Published online May 26, 2020. doi: 10.4252/wjsc.v12.i5.351
Peer-review started: January 12, 2020
First decision: February 25, 2020
Revised: March 27, 2020
Accepted: April 24, 2020
Article in press: April 24, 2020
Published online: May 26, 2020
Mesenchymal stem cells (MSCs) are a heterogeneous population that can be isolated from various tissues, including bone marrow, adipose tissue, umbilical cord blood, and craniofacial tissue. MSCs have attracted increasingly more attention over the years due to their regenerative capacity and function in immunomodulation. The foundation of tissue regeneration is the potential of cells to differentiate into multiple cell lineages and give rise to multiple tissue types. In addition,the immunoregulatory function of MSCs has provided insights into therapeutic treatments for immune-mediated diseases. DNA methylation and demethylation are important epigenetic mechanisms that have been shown to modulate embryonic stem cell maintenance, proliferation, differentiation and apoptosis by activating or suppressing a number of genes. In most studies, DNA hypermethylation is associated with gene suppression, while hypomethylation or demethylation is associated with gene activation. The dynamic balance of DNA methylation and demethylation is required for normal mammalian development and inhibits the onset of abnormal phenotypes. However, the exact role of DNA methylation and demethylation in MSC-based tissue regeneration and immunomodulation requires further investigation. In this review, we discuss how DNA methylation and demethylation function in multi-lineage cell differentiation and immunomodulation of MSCs based on previously published work. Furthermore, we discuss the implications of the role of DNA methylation and demethylation in MSCs for the treatment of metabolic or immune-related diseases.
Core tip: Mesenchymal stem cells (MSCs) harbor the capacity to regenerate diverse tissues and can also perform key immunomodulatory functions. DNA methylation and demethylation are known to modulate stem cell maintenance and differentiation in embryonic stem cells. However, the role of DNA methylation and demethylation in MSC-based tissue regeneration and immunomodulation requires further investigation. In this review, we discuss how DNA methylation and demethylation function in multi-lineage cell differentiation and immunomodulation of MSCs based on previously published work. In addition, we discuss the implications of the role of DNA methylation and demethylation in MSCs for the treatment of metabolic or immune-related diseases.
- Citation: Xin TY, Yu TT, Yang RL. DNA methylation and demethylation link the properties of mesenchymal stem cells: Regeneration and immunomodulation. World J Stem Cells 2020; 12(5): 351-358
- URL: https://www.wjgnet.com/1948-0210/full/v12/i5/351.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i5.351
DNA methylation and demethylation are two vital epigenetic regulatory mechanisms for gene expression. DNA cytosine methylation is a frequently occurring process that is orchestrated by DNA methyltransferases (DNMTs), which generate 5-methylcytosine (5mC)[1]. The methylation process at the 5th cytosine can be reversible, which is called DNA demethylation. This process has received increased attention over recent years. Increasingly more researchers began to identify enzymes that could generate 5-hydroxymethylcytosine (5hmC) from 5mC in mammalian cells. For the first time, in 2009, TET1 was shown to convert 5mC into 5-hmC[2]. Thereafter, all three of the TET family proteins (TET1, TET2, and TET3) were demonstrated to catalyze a similar hydroxymethylation reaction[3]. TET family proteins are also receiving increased attention because of their function in DNA demethylation.
In addition to their function in multi-lineage differentiation and tissue regeneration[4], mesenchymal stem cells (MSCs) also display profound immunomodu-lation capacity via a sophisticated molecular network[5]. DNA methylation and demethylation are known to modulate stem cell maintenance and differentiation by activating or suppressing an array of genes[6]. Previous research on DNA methylation and demethylation has primarily focused on embryonic stem cells and neural systems. Nevertheless, how DNA methylation and demethylation impact MSC function remains elusive. Here, we discuss recent studies concerning the effect of DNA methylation and demethylation on MSC-based regeneration and immunomodulation.
