and subsequent Triton X-100 treatment, displayed a clear/vacant field with no globular structures or HBV-like particles visible using AFM (Figure 7A). Even at higher magnification, no such particles could be captured by AFM (Figure 7B).
To the best of our knowledge, the present study is the first to detect potentially viable HBV from circulating paper currency samples collected from various parts of Kolkata, a metropolitan city in eastern India. Sequences of the HBV-specific PCR products upon BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 15th February 2020) and phylogenetic analysis closely matched with recently circulating strains of HBV genotype D2 previously isolated from eastern Asia including Kolkata[10-15].
Although the potential role of currency in virus transmission is generally thought to be limited[16], it could still play a role in the spread of communicable viral diseases[17]. HBV, the causative agent of a highly infectious and acute/chronic liver disease, has been reported to retain infectivity when stored at approximately 30 °C for at least 6 mo and withstand drying on a surface for around 7 d when present in blood[18]. It can also remain infectious and survive in environmental surfaces for about a week[19]. This information coupled with the observation of intact and potentially viable HBV on paper currencies from Kolkata area in the present study demands attention because people of this region are of varied immune status and often apply saliva on fingertips to count the paper money. If these people have oral lesions (e.g., mouth ulcers) or broken skin surfaces, it is quite probable that HBV particles on the infected currencies may be transmitted into the human subjects. Concurrent handling of paper currency and household materials may also set the stage for transmission of HBV among different family members[20]. This was further strengthened by reports from India indicating that intra-familial horizontal transmission played a more important role in the spread of HBV infection than the sexual mode of HBV transmission[21,22].
The HBV sequences, identified in the present study, closely matched with HBV genotype D2, widely prevalent in various countries including India[10-12]. All these isolates carried S protein mutations previously reported to impair immune-detection and this explains why these isolates were HBsAg-negative by the enzyme immunoassay[23].
Our findings corroborate well with the fact that HBV genotype D2 is most prevalent in India including Kolkata[24,25]. In fact, the HBV S protein sequences retrieved from the currency samples closely matched at the amino acid level (Figure 2) and nucleotide level (Figure 3) with the HBV sequences, surprisingly detected in blood of Pityriasis rosea skin disease patients from Kolkata during 2016, who were screened for HBV in another study in our laboratory. More than 50% of these samples were occult HBV infection (OBI) and carried at least one OBI-signature S protein mutation[23].
HBV spread is silent as most of these infections are going undiagnosed as ELISAs are still the mainstay of virus diagnosis in developing countries like India, and the HBV strains in question are mostly “occult” in nature (i.e. not easily detectable by HBsAg-detection ELISAs).
The HBV DNA copy numbers observed in the five HBV-positive currency samples were higher than the 50% minimum infectious dose of even OBI DNA, which had been estimated at 1049 (117-3441) copies[26]. However, the SYBR-green qPCR method used for the HBV DNA quantification is prone to overestimation of the copy numbers as SYBR-green binds to nonspecific double stranded DNA sequences (e.g., primer dimers) besides the target sequence, producing false positive signals[27,28]. In order to cross-check the calculated HBV DNA copy numbers present in entire 5 mL washings from each banknote, the numbers were back-calculated from the number of particles visible in AFM images (Figure 6). This was done with the assumption that HBV particles were evenly distributed in the circular area over which the spherical drop of the sample adsorbed on the mica sheet. The numbers came to be of the order of 109 copy numbers (total 5 mL washings from the concerned banknotes), which corroborated with PCR-estimated copy number for the said isolates. Conclusively, the HBV particles present on the currency notes were higher than 50% minimum HBV infectious dose, thereby posing the contaminated currencies as an effective source of HBV transmission.
It was also interesting to observe, in this context, that human saliva, often used for enumerating paper currencies in India, can have significantly high level of HBV contamination even to the level of 107/mL virus titre[29,30]. Furthermore, human saliva is often implicated in horizontal mode of HBV transmission besides the common and major routes of HBV transmission, namely, sexual contact, parenteral drug use, transfusion and vertical transmission[31,32].
The HBV genotype D2 strains detected in the currency notes carried several mutations (the trio of T118V⁄A128V double mutation and P127T mutation; M133I) known to impair immunological detection[23]. We were, indeed, unable to detect S antigen with a method (Bio-Rad's Monolisa ELISA) that is known to use an array of different monoclonal antibodies to reduce the number of S protein mutants that are not detected. Still, it was possible to concentrate these “occult” viruses for microscopy using the monoclonal antibody (SAB4700767), due to the fact that the S protein mutations in these viruses possibly did not affect the binding site of the said Mab. The result of the phylogenetic analysis is not surprising since the HBV genotype and lineage detected is the same that was circulating in the city, and this observation, in fact, supports our finding.
