Published online Jun 26, 2020. doi: 10.4252/wjsc.v12.i6.438
Peer-review started: February 29, 2020
First decision: April 25, 2020
Revised: May 13, 2020
Accepted: May 27, 2020
Article in press: May 27, 2020
Published online: June 26, 2020
Mesenchymal stem cells (MSCs) are multipotent stromal cells with great potential for clinical applications. However, little is known about their cell heterogeneity at a single-cell resolution, which severely impedes the development of MSC therapy. In this review, we focus on advances in the identification of novel surface markers and functional subpopulations of MSCs made by single-cell RNA sequencing and discuss their participation in the pathophysiology of stem cells and related diseases. The challenges and future directions of single-cell RNA sequencing in MSCs are also addressed in this review.
Core tip: Mesenchymal stem cells (MSCs) are an important kind of multipotent stroma cells in vivo. Previous studies have focused on large groups of cells. In this review, we focus on advances in the identification of novel surface markers and functional subpopulations of MSCs made by single-cell RNA sequencing and discuss their participation in the pathophysiology of stem cells and related diseases.
- Citation: Zheng G, Xie ZY, Wang P, Wu YF, Shen HY. Recent advances of single-cell RNA sequencing technology in mesenchymal stem cell research. World J Stem Cells 2020; 12(6): 438-447
- URL: https://www.wjgnet.com/1948-0210/full/v12/i6/438.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i6.438
Mesenchymal stem cells (MSCs) are multipotent stromal cells with great potential for clinical applications[1-3]. However, there are many problems that need to be solved before the wide application of MSC therapy. First, MSCs exhibit heterogeneity at multiple levels, for example, among donors and tissue sources and within cell populations[4]. Although the criteria for MSCs were established in 2006 by the International Society of Cell Therapy[5], they need to be improved on the basis of new knowledge. Surface markers for identifying specific groups of MSCs or predicting their potential for cell differentiation remain to be elucidated. MSCs are produced and expanded in vitro to assure the availability of sufficient cell numbers for clinical therapy. Cell morphology changes, proliferation ability decreases, along with other alterations in cell function, and how culture conditions influence MSCs remains unclear. Finally, the efficacy of MSC therapy varies among different clinical studies, and more data are needed to explore the mechanism of immunoregulation and tissue repair[6]. Single-cell sequencing is a powerful tool for characterizing heterogeneous cell populations and identifying novel stem cell types[7-13]. The aims of this review are to emphasize the advances in the identification of novel surface markers and functional subpopulations of MSCs by single-cell RNA sequencing (scRNA-seq) and discuss their participation in the pathophysiology of stem cells and related diseases.
Mesenchymal stem cells are defined as multipotent mesenchymal stromal cells that can be isolated from many adult organs. They were first reported in 1974 by Friedenstein[14] and were described as colony-forming unit fibroblasts. These cells have the capacity to differentiate into mesodermal tissues, such as bone, cartilage, and fat cells[15,16], as well as other tissues, such as myocytes and neural cells[17]. Moreover, the trophic function of MSCs in supporting hematopoietic stem cells (HSCs) is well studied[17]. In preclinical studies, the advantages of suppressing the inflammation and immunoregulation of MSCs have attracted great interest[18,19]. On the basis of these properties, many clinical trials are using MSCs to treat orthopedic diseases, degenerative diseases, and autoimmune diseases affecting single or multiple organs.
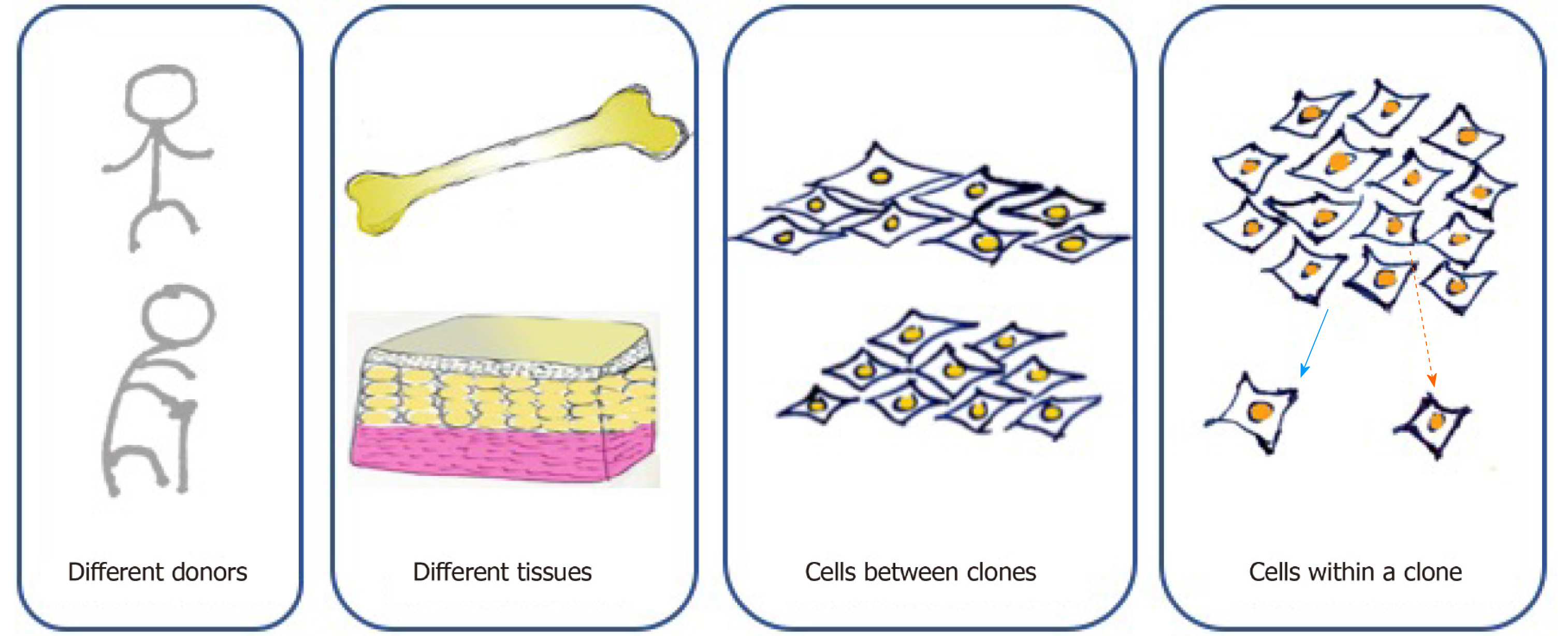
According to the minimal criteria developed by the International Society of Cell Therapy in 2006 for defining MSCs, they must be adherent cells with a spindle-shaped morphology in standard culture conditions; they must express CD105, CD73, and CD90 and lack the expression of CD45, CD34, CD14 or CD11b, CD79alpha or CD19, and HLA-DR surface molecules; and they must be capable of differentiating into osteoblasts, adipocytes, and chondroblasts in vitro[5]. However, cell-to-cell variation exists at multiple levels (Figure 1). First, MSCs can be found in various kinds of connective tissues, even when cultured under standard conditions. MSCs from adipose tissue and bone marrow show different surface markers and differentiation capacities because of distinct biological, chemical, and