Published online Aug 15, 2003. doi: 10.3748/wjg.v9.i8.1734
Revised: January 6, 2003
Accepted: April 1, 2003
Published online: August 15, 2003
AIM: To explore the transcriptional gene expression profiles of HGF/SF-met signaling pathway in colorectal carcinoma to understand mechanisms of the signaling pathway at so gene level.
METHODS: Total RNA was isolated from human colorectal carcinoma cell line LoVo treated with HGF/SF (80 ng/L) for 48 h. Fluorescent probes were prepared from RNA labeled with cy3-dUTP for the control groups and with cy5-dUTP for the HGF/SF-treated groups through reverse-transcription. The probes were mixed and hybridized on the microarray at 60 °C for 15-20 h, then the microarray was scanned by laser scanner (GenePix 4000B). The intensity of each spot and ratios of Cy5/Cy3 were analyzed and finally the differentially expressed genes were selected by GenePix Pro 3.0 software. 6 differential expression genes (3 up-regulated genes and 3 down-regulated genes) were selected randomly and analyzed by β-actin semi-quantitative RT-PCR.
RESULTS: The fluorescent intensities of built-in negative control spots were less than 200, and the fluorescent intensities of positive control spots were more than 5000. Of the 4004 human genes analyzed by microarray, 129 genes (holding 3.22% of the investigated genes) revealed differential expression in HGF/SF-treated groups compared with the control groups, of which 61 genes were up-regulated (holding 1.52% of the investigated genes) and 68 genes were down-regulated (holding 1.70% of the investigated genes), which supplied abundant information about target genes of HGF/SF-met signaling.
CONCLUSION: HGF/SF-met signaling may up-regulate oncogenes, signal transduction genes, apoptosis-related genes, metastasis related genes, and down-regulate a number of genes. The complexity of HGF/SF-met signaling to control the gene expression is revealed as a whole by the gene chip technology.
- Citation: Li XN, Ding YQ, Liu GB. Transcriptional gene expression profiles of HGF/SF-met signaling pathway in colorectal carcinoma. World J Gastroenterol 2003; 9(8): 1734-1738
- URL: https://www.wjgnet.com/1007-9327/full/v9/i8/1734.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i8.1734
HGF/SF-met signaling is a pathway by stimulation of c-met via its ligand hepatocyte growth factor /scatter factor (HGF/SF), leading to a considerable variety of biological and biochemical effects on the cells[1-3]. HGF/SF, a multifunctional cell growth factor, is yielded by interstitial cells such as fibroblast, neuroglial cell, glial cell, fat-storing cell and macrophage[3,4]. It binds to the c-met receptor specifically, inducing a series of conformational changes of signaling proteins by activating JAK[5], phosphatidylinositol 3-kinase (PI-3 K)[6], phospholipase C-γ[7], Raf-1 kinase[8] etc, by which it transmits the signal to the nuclear transcription machines to control certain gene transcription and expressions. Finally it affects the cell proliferation, differentiation, locomotion and other cell functions[9-12]. HGF/SF has been proved to have mitogen, motogen, morphogen activity for almost all epithelial cells, and to promote adhesiveness, invasiveness, motion and metastasis and stimulation for blood vessel growth of cancer cells[8-13]. It was revealed to have anti-cytotoxin and anti-apoptosis for cancer cells in the recent investigation[11,12]. It is evidenced that abnormal activation of HGF/SF-met signaling plays an important or critical causal role in tumor progression and invasion and metastasis. However, little is known about the target genes and gene expression profiles by which HGF/SF exerts biological functions through start-up of transcriptional gene expression.
In the present study, we aimed to explore the transcriptional gene expression profiles by activating HGF/SF-met signaling in colorectal carcinoma cells using the microarray technology by which thousands of genes could be detected at the same time.
Hepatocyte growth factor/scatter factor (HGF/SF), ethidium bromide(EB), and SDS were purchased from Sigma. AMV reverse transcriptase, oligo(dT), dNTP, Taq, and agarose were Promega products. Trypsin, Cy3-dUTP, and Cy5-dUTP were purchased from Amersham Phamacia, and Oligotex mRNA midi kit from Qiagen, and other reagents from Shanghai Biostar Co.
Human colorectal carcinoma cell line LoVo was routinely cultured in RPMI-1640 medium with 10% FCS, incubated at 37 °C with 5% CO2. The well-grown cells at the logarithmic growth phase were treated with HGF/SF (80 ng/L) for 24-48 h in the same medium containing 2.5% FCS. The blank control groups were treated with D-Hanks solution in the same culture condition as HGF/SF-treated groups. Triduplication of the experiments was for the microarray assay.