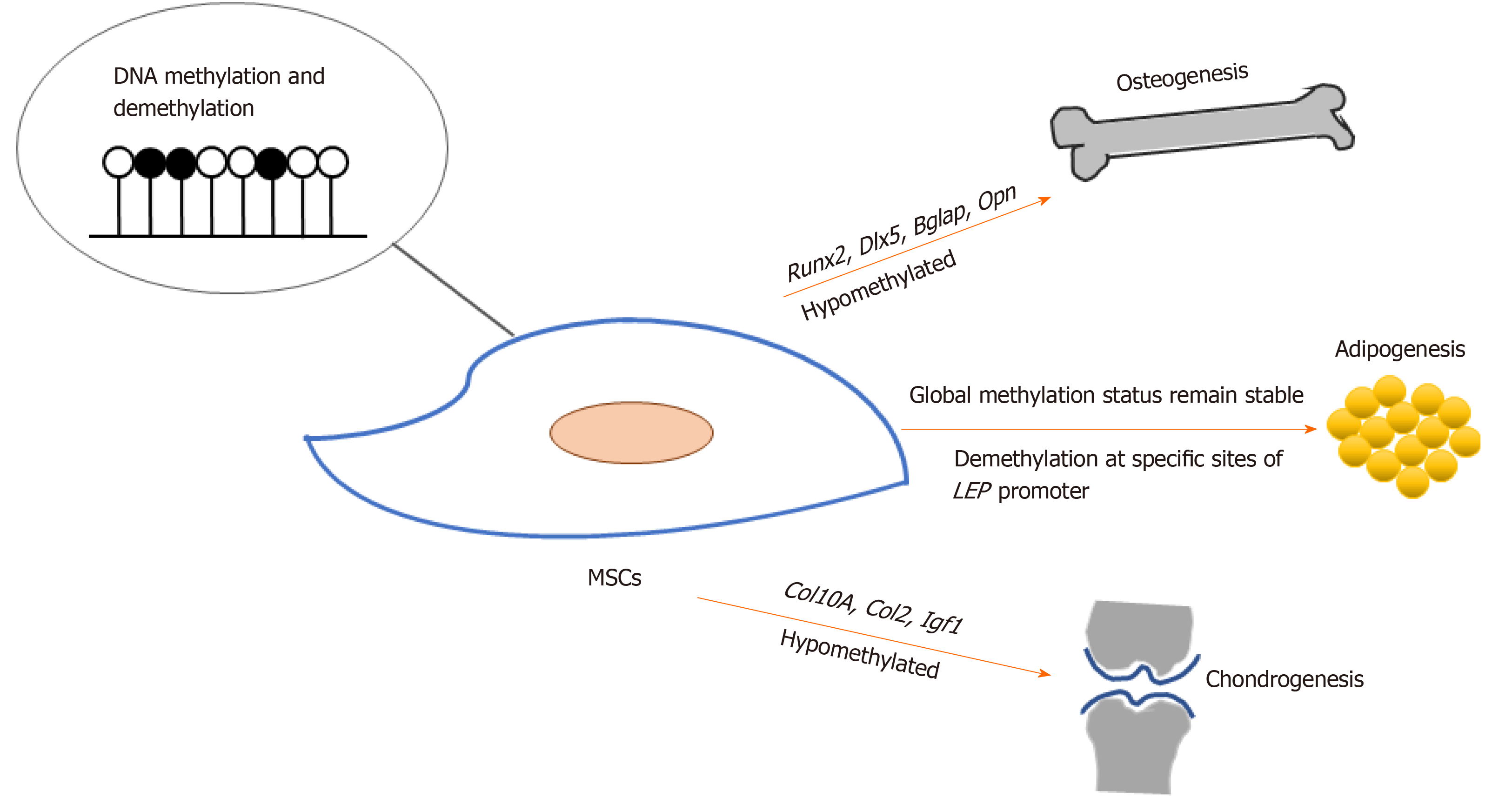
MSCs hold promising potential for regenerative medicine due to their capacity for self-renewal and multi-lineage differentiation into tissue-specific cells, which include osteoblasts, chondrocytes, and adipocytes. During osteogenic differentiation of MSCs, osteogenic-specific genes such as RUNX2, OPN, COX2, ALP, and OCN[7-11], which are regulated by DNA methylation, showed increased expression and decreased DNA methylation. Demethylation was observed at specific CpG regions in the promoters of osteogenic lineage-specific genes, including Runx2, Dlx5, Bglap, and Osterix, during osteogenic differentiation in adipose-derived MSCs (Ad-MSCs). Upon demethylation inhibition, osteogenic gene expression became down-regulated[12]. On the other hand, Daniunaite et al[13] found that genes encoding the main pluripotency factors, such as Nanog and Sox2, showed decreased gene expression along with decreased 5hmC levels during the osteogenic differentiation of Ad-MSCs.
In another study on Ad-MSCs, an age-related decline in proliferation was observed. Ad-MSCs isolated from old donors showed significantly impaired osteogenic differentiation capacity compared to young donors. Furthermore, decreased expression of Nanog, Oct4, and Lin28A and increased expression of Sox2 were observed. A simultaneous decrease of global 5hmC in Ad-MSCs from old donors also occurred. When 5-azacytidine (5-Aza), a DNMT inhibitor, was used to treat Ad-MSCs from old donors, increased global 5hmC and increased TET2 and TET3 expression were observed, which was accompanied by an increase in osteogenic differentiation capacity[14]. These results suggest that global DNA demethylation levels correlate with the osteogenesis capacity of MSCs, and that DNMT inhibitors could down-regulate DNA methylation to improve osteogenesis. Notably, an additional study by Kornicka et al[15] drew similar conclusions.