mechanical stresses in stem cell niches[20]. For MSCs originating from the same tissue, those from different donors present distinct functions due to the influence of age, health condition, and other individual differences[4]. Moreover, MSCs form clones, and cell heterogeneity exists both interclonally and intraclonally. Kuznetsov et al[21] reported that only half of clonal MSC implants differentiated into bone in mice. Extracellular matrix genes and osteogenic transcription factor-related genes show increases in highly osteogenic clones compared to poorly osteogenic clones[22]. MSCs within a clone also present differences in cell morphology and differentiation ability. For instance, cells located at the outer periphery of clones express higher levels of genes (MKI67 and PODXL) related to cell proliferation, while extracellular matrix genes (VCAM1) tend to exhibit higher expression in interior MSCs[23].
To identify cell subsets with specific functions in heterogeneous MSCs, cell surface markers and global molecular signatures have been continuously explored[24]. Single-cell-derived colonies with small, rapidly dividing cells show a high colony-forming efficiency. STRO-1, CD146, and CD271 have been identified as cell surface markers[25,26]. However, markers of proliferation cannot predict the differentiation potential of MSCs, and cell subsets sharing similar surface markers exhibit different chondrogenic differentiation abilities under the same culture conditions[27]. RNA sequencing and microarray analysis have helped to elucidate transcriptional signatures predicting differentiation potential. Osterix and distal-less homeobox5 are the main transcription factors involved in osteoblast differentiation[28], while peroxisome proliferator-activated receptor gamma and CCAAT/enhancer-binding protein alpha are a signature of adipogenic potential[29]. In addition, MSCs with specific surface markers of differentiation potential may present various physiological functions. Studies have proved that CD105+ MSCs exhibit myogenic potential, which could help to repair the infarcted myocardium[30]; however, the differentiation incidence is low in vivo. Other studies have shown that an enhanced paracrine effect could be more important in the myocardial repair of CD105+ MSCs, and the subpopulations need to be further identified[31,32]. CD106 (VCAM-1) is a cell surface protein related to the adhesion of leukocytes to the vascular endothelium[33]. CD106+ MSCs show enhanced multipotency, while their protein expression decreases with extended passaging or differentiation. Moreover, CD106+ MSCs display an immunosuppressive ability by mediating cell-to-cell contact with immune cells, as reported in human placenta[34]. Increasing evidence shows that MSCs contain multiple subsets with specific surface markers. More work is needed to identify these subpopulations and clarify their biological functions.
Cells are the smallest functional unit of organisms. Cellular heterogeneity is a universally acknowledged characteristic of biological tissues, including MSCs. Even in almost pure cell groups, gene expression differs among individual cells because of the variability of the intrinsic regulatory system and extrinsic microenvironment. Single-cell sequencing, which includes single-cell transcriptome-, epigenome-, and genome-sequencing technologies, provides powerful tools for characterizing heterogeneous cell populations[10,13,35]. As scRNA-seq is one of the most widely used of these approaches, this review will mainly discuss the advances of scRNA-seq applications.
Tang et al[36] first reported the methodology for scRNA-seq in 2009, which provides an unbiased view of the transcriptome at a single-cell resolution. Since that time, scRNA-seq strategies have been constantly updated, including methods for cell capture, RNA capture, cDNA amplification, and library establishment and bioinformatics tools for bulk data analyses[37,38]. For example, by using a combination of the one-bead-one-cell droplet approach and a special barcoding strategy, Drop-seq and inDrop systems were developed, which are high-throughput systems that can handle thousands or even tens of thousands of individual cells in a single experimental run[39]. Defining subpopulations within a dataset of MSCs can be achieved by unbiased clustering and differential gene expression analysis[40-42]. More recently, a “pseudotime” trajectory was produced in a reduced-dimension data space, which helps to record the spatial or lineage information of MSCs[35,43]. To date, scRNA-seq has been widely applied in the fields of early embryo development, neuroscience, cancer, and stem cell research.
