Published online May 26, 2020. doi: 10.4252/wjsc.v12.i5.303
Peer-review started: February 27, 2020
First decision: April 7, 2020
Revised: April 19, 2020
Accepted: May 5, 2020
Article in press: May 5, 2020
Published online: May 26, 2020
Autophagy is a highly regulated catabolic process in which superfluous, damaged organelles and other cytoplasmic constituents are delivered to the lysosome for clearance and the generation of macromolecule substrates during basal or stressed conditions. Autophagy is a bimodal process with a context dependent role in the initiation and the development of cancers. For instance, autophagy provides an adaptive response to cancer stem cells to survive metabolic stresses, by influencing disease propagation via modulation of essential signaling pathways or by promoting resistance to chemotherapeutics. Autophagy has been implicated in a cross talk with apoptosis. Understanding the complex interactions provides an opportunity to improve cancer therapy and the clinical outcome for the cancer patients. In this review, we provide a comprehensive view on the current knowledge on autophagy and its role in cancer cells with a particular focus on cancer stem cell homeostasis.
Core tip: Cancer stem cells (CSCs) are a distinct subpopulation in the tumor bulk that are highly plastic, and autophagy has been suggested to modulate their stemness and development during cancer progression. Autophagy is a pro-survival mechanism used by cancer cells to provide bioenergetic substrates. Therefore, dissecting the role of autophagy in cancer propagation can theoretically lead to a more efficient cancer treatment via the modulation of autophagy, in combination with chemotherapeutics to sensitize and target CSCs. This review summarizes the divergent role of autophagy in CSCs and cancer cells and attempts to elucidate the molecular mechanisms involved.
- Citation: Mandhair HK, Arambasic M, Novak U, Radpour R. Molecular modulation of autophagy: New venture to target resistant cancer stem cells. World J Stem Cells 2020; 12(5): 303-322
- URL: https://www.wjgnet.com/1948-0210/full/v12/i5/303.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i5.303
Autophagy (“self-consumption”) is a conserved catabolic process which assists in the clearance of superfluous, damaged organelles and proteins, and contributes in the recycling of the constituents for the maintenance of metabolic homeostasis and as a pro-survival mechanism[1]. Autophagy is further activated by intrinsic and environmental stressors including nutrient deprivation, oxidative stress, cytokine and growth factor deficiency, hypoxia and exposure to infection[2,3]. It can be noted that basal autophagy acts as a quality assurance mechanism in cells and as a source of metabolites[4]. Dysregulation of autophagy is associated with a variety of inflammatory and infectious conditions, as well as neurodegenerative pathologies, ageing and cancer[5].
Autophagy is a highly regulated mechanism that facilitates the deliverance of cytoplasmic components for lysosomal mediated degradation. There are three distinct forms of autophagy, such as microautophagy, chaperone mediated autophagy (CMA) and macroautophagy. Microautophagy is modulated by the direct sequestration of cytosolic cargo causing engulfment, followed by indentation of the lysosome leading to degradation[6]. In comparison, CMA is a prime example of selective autophagy. In this particular pathway, chaperones are utilized targeting specific proteins containing a pentapeptide KFERQ motif sequence. Once engaged this leads to the translocation across the lysosome membrane mediated by lysosome associated membrane protein 2A[7,8]. In contrast, macroautophagy (herein referred to as autophagy) initiates the degradation of intracellular organelles by delivering them to the lysosome by sequestrating sections of the cytoplasm via double membrane vesicles called autophagosomes. The fusion between these two entities not only promotes degradation but also generates bioenergetic substances for recycling. Emerging studies describe the existence of a cross talk between CMA and macroautophagy that promotes a compensatory mechanism under basal and stressed conditions[7,9].
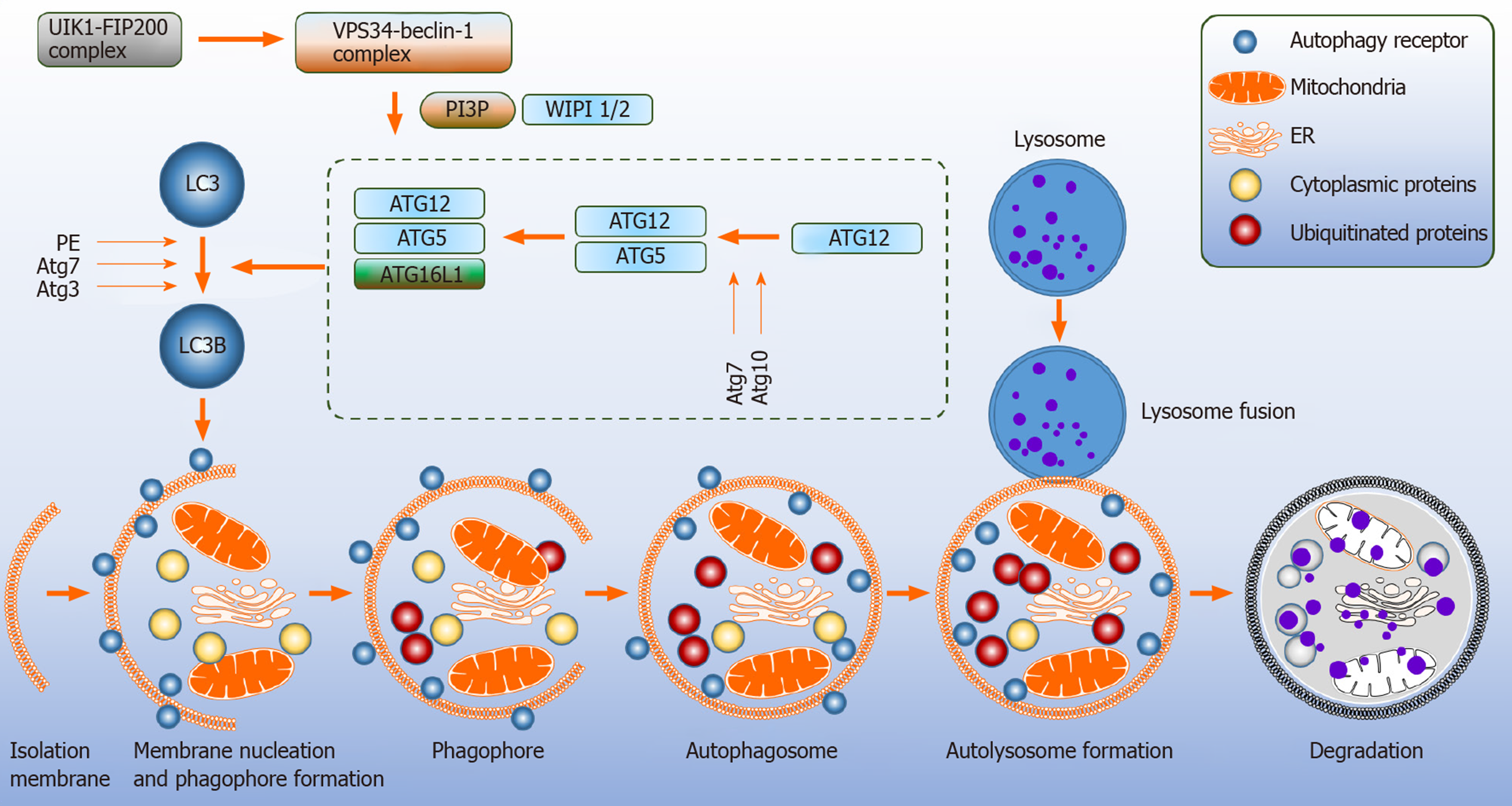
Autophagy relates genes (Atg) are involved in the development and turnover of the autophagosomes. Formation of the autophagosome proceeds through multiple steps that include initiation, nucleation, elongation, maturation and thereafter fusion with the lysosome[8,10]. The mitochondria and the endoplasmic reticulum (ER) are contact sites for the formation of autophagosomes[11,12] (Figure 1).
Upstream signaling pathway such mammalian target of rapamycin (mTOR) is a major negative regulator of autophagy as it senses amino acid availability and initiation of cellular anabolism. 5’-AMP-activated protein kinase (AMPK) is activated during starvation[2]. Under these circumstances, AMPK phosphorylates tuber sclerosis complex 2 which inhibits mTOR[13]. Moreover, Atg7 protein is essential in modulating starvation-induced autophagy as demonstrated in Atg7 conditional knockout mice[14].
The inhibition of mTOR sequentially leads to the activation of pre initiation complex composed of unc-51-like kinase 1 (ULK1) complex, FAK family kinase interacting protein of 200 kDa, Atg13 and Atg101, causing translocation to the membrane, and triggering the initiation step for the assembly of autophagosomes[10]. The ULK1 complex phosphorylates the class III phosphatidylinositol-3-kinase (PI3K) vacuole protein sorting 34 (VPS34) complex; consisting of VPS15, Beclin-1 (BECN1) and Atg14, which stimulates the generation of phosphatidylinositol-3-phospate 3 (PI3P), an essential lipid molecule required for the nucleation step of the phagophore[15-17]. Atg9 positive vesicles on the ER contribute to the nucleation process by interacting with the ULK1 complex[17]. To promote autophagosomes elongation, WD repeat domain phosphoinositide-interacting protein 2 (WIPI-2) and zinc-finger FYVE domain-containing protein 1 are employed for the recruitment of two ubiquitin like systems[16]. Firstly, Atg7 and Atg10 act as E1 like and E2 like enzymes to covalently conjugate Atg12 to Atg5 and then attach to Atg16L[8,18,19]. In the second conjugation pathway, Atg12-Atg5 conjugate serves as an E3 like enzyme, where Atg8 family member LC3 is attached to phosphatidylethanolamine[2,19]. Atg7 and Atg3 mediate this process. Next, the autophagosome matures by membrane bound LC3. NBR1 neighbor of BRAC1 and adaptor protein p62 facilitate in the degradation of misfolded and ubiquitinated substrates by binding to Atg8-LC3[18-20]. The closure of the autophagosome is driven by LC3 causing the Atg12-Atg5-Atg16L complex to dissociate from the autophagosome membrane leaving the lipidated LC3 (LC3B; microtubule-associated proteins 1A/1B light chain 3B) in the autophagosome[16,18]. The degradation of LC3B and p62 are widely accepted markers to measure the autophagic flux.
It should be noted, however, that multiple signaling cascades control autophagy and modify ULK1 and class III PI3K complexes. These include antigen specific receptors (B cell receptor and T cell receptor), CD40 “the co-stimulatory molecule”, Toll like receptors, cytokine receptors and nucleotide-binding oligomerization domain protein 2[2]. The VPS34-BECN1 complex can be inactivated by the anti-apoptotic proteins from the B cell lymphoma-2 (BCL-2) family[16]. Here we have discussed the major canonical pathway that utilizes mTOR (Figure 1).
Autophagy that precedes the formation of autophagosomes without the involvement of the core machinery is referred to as non-canonical autophagy. An example of non-canonical autophagy would be LC3-associated phagocytosis (LAP) which depends on class III PI3K subunit called RUBICON, a negative regulator of autophagy[2,21]. Unlike canonical autophagy, LAP only requires BECN1 and VPS34 as a pre-initiation complex and downstream conjugation of LC3 to generate NADH oxidase 2[22]. LAP-LC3 is associated to autophagosome maturation and facilitating the degradation of engulfed cells. LAP does not respond to nutrient deficiency or intracellular stressors, unlike canonical autophagy. Additionally, the substrates for this process are extracellular entities including Toll like receptor, pattern recognition receptors and dead cells[22]. LAP occurs in multiple immune cells, such as macrophages, dendritic cells (DCs) and epithelial cells[21]. LAP deficiency in cells and animal models trigger exaggerated inflammation[22].
In the canonical form, it is assumed that the generation of PI3P is essential for the process of autophagy. However, Mauthe et al[23] reported resveratrol mediated autophagy did not stimulate PI3P dependent accumulation of WIPI-1 at the autophagosome membrane. This finding was confirmed by PI3P inhibition using wortmannin in combination with resveratrol which led to an increased autophagic flux of LC3B and GFP-LC3 puncta formation. This was promoted in the absence of phagophore formation suggesting an alternative contact site for autophagosome formation. Additionally, the actions of resveratrol were found to be independent of BECN1; however, required Atg7 and Atg5 to induce the LC3 lipidation. It can be concluded that resveratrol induces non-canonical autophagy[23].