The genes for HGEC-40S microarray containing 4096 cDNA spots (including 92 negative and positive control spots) were provided by Shanghai Biostar Co. These genes were amplified by PCR with universal primers and then purified by the routine method[13-15]. The purity of PCR products was supervised by agarose electrophoresis and then the PCR products were dissolved in 3 X SSC solution. The target genes were spotted on siliconlated slides (TeleChem Inc) by using Cartesian 7500 spotting robotics (Cartesian Inc), then hydrated for 2 h, dried for 0.5 h, UV cross-linked by UVP CL-1000 at 65 mJ/cm, and then treated with 0.2% SDS(10min), H2O (10 min) and 0.2% NaBH4 (10 min). Finally the slides were dried again and ready for use.
Total RNA was isolated from human colorectal carcinoma cell line LoVo by the routine method[13-15]. And then they were tested by hot-stabilization experiments and RNA electrophoresis. cDNA probes were prepared through reverse transcription in 50 μl reaction system in reference to Schena et al[15]. The fluorescent probes derived from cDNA reverse-transcripted from RNA were labeled with cy3-dUTP for the control groups and with cy5-dUTP for the HGF/SF-treated groups. The two kinds of probes were mixed equally after ethanol precipitation, then inspissated by vacuum to 50 μl, purified by using S-200 columns, and finally dissolved in hybridizing solution (20 μl 5×SSC+0.2% SDS).
After denatured at 95 °C water bath, the microarray was prehybridized with 6 µl hybridization solutions 1 and 2 (Biostar) at 42 °C for 6 h. The probe mixtures were added on the prehybridized microarray, denatured at 95 °C for 5 min, then inoculated in hybridization cabin at 60 °C for 15-20 h. After the glass cover was removed, the microarray was washed in solutions 1 (2×SSC+0.2% SDS), and 2 (0.1×SSC+0.2% SDS) and 3 (0.1×SSC) for 10 min each, then dried at room temperature.
GenePix 4000B laser scanner (Axon) was used to scan microarray, with laser power 3.72 and photomultiplier tube volts 750. GenePix Pro 3.0 software was used for analysis of intensity of each spot and ratio of Cy5/Cy3 from the acquired images. The intensities of Cy3 and Cy5 were normalized by a coefficient according to the ratio of the located 40 housekeeping genes. The standards for differentially expressed genes were as follows: the ratio of Cy5/Cy3 was more than 4 or less than 0.25, and the intensity value of Cy3 and Cy5 was more than 200.
RT-PCR primers were designed by Omiga 2.0 software with basic reaction parameters as follows: primer length 18-22 bp, primer% GC 40%-60%, primer Tm 55 °C; PCR products length 100-600 bp, PCR products% GC 40%-60%, and PCR products Tm 82.5 °C. The primer sequences were as follows:
CAMK4 (321 bp): 5’GAGACCCTTCTCCAATCC3’ 5’GAACTTCAAAACCCACAGC 3’
SHPG (555 bp): 5’ACATCTCCCCTTTGCTAACG 3’ 5’GCCACAGTACCCTCATAACTCC 3’
Heregulin (424 bp): 5’TGCTCAACAGCAACATCC 3’ 5’TCATACATCTGCCCCTCC 3’
p130 (523 bp): 5’GCACTTCAGTGTTCTAATCG 3’ 5’GGCTATTCTCCTTAATGTACC 3’
DAP-1 (322 bp): 5’ACATGAGACACCACATTCC 3’ 5’ACGACACAGTTGCTGACC 3’
TRAMP (381 bp): 5’ATTCGCAAGAAAAGCACC 3’ 5’GTAGAACGCACTAAGCTGACC 3’
β-actin(206bp): 5’GGCGGCACCACCATGTACCCT 3’ 5’AGGGGCCGGACTCGTCATCATACT 3’
Components of the reverse transcription reaction system were as follows: oligo (dT) 0.5 μl, AMV5x buffer 4 μl, dNTP (10 mM) 1 μl RNasin (20u/ μl) 1 μl, AMV (10u/ μl) 0.5 μl, and DEPC-treated water 12 μl. Components of PCR reaction system were as follows: 10X PCR buffer 2 μl, cDNA 5 μl, dNTP (10 mM) 1 μl, forward primer 1 μl (1 μM), reverse primer 1 μl (1 μM), β-actin forward primer 0.5 μl (0.2 μM), β-actin reverse primer 0.5 μl (0.2 μM), Taq DNA polymerase (2U/ μl) 1 μl, and DEPC-treated water 8 μl, PCR amplification consisted of 35 cycles of denaturation for 60-90 s at 94 °C, annealing for 60-90 s at 56 °C, and extension for 90-120 s at 72 °C. After 35 turns of the cycle, it ended after extension at 72 °C for 7 min. DEPC-treated water was replaced with DNA template for the negative control. 1.5% agarose gel electrophoresis with ethidium bromide was used for the analysis of PCR products. Bandleader 3.0 software was applied to detect the density of bands of PCR products. The value of gene expression was calculated from percent of the ratio of band density of PCR product and the band density of β-actin.