MSCs can differentiate into osteoblasts, adipocytes, and chondrocytes. Specific surface markers and global molecular signatures have been reported in studies based on bulk cells or tissues[4,44]. Novel functional subsets of MSCs have been identified at a single-cell resolution in recent years[7,9,13,45]. Worthley et al[45] identified a subpopulation of skeletal stem cells in mice that express gremlin 1 as osteochondroreticular stem cells in the bone marrow. This subset of cells can generate osteoblasts, chondrocytes, and reticular marrow stromal cells but not adipocytes. Chan et al[46] reported skeletal stem cells (mSSCs) with a similar differentiation potential in mice. Subsequently, using scRNA-seq, Chan et al[47] identified novel surface markers in human bone-derived cells that were similar to mSSCs. PDPN+CD146-CD73+CD164+ cells are likely enriched in human skeletal stem cells (hSSCs) in the bone. The self-renewal ability and multipotency of the cells are maintained, and injury-induced hSSCs (PDPN+CD146-CD73+CD164+) represent a regenerative response to skeletal injury in a human bone xenograft mouse model. In addition, hSSCs and hSSC-derived subsets express a series of potential hematopoiesis-supportive cytokines, including ANGPT1, CSF1, SDF, IL27, IL7, and SCF. Gene expression was also compared among hSSCs, mSSCs, and iPSC-derived hSSCs, and hSSCs isolated from any source were shown to cluster together. These cells are capable of forming the whole skeletal lineage hierarchy, which suggests great potential for use in regenerative medicine. Endosteal and outer periosteal compartments are both important to bone physiology, and each compartment maintains separate pools of cells separated by the bone cortex. Debnath et al[48] identified periosteal stem cells (PSCs) in mice that form bone in an intramembranous manner, which differ from other skeletal MSCs that mediate endochondral ossification. In addition, the discrete existence of PSCs suggests that bone is composed of separate pools of stem cell progenitors that provide a special derived microenvironment for each type of stem cells (Table 1). To some extent, PSCs are specialized for intramembranous bone formation. The identified subsets contribute to fracture healing as well as the modeling of the bone cortex, providing a promising novel clinical target for bone fracture regeneration.
| Ref. | Marker | Cell | Function | Species |
| Worthley et al[45] | Gremlin 1+ | Osteochondroreticular stem cell | Generate osteoblasts, chondrocytes and reticular marrow stromal cells but not adipocytes | Mouse |
| Chan et al[46] | CD45−Ter119−Tie2− AlphaV+Thy−6C3− CD105−CD200+ | Skeletal stem cells (mSSCs) | Bone cartilage and stromal progenitor | Mouse |
| Chan et al[47] | PDPN+CD146-CD73+CD164+ | Skeletal stem cells (hSSCs) | Response to skeletal injury; express potential hematopoiesis-supportive cytokines | Human |
| Debnath et al[48] | CTSK-mGFP+CD200+CD105− | Periosteal stem cells (PSCs) | Form bone in an intramembranous manner | Mouse |
| Scott et al[9] | Hypermethylated in cancer 1 (Hic1) + | Mesenchymal progenitors (MPs) | Provide stage-specific immunomodulation and trophic and mechanical support | Mouse |
| Merrick et al[51] | Dipeptidyl peptidase-4 (DPP4)/CD26+ | Mesenchymal cells | Give rise to ICAM1+ and CD142+ committed preadipocytes that can differentiate into mature adipocytes | Mouse |
| Schwalie et al[52] | CD142+ | Adipose stem cells | Suppress adipocyte formation in a paracrine manner | Mouse |
Tissue-resident mesenchymal progenitors (MPs) are responsible for tissue maintenance and regeneration. Scott et al[9] revealed hypermethylated in cancer 1 (Hic1) to be a marker for MPs. scRNA-seq and ATAC-seq analysis demonstrated that Hic1+ MPs present distinct functions and lineage potential in skeletal muscle regeneration by providing stage-specific immunomodulation and trophic and mechanical support. Moreover, Giordani et al[49] mapped ten mononuclear cell types in mouse muscle by scRNA-seq and illustrated gene signatures and key discriminating markers in each group, which can help to understand more about muscle-resident cell type identities and muscle diseases.
Regarding adipose tissues, Liu et al[50] performed an analysis of adipose-derived mesenchymal stem cells (ADSCs) from three donors by scRNA-seq, and the resulting high-quality dataset is valuable for distinguishing the heterogeneity of cultured ADSCs at a single-cell resolution. Merrick et al[51] found that dipeptidyl peptidase-4/CD26+ mesenchymal cells are highly proliferative, multipotent progenitors that give rise to ICAM1+ and CD142+ committed preadipocytes that can differentiate into mature adipocytes. The in vivo origin of adipose stem cells is currently poorly understood. Schwalie et al[52] identified distinct subsets of adipose stem cells in the stromal vascular fraction of subcutaneous adipose tissue. The CD142+ group was shown to suppress adipocyte formation in a paracrine manner. The potentially key role of adipogenesis-regulatory cells in regulating adipose tissue plasticity is related to metabolic diseases such as type 2 diabetes.
Other studies have identified subpopulations of Col2a1-creER-marked neonatal chondrocytes that behave as transient mesenchymal precursor cells at the growth plate borderline[53]. With the application of scRNA-seq technology, more subsets and specific surface markers of MSCs have been revealed, which helps not only to predict differentiation potential but also to explain the regulatory network under physiological and pathological conditions.
MSCs can modulate both the innate and adaptive immune systems, including effects on neutrophils, macrophages, dendritic cells, natural killer cells, B lymphocytes, and T lymphocytes[19]. For example, MSCs impede B lymphocytes from differentiating into plasma cells as well as secreting immunoglobulins. They can promote the generation of regulatory T cells while inhibiting the differentiation of helper T cells[19]. The immunosuppression function can be executed via direct cell-cell interactions and paracrine actions. Many molecules secreted by MSCs are responsible for immunosuppression, including TGF-b, IL-10, PGE2, IDO, and NO. Although MSCs have been applied to treat several autoimmune diseases, such as Crohn’s disease, rheumatoid arthritis, and systemic lupus erythematosus, the mechanism underlying the immunosuppressive ability of MSCs in vivo is not clear[1,18].