The origin of the autophagosome membrane and the formation of the autophagosome remains unclear[24]. Recently, using freeze fracture replica immunolabelling, WIPI-1 puncta were found to be localized on the ER and Plasma membrane and WIPI-2 was detected close to the Golgi cisternae under starvation induced autophagy, exclusively. These findings suggest that WIPI-1 and WIPI-2 are essential components of the autophagosome and the autophagosome membrane site and formation may potentially originate from the ER, Plasma membrane and the Golgi[25]. Interestingly, the deletion of WIPI-2 in the germinal center (GC) B cells enhanced the autophagic activity, suggesting that B cells derived from the GC have the ability to switch from canonical autophagy upon challenge to non-canonical autophagy to meet their metabolic demands[26].
It is believed that Atg5 and At7 are essential for autophagy. However, recent studies have challenged this notion. Atg5/Atg7 independent non-canonical autophagic pathway have been identified, which are able to form autophagosomes mediated in a Rab9 dependent manner from the trans-Golgi network and late endosomes. Autophagy proteins, such as ULK1 and BECN1 were found to regulate this process independent of LC3[27]. The resulting autophagosomes mature and fuse with the lysosome and undergo cargo clearance[28]. Furthermore, ULK1 dependent/Atg5 independent autophagy has been implicated in the removal of the mitochondria from fetal definitive erythroid cells in vivo[29]. Additionally, ULK1-/- mice models were able to express LC3B under nutrient depleted conditions; indicating the role of ULK1 in the induction of autophagy is dispensable[30]. These reported studies suggest ULK1 is not essential for Atg5/Atg7 dependent canonical autophagy[14,31]. Moreover, ULK1 is upregulated during non-canonical autophagy, and the silencing of ULK1 inhibits this process[27].
It is evident that autophagy participates in catabolism including the breakdown of long-lived proteins, providing bioenergetics material to facilitate in the production of adenosine triphosphate (ATP) and meet the metabolic demands of cells undergoing adverse conditions and rescue them. However, under prolonged metabolic-stressed conditions the pool of bioenergetic substrates will be facilitated to generate ATP dependent apoptosis[32]. Predominately, autophagy has a cytoprotective role. Overall, it can be assumed that autophagy and apoptosis are activated by a common stimulus[19].
Apoptosis “self-killing” is a form of type 1 programmed cell death (PD) and is characterized by the distinct morphological changes causing nuclear condensation (Pyknosis) and fragmentation (Karyorhexis), and membrane blebbing a requisite for the generation of apoptotic bodies (smaller apoptotic cell fragments)[33,34].
Emerging literature indicates a complex network that regulates the interplay between autophagy and apoptosis. This is cell type and stimuli dependent. This dynamic interplay has been described in the following examples: Autophagy and apoptosis can function together in order to induce cell death, autophagy can promote cell survival by antagonizing apoptosis, or autophagy can assist in cell death by activating apoptosis[16,35,36].
Multiple stimuli that can trigger cell death can also induce autophagy. Autophagy as a cytoprotective mechanism is usually induced first, followed by apoptosis[16,37]. Death associated protein kinase (DAPK) signaling is an example when both apoptosis and autophagy are induced either simultaneously or sequentially. Upon stimulation, DAPK phosphorylates BECN1 leading to its dissociation from BCL-2; thus, activating autophagy by binding to VPS34[38,39]. However, activated DAPK is also able to stimulate apoptosis in autophagy deficient conditions[40]. It can be postulated that DAPK regulated autophagy is induced by low levels of stress, however, intense and chronic stress stimuli can initiate apoptosis through DAPK[16].
It has been proposed that autophagy and apoptosis display an inhibitory relationship during the removal of pro-apoptotic proteins in the cytoplasm caused by autophagy, resulting in reduced apoptosis. Caspase-8 activation is a critical step during the extrinsic apoptosis signaling. However, selective autophagy may interfere in an inhibitory manner with the cell-death pathway through the degradation of capsase-8[41]. Furthermore, autophagy can be inhibited by apoptosis via numerous mechanisms; for example, autophagy exhaustion during increased intensity levels of stress. In this condition, degradation of autophagic proteins and caspases activity is reduced. For example, BECN1 inactivation occurs after caspase-mediated cleavage, stimulating the release of pro-apoptotic factors, and resulting in autophagy inhibition and induction of apoptosis[42].
Autophagy dependent cell death is defined as a form of cell death distinct from apoptosis or necrosis that mechanistically depends on the autophagic machinery[43]. It is postulated that the formation of autophagosome, and not degradation, leads to the activation of caspase-8 and the execution of cell death. As reported in mouse embryonic fibroblasts treated with proteasome inhibitor Bortezomib, and pan-sphingosine kinase inhibitor. Pro-caspase- 8 interactions with p62 have been shown to co-localize with the autophagosomes. The surface of the autophagosomes serves as a platform for the maturation of caspase-8 and the initiation of apoptosis[16,43]. Furthermore, the depletion of Atg5 ablated caspase-8 processing in the presence of Bortezomib leading to a significant reduction in cell death[44].
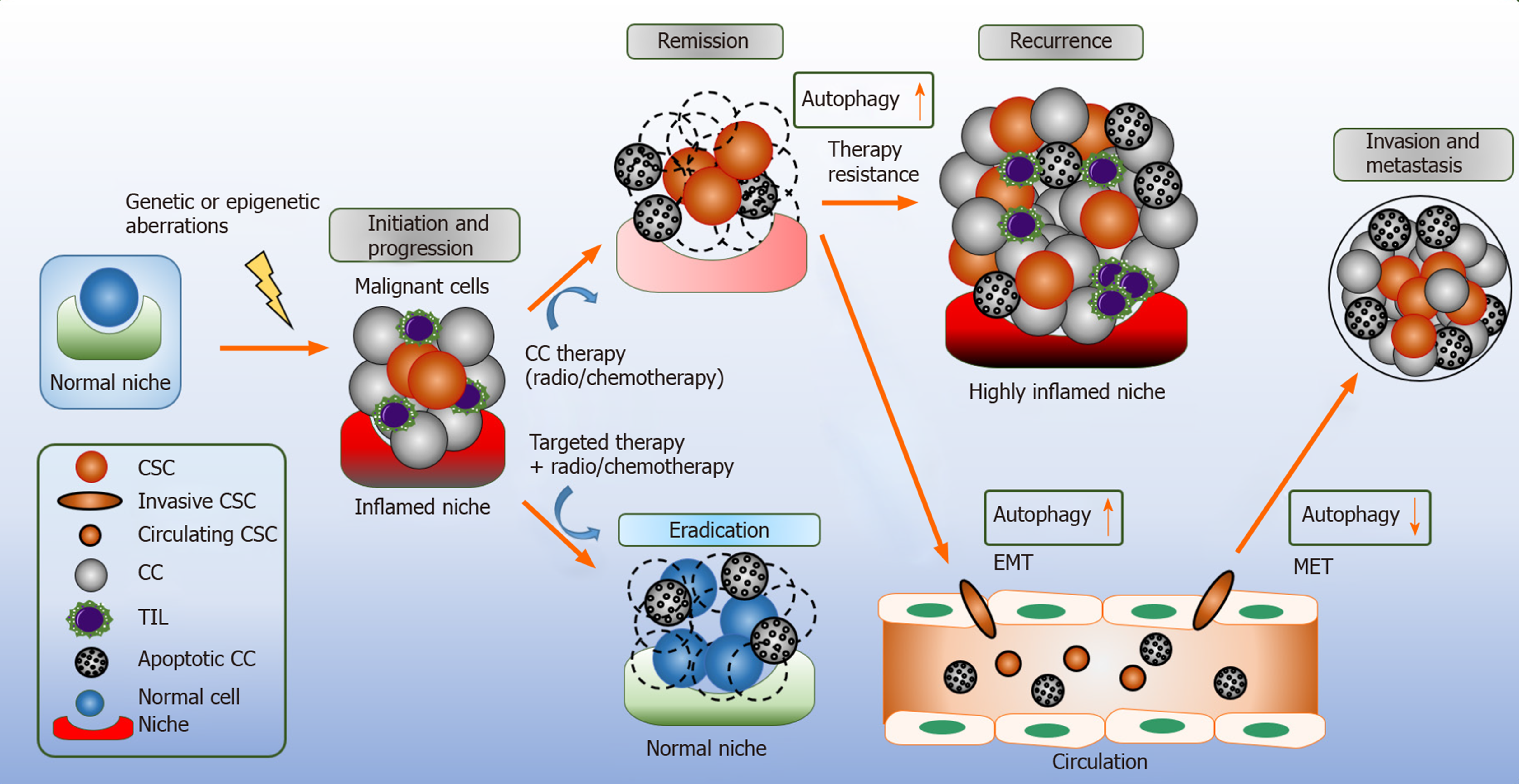
Tumors are derived from heterogeneous cell types. Cancer stem cells (CSCs; also known as tumor initiating cells) are a small subpopulation of cancer cells within the tumor bulk tissue that retain the capacity for self-renewal, disease propagation, and metastasis, which are decisive for tumor recurrences and are therapy resistance[45-47]. In general, stem cells are characterized by their distinct ability to switch their cell cycle profile from quiescent to proliferative behavior in order to maintain their capability for self-renewal and later multi-potency[48]. Similarly, CSCs have the extraordinary capability to self-renew and differentiate rapidly; accumulating mutations and genetic alterations and transmitting these defects to the proliferating progeny, giving rise to tumor heterogeneity conferring to resistance against anti-cancer therapeutics[49].
Similar to normal stem cells, CSCs reside in dynamic microenvironments known as the stem cell niche, this regulates the fate of adult stem cells by providing signals, such as cell-cell contact and secreting mediators to promote CSCs renewal, tumor invasion and metastasis[24,50]. Normal niches are comprised of heterogeneous collection of cells, such as endothelial, fibroblasts, immune cells, perivascular cells, components of the extracellular matrix, cytokines and growth factors[51]. In comparison, the CSCs niche itself is part of the tumor specific microenvironment that remains distinct from the normal niche[52]. During tumor progression to a malignancy, the CSC state in the primary tumor depends crucially on the microenvironment and potentially on the CSC niche itself[53]. Targeting the CSCs niche is the current subject of research as it is a valuable modality for the treatment and prevention of CSCs growth and downstream signaling[52].
The functional characterization of CSCs in multiple studies have clarified that CSCs are the foundation of tumor formation that can survive treatment with conventional therapies and can cause the recurrence of cancer[54,55]. According to the concept of a stem cell, it can be assumed that even a few surviving CSCs after therapy is sufficient to develop a new tumor leading to a relapse. Due to the ability of CSCs to initiate relapse after conventional cancer therapy, they represent a crucial therapeutic target[46]. CSCs were first identified in acute myeloid leukemia (AML); their presence was confirmed by the isolation of AML-initiating cells based on their phenotypical markers[56]. In solid tumors, breast cancer was one of the first to be characterized, which led to the identification of a specific subpopulation of CSCs marked by CD44+CD24-/Low lineage. This tumorigenic population of cells was able to initiate tumor growth in immunosuppressed mice[57,58]. Furthermore, CSCs have been discovered in several solid cancers, such as lung[55], pancreatic[59], colon[60,61], melanoma[62], ovarian[63,64], brain cancers[65,66] and hematological malignancies of both myeloid and lymphoid origin[67-69].
To date, two paradigms: hierarchical and stochastic have been proposed to account for the tumor origin, progression and heterogeneity. In brief, the hierarchical model is based on a concept that tumor cells are hierarchically arranged cell populations and CSCs represent the top of the arrangement. Carcinogenesis proceeds when a healthy normal stem cell escapes regulation and transforms into a stem cell-like phenotype- CSCs. This in turn gives rise to heterogeneity by generating differentiated and quiescent cells whose proliferation capacity is restricted[52,70]. By contrast, in the stochastic model, cancer is derived from a single somatic cell that initiates tumorigenesis and progression. This paradigm partially relies upon the environment in which the cancer cell is located in, but, fundamentally is defined by hyper proliferation and the acquisition of mutational burden during the cell cycle process contributing to clonal expansion[52].