Data for gene expression were analyzed by two sided Student’s t test using SPSS 10.0 software and the significant value was P < 0.05.
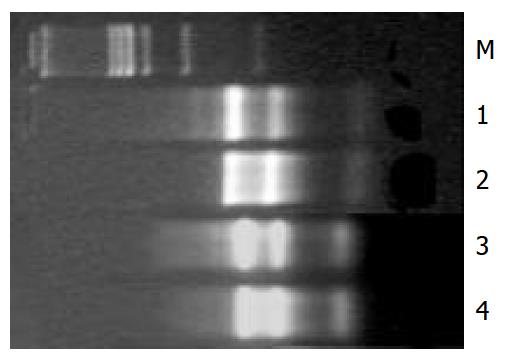
The values of D260/D280 for control and HGF/SF-treated groups were 2.022, 2.103 respectively. After hot stabilization test the 18S and 28S bands of total RNA extracts were as clear as before the test (Figure 1).
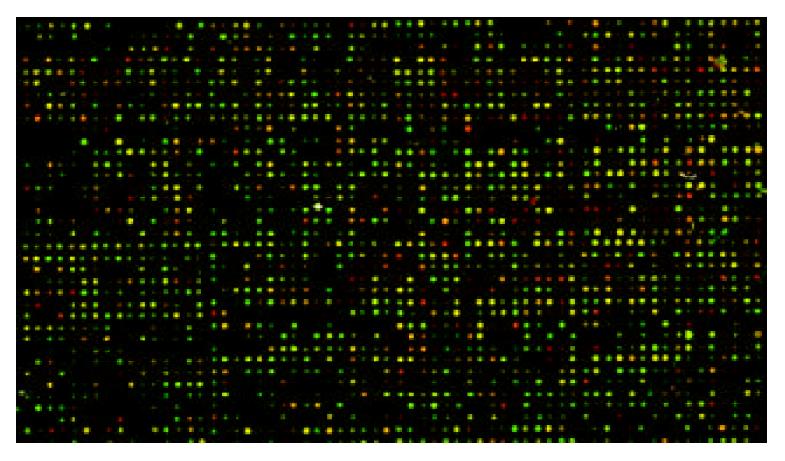
The fluorescent intensities of built-in negative control including rice U2 RNA gene, HCV coat protein gene, spotting solution (without DNA) as blank spots were lower than 200, while the intensities of built-in 40 house-keeping genes as positive control spots were larger than 5000. Normalization coefficient was 1.028. The odds of spot average intensities with background for HGF/SF-treated groups and the control groups were 10.254, 12.856 respectively. The ratio of Cy5/Cy3 by self-check test was 0-1.7, with the average 1.0112.
The scanning images for HGF/SF-treated groups labeled with Cy5 and images for control groups labeled with Cy3 showed lower noise background and appropriate spot intensity of signal with clear circle appearance. The overlaying image for bicolor fluorescence label is shown in Figure 2.
Of the 4004 human genes analyzed by microarray, 129 differential expression genes were identified by the filter standard mentioned above. Among the 129 differential expression genes, 61 genes were up-regulated and 68 genes were down-regulated. The up-regulated genes by HGF/SF-met signaling consisted of cell growth factor genes, cell surface receptor genes, angiogenesis genes, cell cycle positive-regulation genes, calcium-, MAPK signaling-related genes and nuclei-receptor genes, etc. The down-regulated genes by HGF/SF-c-met signaling were cell death correlation receptor genes, transmembrane-4 superfamily, cell cycle negative- regulation genes, calcium-, MAPK signaling-related negative genes, cytoskeleton rearrangement inhibitor, anti-oncogene and protease inhibitor, etc.