In addition, MSCs are capable of supporting the maintenance, expansion, and differentiation of HSCs by producing growth factors, chemokines, interleukins, and extracellular matrix molecules. HSCs cotransplanted with MSCs in vivo ameliorated HSC engraftment and improved hematopoietic function recovery. In addition, MSCs secrete chemokines such as Ang-1 and CXCL12 to promote angiogenesis by recruiting endothelial progenitor cells. They can also produce neurotrophic factors that are important in neurogenesis and neurodegenerative diseases, such as amyotrophic lateral sclerosis and multiple sclerosis. The multipotency of MSCs is considered an important function for tissue regeneration and the treatment of degenerative diseases. However, less than 1% of transplanted MSCs could be found in the host bone of a patient who suffered from severe osteogenesis imperfecta. Similar observations were made in patients with eye diseases who were receiving MSC therapy, and no clear evidence showed MSC engraftment into the retina. Other functions, such as the roles of trophic factors, should also be considered in MSC therapy.
Although the importance of MSCs in bone marrow in supporting HSCs has been recognized since 1974[14], the molecular complexity of this relationship and its response to stress are unclear. Tikhonova et al[54] mapped the transcriptional signatures of bone marrow vascular, perivascular, and osteoblast cells in mice at single-cell resolution and revealed novel cellular subsets and cellular sources of pro-hematopoietic factors in vivo. The vascular notch delta-like ligands (Dll1,4) play critical roles in HSC differentiation and lineage commitment and are downregulated under stress-induced changes. These authors elucidated the cellular architecture of the bone marrow niche and revealed a heterogeneous molecular landscape that responds to stress. Severe et al[24] reported that CD73+ BMSCs contribute to hematopoietic stem and progenitor cell engraftment and acute hematopoietic recovery.
Currently, effective cell surface markers are available to identify hematopoietic cells, and it is still difficult to elucidate the interactions with classical myeloid and lymphoid cells in vivo. Jaitin et al[55] analyzed spleen tissues using scRNA-seq, and single-cell transcriptional states in dendritic cells and hematopoietic cells were considered together to reveal gene-module activity and cell-type heterogeneity in both steady-state conditions and pathogen activation states.
Large numbers of MSCs are needed for clinical therapy, requiring cell expansion ex vivo. Although MSCs are capable of self-renewal and proliferation, these abilities decrease with time when the cells are cultured in vitro along with other functional changes[56]. With cell cycle arrest and the loss of proliferation, some MSCs may undergo senescence[56,57]. Oxidative stress and the dysregulation of regulatory factors associated with differentiation are related to the decreased differentiation potential of senescent MSCs. Bork et al[58] revealed that DNA methylation identified at specific CpG sites is a typical epigenetic signature. When expanded in vitro, MSCs experience genomic DNA damage, and attempts to repair DNA damage seem to promote senescence, which could help defend against cell death. Other studies have shown that the functional decline associated with age can be reversed by manipulating epigenetic factors that are dysregulated. This could shed new light on the epigenetics of cell aging to improve therapeutic potential[57].
Cells cultured in vitro are exposed to various conditions. The properties of the biomaterial interface, such as its topography, stiffness, and chemistry, can lead to transcriptional variations[59-61]. Darnell et al[62] found that the stiffness of hydrogels could regulate cytokine secretion by mouse mesenchymal stem cells. These authors further revealed many stiffness-sensitive genes by RNA-seq, which showed that stiffness can regulate the expression of MSC immunomodulatory markers. In addition, MSC signatures change as cell confluence increases in vitro, and 100% confluent MSCs may exhibit compromised pro-angiogenesis properties[63]. Other studies are building upon the complexity of the niches produced in vitro to create a tissue-like system[59]; however, there is a lack of research at a single-cell resolution regarding how cells sense and respond to the ex vivo culture microenvironment.
Lazarus et al[64] reported the first use of MSCs as a cellular pharmaceutical agent in humans in 1995. Since that time, the number of clinical trials of MSCs has increased worldwide. The niche-like regenerative properties and anti-inflammatory ability of MSCs make them candidates for the treatment of acute tissue injury, chronic degenerative diseases, and inflammatory diseases[1]. Conditional approval for MSC treatment in children with GvHD (graft-versus-host disease) was secured in 2012 in Canada, which was a historic event in the application of MSC therapy in the clinic. In addition, the use of MSCs to treat Crohn’s-related enterocutaneous fistular disease was approved by the European Commission in 2018. Other high-quality clinical trials have focused on heart disease. Bartunek et al[65] performed an MSC trial (NCT00810238) of heart failure secondary to ischemic cardiomyopathy in 2013, which showed improved cardiac outcomes. However, three years later, another phase 3 trial (NCT01768702) of the same MSC therapy for the treatment of chronic advanced ischemic heart failure was performed. No significant difference was found between the MSC-treated and placebo groups from baseline to 39 wk in the primary endpoint. The authors also evaluated ventricular remodeling at 52 wk. The data revealed an inverted U-shaped dose curve with worse outcomes at a higher dose delivery of MSCs[66].