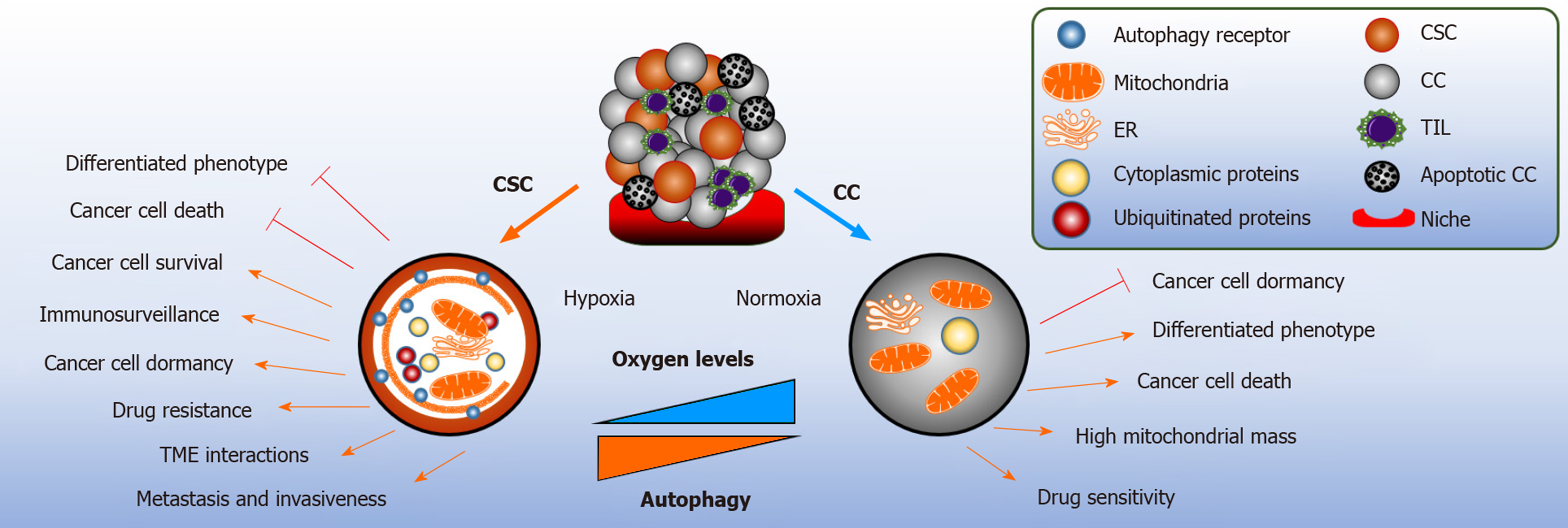
As highlighted earlier in this review, autophagy is a multifaceted pro-survival mechanism. In cancer, the role of autophagy is context dependent. Autophagy elicits tumor suppressing functions during tumor initiation by limiting inflammation, tissue damage, and genome instability by removing damaged mitochondria and reducing oxidative stress[2]. Extracellular stimuli, such as oxidative stress, nutrient depletion, increased metabolism and hypoxia result to disease propagation; thus, demanding autophagy to meet the high metabolic demands by providing recycled bioenergetic substrates to the CSCs, and whilst doing so, implementing its role as a tumor promotor (Figure 2)[71].
It has been proposed that autophagy is associated to CSC maintenance. LC3B gene knockdown in human embryonic stem cells (ESCs) leads to a reduction in pluripotency and due to the accumulation of pluripotency associated proteins suggesting autophagy regulates these proteins[72]. Autophagic flux is upregulated in mammospheres in basal and starvation-induced autophagy and is driven by BECN1 and Atg4A for their survival and expansion. Inhibition of these autophagy genes abolishes the tumor formation[73,74]. Aldehyde dehydrogenase 1-positive (ALDH1+) CSCs isolated from MCF-7 mammospheres presented an increased LC3B dependent autophagic flux with higher rate of p62 degradation compared to the bulk population; indicating an increased synthesis of autophagosomes. In addition, suggesting elevated autophagy is critical for CSCs[74]. Moreover, Antonelli et al[75] reports that ataxia-telangiectasia mutated (ATM) kinase modulates breast CSCs through Atg4C. This was validated in an overexpression study of Atg4C that was assessed in ATM gene silenced cells using shATM; this led to the rescue of mammosphere formation in ATM knockdown cells. These findings correlated with the microarray data of breast cancer samples, however, excluded triple negative tumors[75]. Indeed, these autophagy genes have shown to promote CSC survival and tumorigenicity. RNAi screenings reveal constitutive STAT3 activity is regulated by autophagy and is enriched in the triple negative breast cancer cell lines[73,76]. In those cell lines, Atg7 and BECN1 modulate CD24 expression in CD44+CD24-/low CSC population and secret interleukin 6 (IL-6) through gp130 and JAK-STAT pathway for CSC maintenance[73,77].
MMTV-PyMT is a well-characterized transgenic murine model for breast CSCs tumorigenesis. Yeo et al[78] reported autophagy differentially regulates two distinct breast cancer stem-like cells ALDH1+ and CD29hiCD61+ though EGFR/STAT3 and Tgfβ/Smad. Depletion of FIP200 decreased STAT3 activation by decreasing phosphorylation of EGFR and had consequently impaired the tumor initiating properties of ALDH1+ and CD29HighCD61+ breast CSCs. Autophagy inhibition led to decreased mRNA levels of TGFβ2 and TGFβ3 triggering dysregulation in Smad signaling which is essential for CD29HighCD61+ CSCs[78]. The secretion of IL-6 is autophagy dependent and is mediated through STAT3/JAK2 pathway[77]. From these studies, it can be assumed STAT3 signaling may potentially be an important factor in CSCs transformation.
In general, FOXO transcription factors have been associated in the regulation of cellular homeostasis, stem cell maintenance, ageing and tumor suppression. Mice with somatic deletion of FOXO1, FOXO2 and FOXO4 resulted in thymic lymphomas and hemangiomas[79]. Upregulation of FOXO1 promotes self-renewal of t(8;21) pre leukemia cells in vitro and in vivo, and restricts differentiation of AML cells with t(8;21) translocation; indicating FOXO1 is not a tumor suppressor, however, plays a crucial role in leukemia stem cells (LSCs) maintenance[80]. Absence of FOXO3 has been reported to contribute to the expansion of CSC population as well as increase self-renewal and tumorigenesis in prostate[81], colon[82], and glioblastoma[83] and promote tumor initiation in breast cancer[84]. Recently, it has been proposed that DNA methyltransferase 1 mediates FOXO3a promoter hyper methylation causing downregulation of FOXO3a gene expression in breast CSCs; thus, suppressing CSCs phenotype markers and tumorigenicity[85]. To date, the role of FOXO in CSCs remains controversial. It has been reported FOXOs are implicated in autophagy[86-88]. FOXO3 overexpression studies reveal this gene directly regulates autophagy related genes involved in the autophagosome pre-initiation complex: WIPI-1/2, core initiation complex: ULK1, autophagosome formation and elongation: Atg14, GABARAP, Atg5, Atg10. FOXO3 knockout cells downregulated many of these genes and PINK1 (component of mitophagy) and exhibited poor LC3B lipidation turnover; indicating FOXOs are required to maintain basal autophagy in neural stem and progenitor cells[87]. FOXO3A induced autophagy promotes survival in human pluripotent stem cells[88]. The pro autophagy protein, AMBRA1, modulates the differentiation of regulatory T cells through FOXO3/FOXP3 axis. In the context of immunosurveillance against tumors, AMBRA1 deficiency leads to defective generation of the induced regulatory T cells in lymph nodes of tumor bearing mice; influencing the regulatory T cells function in tumor response[89].
Additional FOXO family members are associated to autophagy. It is reported that FOXA2 knockdown in ovarian CSCs leads to a reduction in the number of spherical clusters of cells, size and the percentage of phenotype surface markers; suggesting FOX2A modulates the ability of self-renewal in vitro[90]. Inhibition of autophagy by Atg5 knockdown, bafilomycin A1 (vacuolar H+ ATPase inhibitor) or chloroquine (CQ; lysosomotropic agent- late stage autophagy inhibitor) repressed FOXA2 expression. FOXA2 overexpression partially rescues these effects; indicating autophagy modulates ovarian CSCs stemness through FOX2A[90]. These studies identify a synergy between FOXOs and autophagy. This relationship promotes CSC stemness and tumorigenesis; however, the mechanisms behind these actions remains unclear and require further elucidation. Though it is noteworthy that the regulatory role of autophagy in CSC is very complex.
The tumor microenvironment (TME) is a critical driver of tumor heterogeneity, encouraging CSCs plasticity, remodeling immune surveillance, and facilitating their metastatic potential and ultimately conferring CSCs resistance to chemotherapy drugs[52,91]. Non neoplastic cells, and their secreted mediators, such as growth factors and the release of cytokines, are found to contribute to the TME[92]. The core regulatory mechanism for oxygen sensing and adaption to hypoxia is hypoxia inducible factor (HIF), in particular HIF-1α and HIF-2α. HIF target genes are able to induce human ESCs markers sufficient to induce pluripotent stem cell inducers: OCT4, NANOG, SOX2, KLF4, MYC and miRNA-302 in multiple cancer cell lines. Similar results were reported in prostate tumors[93]. Hypoxia-Notch1-SOX2 signaling axis has been found to activate ovarian CSCs by stimulating self-renewal capacity and drug resistance[94]. Hypoxia activation and upregulation of HIFs has been implicated in aggressive tumor phenotypes, including breast and glioma CSCs, as a result correlating with co-localization studies of these markers with CSCs markers results to poor survival outcome in cancer patients[95,96].
The integration of autophagy in the cancer stem niche provides metabolic plasticity to CSCs from hypoxic conditions, nutrient limitation and acidosis[92]. Immuno-histochemistry of pancreatic ductal adenocarcinoma (PDA) tissues reveals co-expression of hypoxia, pancreatic CSC markers (CD44, CD24) and autophagy (BECN1 and LC3B). Hypoxia starvation induced autophagy has been demonstrated to increase clonogenicity and migration of PDA-CSCHigh cells and the number of autophagosomes formed[97]. In accordance, CD133+ pancreatic CSCs is dependent on HIF-1α to induce autophagy for stem cell maintenance[98]. Similarly, CD133+ liver CSCs showed higher survival capacity under hypoxic and nutrient deprived conditions[99]. Recently, phosphorylation of EZR at Thr567 residue and activation of PRKCA/PKCα kinase has been suggested to be a responsible candidate for enhanced self-renewal capacity of colorectal CSCs in hypoxia induced autophagy. The blockade of Atg5, BNIP3, BNIP3L, or BECN1 reversed these effects[100]. Limited literature is available to define the exact interplay between hypoxia, autophagy and the maintenance of the TME.
HIF-1α enhances the secretion of TGF-β1-Smad in mesenchymal stem cells (MSC) which facilitates the propagation of CD44+ breast cancer stem-like cells[94], promoting epithelial to mesenchymal transition (EMT)[101]. Autophagy inhibition by Atg5 silencing and CQ treatment, notably enhanced the transcriptional activation of epithelial marker CD24 whilst repressing EMT marker vimentin in response to TGF-1β, dysregulating cellular ability to migrate and invade[102]. In non small cell lung cells, vimentin was downregulated in the presence of TGF-β1 treatment in Atg7 knockdown cells, indicating autophagy positively regulates TGF-β1 in EMT[103]. To the contrary, autophagic targeting of EMT transcription factors, such as Snail and TWIST, through death-effector domain-containing DNA-binding protein-PI3KC3 has been shown to inhibit tumor metastasis and growth in breast cancer[104]. The divergent role of autophagy in EMT has been illiustrated in Figure 2.
Recently, it has been shown that pluripotent transcription factor NANOG, contributes to hypoxia-induced autophagy by directly activating BNIP3L. NANOG promotes resistance to immune mediated actions of cytotoxic T cells[105].