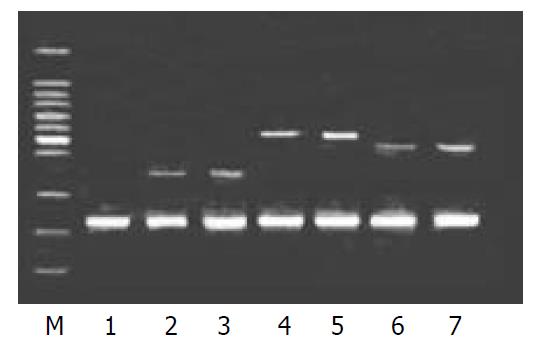
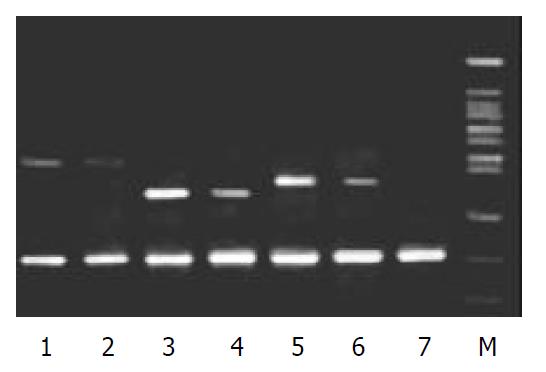
To validate the reliability of microarray results, 6 differential expression genes (3 up-regulated genes and 3 down-regulated genes) were selected randomly and analyzed by β-actin semi-quantitative RT-PCR. Expressions of the 6 differential genes by RT-PCR were confirmed to be consistent with the results of microarray (shown in Figure 3 and Figure 4). The average relative densities of bands detected by Bandlead 3.0 computer program showed significant differences (P < 0.05) between HGF/SF-treated groups and control groups except SHPG, heregulin, and DAP-1 groups.
Gene chip, or DNA chip, including oligonucleotide chip synthesized in situ and cDNA microarray by direct-spot or cDNA array[22-24], is a key technique platform[25-27] in post-genome times for its striking superiority of high-throughput, high-parallelity, high-sensitivity, micromation and automatization. cDNA microarray is a mature and widespread technique. Its stability, reliability and reproducibility have been confirmed by many investigators[28-31]. The microarray experiment in this research was successful and under strict quality control, with mRNA in good quality and no degradation tested by the test of hot-stabilization. In our experiment the built-in positive and negative control showed the qualified results, the scanning images showed lower noise background, and the positive spots indicated high intensity of signal with clear circle appearance. All the parameters such as odds of spot average intensity with background for the HGF/SF-treated group and for the control, normalization coefficient, and ratio of Cy5/Cy3 by self-check test were in accord with theoretic values of successful experiment. The expressions of the 6 differential expression genes randomly selected by RT-PCR were confirmed to be consistent with the results of microarray. These results indicate satisfactory reliability of the microarray experiment.
It has been evidenced that HGF/SF-met signaling plays an important, even a critical role in tumor progression, invasion and metastasis. Hamasuna et al[32] showed that HGF/SF activated membrane type metalloproteinases (MT1-MMP) in dosage-related manner and decreased wild-type metalloproteinases inhibitor TIMP-2 mRNA. They indicated that glioblastoma cells U251 excreted more MMP-2 protein and MMP-2 mRNA expression increased by 2.5 fold by HGF/SF stimulation. Abouader et al[33] argued that HGF/SF-met signaling was a key molecule path for control of glioblastoma cells growth, progression, invasion and metastasis. Recently Webb et al[34] have confirmed that HGF/SF-met-uPA -plasmin pathway exists in some tumor cell lines and is related to invasion, metastasis and other malignant behaviors. Only a few genes can be investigated by the traditional technology of molecular biology and the whole transcriptional gene expression profiles of HGF/ SF-met signaling pathway can hardly be explored. In this paper the transcriptional gene expression profiles of HGF/SF-met signaling pathway in colorectal carcinoma cells were discussed for the first time so as to explore the target genes related to the signaling pathway. The results indicated that HGF/SF-met signaling might activate oncogenes, signal transduction genes, apoptosis-related genes, metastasis related genes and many other genes with up-regulation effects, and meanwhile it might down-regulate a number of genes. It is suggested that a complex signaling-adjusting-gene-expression network may be in existence, and contribute to extensive gene transcription effects and comprehensive biological roles. The complexity of HGF/ SF-met signaling to control the gene expression was revealed as a whole by the gene chip technology, by which it manifested incomparable superiority.