One limitation of our study is that although we think that the popular habit of using saliva in counting paper notes may be related to spread of HBV, we could not rule out several other routes of transmission that could also contribute to the surge of HBV infections in Kolkata in recent years. These include practices such as tattooing, piercing, drugs abuse, sex workers, unregistered medical practices, manicure/pedicure, ritual practices involving needles, knife and other sharp elements and so on[31,32].
Still, it appears that use of saliva in counting currency is a more predominant and generalized habit in the overall Indian population than the aforesaid other practices that expose the population to the risk of HBV transmission. However, many of these factors remain unknown due to the lack of reliable model for virus propagation and experimental infections. Some of these factors include evaluation of infectious dose of the virus by this route, the stability of the virus, the probability of counting money each day (time of exposition) and so on.
We could not demonstrate the “infectivity” of the HBV DNA-positive samples and it was also not feasible to demonstrate experimentally the transmission of HBV via paper notes. However, detection of intact virions in considerable amount on heavily used currency notes sets up the scenario that if people do contract HBV from the currency samples, they are likely to develop hepatitis B, which is difficult to diagnose by routine ELISA. Thus, there is increased risk of silent spread of the infection in the susceptible population.
Presently, the severe acute respiratory syndrome coronavirus 2 (SARS CoV-2), first reported from Wuhan, China in 2019, has emerged as a pandemic and a major health concern worldwide[33]. It has been stated that there is a high risk of transmission of this virus by touching contaminated dry inanimate surfaces like paper, wood, plastic, metal, steel, glass, ceramic, Teflon and so on. It has been already reported that the various strains of SARS CoV like P9 or GVU6109 can persist on the paper surfaces for 24 h to 5 d at room temperature[34]. Thus, the handling of contaminated paper currencies and enumerating them using saliva can pose a substantial risk of transmission of not only hepatitis B virus, as described in this study, but also SARS CoV-2, thereby contributing to its currently observed rapid spread in densely populated countries like India.
To put into perspective the role of paper currency (with HBV contamination) in virus transmission, we propose a model as follows: Infected saliva to finger to paper notes and then contaminated notes to finger to saliva of susceptible human, supposing that many people count the money in this way.
In summary, the presence of highly infectious HBV in commonly circulating currency notes in a populous region, as detected in this study, imposes possible risk of transmission of this pathogen among the general population. This, however, needs further experimental validation as already discussed above. This phenomenon might be contributing to the increasing incidence of HBV infections among the population besides other routes of exposure. Hence, people should be made aware about the unhygienic practices leading to microbial contamination of currencies to reduce the risk of transmission of infectious microbes through currency route.
The recent rise in the incidence of hepatitis B virus (HBV) infections in a densely populated city of eastern India prompted the search. Paper currency is widely used as a mode of transaction for various goods and services irrespective of socio-economic status among the population. Therefore, the chances of microbial contamination specifically in the currencies of lower denominations are higher. The common practice of enumerating currency notes using saliva in Indian subcontinent may be a potential source of horizontal transmission of HBV, especially if there are cuts/bruises on the oral mucous membrane or skin.
The increasing number of cases of HBV infections in eastern India served as the impetus to investigate possible presence of this virus in low denomination paper notes in a densely populated city of India such as Kolkata.
To investigate whether paper currency can serve as a plausible mode of horizontal transmission of HBV infection in areas of high population density.
HBV was detected by performing polymerase chain reactions (PCRs) on nucleic acids extracted from ultracentrifuged washings from paper currencies, followed by nucleotide sequencing for the confirmation of the presence of the virus. Hepatitis B virus surface antigen-enzyme-linked immunosorbent assay (HBsAg-ELISA) was carried on HBV DNA-positive samples to check for the detectability of HBV surface antigen. Atomic force microscopy (AFM) was used for visual confirmation of HBV particles in ultracentrifuged/immunoprecipitated samples from currency paper washings.
Out of all the currency notes screened (n = 70), 7.14% of the samples were found to be contaminated with potentially intact/viable HBV of genotype D2. Atomic force microscopy provided visual confirmation of HBV particles in ultracentrifuged/immunoprecipitated samples from currency paper washings. However, HBV isolates from the currency notes failed to be detected by hepatitis B surface antigen ELISA. Molecular analysis and enzyme immunoassays suggested that the circulating HBV are “occult” in nature (i.e. ELISA-negative but DNA-positive).
Applying saliva on fingers for counting bank notes is a common practice in the Indian subcontinent and many other countries of the world. Paper notes may be a source of “horizontal” transmission of HBV as well as other environmentally stable infectious viruses like severe acute respiratory syndrome coronavirus 2, especially if there are cuts/bruises on the oral mucous membrane or skin. However, it was practically not possible to demonstrate experimentally such transmission. Detection of potentially intact/viable and “occult” HBV on currency notes and in considerable numbers poses potential risk of silent transmission of this virus in densely populated cities like Kolkata.