Bone marrow MSCs (BMMSCs) are a population of multipotent stem cells isolated from bone marrow that harbor the capacity for self-renewal and multi-lineage differentiation. The osteogenic differentiation of BMMSCs is also regulated by dynamic changes, as well as a balance of DNA methylation and demethylation. Bone loss caused by mechanical unloading is partially due to the impaired regeneration capacity of BMMSCs[16]. When mechanical stimuli were rescued, Dnmt3b was released from the Shh gene promoter, thus leading to promoter demethylation and up-regulated gene expression. Hedgehog signal was then activated by Shh, promoting BMMSCs to differentiate into osteoblasts[17]. Yang et al[18] found that in Tet1 and Tet2 double knockout mice, 5hmC levels of the P2rX7 promoter were down-regulated, leading to miR-293a-5p, miR-293b-5p, and miR-293c-5p accumulation, and a decrease in BMMSC osteogenic differentiation capacity. Upon re-activating P2rX7, microRNA secretion from Tet double knockout BMMSCs was increased, thus partly rescuing both the osteopenia phenotype and BMMSC function.
Mechanisms of TET-mediated DNA demethylation in distinct MSCs vary due to their diverse sources. When small hairpin RNA lentiviral vectors were transfected to knock down TET1, the proliferation rate and odontogenic differentiation capacity of human dental pulp stem cells were significantly suppressed. This indicated that TET1 plays an important role in dental pulp repair and regeneration[19]. In another study focusing on human BMMSCs, TET1 recruited other epigenetic modifiers, including SIN3A and EZH2, to inhibit the osteogenic differentiation of BMMSCs in an indirect manner. On the other hand, TET2 was found to directly promote the osteogenic differentiation of BMMSCs[20]. The underlying mechanisms of how the TET family proteins regulate MSC function from distinct sources require further investigation.
Noer et al[21] reported that in undifferentiated Ad-MSCs, the promoters of adipogenic genes, including LEP, PPARγ2, FABP4 and LPL, are hypomethylated, in contrast to myogenic or endothelial genes. During adipogenic differentiation, although specific CpG sites of the LEP promoter undergo demethylation, the global methylation status of LEP, PPARG2, FABP4, and LPL promoters across different Ad-MSC clones remains stable. Yang et al[18] showed that Tet1 and Tet2 small interfering RNA treatment does not alter the adipogenic differentiation capacity of BMMSCs.
Barrand et al[22] showed that in adipose MSCs, the promoter of OCT4 was hypermethylated consistent with its repression. Melzner et al[23] found that the promoter of leptin underwent extreme demethylation (9.4% ± 4.4%) during the maturation of human preadipocytes toward terminally differentiated adipocytes. What’s more, methyl-CpG binding proteins could bind to specific sites in the promoter and repressed leptin expression. Fujiki et al[24] reported that during the differentiation of 3T3-L1 preadipocytes to adipocytes, the hypermethylated PPARγ2 promoter was progressively demethylated, while 5-Aza could increase the expression of PPARγ2, indicating that the methylation of its promoter inhibited the gene expression.
Overall, additional research on the dynamics of DNA methylation and demethylation during adipogenesis from different MSC sources is necessary.
DNA methylation and demethylation status also change during MSC differentiation into chondrocytes. Chondrogenic differentiation of Ad-MSCs and BMMSCs was associated with a < 50% reduction in methylation rates at two specific CpG sites in the COL10A1 gene, and transcription of this gene was strongly induced[25]. Ito et al[26] discovered that 5hmC increased during chondrogenic differentiation of C3H10T1/2, a MSC line, and that TET1 expression was significantly up-regulated. Furthermore, Tet1 knockdown resulted in a marked decrease in the expression of chondrogenesis markers such as Col2 and Col10. In addition, 5hmC in the Igf1 promoter is a preferable binding site for TET proteins in chondrocytes. Additional targets of Tet genes, as well as other enzymes that function in DNA methylation and demethylation, need to be identified in order to reveal the underlying mechanisms of chondrogenic differentiation of MSCs.
Lin et al[27] found that stepwise preconditioning–manipulated BMMSCs showed improved cell proliferation and chondrogenic differentiation potential in vitro and enhanced therapeutic effect on the progression of osteoarthritis in vivo, and one mechanism of that is the reduction in CpG methylation at the promoters of Nanog and Oct4. Pollock et al[28] demonstrated an experimental DMSO-free formulation which could improve post-thaw function of MSCs including chondrogenesis, as DMSO is a strong inducer of demethylation which may affect the potential of MSCs for therapeutic use in treatment of human diseases. These studies reminded us that epigenetic modification of MSCs could be a promising approach to improve their therapeutic effects.
These results regarding DNA methylation and demethylation indicate that hypomethylation of specific genes, such as Runx2, Opn, Dlx5, Osterix, Col2, and Col10, play important roles in multi-lineage differentiation of and tissue regeneration by MSCs (Figure 1).