Mitophagy is the selective degradation of defective mitochondria by autophagy to avoid the accumulation of oxygen species and its association to cell death, senescence and malignant transformation. Mitochondria has a central role in generating ATP derived from oxidative phosphorylation (OXPHOS) and the tricarboxylic acid cycle[2]. Human pancreatic CSCs are primarily reliant upon OXPHOS for energy acquisition, as compared to their counterpart; indicating increased mitochondrial activity contributes to CSC stemness[106]. Similar results were observed in mice exhibiting KRAS gene ablation in pancreatic adenocarcinoma cells[107]. Moreover, KRASG12D mutated pancreatic adenocarcinoma cells have been shown to enter into quiescence in response to oncogene ablation and did not present metabolic stress and induced autophagy. This finding was confirmed by measuring the levels of LC3B by immunoblotting and using flow cytometry to quantify the autophagic flux of KRAS mutated cells stably expressing GFP-LC3; and Bafilomycin A1 treatment rescued the GFP signaling. Interestingly, these cancer cells exhibited stem cell-like phenotype[107]. Increased mitophagy is reported in esophageal squamous cell carcinoma CD44High undergoing EMT; the expression of CD44 is rendered during the inhibition of Parkin dependent mitophagy, resulting to cell death[108]. Hepatic CSCs stemness and self-renewal capacity is maintained by the removal of p53 localized to the mitochondria and removed in a mitophagy dependent manner. In contrast, during the suppression of mitophagy, p53 is phosphorylated by PINK1 and translocated to the nucleus to prevent Oct4, SOX2 and NANOG transcription in the hepatic CSC population. These results suggest that the activity of p53 is regulated by mitophagy to promote hepatocarcinogenesis[94]. In LSCs, the loss of p53 simultaneously activates endogenous KRASG12D mutation inducing aggressive AML phenotype; thus, enabling abnormal growth[109]. Mitophagy is activated in LSCs by the constitutive activity of AMPK and FIS1; preventing differentiation via GSK3 downstream mechanism and promotes stemness. Inhibition of AMPK-FIS1 axis results to suppression of proliferation and induction of differentiation[110].
Bcl-2 binds directly to BECN1 and plays a vital role in the development and differentiation of normal B cells to inhibit autophagy[111-113]. In accordance, immuno-histochemistry studies of patients with diffuse large B cell lymphomas (DLBCL) revealed that increased BECN1 levels with reduced levels of Bcl-2 correlated favorably to the clinical survival outcome with better response to the first line treatment of R-CHOP[114,115]. The incidence of breast, ovarian and prostate cancer is higher in 40%-75% patients with monoallelic deletions of BECN1 gene. Furthermore, in mice with heterozygous deletion of BECN1 predisposed them to spontaneous malignancies including DLBCL, suggesting BECN1 is a haplo-insufficient tumor suppressor gene[116]. Similar findings were reported in the incidence of pre-B acute lymphoblastic lymphoma with elevated expressions of programmed death ligand 1 (PD-L1) and IL-10[117]. A study led by Bertolo et al[118], suggests constitutive suppression of autophagy responses in BCL-6 driven GC-derived lymphomas, including DLBCL contribute to lymphomagenesis. In mice, the homozygote deletion of BECN1 results to embryonic lethality, in comparison BECN1 heterozygous deletion leads to the establishment of spontaneous tumors and defective autophagy; however, did not impair apoptosis[119].
Enhanced autophagic flux has been attributed significantly to metastatic tumorigenesis and immunosuppression related chemoresistance. In ex vivo lung cancer cells, CQ augments carboplatin treatment by sensitizing the lung cancer drug resistant cells and non-resistant cells by limiting the proliferation status and providing synergistic effects with carboplatin to induce apoptosis. These findings corroborated with the decreased LC3B level and BECN1 protein expressions suggesting a decrease in the formation of autophagosomes. The administration of CQ in drug resistant cancer cells, strikingly reduced the drug resistant proteins: MDR1, MRP1 and ABCG2 and mRNA reduction of MRP1 and ABCC2. The combination treatment of CQ and Carboplatin significantly reduced both the protein and the mRNA levels. Furthermore, this decreased the expression of PD-L1 suggesting autophagy has a role in modulating of PD-L1 in cancer evasion and immunosuppression. Interestingly, the combination treatment promoted the infiltration of CD4+, FOXP3+ tumor infiltrating lymphocytes (TILs) indicating autophagy inhibition with carboplatin could mediate lymphocyte infiltration in the tumor and upregulate only specific expression of TILs, leading to immune system activation[120].
Cancer progression leads to metastatic growth resulting to a majority of cancer related deaths[121]. In many cases, dissemination of tumor cells (DTCs) has already occurred in patients at diagnosis. It is challenging to detect DTCs at secondary sites, as they may have entered into dormancy and become refractory to therapeutic targets[122]. The divergent characteristics of DTCs have emphasized the need to improve this phenomenon. It is postulated that autophagy is activated during the seeding process of DTCs at secondary sites providing an adaptive response to nutrient depletion and environmental stress[123]. For example, the tumor suppressor gene ARHI (RAS homologue) is downregulated in 60% of ovarian cancer cases. Studies in ovarian cancer cell lines revealed autophagy induction is mediated by ARHI as it inhibits PI3K-mTOR signaling. This is corroborated by co-localization staining of Atg4 and LC3B in autophagosomes suggesting ARHI facilitates the autophagosome formation through this signaling. Xenograft model expressing SKOv3-ARHI cells supplemented with ARHI by doxycycline repressed tumor growth, however, the withdrawal of ARHI after 32 or 42 d stimulated rapid tumor growth, indicating that the cancer cells, in particular, CSCs remained viable and dormant during latency. Autophagy inhibition by CQ in this model confirms dormancy requires ARHI mediated autophagy[124]. Accordingly, Atg7 is essential for the reduction of lung metastatic burden utilizing a non-canonical autophagy pathway independent of BECN1[125]. In contrast, recent microarray analysis of CSCs in breast cancer patients revealed the expression of 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (Pfkfb3), which correlated with an aggressive cancer phenotype coupled with self-renewal ability and metastasis potential. Dormant breast cancer cells display Pfkfb3LowAutophagyHigh phenotype with elevated levels of LC3B and p62. In contrast, the metastatic breast cancer cells which exhibited Pfkfb3HighAutophagyLow; suggesting the status of autophagy changes during the phenotypic transition. Knockdown of Atg3, Atg7 or p62 genes promoted the proliferation and outgrowth restoring the upregulated expression of Pfkfb3 in dormant breast CSCs. The ablation of autophagy related genes gained CD49fHigh/CD24Low phenotype with increased stemness signature in CSCs. These findings reveal autophagy activation could function to prolong the overall survival of patients by promoting permanent dormancy of CSCs. Additionally, Pfkfb3 protein was found to directly interact with ubiquitin binding domain of p62, suggesting its role as a substrate. Moreover, inactivation of autophagy can facilitate dormancy of breast CSCs to metastatic lesions by stabilizing Pfkfb3 gene expressions via p62[126]. These studies highlight the poorly understood role of autophagy during dormancy in breast CSCs, in which targeting autophagy enables the sensitization of CSCs to chemotherapy by eliminating the adaptive response to autophagy[124,125]. Though it is noteworthy, CSCs are heterogeneous and disease specificity adds complexity to the matter[126].
Autophagy demonstrates tumor-suppressing actions in early cancer initiation; but fundamentally provides adaptive responses – an advantage to cancer cells and CSC during cellular stress. It remains an open question whether to stimulate or to inhibit autophagy in cancer, specifically, in combination with anti-cancer therapeutics. Autophagy inhibition may provide a reasonable rational to be used; as multiple tumors stimulate autophagy as a source of nutrient replenishment for their increased metabolic demands, survival and disease propagation (Figure 3)[127]. Hydro-xychloroquine (HCQ) is an FDA approved drug, with the capability to suppress autophagy at a later stage by inhibiting lysosome acidification and due to these functions it has been used in numerous early phase clinical trials[128]. Meta-analysis data reveals autophagy inhibition based treatment leads to a better therapeutic response as compared to chemotherapy or radiation in the absence of autophagy; suggesting this may provide a new therapeutic strategy for anti-tumoral therapy[129]. However, the activation of autophagy may potentially hold a beneficial role as an anticancer therapy. For example, tat-BECN1 peptide was shown to induce autophagy in HER2-positive breast cancer xenografts and prevented tumor growth[131].
In hindsight, CSCs and their counterparts have a unique and complex interaction within the tumor niche which challenges the opportunity to target autophagy directly[132]. Several studies indicate the beneficial impact of combination treatments of chemotherapeutics with autophagy modulators. For instance, the combination of autophagy modulators with chemotherapy showed to stimulate of CD8+ T cell-dependent anticancer immune responses leading to tumor sensitization and cancer cell growth reduction (Figure 2)[133].
CSCs are highly tumorigenic and contribute to cancer relapse due to their ability to self-renew and differentiate into heterogeneous cancer cell lineages. Their resilience is demonstrated in the treatment of chemotherapy and radiation therapy[57]. In addition, CSCs are able to remain in a quiescent state and cultivate their ability to become resistant by gaining adaption to their environment[123]. For example, in castration resistant prostate cancer it has been shown that autophagy is induced during Docetaxel treatment and STAT3 contributes to cancer cell survival[134]; suggesting it is important to target autophagy directly or as a combination treatment to sensitize cancer cells.
It should be highlighted that CQ and HCQ exert anti-tumor effects in combination with anti-cancer treatments in clinical trials[135]. In PDA the combination of Gemcitabine with HCQ was assessed[136], this was also evaluated in studies of breast cancer and irradiation[137], or in combination with the autophagy inducer Temsirolimus in patients with various solid cancers including melanoma[138]. In preclinical in vitro models of breast cancer, similar results were reported[139]. These findings suggest that autophagy inhibition and activation are promising methods to elicit the sensitization of CSCs to chemotherapy. Moreover, it can be concluded that metastatic cells are preferentially vulnerable to lysosomal inhibition; however, it would be important to assess if these metastatic cancer cells express stem cell-like phenotypical features[140]. For example, autophagy inhibition in breast CSCs expressing Pfkfb3 were found to promote tumor metastasis[126]; suggesting therapeutic strategies involving autophagy modulation in treating CSCs, also depends on the cancer phenotype. As mentioned above, CQ and HCQ have been used as late stage autophagy inhibitors in numerous studies. However, the development of newer generation of lysosome inhibitors are more selective and potent which have been introduced, including Lys05 (analogue of CQ) and dimeric quinacrine (DQ661) - a derivative of Lys05. Both are specific in targeting the lysosome and causing impairment of palmitoyl-protein thioesterase activity by impairing mTOR signaling pathway[141]. Lys05 is a potent autophagy inhibitor in comparison to HCQ. Lys05 has shown to decrease the number of LSCs in vitro by promoting their maturation; similar results were seen in patient-derived samples[142]. DQ661 is effective in targeting cancer paradigms of melanoma, colon cancer and PDA by repressing growth and inhibiting autophagy[141]. Inhibitor of V-ATPase called Concanamycin A, protease inhibitor E64d and pepstatin A, have also been introduced[143]. These autophagy modulators are providing an opportunity to explore different combination treatments in different cancer types. Moreover, these lysosomotropic targets are deemed to be effective in bulk autophagy degradation, in comparison to selective autophagy, such as mitophagy[2]; in such instances early stage autophagy inhibitors would be considered to be more beneficial. Early stage autophagy inhibitors could target the initiation of autophagy, for example PIK-III (Vps34 inhibitor)[144], MRT68921, SBI-0206965 (ULK inhibitors)[145,146] and SAR405 (PIK3C3/Vps34 inhibitor)[147]. Interestingly, SAR405 and Everolimus (an autophagy inducers) demonstrate significant synergism in renal tumor cells by reducing cancer cell proliferation[147]. Additionally, early stage autophagy inhibitors would be a strategic method to target tumors grown in oxygenated environments, as they use OXPHOS as an alternative source of metabolism.
Autophagy is an adaptive mechanism modulating the TME surrounding CSCs. Several studies defined CSCs inducing autophagy in the TME to support their stemness and cancer propagation by activating the autophagic machinery under nutrient depleted and hypoxic conditions, for example in breast cancer[72-74]. By these actions, autophagy can initiate the development of an aggressive cancer phenotype and develop resistance to cell death. Further investigations are needed to explore the role of autophagy in these cells within the tumor niche in order to tackle the protective surroundings of the TME.
Oncolytic viruses (OVs) therapy is an emerging anti-cancer treatment capable of efficiently killing CSCs and cancer cells in several tumor types[148]. The most commonly used OVs include adenoviruses, herpes simplex virus, measles virus, reovirus, Newcastle disease virus and adenovirus serotype 5[149]. OVs retain the capability to infect, replicate and integrate into tumor cells and potentially in their immunosuppressed TME. Malignant cells overexpressing certain virus receptors, including coxsackie-adenovirus receptor[150,151], CD155[152], CD46[153] and laminin[154] are targeted by OVs. Several studies revealed that autophagy facilitates immunogenic cell death via stimulating the release of pathogen associated molecular pattern and damaged associated molecular pattern and initiating their responses in the TME[155]. These responses activate the secretion of ATP from the tumor cells promoting the stimulation of antigen presenting cells, such as DCs to elicit antigens on major histocompatibility I and II molecules which stimulate T cells[13,156,157]. Consequently, pro inflammatory cytokines, including type I interferons induce the stimulator of interferon genes signaling in DCs, further benefiting anti-tumoral T cell responses[158]. In the context of autophagy, OVs employ strategic methods to survive and propagate within the cancer cells by perturbing the core autophagic machinery[159,160].