Heavily used paper currency may play a potential role in transmission of infectious viruses like HBV. The present study puts forward a model of horizontal HBV transmission from infected saliva to finger to paper currencies and then from contaminated bank notes to finger to saliva of susceptible humans, especially in places where people have the habit of using saliva for counting bank notes.
Biswas S and Supekar R acknowledge Academy of Scientific and Innovative Research, New Delhi, India. Roy S acknowledges UGC for his SRF. Authors would also like to thank T. Muruganandan (Technical Officer at Council of Scientific and Industrial Research-Indian Institute of Chemical Biology) for his expert help with the atomic force microscopy imaging.
Manuscript source: Invited manuscript
Specialty type: Virology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Cui J, Lei YC, Sergi C S-Editor: Wang DM L-Editor: Filipodia P-Editor: Wang LL
| 1. | Emikpe OB, Oyero OG. In vitro antibiotics sensitivity pattern of some bacteria isolated from Nigerian currency. Res J Biol Sci. 2007;2:209-211. [Cited in This Article: ] |
| 2. | Ahmed MS, Parveen S, Nasreen T, Feroza B. Evaluation of the microbial contamination of Bangladesh paper currency notes (Taka) in circulation. Adv Biol Res. 2010;5:266-271. [Cited in This Article: ] |
| 3. | Sharma S, Sumbali G. Contaminated money in circulation: a review. Int J Recent Sci Res. 2014;5:1533-1540. [Cited in This Article: ] |
| 4. | Girma G, Ketema T, Bacha K. Microbial load and safety of paper currencies from some food vendors in Jimma Town, Southwest Ethiopia. BMC Res Notes. 2014;7:843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Angelakis E, Azhar EI, Bibi F, Yasir M, Al-Ghamdi AK, Ashshi AM, Elshemi AG, Raoult D. Paper money and coins as potential vectors of transmissible disease. Future Microbiol. 2014;9:249-261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Bhattacharya P, Chandra PK, Datta S, Banerjee A, Chakraborty S, Rajendran K, Basu SK, Bhattacharya SK, Chakravarty R. Significant increase in HBV, HCV, HIV and syphilis infections among blood donors in West Bengal, Eastern India 2004-2005: exploratory screening reveals high frequency of occult HBV infection. World J Gastroenterol. 2007;13:3730-3733. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 51] [Cited by in F6Publishing: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Candotti D, Lin CK, Belkhiri D, Sakuldamrongpanich T, Biswas S, Lin S, Teo D, Ayob Y, Allain JP. Occult hepatitis B infection in blood donors from South East Asia: molecular characterisation and potential mechanisms of occurrence. Gut. 2012;61:1744-1753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 8. | Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870-1874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27225] [Cited by in F6Publishing: 26589] [Article Influence: 3323.6] [Reference Citation Analysis (0)] |
| 9. | Zhao Q, Wang Y, Freed D, Fu TM, Gimenez JA, Sitrin RD, Washabaugh MW. Maturation of recombinant hepatitis B virus surface antigen particles. Hum Vaccin. 2006;2:174-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Datta S, Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Roy G, Das S, Das K, Gupta S, Basu K, Basu A, Datta S, Chowdhury A, Banerjee S. Novel point and combo-mutations in the genome of hepatitis B virus-genotype D: characterization and impact on liver disease progression to hepatocellular carcinoma. PLoS One. 2014;9:e110012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 11. | Mondal RK, Khatun M, Ghosh S, Banerjee P, Datta S, Sarkar S, Saha B, Santra A, Banerjee S, Chowdhury A, Datta S. Immune-driven adaptation of hepatitis B virus genotype D involves preferential alteration in B-cell epitopes and replicative attenuation--an insight from human immunodeficiency virus/hepatitis B virus coinfection. Clin Microbiol Infect 2015; 21: 710.e11-710. e20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 10] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Ghosh S, Mondal RK, Banerjee P, Nandi M, Sarkar S, Das K, Santra A, Banerjee S, Chowdhury A, Datta S. Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin Microbiol Infect. 2012;18:E412-E418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Chakravarty R, Pal A. Insights into human immunodeficiency virus-hepatitis B virus co-infection in India. World J Virol. 2015;4:255-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 14. | Thai H, Campo DS, Lara J, Dimitrova Z, Ramachandran S, Xia G, Ganova-Raeva L, Teo CG, Lok A, Khudyakov Y. Convergence and coevolution of hepatitis B virus drug resistance. Nat Commun. 2012;3:789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Ghosh S, Banerjee P, Deny P, Mondal RK, Nandi M, Roychoudhury A, Das K, Banerjee S, Santra A, Zoulim F, Chowdhury A, Datta S. New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013;20:209-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Alemu A. Microbial Contamination of Currency Notes and Coins in Circulation: A Potential Public Health Hazard. Biomed Biotechnol. 2014; 2:46-53. [DOI] [Cited in This Article: ] |