Cardiogenic differentiation is another important property of MSCs, and stem cell therapy for cardiovascular diseases is now in clinical trial[29]. Bhuvanalakshmi et al[30] found that in differentiated cardiomyocytes from MSCs, six out of the ten CpG islands of the promoter regions of Nkx2.5, the early cardiac gene, underwent demethylation. What’s more, the CpG promoter demethylation of sFRP4, a Wnt antagonist, was also observed. This result is consistent with the previous findings that 5-Aza treatment of BMMSCs inhibited the ventricular scar from thinning and expanding, minimized left ventricular chamber dilatation, and thus improved myocardial function[31]. Antonitsis et al[32] treated hBMMSCs with 5-Aza in vitro to induce them to differentiate towards a cardiomyogenic lineage. Nakatsuka et al[33] also used 5-Aza to investigate the myogenic differentiation potential of mouse dental pulp stem cells. DNA demethylation induced by 5-Aza and forced expression of Myod1 upregulated the muscle-specific transcriptional factors such as Myogenin and Pax7.
Aside from tissue regeneration, MSCs play an important role in immunomodulation, which may prove critical for treating a variety of immune diseases such as colitis, arthritis, and systemic lupus erythematosus[34-36]. Immunomodulation by MSCs relates to the secretion of extracellular matrix proteins[37] as well as a variety of cytokines including IL-2, IL-4, IL-10, TNF-α, and INF-γ[38-40]. MSC immunoregulation can also occur through cellular contacts[40-42]. B cell proliferation was found to be inhibited by human MSCs, not through the induction of apoptosis but through G0/G1 cell cycle arrest[43]. MSCs may suppress T cell proliferation, cytokine release, cytotoxicity, and Th1/Th2 balance[44,45].
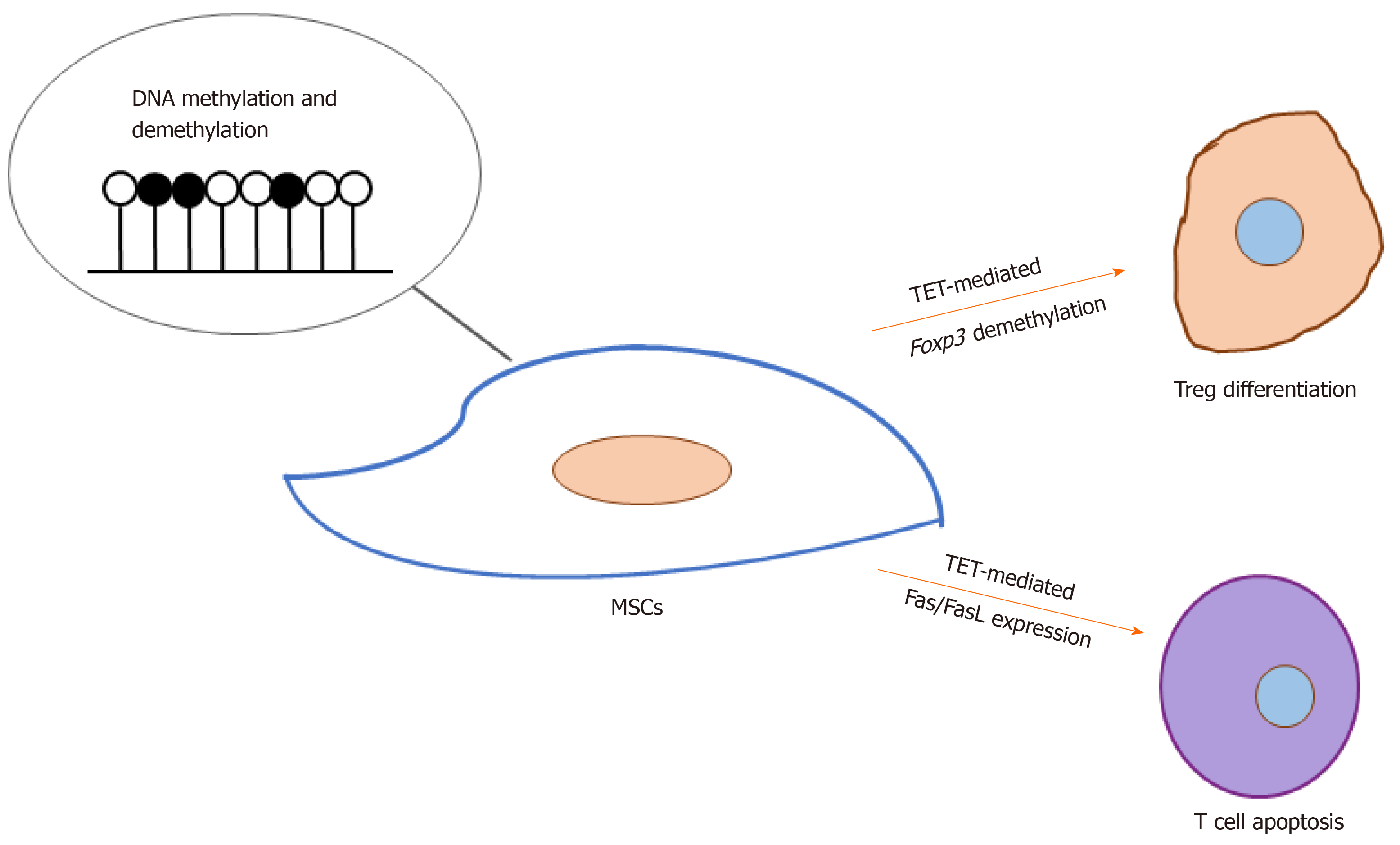
Of late, how DNA methylation and demethylation regulate MSC-induced immunomodulation has received increasingly greater attention. Yang et al[46] found that Tet1- and Tet2-mediated Foxp3 demethylation plays a significant role in the differentiation of regulatory T cells as well as the maintenance of immune homeostasis. Khosravi et al[47] reported that MSCs could enhance the demethylation of the Treg-specific demethylated region upon cell-cell contact, and MSC-based induction of regulatory T cells is associated with direct modifications of the RUNX complex genes (RUNX1, RUNX3, and CBFB). Yu et al[48] found that the down-regulation of both TET1 and TET2 leads to hypermethylation of the DKK-1 promoter, which leads to activation of the Wnt/β-catenin signaling pathway and thus up-regulates Fas ligand (the FasL