Autophagy can either be promoted or inhibited during oncolytic adenovirus therapy[161]. The expression of adenovirus oncoprotein triggers the upregulation of Atg1, Atg5 and LC3 proteins[162]. Leukemic cells treated with oncolytic adenovirus encoding BECN1 (SG511-BECN1) significantly induced autophagic cell death in vitro. Similarly, primary blasts isolated from chronic myelocytic leukemia patients with Imatinib resistance and AML patients with relapse disease treated with SC511-BECN1 showed an increase in BECN1 expression and LC3B accumulation. This led to significantly reduction of colony formation in comparison to SG511 control[163]. Interestingly, combination treatment of SG511-BECN1 and Doxorubicin is highly synergistic in chronic myelocytic leukemia cell lines leading to significant cancer cell death. Increased levels of BECN1 and LC3B proteins were observed in comparison with normal mononuclear cells; suggesting the combination of SG511-BECN1 and Doxorubicin elicits synergistic effects in an autophagy dependent manner[164].
In liver CSCs, oncolytic virus expressing tumor suppressor gene, TSLC1, and specifically targeting Wnt signaling, promoted the generation of autophagosomes. This was confirmed by the upregulation of BECN1 and accumulation of total LC3 and led to the reduction of p62 and Survivin. This resulted in cell death in an autophagy dependent manner. The inhibition of autophagy by CQ induced the accumulation of total LC3 and p62, this in turn promoted the survival of the liver CSCs. The hepatic xenograft models treated with this adenovirus induced apoptosis and inhibited tumor metastasis resulting in an improved survival outcome[165]. It has been proposed autophagy activators, such as Rapamycin or Temozolomide synergistically sensitize tumor cells to adenovirus by stimulating autophagy, without modifying the viral replication; thus, inducing autophagy dependent cell death as an antitumor mechanism[166]. In addition, the adenovirus E4 protein suppresses autophagy by activating mTOR signaling and inhibiting ULK1 activity[161].
The clinical development of immune checkpoint inhibitors (ICIs) is an emerging treatment modality for the reversal of TILs dysregulation phenotype, thereby imposing antitumor responses. Different immune checkpoints, such as T lymphocyte antigen-4 (CTLA-4), PD-1 and PD-L1 could be clinically targeted using ICIs[167].
It is reported that PD-L1 expression on melanoma and ovarian cancer cells elicits tumor growth mainly via Akt-mTOR regulated autophagy; this data corroborated with a comparative microarray analysis. Moreover, melanoma PD-L1High expressing tumors demonstrated increased sensitivity to CQ; thus, limiting proliferation in vitro and in vivo[168]. RNA sequencing data in PD-L1 positive glioma cells promoted cancer invasion in starvation induced autophagy, utilizing the Akt-F-Actin signaling[169]. In gastric cancers the knockdown of Atg5 and Atg7 genes inhibited LC3B formation, leading to the upregulation of PD-L1 by the activation of NF-Kb pathway[170]. These accumulating studies confirm intrinsic PD-L1 functions through the activation of Akt-mTOR pathway, however, the mechanisms by which PD-L1 transduces signals remains unknown. The identification of these targets may potentially lead to targeted combinational treatments using autophagic agents. Recently, it is reported that Sigma1 promotes the degradation of PD-L1 using selective autophagy and ablates the functional interaction of PD-1 and PD-L1 in co-cultures of T cells and tumor cells[171]. In accordance, targeting cancer cells expressing CD274 with PD-L1/PD-1 inhibitors can stimulate autophagy and promote sensitization of cancer cells when combined with autophagy inhibitors[172].
CTLA-4 inhibitor is an effective ICI in a subset of patients with metastatic melanoma. In a small cohort of melanoma patients, a subcluster of MAGE-A cancer germline antigens, were found to be overexpressed causing resistance to CTLA-4 inhibition, but not PD-1. Tissue microarray data revealed that the LC3B expression in MAGE-A+ tumors was significantly attenuated as compared to MAGE-A- tumors. Moreover, immunohistochemistry data indicated MAGE-A and damaged associated molecular pattern protein high-mobility group box 1 (HMGB-1) were mutually expressed in the clinical samples. In vitro ubiquitination screening confirmed that autophagy was suppressed by the MAGE-TRIM28 ubiquitin ligase complex[173]. HMGB-1 is a pro autophagic protein that directly interacts with BECN1 by displacing BCL-2; thus, sustaining autophagy and promoting cellular survival[174]. The secretion of HMGB-1 mediates the priming of immune adaptive response[175]. To overcome CTLA-4 therapy resistance in melanoma patients, the induction of autophagy may potentially be relevant in enhancing the effect of CTLA-4 inhibitors; thus, minimizing tumor immune tolerance. Combining CTLA-4 inhibition with Rapamycin in vivo during CD8+ T cell priming, led to an increase of Ag-specific memory CD8+ T cells and enhanced their function, which in turn, resulted to tumor growth reduction, rapid bacterial clearance and mediated cytokine production[176]. Taking these findings into consideration, the induction of autophagy would reinstate the CTLA-4 expression and its suppressive functions, thereby, eliciting antitumoral activity.
New therapeutic concepts are needed to improve the prognosis of cancer patients. One possible starting point is the tumor-specific metabolism of cancer cells. Autophagy is a catabolic recycling process exciting different forms of cancer cells and CSCs. In general, CSC maintenance and the development of an aggressive cancer phenotype have strongly been correlated to autophagy. In cancer, the role of autophagy is context dependent as it demonstrates functions both as a tumor suppressor during tumor initiation and as a pro-survival mechanism during cancer propagation by facilitating CSCs and cancer cells adaptive responses during metabolic stresses and dormancy.
Targeting autophagy could potentially represent a promising therapeutic target for preventing the aggressive and resistance cancer phenotypes. There is convincing evidence that the inhibition of autophagy in cancer cells, and specifically in CSCs, augments cytotoxicity leading to antitumoral effects under certain conditions. Therefore, we can expect valuable knowledge regarding suitable autophagy-associated biomarkers in tumor cells and new therapeutic approaches that are specifically directed against autophagy-dependent pathways in cancer cells or CSCs. Additionally, it is increasing evident that autophagy is involved in the maintenance of immune cell homeostasis, activation and function in the TME. However, limited studies are available to interpret whether autophagy enhancement or inhibition my support the effects of immunotherapy. Several additional preclinical studies are necessary to identify them, specifically, in a context dependent manner. This would represent an important step in the direction of improved and individualized cancer therapy.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: Switzerland
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cenciarelli C, Wang YG S-Editor: Dou Y L-Editor: A E-Editor: Liu JH
| 1. | Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741-752. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2430] [Cited by in F6Publishing: 2658] [Article Influence: 156.4] [Reference Citation Analysis (0)] |
| 2. | Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat Rev Immunol. 2019;19:170-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 211] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 3. | Nazio F, Bordi M, Cianfanelli V, Locatelli F, Cecconi F. Autophagy and cancer stem cells: molecular mechanisms and therapeutic applications. Cell Death Differ. 2019;26:690-702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 237] [Cited by in F6Publishing: 236] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 4. | Loukil A, Zonca M, Rebouissou C, Baldin V, Coux O, Biard-Piechaczyk M, Blanchard JM, Peter M. High-resolution live-cell imaging reveals novel cyclin A2 degradation foci involving autophagy. J Cell Sci. 2014;127:2145-2150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Djavaheri-Mergny M, Giuriato S, Tschan MP, Humbert M. Therapeutic Modulation of Autophagy in Leukaemia and Lymphoma. Cells. 2019;8:103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 6. | Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811-1836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1076] [Article Influence: 153.7] [Reference Citation Analysis (0)] |
| 7. | Massey AC, Kaushik S, Sovak G, Kiffin R, Cuervo AM. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci USA. 2006;103:5805-5810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 400] [Cited by in F6Publishing: 396] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 8. | Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1382] [Cited by in F6Publishing: 1664] [Article Influence: 237.7] [Reference Citation Analysis (0)] |
| 9. | Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179-2192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 240] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 10. | Goldsmith J, Levine B, Debnath J. Autophagy and cancer metabolism. Methods Enzymol. 2014;542:25-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 101] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 11. | Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759-774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1033] [Cited by in F6Publishing: 968] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 12. | Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1168] [Cited by in F6Publishing: 1242] [Article Influence: 112.9] [Reference Citation Analysis (0)] |
| 13. | Doherty J, Baehrecke EH. Life, death and autophagy. Nat Cell Biol. 2018;20:1110-1117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 448] [Article Influence: 74.7] [Reference Citation Analysis (0)] |
| 14. | Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425-434. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1755] [Cited by in F6Publishing: 1827] [Article Influence: 96.2] [Reference Citation Analysis (0)] |
| 15. | Jiang X, Bao Y, Liu H, Kou X, Zhang Z, Sun F, Qian Z, Lin Z, Li X, Liu X, Jiang L, Yang Y. VPS34 stimulation of p62 phosphorylation for cancer progression. Oncogene. 2017;36:6850-6862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 16. | Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1403] [Cited by in F6Publishing: 1579] [Article Influence: 157.9] [Reference Citation Analysis (0)] |
| 17. | Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, Ahmed Q, Domart MC, Collinson L, Ktistakis NT. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016;7:12420. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 205] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 18. | Onorati AV, Dyczynski M, Ojha R, Amaravadi RK. Targeting autophagy in cancer. Cancer. 2018;124:3307-3318. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 455] [Cited by in F6Publishing: 439] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 19. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2401] [Cited by in F6Publishing: 2580] [Article Influence: 184.3] [Reference Citation Analysis (0)] |
| 20. | Turco E, Witt M, Abert C, Bock-Bierbaum T, Su MY, Trapannone R, Sztacho M, Danieli A, Shi X, Zaffagnini G, Gamper A, Schuschnig M, Fracchiolla D, Bernklau D, Romanov J, Hartl M, Hurley JH, Daumke O, Martens S. FIP200 Claw Domain Binding to p62 Promotes Autophagosome Formation at Ubiquitin Condensates. Mol Cell. 2019;74:330-346.e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 193] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 21. | Cunha LD, Yang M, Carter R, Guy C, Harris L, Crawford JC, Quarato G, Boada-Romero E, Kalkavan H, Johnson MDL, Natarajan S, Turnis ME, Finkelstein D, Opferman JT, Gawad C, Green DR. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell. 2018;175:429-441.e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 205] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 22. | Wong SW, Sil P, Martinez J. Rubicon: LC3-associated phagocytosis and beyond. FEBS J. 2018;285:1379-1388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 23. | Mauthe M, Jacob A, Freiberger S, Hentschel K, Stierhof YD, Codogno P, Proikas-Cezanne T. Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy. 2011;7:1448-1461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 24. | Longatti A, Tooze SA. Vesicular trafficking and autophagosome formation. Cell Death Differ. 2009;16:956-965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 162] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 25. | Proikas-Cezanne T, Robenek H. Freeze-fracture replica immunolabelling reveals human WIPI-1 and WIPI-2 as membrane proteins of autophagosomes. J Cell Mol Med. 2011;15:2007-2010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 26. | Martinez-Martin N, Maldonado P, Gasparrini F, Frederico B, Aggarwal S, Gaya M, Tsui C, Burbage M, Keppler SJ, Montaner B, Jefferies HB, Nair U, Zhao YG, Domart MC, Collinson L, Bruckbauer A, Tooze SA, Batista FD. A switch from canonical to noncanonical autophagy shapes B cell responses. Science. 2017;355:641-647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 27. | Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 801] [Cited by in F6Publishing: 856] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 28. | Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13:7-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 418] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 29. | Honda S, Arakawa S, Nishida Y, Yamaguchi H, Ishii E, Shimizu S. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun. 2014;5:4004. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 30. | Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493-1502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 411] [Cited by in F6Publishing: 428] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 31. | Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032-1036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2154] [Cited by in F6Publishing: 2215] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 32. | Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy. 2013;9:1270-1285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 33. | González-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquère S, Eskelinen EL, Pierron G, Saftig P, Kroemer G. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005;118:3091-3102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 397] [Cited by in F6Publishing: 384] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 34. | Poon IK, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 732] [Cited by in F6Publishing: 920] [Article Influence: 92.0] [Reference Citation Analysis (0)] |