| 17. | Datta S. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. Virol J. 2008;5:156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 100] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 18. | Hollinger FB, Liang TJ. Hepatitis B Virus Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2001: 2971-3036. [Cited in This Article: ] |
| 19. | Bond WW, Favero MS, Petersen NJ, Gravelle CR, Ebert JW, Maynard JE. Survival of hepatitis B virus after drying and storage for one week. Lancet. 1981;1:550-551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 241] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 20. | Goh KT, Ding JL, Monteiro EH, Oon CJ. Hepatitis B infection in households of acute cases. J Epidemiol Community Health. 1985;39:123-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Chakravarty R, Chowdhury A, Chaudhuri S, Santra A, Neogi M, Rajendran K, Panda CK, Chakravarty M. Hepatitis B infection in Eastern Indian families: need for screening of adult siblings and mothers of adult index cases. Public Health. 2005;119:647-654. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 22. | Kavitha R, Kumar K S, Sandesh K, Ramachandran T M, Varghese T. Intrafamilial occurrence of hepatitis B virus (HBV) infection and the profile of liver disease in close relatives of patients with HBV infection. Hepatitis B Annual. 2011;8:4-16. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 23. | De A, Roy S, Sukla S, Ansari A, Biswas S. Occult Hepatitis B Virus Infections (Often with Human Herpesvirus 7 Co-Infection) Detected in Pityriasis rosea Patients: A Pilot Study. Indian J Dermatol. 2017;62:598-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 24. | Ismail AM, Puhazhenthi KS, Sivakumar J, Eapen CE, Kannangai R, Abraham P. Molecular epidemiology and genetic characterization of hepatitis B virus in the Indian subcontinent. Int J Infect Dis. 2014;20:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Saha D, Pal A, Biswas A, Panigrahi R, Sarkar N, Das D, Sarkar J, Guha SK, Saha B, Chakrabarti S, Chakravarty R. Molecular characterization of HBV strains circulating among the treatment-naive HIV/HBV co-infected patients of eastern India. PLoS One. 2014;9:e90432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 26. | Allain JP, Mihaljevic I, Gonzalez-Fraile MI, Gubbe K, Holm-Harritshøj L, Garcia JM, Brojer E, Erikstrup C, Saniewski M, Wernish L, Bianco L, Ullum H, Candotti D, Lelie N, Gerlich WH, Chudy M. Infectivity of blood products from donors with occult hepatitis B virus infection. Transfusion. 2013;53:1405-1415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 109] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 27. | Cao H, Shockey JM. Comparison of TaqMan and SYBR Green qPCR methods for quantitative gene expression in tung tree tissues. J Agric Food Chem. 2012;60:12296-12303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 28. | Hou Y, Zhang H, Miranda L, Lin S. Serious overestimation in quantitative PCR by circular (supercoiled) plasmid standard: microalgal pcna as the model gene. PLoS One. 2010;5:e9545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 29. | Jenison SA, Lemon SM, Baker LN, Newbold JE. Quantitative analysis of hepatitis B virus DNA in saliva and semen of chronically infected homosexual men. J Infect Dis. 1987;156:299-307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 74] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | van der Eijk AA, Niesters HG, Götz HM, Janssen HL, Schalm SW, Osterhaus AD, de Man RA. Paired measurements of quantitative hepatitis B virus DNA in saliva and serum of chronic hepatitis B patients: implications for saliva as infectious agent. J Clin Virol. 2004;29:92-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 34] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 31. | Marie-Cardine A, Mouterde O, Dubuisson S, Buffet-Janvresse C, Mallet E. Salivary transmission in an intrafamilial cluster of hepatitis B. J Pediatr Gastroenterol Nutr. 2002;34:227-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Williams I, Smith MG, Sinha D, Kernan D, Minor-Babin G, Garcia E, Robertson BH, Di Pentima R, Shapiro CN. Hepatitis B virus transmission in an elementary school setting. JAMA. 1997;278:2167-2169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | van Doremalen N, Bushmaker T, Morris DH, Holbrook MG, Gamble A, Williamson BN, Tamin A, Harcourt JL, Thornburg NJ, Gerber SI, Lloyd-Smith JO, de Wit E, Munster VJ. Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1. N Engl J Med. 2020;382:1564-1567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5894] [Cited by in F6Publishing: 5442] [Article Influence: 1360.5] [Reference Citation Analysis (0)] |
| 34. | Kampf G, Todt D, Pfaender S, Steinmann E. Corrigendum to "Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents" [J Hosp Infect 104 (2020) 246-251]. J Hosp Infect. 2020;Online ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2034] [Cited by in F6Publishing: 1892] [Article Influence: 473.0] [Reference Citation Analysis (0)] |