gene) expression in periodontal ligament stem cells. This in turn enhances their immunomodulatory ability, which is demonstrated by their elevated capacity to induce T cell apoptosis. Taken together, these results demonstrate a significant role for TET-mediated DNA demethylation in MSC-based immunomodulation (Figure 2). Nevertheless, further investigations are required to reveal whether the methylation of MSCs is involved in regulation of other immune cells such as macrophages and natural killer cells and the underlying mechanisms.
As previously mentioned, DNA methylation participates in the regulation of gene expression, which may contribute to metabolic diseases when there is an imbalance in DNA methylation vs demethylation. García-Ibarbia et al[49] compared bone tissue samples from patients with osteoporotic hip fractures and osteoarthritis. Their results showed that Wnt pathway activity is reduced in patients with hip fractures compared with those with osteoarthritis. Additionally, six genes, including FZD10, TBL1X, CSNK1E, SFRP4, CSNK1A1L, and WNT8A, showed significantly different methylation rates between both groups. FZD10, CSNK1E, TBL1X, and SFRP4 are hypermethylated in osteoarthritis, while WNT8A and CSNK1A1L are hypomethylated compared with fractures. This result may help explain the distinctions in Wnt pathway activity between the two groups. MSCs from spinal ligaments with ectopic ossification largely differentiated into osteogenic lineage. Chiba et al[50] found that MSCs isolated from the spinal ligaments of ossification from yellow ligament patients showed higher expression of GDNF and Wnt5a, which are hypomethylated compared with the control group. This result indicates that osteogenic features of MSCs from patients with ossification of the yellow ligaments are promoted by GDNF and Wnt5a demethylation.
In 2002, Bartholomew et al[51] first reported that MSCs harbored immuno-suppressive functions by demonstrating their ability to inhibit a mixed lymphocyte response in vitro as well as prevent rejection in a baboon skin allograft model in vivo. The immunosuppressive capacities of MSCs have therein provided new therapeutic insights into immune-mediated disease treatments. Centeno et al[52] reported that autologous MSCs and physiologic doses of dexamethasone could increase meniscus volume of the human knee. In addition, MSCs can relieve symptoms of multiple sclerosis, multiple system atrophy, and amyotrophic lateral sclerosis in varying degrees[53,54]. How DNA methylation and demethylation function in MSC therapy for immunological diseases necessitates further exploration.