Of the 4004 human genes analyzed by microarray (totally 4096 spots, subtracted 92 built-in positive and negative control spots), 129 genes (holding 3.22% of the investigated genes) revealed differential expression in the HGF/SF-treated groups compared with the control groups, which supplied abundant information about target genes of HGF/SF-met signaling. Among the 129 differential expression genes, 61 genes were up-regulated (holding 1.52% of the investigated genes), and 68 genes were down-regulated (holding 1.70% of the investigated genes). The expressions of some differential genes analyzed by microarray, for instance, collagenase IV[35], catenin[36], were consistent with the results reported in literatures that they were uncovered by traditional technology. Still a number of genes with differential expression not reported previously were found in our investigation. Some of them are genes with unknown function. Some genes with elevated expression by HGF/SF stimulation not reported previously are listed as following: Syndeans-2, calmodulin-dependent protein kinase IV(CaMK4), cadherein 8, protocadherin, calcium-dependent serine protein kinase (CASK), human keractnocyte growth factor(KGF), Heregulin, angiogenin, etc. In a word, the up-regulated genes by HGF/SF stimulation usually belong to cell growth factor genes, cell surface receptor genes, angiogenesis genes, cell cycle positive-regulation genes, calcium-, MAPK signaling-related genes, transcription factor, cytoskeleton regulation genes, and nuclei-receptor genes. These up-regulation genes stimulating cell growth, promoting cell transformation, resisting apoptosis, facilitating cell locomotion, accelerating signal conduction, and promoting vascularization and extracellular matrix lysis[37-44], may provide a foundation for mitogen, motogen, and morphogen activity in HGF/SF-met signaling pathway further.
Versatile effects on down-regulated gene expression by HGF/SF-met signaling have been found in this microarray assay. The genes of reduced expression by HGF/SF stimulation not reported previously include dual-specificity phosphatase 6 (DUSP6), SPINT2 (serine protease inhibitor, Kunitz-type 2, hepatocyte growth factor activator inhibitor type 2), TIMP1(tissue inhibitor of metalloproteinases 1), elastase inhibitive factor, TAPA1(Target of antiproliferative antibody 1, CD81), MRP-1 (motility-related protein 1, CD9), PRB2/p130, pRB2/p107, TRAMP (TNF receptor-related apoptosis mediating protein, DR3), WD40, DAP-1, etc. On the whole, the adjustability of multi-aspects and multi-targets in down-regulation of gene expression in colorectal carcinoma cells by HGF/SF-met signaling was revealed by microarray assay. The down-regulated genes by HGF/SF stimulation usually belong to cell death correlated receptor genes, transmembrane-4 superfamily, cell cycle negative-regulation genes, calcium and MAPK signaling-related negative genes, cytoskeleton rearrangement inhibitors, anti-oncogene and protease inhibitors. These genes are responsible for the negative control of cell function, which makes reinforcement and maintenance in stimulation of cell growth, promotion of cell transformation, resistance to apoptosis, facilitation for cell locomotion, acceleration of signal transduction, promotion of vascularization and lysis of extracellular matrix[45-56]. These are another annotation for mitogen, motogen, and morphogen activity of HGF/SF-met signaling pathway in tumor progression, also the first confirmation of pluripotency and versatility in regulation of gene expression by HGF/SF-met signaling.
The transcriptional gene expression profiles induced by HGF/SF-met signaling pathway in the colorectal carcinoma cells were described by cDNA microarray technology, and meanwhile the complexity and pluripotency of regulation in gene expression were also revealed for the first time. A majority of the genes with differential expression can be explained by the existing scientific theories, although some of them have unknown-function and a few of them cannot be explained reasonably for the time being, which deserve further investigation.