Although MSCs are effective for treating some cardiac patients, the underlying molecular mechanism is poorly understood. Lescroart et al[67] revealed that the population of Mesp1 cardiovascular progenitors are the progenitors of distinct cell lineages and regions of the heart, identifying molecular characteristics in the early stage of mouse gastrulation. In addition to cardiomyocytes, interstitial cells, including fibroblasts and vascular and immune cells, are also important for heart repair. Farbehi et al[68] identified more than 30 populations, representing nine cell lineages. The novel fibroblast lineage trajectory observed under both sham and myocardial infarction and in myofibroblasts leads to a uniquely activated cell state. To evaluate whether novel cardiovascular progenitors can differentiate into cardiomyocytes, scRNA-seq was used, and laminin-221 was identified as the most likely cardiac laminin that was highly expressed during development. Moreover, cells coated with laminin-221 could be reproducibly differentiated at different time points[69]. Future sc-RNA-seq studies will help to identify disease-related phenotypes and the transition trajectories of cells during disease development.
scRNA-seq has been applied and improved for more than ten years since 2009; it was highlighted as the “Method of the Year” in 2013[70] and has recently become a routine approach for investigating cell heterogeneity. The challenges and limitations of the technology should also be considered. Eleven grand challenges in single-cell data science were reviewed by Lahnemann et al[71]. For example, scRNA-seq data may show zero results, where a given gene has no unique molecular identifiers or reads mapping to it in a given cell. Such sparsity of scRNA-seq results can affect downstream analyses, which necessitates further methodological development. Regarding the application of MSCs, the lack of information other than transcript levels could lead to inaccurate analysis. Then, although novel cell surface markers and cell subpopulations may be identified by scRNA-seq, few interactions with surrounding cells and organisms can be demonstrated.
In the future, more efforts are needed to explore methods that provide multimodal data[35]. Such methods may include the simultaneous profiling of multiple types of molecular data within a single cell, for instance, scRNA-seq coupled with DNA sequence or protein information. Other types of technologies may allow the detection and analysis of different kinds of data together. If the combination of multiple types of single-cell data across different experiments or experimental conditions can be achieved, we will obtain new information about MSCs from a transcriptome-centric perspective, which will undoubtedly improve the efficiency of MSC therapy.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tanabe S S-Editor: Wang J L-Editor: Wang TQ E-Editor: Xing YX
| 1. | Galipeau J, Sensébé L. Mesenchymal Stromal Cells: Clinical Challenges and Therapeutic Opportunities. Cell Stem Cell. 2018;22:824-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1009] [Cited by in F6Publishing: 1011] [Article Influence: 168.5] [Reference Citation Analysis (0)] |
| 2. | Naji A, Eitoku M, Favier B, Deschaseaux F, Rouas-Freiss N, Suganuma N. Biological functions of mesenchymal stem cells and clinical implications. Cell Mol Life Sci. 2019;76:3323-3348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 263] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 3. | Phinney DG, Galipeau J, Krampera M, Martin I, Shi Y, Sensebe L. MSCs: science and trials. Nat Med. 2013;19:812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 4. | McLeod CM, Mauck RL. On the origin and impact of mesenchymal stem cell heterogeneity: new insights and emerging tools for single cell analysis. Eur Cell Mater. 2017;34:217-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 118] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 5. | Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11055] [Cited by in F6Publishing: 11776] [Article Influence: 692.7] [Reference Citation Analysis (1)] |
| 6. | Shi Y, Hu G, Su J, Li W, Chen Q, Shou P, Xu C, Chen X, Huang Y, Zhu Z, Huang X, Han X, Xie N, Ren G. Mesenchymal stem cells: a new strategy for immunosuppression and tissue repair. Cell Res. 2010;20:510-518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 380] [Cited by in F6Publishing: 410] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 7. | Kumar P, Tan Y, Cahan P. Understanding development and stem cells using single cell-based analyses of gene expression. Development. 2017;144:17-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 8. | LeBleu VS, Neilson EG. Origin and functional heterogeneity of fibroblasts. FASEB J. 2020;34:3519-3536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 116] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 9. | Scott RW, Arostegui M, Schweitzer R, Rossi FMV, Underhill TM. Hic1 Defines Quiescent Mesenchymal Progenitor Subpopulations with Distinct Functions and Fates in Skeletal Muscle Regeneration. Cell Stem Cell. 2019;25:797-813.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 112] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 10. | Tanay A, Regev A. Scaling single-cell genomics from phenomenology to mechanism. Nature. 2017;541:331-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 490] [Cited by in F6Publishing: 449] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 11. | Xiong X, Kuang H, Liu T, Lin JD. A Single-Cell Perspective of the Mammalian Liver in Health and Disease. Hepatology. 2020;71:1467-1473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 12. | Wu H, Humphreys BD. The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int. 2017;92:1334-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Wen L, Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016;17:71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 117] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 14. | Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17:331-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 998] [Cited by in F6Publishing: 909] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 15. | Collas P. Programming differentiation potential in mesenchymal stem cells. Epigenetics. 2010;5:476-482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Barry F, Murphy M. Mesenchymal stem cells in joint disease and repair. Nat Rev Rheumatol. 2013;9:584-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 279] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 17. | Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2360] [Cited by in F6Publishing: 2512] [Article Influence: 167.5] [Reference Citation Analysis (0)] |