Although a wealth of research has investigated MSC therapy, including hundreds of MSC-based clinical trials that have been administered, the mechanisms that underlie the multiple distinct MSC functions remain elusive. This review sheds light on the roles that DNA methylation and demethylation play in regulating MSC-based regeneration and immunomodulation, although it is possible that we overlooked a few studies due to our literature resource limitations. However, the precise function of DNA methylation and demethylation in different MSC types, as well as the associated underlying mechanisms, remain to be thoroughly investigated. This knowledge would inform the development of novel approaches for enhancing MSC-based tissue regenerative and immune therapies.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Fatkhudinov T, Scarfì S S-Editor: Dou Y L-Editor: Wang TQ E-Editor: Xing YX
| 1. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5074] [Cited by in F6Publishing: 4679] [Article Influence: 212.7] [Reference Citation Analysis (0)] |
| 2. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4143] [Cited by in F6Publishing: 4127] [Article Influence: 275.1] [Reference Citation Analysis (0)] |
| 3. | Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129-1133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1873] [Cited by in F6Publishing: 1865] [Article Influence: 133.2] [Reference Citation Analysis (0)] |
| 4. | Friedenstein AJ, Chailakhyan RK, Latsinik NV, Panasyuk AF, Keiliss-Borok IV. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17:331-340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 998] [Cited by in F6Publishing: 903] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 5. | Akiyama K, Chen C, Wang D, Xu X, Qu C, Yamaza T, Cai T, Chen W, Sun L, Shi S. Mesenchymal-stem-cell-induced immunoregulation involves FAS-ligand-/FAS-mediated T cell apoptosis. Cell Stem Cell. 2012;10:544-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 486] [Cited by in F6Publishing: 527] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 6. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7446] [Cited by in F6Publishing: 7317] [Article Influence: 430.4] [Reference Citation Analysis (0)] |
| 7. | Hu X, Zhang X, Dai L, Zhu J, Jia Z, Wang W, Zhou C, Ao Y. Histone deacetylase inhibitor trichostatin A promotes the osteogenic differentiation of rat adipose-derived stem cells by altering the epigenetic modifications on Runx2 promoter in a BMP signaling-dependent manner. Stem Cells Dev. 2013;22:248-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 8. | Chen JC, Chua M, Bellon RB, Jacobs CR. Epigenetic changes during mechanically induced osteogenic lineage commitment. J Biomech Eng. 2015;137:020902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Arnsdorf EJ, Tummala P, Castillo AB, Zhang F, Jacobs CR. The epigenetic mechanism of mechanically induced osteogenic differentiation. J Biomech. 2010;43:2881-2886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 10. | Villagra A, Gutiérrez J, Paredes R, Sierra J, Puchi M, Imschenetzky M, Wijnen Av Av, Lian J, Stein G, Stein J, Montecino M. Reduced CpG methylation is associated with transcriptional activation of the bone-specific rat osteocalcin gene in osteoblasts. J Cell Biochem. 2002;85:112-122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 79] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Delgado-Calle J, Sañudo C, Sánchez-Verde L, García-Renedo RJ, Arozamena J, Riancho JA. Epigenetic regulation of alkaline phosphatase in human cells of the osteoblastic lineage. Bone. 2011;49:830-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 12. | Zhang RP, Shao JZ, Xiang LX. GADD45A protein plays an essential role in active DNA demethylation during terminal osteogenic differentiation of adipose-derived mesenchymal stem cells. J Biol Chem. 2011;286:41083-41094. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 13. | Daniunaite K, Serenaite I, Misgirdaite R, Gordevicius J, Unguryte A, Fleury-Cappellesso S, Bernotiene E, Jarmalaite S. Epigenetic regulation of human adipose-derived stem cells differentiation. Mol Cell Biochem. 2015;410:111-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Yan X, Ehnert S, Culmes M, Bachmann A, Seeliger C, Schyschka L, Wang Z, Rahmanian-Schwarz A, Stöckle U, De Sousa PA, Pelisek J, Nussler AK. 5-azacytidine improves the osteogenic differentiation potential of aged human adipose-derived mesenchymal stem cells by DNA demethylation. PLoS One. 2014;9:e90846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 15. | Kornicka K, Marycz K, Marędziak M, Tomaszewski KA, Nicpoń J. The effects of the DNA methyltranfserases inhibitor 5-Azacitidine on ageing, oxidative stress and DNA methylation of adipose derived stem cells. J Cell Mol Med. 2017;21:387-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 16. | Eimori K, Endo N, Uchiyama S, Takahashi Y, Kawashima H, Watanabe K. Disrupted Bone Metabolism in Long-Term Bedridden Patients. PLoS One. 2016;11:e0156991. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Wang C, Shan S, Wang C, Wang J, Li J, Hu G, Dai K, Li Q, Zhang X. Mechanical stimulation promote the osteogenic differentiation of bone marrow stromal cells through epigenetic regulation of Sonic Hedgehog. Exp Cell Res. 2017;352:346-356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Yang R, Yu T, Kou X, Gao X, Chen C, Liu D, Zhou Y, Shi S. Tet1 and Tet2 maintain mesenchymal stem cell homeostasis via demethylation of the P2rX7 promoter. Nat Commun. 2018;9:2143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 19. | Rao LJ, Yi BC, Li QM, Xu Q. TET1 knockdown inhibits the odontogenic differentiation potential of human dental pulp cells. Int J Oral Sci. 2016;8:110-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Cakouros D, Hemming S, Gronthos K, Liu R, Zannettino A, Shi S, Gronthos S. Specific functions of TET1 and TET2 in regulating mesenchymal cell lineage determination. Epigenetics Chromatin. 2019;12:3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Noer A, Sørensen AL, Boquest AC, Collas P. Stable CpG hypomethylation of adipogenic promoters in freshly isolated, cultured, and differentiated mesenchymal stem cells from adipose tissue. Mol Biol Cell. 2006;17:3543-3556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 120] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Barrand S, Collas P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem Biophys Res Commun. 2010;391:762-767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Melzner I, Scott V, Dorsch K, Fischer P, Wabitsch M, Brüderlein S, Hasel C, Möller P. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. J Biol Chem. 2002;277:45420-45427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 147] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 24. | Fujiki K, Kano F, Shiota K, Murata M. Expression of the peroxisome proliferator activated receptor gamma gene is repressed by DNA methylation in visceral adipose tissue of mouse models of diabetes. BMC Biol. 2009;7:38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 186] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 25. | Zimmermann P, Boeuf S, Dickhut A, Boehmer S, Olek S, Richter W. Correlation of COL10A1 induction during chondrogenesis of mesenchymal stem cells with demethylation of two CpG sites in the COL10A1 promoter. Arthritis Rheum. 2008;58:2743-2753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 73] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 26. | Ito R, Shimada H, Yazawa K, Sato I, Imai Y, Sugawara A, Yokoyama A. Hydroxylation of methylated DNA by TET1 in chondrocyte differentiation of C3H10T1/2 cells. Biochem Biophys Rep. 2016;5:134-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Lin S, Lee WYW, Xu L, Wang Y, Chen Y, Ho KKW, Qin L, Jiang X, Cui L, Li G. Stepwise preconditioning enhances mesenchymal stem cell-based cartilage regeneration through epigenetic modification. Osteoarthritis Cartilage. 2017;25:1541-1550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 28. | Pollock K, Samsonraj RM, Dudakovic A, Thaler R, Stumbras A, McKenna DH, Dosa PI, van Wijnen AJ, Hubel A. Improved Post-Thaw Function and Epigenetic Changes in Mesenchymal Stromal Cells Cryopreserved Using Multicomponent Osmolyte Solutions. Stem Cells Dev. 2017;26:828-842. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Fisher SA, Zhang H, Doree C, Mathur A, Martin-Rendon E. Stem cell treatment for acute myocardial infarction. Cochrane Systematic Reviews. 2015;. [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 30. | Bhuvanalakshmi G, Arfuso F, Kumar AP, Dharmarajan A, Warrier S. Epigenetic reprogramming converts human Wharton's jelly mesenchymal stem cells into functional cardiomyocytes by differential regulation of Wnt mediators. Stem Cell Res Ther. 2017;8:185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 31. | Tomita S, Li RK, Weisel RD, Mickle DA, Kim EJ, Sakai T, Jia ZQ. Autologous transplantation of bone marrow cells improves damaged heart function. Circulation. 1999;100:II247-II256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 431] [Cited by in F6Publishing: 538] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 32. | Antonitsis P, Ioannidou-Papagiannaki E, Kaidoglou A, Charokopos N, Kalogeridis A, Kouzi-Koliakou K, Kyriakopoulou I, Klonizakis I, Papakonstantinou C. Cardiomyogenic potential of human adult bone marrow mesenchymal stem cells in vitro. Thorac Cardiovasc Surg. 2008;56:77-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Nakatsuka R, Nozaki T, Uemura Y, Matsuoka Y, Sasaki Y, Shinohara M, Ohura K, Sonoda Y. 5-Aza-2'-deoxycytidine treatment induces skeletal myogenic differentiation of mouse dental pulp stem cells. Arch Oral Biol. 2010;55:350-357. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 34. | Regmi S, Pathak S, Kim JO, Yong CS, Jeong JH. Mesenchymal stem cell therapy for the treatment of inflammatory diseases: Challenges, opportunities, and future perspectives. Eur J Cell Biol. 2019;98:151041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 160] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 35. | Duijvestein M, Vos AC, Roelofs H, Wildenberg ME, Wendrich BB, Verspaget HW, Kooy-Winkelaar EM, Koning F, Zwaginga JJ, Fidder HH, Verhaar AP, Fibbe WE, van den Brink GR, Hommes DW. Autologous bone marrow-derived mesenchymal stromal cell treatment for refractory luminal Crohn's disease: results of a phase I study. Gut. 2010;59:1662-1669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 471] [Cited by in F6Publishing: 448] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 36. | Tang X, Li W, Wen X, Zhang Z, Chen W, Yao G, Chen H, Wang D, Shi S, Sun L. Transplantation of dental tissue-derived mesenchymal stem cells ameliorates nephritis in lupus mice. Ann Transl Med. 2019;7:132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Wight TN, Kinsella MG, Keating A, Singer JW. Proteoglycans in human long-term bone marrow cultures: biochemical and ultrastructural analyses. Blood. 1986;67:1333-1343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 38. | Horwitz EM, Dominici M. How do mesenchymal stromal cells exert their therapeutic benefit? Cytotherapy. 2008;10:771-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 39. | Ben-Ami E, Miller A, Berrih-Aknin S. T cells from autoimmune patients display reduced sensitivity to immunoregulation by mesenchymal stem cells: role of IL-2. Autoimmun Rev. 2014;13:187-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood. 2005;105:1815-1822. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3271] [Cited by in F6Publishing: 3170] [Article Influence: 158.5] [Reference Citation Analysis (0)] |