Edited by Zhao P and Wang XL
| 1. | Stuart KA, Riordan SM, Lidder S, Crostella L, Williams R, Skouteris GG. Hepatocyte growth factor/scatter factor-induced intracellular signalling. Int J Exp Pathol. 2000;81:17-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 94] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 2. | Ueoka Y, Kato K, Kuriaki Y, Horiuchi S, Terao Y, Nishida J, Ueno H, Wake N. Hepatocyte growth factor modulates motility and invasiveness of ovarian carcinomas via Ras-mediated pathway. Br J Cancer. 2000;82:891-899. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Delehedde M, Sergeant N, Lyon M, Rudland PS, Fernig DG. Hepatocyte growth factor/scatter factor stimulates migration of rat mammary fibroblasts through both mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt pathways. Eur J Biochem. 2001;268:4423-4429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 4. | Liu ZX, Nickel CH, Cantley LG. HGF promotes adhesion of ATP-depleted renal tubular epithelial cells in a MAPK-dependent manner. Am J Physiol Renal Physiol. 2001;281:F62-F70. [PubMed] [Cited in This Article: ] |
| 5. | Rodrigues GA, Park M, Schlessinger J. Activation of the JNK pathway is essential for transformation by the Met oncogene. EMBO J. 1997;16:2634-2645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 144] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Awasthi V, King RJ. PKC, p42/p44 MAPK, and p38 MAPK are required for HGF-induced proliferation of H441 cells. Am J Physiol Lung Cell Mol Physiol. 2000;279:L942-L949. [PubMed] [Cited in This Article: ] |
| 7. | Day RM, Cioce V, Breckenridge D, Castagnino P, Bottaro DP. Differential signaling by alternative HGF isoforms through c-Met: activation of both MAP kinase and PI 3-kinase pathways is insufficient for mitogenesis. Oncogene. 1999;18:3399-3406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 69] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 8. | Abounader R, Ranganathan S, Kim BY, Nichols C, Laterra J. Signaling pathways in the induction of c-met receptor expression by its ligand scatter factor/hepatocyte growth factor in human glioblastoma. J Neurochem. 2001;76:1497-1508. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 43] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 9. | Sipeki S, Bander E, Buday L, Farkas G, Bácsy E, Ways DK, Faragó A. Phosphatidylinositol 3-kinase contributes to Erk1/Erk2 MAP kinase activation associated with hepatocyte growth factor-induced cell scattering. Cell Signal. 1999;11:885-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Wong AS, Leung PC, Auersperg N. Hepatocyte growth factor promotes in vitro scattering and morphogenesis of human cervical carcinoma cells. Gynecol Oncol. 2000;78:158-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Hiscox S, Jiang WG. Association of the HGF/SF receptor, c-met, with the cell-surface adhesion molecule, E-cadherin, and catenins in human tumor cells. Biochem Biophys Res Commun. 1999;261:406-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 12. | Kitamura S, Kondo S, Shinomura Y, Kanayama S, Miyazaki Y, Kiyohara T, Hiraoka S, Matsuzawa Y. Met/HGF receptor modulates bcl-w expression and inhibits apoptosis in human colorectal cancers. Br J Cancer. 2000;83:668-673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 13. | Luo YQ, Wu MC, Cong WM. Gene expression of hepatocyte growth factor and its receptor in HCC and nontumorous liver tissues. World J Gastroenterol. 1999;5:119-121. [PubMed] [Cited in This Article: ] |
| 14. | Liu LX, Jiang HC, Liu ZH, Zhou J, Zhang WH, Zhu AL, Wang XQ, Wu M. Integrin gene expression profiles of human hepatocellular carcinoma. World J Gastroenterol. 2002;8:631-637. [PubMed] [Cited in This Article: ] |
| 15. | Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci U S A. 1996;93:10614-10619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1087] [Cited by in F6Publishing: 1161] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 16. | Heller RA, Schena M, Chai A, Shalon D, Bedilion T, Gilmore J, Woolley DE, Davis RW. Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc Natl Acad Sci U S A. 1997;94:2150-2155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 509] [Cited by in F6Publishing: 545] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 17. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6477] [Cited by in F6Publishing: 5032] [Article Influence: 173.5] [Reference Citation Analysis (0)] |