| 18. | Castro-Manrreza ME, Montesinos JJ. Immunoregulation by mesenchymal stem cells: biological aspects and clinical applications. J Immunol Res. 2015;2015:394917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 265] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 19. | Wang Y, Chen X, Cao W, Shi Y. Plasticity of mesenchymal stem cells in immunomodulation: pathological and therapeutic implications. Nat Immunol. 2014;15:1009-1016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 830] [Cited by in F6Publishing: 949] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 20. | Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1411] [Cited by in F6Publishing: 1346] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 21. | Kuznetsov SA, Mankani MH, Bianco P, Robey PG. Enumeration of the colony-forming units-fibroblast from mouse and human bone marrow in normal and pathological conditions. Stem Cell Res. 2009;2:83-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | Elsafadi M, Manikandan M, Atteya M, Hashmi JA, Iqbal Z, Aldahmash A, Alfayez M, Kassem M, Mahmood A. Characterization of Cellular and Molecular Heterogeneity of Bone Marrow Stromal Cells. Stem Cells Int. 2016;2016:9378081. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Ylöstalo J, Bazhanov N, Prockop DJ. Reversible commitment to differentiation by human multipotent stromal cells in single-cell-derived colonies. Exp Hematol. 2008;36:1390-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 24. | Severe N, Karabacak NM, Gustafsson K, Baryawno N, Courties G, Kfoury Y, Kokkaliaris KD, Rhee C, Lee D, Scadden EW, Garcia-Robledo JE, Brouse T, Nahrendorf M, Toner M, Scadden DT. Stress-Induced Changes in Bone Marrow Stromal Cell Populations Revealed through Single-Cell Protein Expression Mapping. Cell Stem Cell. 2019;25:570-583.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 25. | Ning H, Lin G, Lue TF, Lin CS. Mesenchymal stem cell marker Stro-1 is a 75 kd endothelial antigen. Biochem Biophys Res Commun. 2011;413:353-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 26. | Simmons PJ, Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55-62. [PubMed] [Cited in This Article: ] |
| 27. | Dickinson SC, Sutton CA, Brady K, Salerno A, Katopodi T, Williams RL, West CC, Evseenko D, Wu L, Pang S, Ferro de Godoy R, Goodship AE, Péault B, Blom AW, Kafienah W, Hollander AP. The Wnt5a Receptor, Receptor Tyrosine Kinase-Like Orphan Receptor 2, Is a Predictive Cell Surface Marker of Human Mesenchymal Stem Cells with an Enhanced Capacity for Chondrogenic Differentiation. Stem Cells. 2017;35:2280-2291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 28. | Lee MH, Kwon TG, Park HS, Wozney JM, Ryoo HM. BMP-2-induced Osterix expression is mediated by Dlx5 but is independent of Runx2. Biochem Biophys Res Commun. 2003;309:689-694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 319] [Cited by in F6Publishing: 308] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 29. | Rosen ED, Hsu CH, Wang X, Sakai S, Freeman MW, Gonzalez FJ, Spiegelman BM. C/EBPalpha induces adipogenesis through PPARgamma: a unified pathway. Genes Dev. 2002;16:22-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1124] [Cited by in F6Publishing: 1061] [Article Influence: 48.2] [Reference Citation Analysis (0)] |
| 30. | Vilahur G, Oñate B, Cubedo J, Béjar MT, Arderiu G, Peña E, Casaní L, Gutiérrez M, Capdevila A, Pons-Lladó G, Carreras F, Hidalgo A, Badimon L. Allogenic adipose-derived stem cell therapy overcomes ischemia-induced microvessel rarefaction in the myocardium: systems biology study. Stem Cell Res Ther. 2017;8:52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 31. | Silva GV, Litovsky S, Assad JA, Sousa AL, Martin BJ, Vela D, Coulter SC, Lin J, Ober J, Vaughn WK, Branco RV, Oliveira EM, He R, Geng YJ, Willerson JT, Perin EC. Mesenchymal stem cells differentiate into an endothelial phenotype, enhance vascular density, and improve heart function in a canine chronic ischemia model. Circulation. 2005;111:150-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 554] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 32. | Noiseux N, Gnecchi M, Lopez-Ilasaca M, Zhang L, Solomon SD, Deb A, Dzau VJ, Pratt RE. Mesenchymal stem cells overexpressing Akt dramatically repair infarcted myocardium and improve cardiac function despite infrequent cellular fusion or differentiation. Mol Ther. 2006;14:840-850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 370] [Cited by in F6Publishing: 324] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 33. | Alon R, Kassner PD, Carr MW, Finger EB, Hemler ME, Springer TA. The integrin VLA-4 supports tethering and rolling in flow on VCAM-1. J Cell Biol. 1995;128:1243-1253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 526] [Cited by in F6Publishing: 558] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 34. | Ren G, Zhao X, Zhang L, Zhang J, L'Huillier A, Ling W, Roberts AI, Le AD, Shi S, Shao C, Shi Y. Inflammatory cytokine-induced intercellular adhesion molecule-1 and vascular cell adhesion molecule-1 in mesenchymal stem cells are critical for immunosuppression. J Immunol. 2010;184:2321-2328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 483] [Cited by in F6Publishing: 461] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 35. | Stuart T, Satija R. Integrative single-cell analysis. Nat Rev Genet. 2019;20:257-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 604] [Cited by in F6Publishing: 688] [Article Influence: 137.6] [Reference Citation Analysis (0)] |