| 35. | Oral O, Akkoc Y, Bayraktar O, Gozuacik D. Physiological and pathological significance of the molecular cross-talk between autophagy and apoptosis. Histol Histopathol. 2016;31:479-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 47] [Reference Citation Analysis (0)] |
| 36. | Nikoletopoulou V, Markaki M, Palikaras K, Tavernarakis N. Crosstalk between apoptosis, necrosis and autophagy. Biochim Biophys Acta. 2013;1833:3448-3459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 844] [Cited by in F6Publishing: 936] [Article Influence: 85.1] [Reference Citation Analysis (0)] |
| 37. | Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol. 2004;6:1221-1228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1017] [Cited by in F6Publishing: 1025] [Article Influence: 51.3] [Reference Citation Analysis (0)] |
| 38. | Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285-292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 441] [Cited by in F6Publishing: 463] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 39. | Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012;19:788-797. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 40. | Gandesiri M, Chakilam S, Ivanovska J, Benderska N, Ocker M, Di Fazio P, Feoktistova M, Gali-Muhtasib H, Rave-Fränk M, Prante O, Christiansen H, Leverkus M, Hartmann A, Schneider-Stock R. DAPK plays an important role in panobinostat-induced autophagy and commits cells to apoptosis under autophagy deficient conditions. Apoptosis. 2012;17:1300-1315. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 41. | Hou W, Han J, Lu C, Goldstein LA, Rabinowich H. Autophagic degradation of active caspase-8: a crosstalk mechanism between autophagy and apoptosis. Autophagy. 2010;6:891-900. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 272] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 42. | Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, Agostinis P, Vanden Berghe T, Lippens S, Vandenabeele P. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010;1:e18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 466] [Cited by in F6Publishing: 490] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 43. | Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, Annicchiarico-Petruzzelli M, Antonov AV, Arama E, Baehrecke EH, Barlev NA, Bazan NG, Bernassola F, Bertrand MJM, Bianchi K, Blagosklonny MV, Blomgren K, Borner C, Boya P, Brenner C, Campanella M, Candi E, Carmona-Gutierrez D, Cecconi F, Chan FK, Chandel NS, Cheng EH, Chipuk JE, Cidlowski JA, Ciechanover A, Cohen GM, Conrad M, Cubillos-Ruiz JR, Czabotar PE, D'Angiolella V, Dawson TM, Dawson VL, De Laurenzi V, De Maria R, Debatin KM, DeBerardinis RJ, Deshmukh M, Di Daniele N, Di Virgilio F, Dixit VM, Dixon SJ, Duckett CS, Dynlacht BD, El-Deiry WS, Elrod JW, Fimia GM, Fulda S, García-Sáez AJ, Garg AD, Garrido C, Gavathiotis E, Golstein P, Gottlieb E, Green DR, Greene LA, Gronemeyer H, Gross A, Hajnoczky G, Hardwick JM, Harris IS, Hengartner MO, Hetz C, Ichijo H, Jäättelä M, Joseph B, Jost PJ, Juin PP, Kaiser WJ, Karin M, Kaufmann T, Kepp O, Kimchi A, Kitsis RN, Klionsky DJ, Knight RA, Kumar S, Lee SW, Lemasters JJ, Levine B, Linkermann A, Lipton SA, Lockshin RA, López-Otín C, Lowe SW, Luedde T, Lugli E, MacFarlane M, Madeo F, Malewicz M, Malorni W, Manic G, Marine JC, Martin SJ, Martinou JC, Medema JP, Mehlen P, Meier P, Melino S, Miao EA, Molkentin JD, Moll UM, Muñoz-Pinedo C, Nagata S, Nuñez G, Oberst A, Oren M, Overholtzer M, Pagano M, Panaretakis T, Pasparakis M, Penninger JM, Pereira DM, Pervaiz S, Peter ME, Piacentini M, Pinton P, Prehn JHM, Puthalakath H, Rabinovich GA, Rehm M, Rizzuto R, Rodrigues CMP, Rubinsztein DC, Rudel T, Ryan KM, Sayan E, Scorrano L, Shao F, Shi Y, Silke J, Simon HU, Sistigu A, Stockwell BR, Strasser A, Szabadkai G, Tait SWG, Tang D, Tavernarakis N, Thorburn A, Tsujimoto Y, Turk B, Vanden Berghe T, Vandenabeele P, Vander Heiden MG, Villunger A, Virgin HW, Vousden KH, Vucic D, Wagner EF, Walczak H, Wallach D, Wang Y, Wells JA, Wood W, Yuan J, Zakeri Z, Zhivotovsky B, Zitvogel L, Melino G, Kroemer G. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018;25:486-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3672] [Cited by in F6Publishing: 3523] [Article Influence: 587.2] [Reference Citation Analysis (0)] |
| 44. | Laussmann MA, Passante E, Düssmann H, Rauen JA, Würstle ML, Delgado ME, Devocelle M, Prehn JH, Rehm M. Proteasome inhibition can induce an autophagy-dependent apical activation of caspase-8. Cell Death Differ. 2011;18:1584-1597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 45. | Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 943] [Cited by in F6Publishing: 1018] [Article Influence: 78.3] [Reference Citation Analysis (0)] |
| 46. | Radpour R. Tracing and targeting cancer stem cells: New venture for personalized molecular cancer therapy. World J Stem Cells. 2017;9:169-178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 47. | Radpour R, Forouharkhou F. Single-cell analysis of tumors: Creating new value for molecular biomarker discovery of cancer stem cells and tumor-infiltrating immune cells. World J Stem Cells. 2018;10:160-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 48. | Takeishi S, Nakayama KI. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br J Cancer. 2014;111:1054-1059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 49. | Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253-1270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1841] [Cited by in F6Publishing: 1866] [Article Influence: 88.9] [Reference Citation Analysis (0)] |
| 50. | Hatina J. The dynamics of cancer stem cells. Neoplasma. 2012;59:700-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011;121:3804-3809. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 438] [Cited by in F6Publishing: 453] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 52. | Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell. 2015;16:225-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 883] [Cited by in F6Publishing: 1021] [Article Influence: 127.6] [Reference Citation Analysis (0)] |
| 53. | Fessler E, Dijkgraaf FE, De Sousa E Melo F, Medema JP. Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett. 2013;341:97-104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 54. | Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010-1012. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 718] [Cited by in F6Publishing: 699] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 55. | Suresh R, Ali S, Ahmad A, Philip PA, Sarkar FH. The Role of Cancer Stem Cells in Recurrent and Drug-Resistant Lung Cancer. Adv Exp Med Biol. 2016;890:57-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 56. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3316] [Cited by in F6Publishing: 3222] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 57. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7830] [Cited by in F6Publishing: 7469] [Article Influence: 355.7] [Reference Citation Analysis (0)] |
| 58. | Radpour R, Barekati Z, Kohler C, Holzgreve W, Zhong XY. New trends in molecular biomarker discovery for breast cancer. Genet Test Mol Biomarkers. 2009;13:565-571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 59. | Wang VM, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, Frith D, Carvalho J, Barry DJ, Snijders AP, Herbert E, Nye EL, MacRae JI, Behrens A. CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol. 2019;21:1425-1435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 60. | O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2977] [Cited by in F6Publishing: 2949] [Article Influence: 163.8] [Reference Citation Analysis (0)] |
| 61. | Aghagolzadeh P, Radpour R. New trends in molecular and cellular biomarker discovery for colorectal cancer. World J Gastroenterol. 2016;22:5678-5693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 51] [Cited by in F6Publishing: 53] [Article Influence: 6.6] [Reference Citation Analysis (1)] |
| 62. | Du Y, Shao H, Moller M, Prokupets R, Tse YT, Liu ZJ. Intracellular Notch1 Signaling in Cancer-Associated Fibroblasts Dictates the Plasticity and Stemness of Melanoma Stem/Initiating Cells. Stem Cells. 2019;37:865-875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 63. | Parte SC, Batra SK, Kakar SS. Characterization of stem cell and cancer stem cell populations in ovary and ovarian tumors. J Ovarian Res. 2018;11:69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 64. | Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008;68:4311-4320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 911] [Cited by in F6Publishing: 965] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 65. | Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5422] [Cited by in F6Publishing: 5376] [Article Influence: 268.8] [Reference Citation Analysis (0)] |
| 66. | Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 335] [Cited by in F6Publishing: 367] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 67. | Riether C, Schürch CM, Flury C, Hinterbrandner M, Drück L, Huguenin AL, Baerlocher GM, Radpour R, Ochsenbein AF. Tyrosine kinase inhibitor-induced CD70 expression mediates drug resistance in leukemia stem cells by activating Wnt signaling. Sci Transl Med. 2015;7:298ra119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 68. | Riether C, Schürch CM, Bührer ED, Hinterbrandner M, Huguenin AL, Hoepner S, Zlobec I, Pabst T, Radpour R, Ochsenbein AF. CD70/CD27 signaling promotes blast stemness and is a viable therapeutic target in acute myeloid leukemia. J Exp Med. 2017;214:359-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 112] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 69. | Radpour R, Riether C, Simillion C, Höpner S, Bruggmann R, Ochsenbein AF. CD8+ T cells expand stem and progenitor cells in favorable but not adverse risk acute myeloid leukemia. Leukemia. 2019;33:2379-2392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 70. | Hernández-Camarero P, Jiménez G, López-Ruiz E, Barungi S, Marchal JA, Perán M. Revisiting the dynamic cancer stem cell model: Importance of tumour edges. Crit Rev Oncol Hematol. 2018;131:35-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 71. | El Hout M, Cosialls E, Mehrpour M, Hamaï A. Crosstalk between autophagy and metabolic regulation of cancer stem cells. Mol Cancer. 2020;19:27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 72. | Cho YH, Han KM, Kim D, Lee J, Lee SH, Choi KW, Kim J, Han YM. Autophagy regulates homeostasis of pluripotency-associated proteins in hESCs. Stem Cells. 2014;32:424-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 73. | Wolf J, Dewi DL, Fredebohm J, Müller-Decker K, Flechtenmacher C, Hoheisel JD, Boettcher M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013;15:R109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 74. | Gong C, Bauvy C, Tonelli G, Yue W, Deloménie C, Nicolas V, Zhu Y, Domergue V, Marin-Esteban V, Tharinger H, Delbos L, Gary-Gouy H, Morel AP, Ghavami S, Song E, Codogno P, Mehrpour M. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261-2272, 2272e.1-2272e.11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 260] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 75. | Antonelli M, Strappazzon F, Arisi I, Brandi R, D'Onofrio M, Sambucci M, Manic G, Vitale I, Barilà D, Stagni V. ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget. 2017;8:21692-21709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 76. | Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, Hansen RJ, Morgan MJ, Porter CC, Gustafson DL, Thorburn A. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer Res. 2014;74:2579-2590. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 138] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 77. | Maycotte P, Jones KL, Goodall ML, Thorburn J, Thorburn A. Autophagy Supports Breast Cancer Stem Cell Maintenance by Regulating IL6 Secretion. Mol Cancer Res. 2015;13:651-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 130] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 78. | Yeo SK, Wen J, Chen S, Guan JL. Autophagy Differentially Regulates Distinct Breast Cancer Stem-like Cells in Murine Models via EGFR/Stat3 and Tgfβ/Smad Signaling. Cancer Res. 2016;76:3397-3410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 79. | Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 788] [Cited by in F6Publishing: 840] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 80. | Lin S, Ptasinska A, Chen X, Shrestha M, Assi SA, Chin PS, Imperato MR, Aronow BJ, Zhang J, Weirauch MT, Bonifer C, Mulloy JC. A FOXO1-induced oncogenic network defines the AML1-ETO preleukemic program. Blood. 2017;130:1213-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 81. | Dubrovska A, Kim S, Salamone RJ, Walker JR, Maira SM, García-Echeverría C, Schultz PG, Reddy VA. The role of PTEN/Akt/PI3K signaling in the maintenance and viability of prostate cancer stem-like cell populations. Proc Natl Acad Sci USA. 2009;106:268-273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 415] [Cited by in F6Publishing: 433] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 82. | Penrose HM, Cable C, Heller S, Ungerleider N, Nakhoul H, Baddoo M, Hartono AB, Lee SB, Burow ME, Flemington EF, Crawford SE, Savkovic SD. Loss of Forkhead Box O3 Facilitates Inflammatory Colon Cancer: Transcriptome Profiling of the Immune Landscape and Novel Targets. Cell Mol Gastroenterol Hepatol. 