| 18. | Shirota Y, Kaneko S, Honda M, Kawai HF, Kobayashi K. Identification of differentially expressed genes in hepatocellular carcinoma with cDNA microarrays. Hepatology. 2001;33:832-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 124] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 19. | Raouf A, Seth A. Discovery of osteoblast-associated genes using cDNA microarrays. Bone. 2002;30:463-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 68] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Verhofstede C, Fransen K, Marissens D, Verhelst R, van der Groen G, Lauwers S, Zissis G, Plum J. Isolation of HIV-1 RNA from plasma: evaluation of eight different extraction methods. J Virol Methods. 1996;60:155-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Mannhalter C, Koizar D, Mitterbauer G. Evaluation of RNA isolation methods and reference genes for RT-PCR analyses of rare target RNA. Clin Chem Lab Med. 2000;38:171-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | Orr MS, Scherf U. Large-scale gene expression analysis in molecular target discovery. Leukemia. 2002;16:473-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 23. | Bashyam MD. Understanding cancer metastasis: an urgent need for using differential gene expression analysis. Cancer. 2002;94:1821-1829. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 24. | Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J Clin Oncol. 2002;20:1932-1941. [PubMed] [Cited in This Article: ] |
| 25. | Lakhani SR, Ashworth A. Microarray and histopathological analysis of tumours: the future and the past. Nat Rev Cancer. 2001;1:151-157. [PubMed] [Cited in This Article: ] |
| 26. | Bertucci F, Houlgatte R, Nguyen C, Viens P, Jordan BR, Birnbaum D. Gene expression profiling of cancer by use of DNA arrays: how far from the clinic. Lancet Oncol. 2001;2:674-682. [PubMed] [Cited in This Article: ] |
| 27. | Balmain A. Cancer genetics: from Boveri and Mendel to microarrays. Nat Rev Cancer. 2001;1:77-82. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 83] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 28. | Xu S, Mou H, Lü G, Zhu C, Yang Z, Gao Y, Lou H, Liu X, Cheng Y, Yang W. Gene expression profile differences in high and low metastatic human ovarian cancer cell lines by gene chip. Chin Med J (Engl). 2002;115:36-41. [PubMed] [Cited in This Article: ] |
| 29. | Lu T, Liu J, LeCluyse EL, Zhou YS, Cheng ML, Waalkes MP. Application of cDNA microarray to the study of arsenic-induced liver diseases in the population of Guizhou, China. Toxicol Sci. 2001;59:185-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 30. | Shen XZ, Chow JF, Koo MW, Cho CH. Gene expression profiles in gastric mucosa of sleep deprivation rats. World J Gastroenterol. 2000;6:754-758. [PubMed] [Cited in This Article: ] |
| 31. | Cheung ST, Chen X, Guan XY, Wong SY, Tai LS, Ng IO, So S, Fan ST. Identify metastasis-associated genes in hepatocellular carcinoma through clonality delineation for multinodular tumor. Cancer Res. 2002;62:4711-4721. [PubMed] [Cited in This Article: ] |
| 32. | Hamasuna R, Kataoka H, Moriyama T, Itoh H, Seiki M, Koono M. Regulation of matrix metalloproteinase-2 (MMP-2) by hepatocyte growth factor/scatter factor (HGF/SF) in human glioma cells: HGF/SF enhances MMP-2 expression and activation accompanying up-regulation of membrane type-1 MMP. Int J Cancer. 1999;82:274-281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 33. | Abounader R, Ranganathan S, Lal B, Fielding K, Book A, Dietz H, Burger P, Laterra J. Reversion of human glioblastoma malignancy by U1 small nuclear RNA/ribozyme targeting of scatter factor/hepatocyte growth factor and c-met expression. J Natl Cancer Inst. 1999;91:1548-1556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 137] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 34. | Webb CP, Hose CD, Koochekpour S, Jeffers M, Oskarsson M, Sausville E, Monks A, Vande Woude GF. The geldanamycins are potent inhibitors of the hepatocyte growth factor/scatter factor-met-urokinase plasminogen activator-plasmin proteolytic network. Cancer Res. 2000;60:342-349. [PubMed] [Cited in This Article: ] |
| 35. | Nabeshima K, Inoue T, Shimao Y, Okada Y, Itoh Y, Seiki M, Koono M. Front-cell-specific expression of membrane-type 1 matrix metalloproteinase and gelatinase A during cohort migration of colon carcinoma cells induced by hepatocyte growth factor/scatter factor. Cancer Res. 2000;60:3364-3369. [PubMed] [Cited in This Article: ] |
| 36. | Hiscox S, Jiang WG. Hepatocyte growth factor/scatter factor disrupts epithelial tumour cell-cell adhesion: involvement of beta-catenin. Anticancer Res. 1999;19:509-517. [PubMed] [Cited in This Article: ] |