| 36. | Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1968] [Cited by in F6Publishing: 2082] [Article Influence: 138.8] [Reference Citation Analysis (0)] |
| 37. | Picelli S, Björklund ÅK, Faridani OR, Sagasser S, Winberg G, Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods. 2013;10:1096-1098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1535] [Cited by in F6Publishing: 1468] [Article Influence: 133.5] [Reference Citation Analysis (0)] |
| 38. | Ramsköld D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777-782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1078] [Cited by in F6Publishing: 1068] [Article Influence: 97.1] [Reference Citation Analysis (0)] |
| 39. | Ziegenhain C, Vieth B, Parekh S, Reinius B, Guillaumet-Adkins A, Smets M, Leonhardt H, Heyn H, Hellmann I, Enard W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol Cell. 2017;65:631-643.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 855] [Cited by in F6Publishing: 817] [Article Influence: 116.7] [Reference Citation Analysis (0)] |
| 40. | Campbell BB, Light N, Fabrizio D, Zatzman M, Fuligni F, de Borja R, Davidson S, Edwards M, Elvin JA, Hodel KP, Zahurancik WJ, Suo Z, Lipman T, Wimmer K, Kratz CP, Bowers DC, Laetsch TW, Dunn GP, Johanns TM, Grimmer MR, Smirnov IV, Larouche V, Samuel D, Bronsema A, Osborn M, Stearns D, Raman P, Cole KA, Storm PB, Yalon M, Opocher E, Mason G, Thomas GA, Sabel M, George B, Ziegler DS, Lindhorst S, Issai VM, Constantini S, Toledano H, Elhasid R, Farah R, Dvir R, Dirks P, Huang A, Galati MA, Chung J, Ramaswamy V, Irwin MS, Aronson M, Durno C, Taylor MD, Rechavi G, Maris JM, Bouffet E, Hawkins C, Costello JF, Meyn MS, Pursell ZF, Malkin D, Tabori U, Shlien A. Comprehensive Analysis of Hypermutation in Human Cancer. Cell. 2017;171:1042-1056.e10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 529] [Cited by in F6Publishing: 509] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 41. | Kornblau SM, Ruvolo PP, Wang RY, Battula VL, Shpall EJ, Ruvolo VR, McQueen T, Qui Y, Zeng Z, Pierce S, Jacamo R, Yoo SY, Le PM, Sun J, Hail N Jr, Konopleva M, Andreeff M. Distinct protein signatures of acute myeloid leukemia bone marrow-derived stromal cells are prognostic for patient survival. Haematologica. 2018;103:810-821. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 42. | Miragaia RJ, Gomes T, Chomka A, Jardine L, Riedel A, Hegazy AN, Whibley N, Tucci A, Chen X, Lindeman I, Emerton G, Krausgruber T, Shields J, Haniffa M, Powrie F, Teichmann SA. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity. 2019;50:493-504.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 335] [Cited by in F6Publishing: 283] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 43. | Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1270] [Cited by in F6Publishing: 1086] [Article Influence: 181.0] [Reference Citation Analysis (0)] |
| 44. | Mo M, Wang S, Zhou Y, Li H, Wu Y. Mesenchymal stem cell subpopulations: phenotype, property and therapeutic potential. Cell Mol Life Sci. 2016;73:3311-3321. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 45. | Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, Levin D, Schwartz MG, Uygur A, Hayakawa Y, Gross S, Renz BW, Setlik W, Martinez AN, Chen X, Nizami S, Lee HG, Kang HP, Caldwell JM, Asfaha S, Westphalen CB, Graham T, Jin G, Nagar K, Wang H, Kheirbek MA, Kolhe A, Carpenter J, Glaire M, Nair A, Renders S, Manieri N, Muthupalani S, Fox JG, Reichert M, Giraud AS, Schwabe RF, Pradere JP, Walton K, Prakash A, Gumucio D, Rustgi AK, Stappenbeck TS, Friedman RA, Gershon MD, Sims P, Grikscheit T, Lee FY, Karsenty G, Mukherjee S, Wang TC. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 470] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 46. | Chan CK, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, Tevlin R, Seita J, Vincent-Tompkins J, Wearda T, Lu WJ, Senarath-Yapa K, Chung MT, Marecic O, Tran M, Yan KS, Upton R, Walmsley GG, Lee AS, Sahoo D, Kuo CJ, Weissman IL, Longaker MT. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 410] [Cited by in F6Publishing: 493] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 47. | Chan CKF, Gulati GS, Sinha R, Tompkins JV, Lopez M, Carter AC, Ransom RC, Reinisch A, Wearda T, Murphy M, Brewer RE, Koepke LS, Marecic O, Manjunath A, Seo EY, Leavitt T, Lu WJ, Nguyen A, Conley SD, Salhotra A, Ambrosi TH, Borrelli MR, Siebel T, Chan K, Schallmoser K, Seita J, Sahoo D, Goodnough H, Bishop J, Gardner M, Majeti R, Wan DC, Goodman S, Weissman IL, Chang HY, Longaker MT. Identification of the Human Skeletal Stem Cell. Cell. 2018;175:43-56.e21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 349] [Article Influence: 69.8] [Reference Citation Analysis (0)] |
| 48. | Debnath S, Yallowitz AR, McCormick J, Lalani S, Zhang T, Xu R, Li N, Liu Y, Yang YS, Eiseman M, Shim JH, Hameed M, Healey JH, Bostrom MP, Landau DA, Greenblatt MB. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562:133-139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 348] [Article Influence: 58.0] [Reference Citation Analysis (0)] |
| 49. | Giordani L, He GJ, Negroni E, Sakai H, Law JYC, Siu MM, Wan R, Corneau A, Tajbakhsh S, Cheung TH, Le Grand F. High-Dimensional Single-Cell Cartography Reveals Novel Skeletal Muscle-Resident Cell Populations. Mol Cell. 2019;74:609-621.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 201] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 50. | Liu X, Xiang Q, Xu F, Huang J, Yu N, Zhang Q, Long X, Zhou Z. Single-cell RNA-seq of cultured human adipose-derived mesenchymal stem cells. Sci Data. 2019;6:190031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 47] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 51. | Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, Seale P. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364:eaav2501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 343] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 52. | Schwalie PC, Dong H, Zachara M, Russeil J, Alpern D, Akchiche N, Caprara C, Sun W, Schlaudraff KU, Soldati G, Wolfrum C, Deplancke B. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559:103-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 274] [Article Influence: 45.7] [Reference Citation Analysis (2)] |