| 41. | Meisel R, Zibert A, Laryea M, Göbel U, Däubener W, Dilloo D. Human bone marrow stromal cells inhibit allogeneic T-cell responses by indoleamine 2,3-dioxygenase-mediated tryptophan degradation. Blood. 2004;103:4619-4621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1243] [Cited by in F6Publishing: 1217] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 42. | Jiang XX, Zhang Y, Liu B, Zhang SX, Wu Y, Yu XD, Mao N. Human mesenchymal stem cells inhibit differentiation and function of monocyte-derived dendritic cells. Blood. 2005;105:4120-4126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 963] [Cited by in F6Publishing: 954] [Article Influence: 50.2] [Reference Citation Analysis (0)] |
| 43. | Corcione A, Benvenuto F, Ferretti E, Giunti D, Cappiello V, Cazzanti F, Risso M, Gualandi F, Mancardi GL, Pistoia V, Uccelli A. Human mesenchymal stem cells modulate B-cell functions. Blood. 2006;107:367-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1263] [Cited by in F6Publishing: 1235] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 44. | Puissant B, Barreau C, Bourin P, Clavel C, Corre J, Bousquet C, Taureau C, Cousin B, Abbal M, Laharrague P, Penicaud L, Casteilla L, Blancher A. Immunomodulatory effect of human adipose tissue-derived adult stem cells: comparison with bone marrow mesenchymal stem cells. Br J Haematol. 2005;129:118-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 685] [Cited by in F6Publishing: 663] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 45. | Selmani Z, Naji A, Zidi I, Favier B, Gaiffe E, Obert L, Borg C, Saas P, Tiberghien P, Rouas-Freiss N, Carosella ED, Deschaseaux F. Human leukocyte antigen-G5 secretion by human mesenchymal stem cells is required to suppress T lymphocyte and natural killer function and to induce CD4+CD25highFOXP3+ regulatory T cells. Stem Cells. 2008;26:212-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 762] [Cited by in F6Publishing: 802] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 46. | Yang R, Qu C, Zhou Y, Konkel JE, Shi S, Liu Y, Chen C, Liu S, Liu D, Chen Y, Zandi E, Chen W, Zhou Y, Shi S. Hydrogen Sulfide Promotes Tet1- and Tet2-Mediated Foxp3 Demethylation to Drive Regulatory T Cell Differentiation and Maintain Immune Homeostasis. Immunity. 2015;43:251-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 254] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 47. | Khosravi M, Bidmeshkipour A, Cohen JL, Moravej A, Hojjat-Assari S, Naserian S, Karimi MH. Induction of CD4+CD25+FOXP3+ regulatory T cells by mesenchymal stem cells is associated with modulation of ubiquitination factors and TSDR demethylation. Stem Cell Res Ther. 2018;9:273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 48. | Yu T, Liu D, Zhang T, Zhou Y, Shi S, Yang R. Inhibition of Tet1- and Tet2-mediated DNA demethylation promotes immunomodulation of periodontal ligament stem cells. Cell Death Dis. 2019;10:780. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 49. | García-Ibarbia C, Delgado-Calle J, Casafont I, Velasco J, Arozamena J, Pérez-Núñez MI, Alonso MA, Berciano MT, Ortiz F, Pérez-Castrillón JL, Fernández AF, Fraga MF, Zarrabeitia MT, Riancho JA. Contribution of genetic and epigenetic mechanisms to Wnt pathway activity in prevalent skeletal disorders. Gene. 2013;532:165-172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 50. | Chiba N, Furukawa K, Takayama S, Asari T, Chin S, Harada Y, Kumagai G, Wada K, Tanaka T, Ono A, Motomura S, Murakami M, Ishibashi Y. Decreased DNA methylation in the promoter region of the WNT5A and GDNF genes may promote the osteogenicity of mesenchymal stem cells from patients with ossified spinal ligaments. J Pharmacol Sci. 2015;127:467-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 51. | Bartholomew A, Sturgeon C, Siatskas M, Ferrer K, McIntosh K, Patil S, Hardy W, Devine S, Ucker D, Deans R, Moseley A, Hoffman R. Mesenchymal stem cells suppress lymphocyte proliferation in vitro and prolong skin graft survival in vivo. Exp Hematol. 2002;30:42-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1730] [Cited by in F6Publishing: 1613] [Article Influence: 73.3] [Reference Citation Analysis (0)] |
| 52. | Centeno CJ, Busse D, Kisiday J, Keohan C, Freeman M, Karli D. Regeneration of meniscus cartilage in a knee treated with percutaneously implanted autologous mesenchymal stem cells. Med Hypotheses. 2008;71:900-908. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 53. | Bai L, Lennon DP, Caplan AI, DeChant A, Hecker J, Kranso J, Zaremba A, Miller RH. Hepatocyte growth factor mediates mesenchymal stem cell–induced recovery in multiple sclerosis models. Nat Neurosci. 2012;15:862-870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 311] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 54. | Lee PH, Kim JW, Bang OY, Ahn YH, Joo IS, Huh K. Autologous mesenchymal stem cell therapy delays the progression of neurological deficits in patients with multiple system atrophy. Clin Pharmacol Ther. 2008;83:723-730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 181] [Article Influence: 10.6] [Reference Citation Analysis (0)] |