2019;7:391-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 83. | Sunayama J, Sato A, Matsuda K, Tachibana K, Watanabe E, Seino S, Suzuki K, Narita Y, Shibui S, Sakurada K, Kayama T, Tomiyama A, Kitanaka C. FoxO3a functions as a key integrator of cellular signals that control glioblastoma stem-like cell differentiation and tumorigenicity. Stem Cells. 2011;29:1327-1337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 84. | Smit L, Berns K, Spence K, Ryder WD, Zeps N, Madiredjo M, Beijersbergen R, Bernards R, Clarke RB. An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget. 2016;7:2596-2610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 85. | Liu H, Song Y, Qiu H, Liu Y, Luo K, Yi Y, Jiang G, Lu M, Zhang Z, Yin J, Zeng S, Chen X, Deng M, Jia X, Gu Y, Chen D, Zheng G, He Z. Downregulation of FOXO3a by DNMT1 promotes breast cancer stem cell properties and tumorigenesis. Cell Death Differ. 2020;27:966-983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 86. | Schäffner I, Minakaki G, Khan MA, Balta EA, Schlötzer-Schrehardt U, Schwarz TJ, Beckervordersandforth R, Winner B, Webb AE, DePinho RA, Paik J, Wurst W, Klucken J, Lie DC. FoxO Function Is Essential for Maintenance of Autophagic Flux and Neuronal Morphogenesis in Adult Neurogenesis. Neuron. 2018;99:1188-1203.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 87. | Audesse AJ, Dhakal S, Hassell LA, Gardell Z, Nemtsova Y, Webb AE. FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet. 2019;15:e1008097. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 88. | Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, Malhotra R, Debnath J, Passegué E. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 428] [Cited by in F6Publishing: 453] [Article Influence: 41.2] [Reference Citation Analysis (1)] |
| 89. | Becher J, Simula L, Volpe E, Procaccini C, La Rocca C, D'Acunzo P, Cianfanelli V, Strappazzon F, Caruana I, Nazio F, Weber G, Gigantino V, Botti G, Ciccosanti F, Borsellino G, Campello S, Mandolesi G, De Bardi M, Fimia GM, D'Amelio M, Ruffini F, Furlan R, Centonze D, Martino G, Braghetta P, Chrisam M, Bonaldo P, Matarese G, Locatelli F, Battistini L, Cecconi F. AMBRA1 Controls Regulatory T-Cell Differentiation and Homeostasis Upstream of the FOXO3-FOXP3 Axis. Dev Cell. 2018;47:592-607.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 90. | Peng Q, Qin J, Zhang Y, Cheng X, Wang X, Lu W, Xie X, Zhang S. Autophagy maintains the stemness of ovarian cancer stem cells by FOXA2. J Exp Clin Cancer Res. 2017;36:171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 91. | Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598-611. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1411] [Cited by in F6Publishing: 1346] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 92. | Kimmelman AC, White E. Autophagy and Tumor Metabolism. Cell Metab. 2017;25:1037-1043. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 585] [Cited by in F6Publishing: 568] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 93. | Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, Tewari M, Liu A, Vessella R, Rostomily R, Born D, Horwitz M, Ware C, Blau CA, Cleary MA, Rich JN, Ruohola-Baker H. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640-4652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 392] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 94. | Seo EJ, Kim DK, Jang IH, Choi EJ, Shin SH, Lee SI, Kwon SM, Kim KH, Suh DS, Kim JH. Hypoxia-NOTCH1-SOX2 signaling is important for maintaining cancer stem cells in ovarian cancer. Oncotarget. 2016;7:55624-55638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 95. | Kitajima S, Lee KL, Fujioka M, Sun W, You J, Chia GS, Wanibuchi H, Tomita S, Araki M, Kato H, Poellinger L. Hypoxia-inducible factor-2 alpha up-regulates CD70 under hypoxia and enhances anchorage-independent growth and aggressiveness in cancer cells. Oncotarget. 2018;9:19123-19135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 96. | Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, Hjelmeland AB, Rich JN. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 994] [Cited by in F6Publishing: 972] [Article Influence: 64.8] [Reference Citation Analysis (0)] |
| 97. | Rausch V, Liu L, Apel A, Rettig T, Gladkich J, Labsch S, Kallifatidis G, Kaczorowski A, Groth A, Gross W, Gebhard MM, Schemmer P, Werner J, Salnikov AV, Zentgraf H, Büchler MW, Herr I. Autophagy mediates survival of pancreatic tumour-initiating cells in a hypoxic microenvironment. J Pathol. 2012;227:325-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 98. | Zhu H, Wang D, Zhang L, Xie X, Wu Y, Liu Y, Shao G, Su Z. Upregulation of autophagy by hypoxia-inducible factor-1α promotes EMT and metastatic ability of CD133+ pancreatic cancer stem-like cells during intermittent hypoxia. Oncol Rep. 2014;32:935-942. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 100] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 99. | Song YJ, Zhang SS, Guo XL, Sun K, Han ZP, Li R, Zhao QD, Deng WJ, Xie XQ, Zhang JW, Wu MC, Wei LX. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett. 2013;339:70-81. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 100. | Qureshi-Baig K, Kuhn D, Viry E, Pozdeev VI, Schmitz M, Rodriguez F, Ullmann P, Koncina E, Nurmik M, Frasquilho S, Nazarov PV, Zuegel N, Boulmont M, Karapetyan Y, Antunes L, Val D, Mittelbronn M, Janji B, Haan S, Letellier E. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy. 2019;1-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 101. | Park SJ, Kim JG, Kim ND, Yang K, Shim JW, Heo K. Estradiol, TGF-β1 and hypoxia promote breast cancer stemness and EMT-mediated breast cancer migration. Oncol Lett. 2016;11:1895-1902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 102. | Cufí S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA. Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle. 2011;10:3871-3885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 145] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 103. | Alizadeh J, Glogowska A, Thliveris J, Kalantari F, Shojaei S, Hombach-Klonisch S, Klonisch T, Ghavami S. Autophagy modulates transforming growth factor beta 1 induced epithelial to mesenchymal transition in non-small cell lung cancer cells. Biochim Biophys Acta Mol Cell Res. 2018;1865:749-768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 66] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 104. | Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H, Yan J, Lv X, Chen X, Hu ZW. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012;72:3238-3250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 105. | Hasmim M, Janji B, Khaled M, Noman MZ, Louache F, Bordereaux D, Abderamane A, Baud V, Mami-Chouaib F, Chouaib S. Cutting Edge: NANOG Activates Autophagy under Hypoxic Stress by Binding to BNIP3L Promoter. J Immunol. 2017;198:1423-1428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 106. | Sancho P, Burgos-Ramos E, Tavera A, Bou Kheir T, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, Graña O, Viera CR, Yuneva M, Sainz B, Heeschen C. MYC/PGC-1α Balance Determines the Metabolic Phenotype and Plasticity of Pancreatic Cancer Stem Cells. Cell Metab. 2015;22:590-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 501] [Article Influence: 55.7] [Reference Citation Analysis (0)] |
| 107. | Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sánchez N, Marchesini M, Carugo A, Green T, Seth S, Giuliani V, Kost-Alimova M, Muller F, Colla S, Nezi L, Genovese G, Deem AK, Kapoor A, Yao W, Brunetto E, Kang Y, Yuan M, Asara JM, Wang YA, Heffernan TP, Kimmelman AC, Wang H, Fleming JB, Cantley LC, DePinho RA, Draetta GF. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628-632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 887] [Cited by in F6Publishing: 878] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 108. | Whelan KA, Chandramouleeswaran PM, Tanaka K, Natsuizaka M, Guha M, Srinivasan S, Darling DS, Kita Y, Natsugoe S, Winkler JD, Klein-Szanto AJ, Amaravadi RK, Avadhani NG, Rustgi AK, Nakagawa H. Autophagy supports generation of cells with high CD44 expression via modulation of oxidative stress and Parkin-mediated mitochondrial clearance. Oncogene. 2017;36:4843-4858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 109. | Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, Shannon K, Lowe SW. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24:1389-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 129] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 110. | Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, Kim H, Tan AC, Myers JR, Ashton JM, Neff T, Pollyea DA, Smith CA, Jordan CT. AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell. 2018;23:86-100.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 159] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 111. | Reed JC. Bcl-2-family proteins and hematologic malignancies: history and future prospects. Blood. 2008;111:3322-3330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 232] [Cited by in F6Publishing: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 112. | Merino R, Ding L, Veis DJ, Korsmeyer SJ, Nuñez G. Developmental regulation of the Bcl-2 protein and susceptibility to cell death in B lymphocytes. EMBO J. 1994;13:683-691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 231] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 113. | Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927-939. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2607] [Cited by in F6Publishing: 2697] [Article Influence: 141.9] [Reference Citation Analysis (0)] |
| 114. | Nicotra G, Mercalli F, Peracchio C, Castino R, Follo C, Valente G, Isidoro C. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol. 2010;23:937-950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 115. | Huang JJ, Zhu YJ, Lin TY, Jiang WQ, Huang HQ, Li ZM. Beclin 1 expression predicts favorable clinical outcome in patients with diffuse large B-cell lymphoma treated with R-CHOP. Hum Pathol. 2011;42:1459-1466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 116. | Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809-1820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1582] [Cited by in F6Publishing: 1768] [Article Influence: 84.2] [Reference Citation Analysis (0)] |
| 117. | Tan P, He L, Xing C, Mao J, Yu X, Zhu M, Diao L, Han L, Zhou Y, You MJ, Wang HY, Wang RF. Myeloid loss of Beclin 1 promotes PD-L1hi precursor B cell lymphoma development. J Clin Invest. 2019;129:5261-5277. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 118. | Bertolo C, Roa S, Sagardoy A, Mena-Varas M, Robles EF, Martinez-Ferrandis JI, Sagaert X, Tousseyn T, Orta A, Lossos IS, Amar S, Natkunam Y, Briones J, Melnick A, Malumbres R, Martinez-Climent JA. LITAF, a BCL6 target gene, regulates autophagy in mature B-cell lymphomas. Br J Haematol. 2013;162:621-630. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 119. | Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci USA. 2003;100:15077-15082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1534] [Cited by in F6Publishing: 1637] [Article Influence: 78.0] [Reference Citation Analysis (0)] |
| 120. | Zarogoulidis P, Petanidis S, Domvri K, Kioseoglou E, Anestakis D, Freitag L, Zarogoulidis K, Hohenforst-Schmidt W, Eberhardt W. Autophagy inhibition upregulates CD4+ tumor infiltrating lymphocyte expression via miR-155 regulation and TRAIL activation. Mol Oncol. 2016;10:1516-1531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 121. | Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7:834-846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1143] [Cited by in F6Publishing: 1071] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 122. | Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14:611-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 743] [Cited by in F6Publishing: 761] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 123. | Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247:708-718. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 234] [Article Influence: 46.8] [Reference Citation Analysis (0)] |