| 37. | Contreras HR, Fabre M, Granés F, Casaroli-Marano R, Rocamora N, Herreros AG, Reina M, Vilaró S. Syndecan-2 expression in colorectal cancer-derived HT-29 M6 epithelial cells induces a migratory phenotype. Biochem Biophys Res Commun. 2001;286:742-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 38. | Munesue S, Kusano Y, Oguri K, Itano N, Yoshitomi Y, Nakanishi H, Yamashina I, Okayama M. The role of syndecan-2 in regulation of actin-cytoskeletal organization of Lewis lung carcinoma-derived metastatic clones. Biochem J. 2002;363:201-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 39. | Tamura N, Tai Y, Sugimoto K, Kobayashi R, Konishi R, Nishioka M, Masaki T, Nagahata S, Tokuda M. Enhanced expression and activation of Ca(2+)/calmodulin-dependent protein kinase IV in hepatocellular carcinoma. Cancer. 2000;89:1910-1916. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 40. | Williams CL, Phelps SH, Porter RA. Expression of Ca2+/calmodulin-dependent protein kinase types II and IV, and reduced DNA synthesis due to the Ca2+/calmodulin-dependent protein kinase inhibitor KN-62 (1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-L-tyrosyl]-4-phenyl piperazine) in small cell lung carcinoma. Biochem Pharmacol. 1996;51:707-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 41. | Wilson SE, Weng J, Chwang EL, Gollahon L, Leitch AM, Shay JW. Hepatocyte growth factor (HGF), keratinocyte growth factor (KGF), and their receptors in human breast cells and tissues: alternative receptors. Cell Mol Biol Res. 1994;40:337-350. [PubMed] [Cited in This Article: ] |
| 42. | Hijazi MM, Thompson EW, Tang C, Coopman P, Torri JA, Yang D, Mueller SC, Lupu R. Heregulin regulates the actin cytoskeleton and promotes invasive properties in breast cancer cell lines. Int J Oncol. 2000;17:629-641. [PubMed] [Cited in This Article: ] |
| 43. | Etoh T, Shibuta K, Barnard GF, Kitano S, Mori M. Angiogenin expression in human colorectal cancer: the role of focal macrophage infiltration. Clin Cancer Res. 2000;6:3545-3551. [PubMed] [Cited in This Article: ] |
| 44. | Lixin R, Efthymiadis A, Henderson B, Jans DA. Novel properties of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun. 2001;284:185-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 45. | Rao CN, Lakka SS, Kin Y, Konduri SD, Fuller GN, Mohanam S, Rao JS. Expression of tissue factor pathway inhibitor 2 inversely correlates during the progression of human gliomas. Clin Cancer Res. 2001;7:570-576. [PubMed] [Cited in This Article: ] |
| 46. | Kataoka H, Uchino H, Denda K, Kitamura N, Itoh H, Tsubouchi H, Nabeshima K, Koono M. Evaluation of hepatocyte growth factor activator inhibitor expression in normal and malignant colonic mucosa. Cancer Lett. 1998;128:219-227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 47. | Longo N, Yáñez-Mó M, Mittelbrunn M, de la Rosa G, Muñoz ML, Sánchez-Madrid F, Sánchez-Mateos P. Regulatory role of tetraspanin CD9 in tumor-endothelial cell interaction during transendothelial invasion of melanoma cells. Blood. 2001;98:3717-3726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 90] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Hashida H, Takabayashi A, Tokuhara T, Taki T, Kondo K, Kohno N, Yamaoka Y, Miyake M. Integrin alpha3 expression as a prognostic factor in colon cancer: association with MRP-1/CD9 and KAI1/CD82. Int J Cancer. 2002;97:518-525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 49. | Luo D, Liu QF, Gove C, Naomov N, Su JJ, Williams R. Analysis of N-ras gene mutation and p53 gene expression in human hepatocellular carcinomas. World J Gastroenterol. 1998;4:97-99. [PubMed] [Cited in This Article: ] |
| 50. | Wang L, Lu W, Chen YG, Zhou XM, Gu JR. Comparison of gene expression between normal colon mucosa and colon carcinoma by means of messenger RNA differential display. World J Gastroenterol. 1999;5:533-534. [PubMed] [Cited in This Article: ] |
| 51. | Sun BH, Zhang J, Wang BJ, Zhao XP, Wang YK, Yu ZQ, Yang DL, Hao LJ. Analysis of in vivo patterns of caspase 3 gene expression in primary hepatocellular carcinoma and its relationship to p21(WAF1) expression and hepatic apoptosis. World J Gastroenterol. 2000;6:356-360. [PubMed] [Cited in This Article: ] |
| 52. | Wu BP, Zhang YL, Zhou DY, Gao CF, Lai ZS. Microsatellite instability, MMR gene expression and proliferation kinetics in colorectal cancer with famillial predisposition. World J Gastroenterol. 2000;6:902-905. [PubMed] [Cited in This Article: ] |
| 53. | Berditchevski F, Odintsova E. Characterization of integrin-tetraspanin adhesion complexes: role of tetraspanins in integrin signaling. J Cell Biol. 1999;146:477-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 235] [Cited by in F6Publishing: 234] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 54. | Tanaka N, Ogi K, Odajima T, Dehari H, Yamada S, Sonoda T, Kohama G. pRb2/p130 protein expression is correlated with clinicopathologic findings in patients with oral squamous cell carcinoma. Cancer. 2001;92:2117-2125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 55. | Paggi MG, Giordano A. Who is the boss in the retinoblastoma family The point of view of Rb2/p130, the little brother. Cancer Res. 2001;61:4651-4654. [PubMed] [Cited in This Article: ] |
| 56. | Classon M, Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264:135-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 189] [Article Influence: 8.2] [Reference Citation Analysis (0)] |