| 53. | Mizuhashi K, Nagata M, Matsushita Y, Ono W, Ono N. Growth Plate Borderline Chondrocytes Behave as Transient Mesenchymal Precursor Cells. J Bone Miner Res. 2019;34:1387-1392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 36] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 54. | Tikhonova AN, Dolgalev I, Hu H, Sivaraj KK, Hoxha E, Cuesta-Domínguez Á, Pinho S, Akhmetzyanova I, Gao J, Witkowski M, Guillamot M, Gutkin MC, Zhang Y, Marier C, Diefenbach C, Kousteni S, Heguy A, Zhong H, Fooksman DR, Butler JM, Economides A, Frenette PS, Adams RH, Satija R, Tsirigos A, Aifantis I. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 404] [Cited by in F6Publishing: 515] [Article Influence: 103.0] [Reference Citation Analysis (0)] |
| 55. | Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776-779. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1265] [Cited by in F6Publishing: 1205] [Article Influence: 120.5] [Reference Citation Analysis (0)] |
| 56. | Turinetto V, Vitale E, Giachino C. Senescence in Human Mesenchymal Stem Cells: Functional Changes and Implications in Stem Cell-Based Therapy. Int J Mol Sci. 2016;17:1164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 312] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 57. | Chen D, Kerr C. The Epigenetics of Stem Cell Aging Comes of Age. Trends Cell Biol. 2019;29:563-568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 58. | Bork S, Pfister S, Witt H, Horn P, Korn B, Ho AD, Wagner W. DNA methylation pattern changes upon long-term culture and aging of human mesenchymal stromal cells. Aging Cell. 2010;9:54-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 312] [Cited by in F6Publishing: 314] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 59. | Donnelly H, Salmeron-Sanchez M, Dalby MJ. Designing stem cell niches for differentiation and self-renewal. J R Soc Interface. 2018;15:20180388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 60. | Barradas AM, Monticone V, Hulsman M, Danoux C, Fernandes H, Tahmasebi Birgani Z, Barrère-de Groot F, Yuan H, Reinders M, Habibovic P, van Blitterswijk C, de Boer J. Molecular mechanisms of biomaterial-driven osteogenic differentiation in human mesenchymal stromal cells. Integr Biol (Camb). 2013;5:920-931. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 61. | Fahy N, Alini M, Stoddart MJ. Mechanical stimulation of mesenchymal stem cells: Implications for cartilage tissue engineering. J Orthop Res. 2018;36:52-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 62. | Darnell M, O'Neil A, Mao A, Gu L, Rubin LL, Mooney DJ. Material microenvironmental properties couple to induce distinct transcriptional programs in mammalian stem cells. Proc Natl Acad Sci U S A. 2018;115:E8368-E8377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 71] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 63. | Ren J, Wang H, Tran K, Civini S, Jin P, Castiello L, Feng J, Kuznetsov SA, Robey PG, Sabatino M, Stroncek DF. Human bone marrow stromal cell confluence: effects on cell characteristics and methods of assessment. Cytotherapy. 2015;17:897-911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 64. | Lazarus HM, Haynesworth SE, Gerson SL, Rosenthal NS, Caplan AI. Ex vivo expansion and subsequent infusion of human bone marrow-derived stromal progenitor cells (mesenchymal progenitor cells): implications for therapeutic use. Bone Marrow Transplant. 1995;16:557-564. [PubMed] [Cited in This Article: ] |
| 65. | Bartunek J, Behfar A, Dolatabadi D, Vanderheyden M, Ostojic M, Dens J, El Nakadi B, Banovic M, Beleslin B, Vrolix M, Legrand V, Vrints C, Vanoverschelde JL, Crespo-Diaz R, Homsy C, Tendera M, Waldman S, Wijns W, Terzic A. Cardiopoietic stem cell therapy in heart failure: the C-CURE (Cardiopoietic stem Cell therapy in heart failURE) multicenter randomized trial with lineage-specified biologics. J Am Coll Cardiol. 2013;61:2329-2338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 355] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 66. | Bartunek J, Davison B, Sherman W, Povsic T, Henry TD, Gersh B, Metra M, Filippatos G, Hajjar R, Behfar A, Homsy C, Cotter G, Wijns W, Tendera M, Terzic A. Congestive Heart Failure Cardiopoietic Regenerative Therapy (CHART-1) trial design. Eur J Heart Fail. 2016;18:160-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 67. | Lescroart F, Wang X, Lin X, Swedlund B, Gargouri S, Sànchez-Dànes A, Moignard V, Dubois C, Paulissen C, Kinston S, Göttgens B, Blanpain C. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science. 2018;359:1177-1181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 68. | Farbehi N, Patrick R, Dorison A, Xaymardan M, Janbandhu V, Wystub-Lis K, Ho JW, Nordon RE, Harvey RP. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8:e43882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 301] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 69. | Yap L, Wang JW, Moreno-Moral A, Chong LY, Sun Y, Harmston N, Wang X, Chong SY, Öhman MK, Wei H, Bunte R, Gosh S, Cook S, Hovatta O, de Kleijn DPV, Petretto E, Tryggvason K. In Vivo Generation of Post-infarct Human Cardiac Muscle by Laminin-Promoted Cardiovascular Progenitors. Cell Rep. 2019;26:3231-3245.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 70. | Method of the year 2013. Nat Methods. 2014;11:1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 71. | Lähnemann D, Köster J, Szczurek E, McCarthy DJ, Hicks SC, Robinson MD, Vallejos CA, Campbell KR, Beerenwinkel N, Mahfouz A, Pinello L, Skums P, Stamatakis A, Attolini CS, Aparicio S, Baaijens J, Balvert M, Barbanson B, Cappuccio A, Corleone G, Dutilh BE, Florescu M, Guryev V, Holmer R, Jahn K, Lobo TJ, Keizer EM, Khatri I, Kielbasa SM, Korbel JO, Kozlov AM, Kuo TH, Lelieveldt BPF, Mandoiu II, Marioni JC, Marschall T, Mölder F, Niknejad A, Raczkowski L, Reinders M, Ridder J, Saliba AE, Somarakis A, Stegle O, Theis FJ, Yang H, Zelikovsky A, McHardy AC, Raphael BJ, Shah SP, Schönhuth A. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 666] [Cited by in F6Publishing: 520] [Article Influence: 130.0] [Reference Citation Analysis (1)] |