| 124. | Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, Liao WS, Bast RC. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917-3929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 223] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 125. | Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun. 2018;9:1944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 261] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 126. | La Belle Flynn A, Calhoun BC, Sharma A, Chang JC, Almasan A, Schiemann WP. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat Commun. 2019;10:3668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 127. | Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res. 2014;24:42-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 475] [Cited by in F6Publishing: 504] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 128. | Pérez-Hernández M, Arias A, Martínez-García D, Pérez-Tomás R, Quesada R, Soto-Cerrato V. Targeting Autophagy for Cancer Treatment and Tumor Chemosensitization. Cancers (Basel). 2019;11:1599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 129. | Xu R, Ji Z, Xu C, Zhu J. The clinical value of using chloroquine or hydroxychloroquine as autophagy inhibitors in the treatment of cancers: A systematic review and meta-analysis. Medicine (Baltimore). 2018;97:e12912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 101] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 130. | Jones TM, Espitia C, Wang W, Nawrocki ST, Carew JS. Moving beyond hydroxychloroquine: the novel lysosomal autophagy inhibitor ROC-325 shows significant potential in preclinical studies. Cancer Commun (Lond). 2019;39:72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 131. | Vega-Rubín-de-Celis S, Zou Z, Fernández ÁF, Ci B, Kim M, Xiao G, Xie Y, Levine B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci USA. 2018;115:4176-4181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 132. | Yoshida GJ. Therapeutic strategies of drug repositioning targeting autophagy to induce cancer cell death: from pathophysiology to treatment. J Hematol Oncol. 2017;10:67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 164] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 133. | Pietrocola F, Pol J, Vacchelli E, Baracco EE, Levesque S, Castoldi F, Maiuri MC, Madeo F, Kroemer G. Autophagy induction for the treatment of cancer. Autophagy. 2016;12:1962-1964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 134. | Hu F, Zhao Y, Yu Y, Fang JM, Cui R, Liu ZQ, Guo XL, Xu Q. Docetaxel-mediated autophagy promotes chemoresistance in castration-resistant prostate cancer cells by inhibiting STAT3. Cancer Lett. 2018;416:24-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 135. | Verbaanderd C, Maes H, Schaaf MB, Sukhatme VP, Pantziarka P, Sukhatme V, Agostinis P, Bouche G. Repurposing Drugs in Oncology (ReDO)-chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience. 2017;11:781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 163] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 136. | Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, Singhi AD, Bao P, Bartlett DL, Liotta LA, Espina V, Loughran P, Lotze MT, Zeh HJ. Safety and Biologic Response of Pre-operative Autophagy Inhibition in Combination with Gemcitabine in Patients with Pancreatic Adenocarcinoma. Ann Surg Oncol. 2015;22:4402-4410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 168] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 137. | Rojas-Puentes LL, Gonzalez-Pinedo M, Crismatt A, Ortega-Gomez A, Gamboa-Vignolle C, Nuñez-Gomez R, Dorantes-Gallareta Y, Arce-Salinas C, Arrieta O. Phase II randomized, double-blind, placebo-controlled study of whole-brain irradiation with concomitant chloroquine for brain metastases. Radiat Oncol. 2013;8:209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 138. | Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, Tan KS, Heitjan DF, DeMichele AM, Vaughn DJ, Redlinger M, Alavi A, Kaiser J, Pontiggia L, Davis LE, O'Dwyer PJ, Amaravadi RK. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1391-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 328] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 139. | Liang DH, Choi DS, Ensor JE, Kaipparettu BA, Bass BL, Chang JC. The autophagy inhibitor chloroquine targets cancer stem cells in triple negative breast cancer by inducing mitochondrial damage and impairing DNA break repair. Cancer Lett. 2016;376:249-258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 140. | Morgan MJ, Fitzwalter BE, Owens CR, Powers RK, Sottnik JL, Gamez G, Costello JC, Theodorescu D, Thorburn A. Metastatic cells are preferentially vulnerable to lysosomal inhibition. Proc Natl Acad Sci USA. 2018;115:E8479-E8488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 141. | Rebecca VW, Nicastri MC, McLaughlin N, Fennelly C, McAfee Q, Ronghe A, Nofal M, Lim CY, Witze E, Chude CI, Zhang G, Alicea GM, Piao S, Murugan S, Ojha R, Levi SM, Wei Z, Barber-Rotenberg JS, Murphy ME, Mills GB, Lu Y, Rabinowitz J, Marmorstein R, Liu Q, Liu S, Xu X, Herlyn M, Zoncu R, Brady DC, Speicher DW, Winkler JD, Amaravadi RK. A Unified Approach to Targeting the Lysosome's Degradative and Growth Signaling Roles. Cancer Discov. 2017;7:1266-1283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 131] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 142. | Baquero P, Dawson A, Mukhopadhyay A, Kuntz EM, Mitchell R, Olivares O, Ianniciello A, Scott MT, Dunn K, Nicastri MC, Winkler JD, Michie AM, Ryan KM, Halsey C, Gottlieb E, Keaney EP, Murphy LO, Amaravadi RK, Holyoake TL, Helgason GV. Targeting quiescent leukemic stem cells using second generation autophagy inhibitors. Leukemia. 2019;33:981-994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 143. | Yang YP, Hu LF, Zheng HF, Mao CJ, Hu WD, Xiong KP, Wang F, Liu CF. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin. 2013;34:625-635. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 268] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 144. | Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, Cantwell J, Luu C, Cornella-Taracido I, Harrington E, Fekkes P, Lei H, Fang Q, Digan ME, Burdick D, Powers AF, Helliwell SB, D'Aquin S, Bastien J, Wang H, Wiederschain D, Kuerth J, Bergman P, Schwalb D, Thomas J, Ugwonali S, Harbinski F, Tallarico J, Wilson CJ, Myer VE, Porter JA, Bussiere DE, Finan PM, Labow MA, Mao X, Hamann LG, Manning BD, Valdez RA, Nicholson T, Schirle M, Knapp MS, Keaney EP, Murphy LO. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014;16:1069-1079. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 395] [Cited by in F6Publishing: 489] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 145. | Petherick KJ, Conway OJ, Mpamhanga C, Osborne SA, Kamal A, Saxty B, Ganley IG. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem. 2015;290:11376-11383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 146. | Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, Lou HJ, Raveendra-Panickar D, Yang CC, Sheffler DJ, Teriete P, Asara JM, Turk BE, Cosford ND, Shaw RJ. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015;59:285-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 450] [Cited by in F6Publishing: 500] [Article Influence: 55.6] [Reference Citation Analysis (0)] |
| 147. | Pasquier B. SAR405, a PIK3C3/Vps34 inhibitor that prevents autophagy and synergizes with MTOR inhibition in tumor cells. Autophagy. 2015;11:725-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 104] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 148. | Chaurasiya S, Chen NG, Warner SG. Oncolytic Virotherapy versus Cancer Stem Cells: A Review of Approaches and Mechanisms. Cancers (Basel). 2018;10:124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 149. | Russell SJ, Peng KW, Bell JC. Oncolytic virotherapy. Nat Biotechnol. 2012;30:658-670. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 906] [Cited by in F6Publishing: 996] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 150. | Martin TA, Watkins G, Jiang WG. The Coxsackie-adenovirus receptor has elevated expression in human breast cancer. Clin Exp Med. 2005;5:122-128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 151. | Kim JS, Lee SH, Cho YS, Choi JJ, Kim YH, Lee JH. Enhancement of the adenoviral sensitivity of human ovarian cancer cells by transient expression of coxsackievirus and adenovirus receptor (CAR). Gynecol Oncol. 2002;85:260-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 31] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 152. | Masson D, Jarry A, Baury B, Blanchardie P, Laboisse C, Lustenberger P, Denis MG. Overexpression of the CD155 gene in human colorectal carcinoma. Gut. 2001;49:236-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 220] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 153. | Anderson BD, Nakamura T, Russell SJ, Peng KW. High CD46 receptor density determines preferential killing of tumor cells by oncolytic measles virus. Cancer Res. 2004;64:4919-4926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 239] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 154. | Sanjuán X, Fernández PL, Miquel R, Muñoz J, Castronovo V, Ménard S, Palacín A, Cardesa A, Campo E. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. J Pathol. 1996;179:376-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 155. | Russell SJ, Barber GN. Oncolytic Viruses as Antigen-Agnostic Cancer Vaccines. Cancer Cell. 2018;33:599-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 152] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 156. | Merkley SD, Chock CJ, Yang XO, Harris J, Castillo EF. Modulating T Cell Responses via Autophagy: The Intrinsic Influence Controlling the Function of Both Antigen-Presenting Cells and T Cells. Front Immunol. 2018;9:2914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 157. | Zhou J, Wang G, Chen Y, Wang H, Hua Y, Cai Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J Cell Mol Med. 2019;23:4854-4865. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 243] [Cited by in F6Publishing: 384] [Article Influence: 76.8] [Reference Citation Analysis (0)] |
| 158. | Ma J, Ramachandran M, Jin C, Quijano-Rubio C, Martikainen M, Yu D, Essand M. Characterization of virus-mediated immunogenic cancer cell death and the consequences for oncolytic virus-based immunotherapy of cancer. Cell Death Dis. 2020;11:48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 159. | Dong X, Levine B. Autophagy and viruses: adversaries or allies? J Innate Immun. 2013;5:480-493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 160. | Beljanski V, Chiang C, Hiscott J. The intersection between viral oncolysis, drug resistance, and autophagy. Biol Chem. 2015;396:1269-1280. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 161. | Tazawa H, Kuroda S, Hasei J, Kagawa S, Fujiwara T. Impact of Autophagy in Oncolytic Adenoviral Therapy for Cancer. Int J Mol Sci. 2017;18:1479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 162. | Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860-4864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 199] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 163. | Tong Y, You L, Liu H, Li L, Meng H, Qian Q, Qian W. Potent antitumor activity of oncolytic adenovirus expressing Beclin-1 via induction of autophagic cell death in leukemia. Oncotarget. 2013;4:860-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 164. | Li L, You LS, Mao LP, Jin SH, Chen XH, Qian WB. Combing oncolytic adenovirus expressing Beclin-1 with chemotherapy agent doxorubicin synergistically enhances cytotoxicity in human CML cells in vitro. Acta Pharmacol Sin. 2018;39:251-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 165. | Zhang J, Lai W, Li Q, Yu Y, Jin J, Guo W, Zhou X, Liu X, Wang Y. A novel oncolytic adenovirus targeting Wnt signaling effectively inhibits cancer-stem like cell growth via metastasis, apoptosis and autophagy in HCC models. Biochem Biophys Res Commun. 2017;491:469-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 166. | Yokoyama T, Iwado E, Kondo Y, Aoki H, Hayashi Y, Georgescu MM, Sawaya R, Hess KR, Mills GB, Kawamura H, Hashimoto Y, Urata Y, Fujiwara T, Kondo S. Autophagy-inducing agents augment the antitumor effect of telerase-selve oncolytic adenovirus OBP-405 on glioblastoma cells. Gene Ther. 2008;15:1233-1239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 167. | Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50:1-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1293] [Cited by in F6Publishing: 1235] [Article Influence: 205.8] [Reference Citation Analysis (0)] |
| 168. | Clark CA, Gupta HB, Sareddy G, Pandeswara S, Lao S, Yuan B, Drerup JM, Padron A, Conejo-Garcia J, Murthy K, Liu Y, Turk MJ, Thedieck K, Hurez V, Li R, Vadlamudi R, Curiel TJ. Tumor-Intrinsic PD-L1 Signals Regulate Cell Growth, Pathogenesis, and Autophagy in Ovarian Cancer and Melanoma. Cancer Res. 2016;76:6964-6974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 265] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 169. | Chen RQ, Xu XH, Liu F, Li CY, Li YJ, Li XR, Jiang GY, Hu F, Liu D, Pan F, Qiu XY, Chen XQ. The Binding of PD-L1 and Akt Facilitates Glioma Cell Invasion Upon Starvation via Akt/Autophagy/F-Actin Signaling. Front Oncol. 2019;9:1347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 170. | Wang X, Wu WKK, Gao J, Li Z, Dong B, Lin X, Li Y, Li Y, Gong J, Qi C, Peng Z, Yu J, Shen L. Autophagy inhibition enhances PD-L1 expression in gastric cancer. J Exp Clin Cancer Res. 2019;38:140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 96] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 171. | Maher CM, Thomas JD, Haas DA, Longen CG, Oyer HM, Tong JY, Kim FJ. Small-Molecule Sigma1 Modulator Induces Autophagic Degradation of PD-L1. Mol Cancer Res. 2018;16:243-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 172. | Clark CA, Gupta HB, Curiel TJ. Tumor cell-intrinsic CD274/PD-L1: A novel metabolic balancing act with clinical potential. Autophagy. 2017;13:987-988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 173. | Shukla SA, Bachireddy P, Schilling B, Galonska C, Zhan Q, Bango C, Langer R, Lee PC, Gusenleitner D, Keskin DB, Babadi M, Mohammad A, Gnirke A, Clement K, Cartun ZJ, Van Allen EM, Miao D, Huang Y, Snyder A, Merghoub T, Wolchok JD, Garraway LA, Meissner A, Weber JS, Hacohen N, Neuberg D, Potts PR, Murphy GF, Lian CG, Schadendorf D, Hodi FS, Wu CJ. Cancer-Germline Antigen Expression Discriminates Clinical Outcome to CTLA-4 Blockade. Cell. 2018;173:624-633.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 102] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 174. | Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010;190:881-892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 646] [Cited by in F6Publishing: 723] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 175. | Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira JP, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, André F, Delaloge S, Tursz T, Kroemer G, Zitvogel L. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050-1059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2084] [Cited by in F6Publishing: 2286] [Article Influence: 134.5] [Reference Citation Analysis (0)] |
| 176. | Pedicord VA, Cross JR, Montalvo-Ortiz W, Miller ML, Allison JP. Friends not foes: CTLA-4 blockade and mTOR inhibition cooperate during CD8+ T cell priming to promote memory formation and metabolic readiness. J Immunol. 2015;194:2089-2098. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |