Published online Nov 15, 2003. doi: 10.3748/wjg.v9.i11.2419
Revised: May 27, 2003
Accepted: June 4, 2003
Published online: November 15, 2003
AIM: To construct subtracted cDNA libraries of human vascular endothelial cells (VECs) related to gastrocarcinoma using suppression substractive hybridization (SSH) and to analyze cDNA libraries of gastrocarcinoma and VECs in Cancer Gene Anatomy Project (CGAP) database.
METHODS: Human VECs related to gastric adenocarcinoma and corresponding normal tissue were separated by magnetic beads coupled with antibody CD31 (Dynabeads CD31). A few amount of total RNA were synthesized and amplified by SMARTTM PCR cDNA Synthesis Kit. Then, using SSH and T/A cloning techniques, cDNA fragments of differentially expressed genes in human VECs of gastric adenocarcinoma were inserted into JM109 bacteria. One hundred positive bacteria clones were randomly picked and identified by colony PCR method. To analyze cDNA libraries of gastrocarcinoma and VECs in CGAP database, the tools of Library Finder, cDNA xProfiler, Digital GENE Expression Displayer (DGED), and Digital Differential Display (DDD) were used.
RESULTS: Forward and reverse subtraction cDNA libraries of human VECs related to gastrocarcinoma were constructed successfully with SSH and T/A cloning techniques. Analysis of CGAP database indicated that no appropriate library of VECs related to carcinoma was constructed.
CONCLUSION: Construction of subtraction cDNA libraries of human VECs related to gastrocarcinoma was successful and necessary, which laid a foundation for screening and cloning new and specific genes of VECs related to gastrocarcinoma.
- Citation: Liu YB, Wei ZX, Li L, Li HS, Chen H, Li XW. Construction and analysis of SSH cDNA library of human vascular endothelial cells related to gastrocarcinoma. World J Gastroenterol 2003; 9(11): 2419-2423
- URL: https://www.wjgnet.com/1007-9327/full/v9/i11/2419.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i11.2419
The microvascular system is indispensable throughout the full life span of an organism. Malformation and/or malfunction of VECs contribute to numerous human pathologies including various congenital abnormalities, arteriosclerosis, benign tumors, and cancer. Angiogenesis, the development of new blood vessels by sprouting from the preexisting vasculature, plays an important role in a number of physiological and pathological processes. Stimulated growth or strengthening of so-called collateral vascular branches might circumvent areas with obstructed blood flow in the case of stroke or coronary heart disease. On the other hand, there is substantial evidence suggesting that inhibition of tumor vascular rise might slow down, stop or eventually even reverse tumor growth and could thus become an important part of cancer therapy. The growth of micro vessel accompanying tumor development has, in particular, aroused greater interest and helped improve our understanding of the central role of Vices. Suppression subtractive hybridization (SSH), a PCR-based coda subtracted method, is an important method to reach this aim[1-4]. Because it still needs a lot of initiating RNA, bulk tissues (normal and cancerous) instead of individual cells are routinely used in the analysis. In recent two years, many articles have been publicized employing this method using various tissues[5,6] or cell lines[7,8]. However, bulk tissue contains many different cell types, which will burden the latter work for identifying special genes, and cell lines are different from cells in vivo especially for disease cells. With the introduction of Laser Capture Microdissection (LCM), the quantity and purity of RNA from either malignant or benign individual cells invariably yield more reliable and comprehensive experimental results[9-12]. But cell selection and extraction by LCM is currently a manual process, and at most only a few thousand cells can be extracted in an amount of time necessary to limit RNA degradation, and the machine is too expensive for average laboratory. Gastrocarcinoma, one of the most common human malignant tumors, ranks worldwide as the first leading cause of cancer-related mortality. A great deal of articles have been publicized about differentially expressed genes in normal and tumor gastric tissues[13-15]. Some researchers used cells separated from tissues digested with collagenase or cultured cells as material[16-18]. However, with cell culture even primary cell culture, the expressed gene map may change a lot. Likewise, in cells treated with collagenase at 37 °C for 30 min for separating the target cells from tissue, the specially expressed genes may lose before disposal or new genes may appear after disposal. The Cancer Gene Anatomy Project (CGAP) database of the National Cancer Institute has thousands of expressed sequences, derived from diverse normal and tumor cDNA libraries, but has no appropriate VECs cDNA library related to tumor. The aim of the present study was to select all genes specially expressed in VECs related to gastrocarcinoma rather than in normal gastric tissue. Using VECs mechanically separated by Dynabeads CD31 as materials without collagenase and culture, we built a cDNA library through SSH method, thus paved the way for further research on changes of VECs in gastrocarcinoma.
The resected specimens about 2 cm3 from gastric adenocarcinoma confirmed pathologically and from normal gastric tissue 7 cm away from the edge of the adenocarcinoma in same patients who were admitted to Henan Provincial Hospital were put into RNA protecting solution (RNAlaterTM, Ambion Company) after being rinsed with PBS immediately after resection.
Separation of ECs with Dynabeads CD31 About 1 cm3 of tissue cut from tissues stabilized by RNAlater was put into a mortar with a little RNAlater RNA Stabilization Reagent, and then was scissored and ground with scissors and pestle. To make cell suspension, 1 mL RNAlater RNA Stabilization Reagent was added to it, then filtered through a sterile 80-μm-nylon filter. 25 μL washed Dynabeads® CD31 beads were added to the suspension for 30 min at 2-8 °C. For identification, slide was made with one drop of this mixture and stained with hematoxylin and eosin (H&E). Then bead-bound cells were separated in a magnetic device (Dynal MPC®).
Rnaeasy Mini Kit and Rnase-Free Dnase set (QIAGEN Company) were used to extract the total RNA from bead-bound cells. RNA purified by Rnaeasy Column was analyzed for integrity and size by formaldehyde agarose gel electrophoresis and quantification and purity of RNA by OD value.
About 100 ng total RNA was used to synthesize the first strand of cDNA with SMARTTM PCR cDNA Synthesis Kit (Clontech Company), then amplified by LD-PCR with 15, 18, 21, 24, 27 cycles separately and analyzed through 1.2% agarose gel electrophoresis in order to get the perfect cycle number with which we harvested a suitable amount rather than a superfluous one to build the library. Placental total RNA was performed as control. CHROMA SPIN-100 Column was used to purify the cDNA.
SampleI, cDNA from VECs of gastrocarcinoma, and sample II, cDNA from VECs of normal stomach tissue, and sample III, cDNA from VECs of placental tissue were treated with enzyme RsaI respectively. From each sample, 10-mL solutions was taken before digestion and 1 h, 3 h and 3.5 h after digestion, and 1.2% agarose gel electrophoresis was performed for identification of digestion efficiency. Digested products were purified by QIAquick PCR purification Kit (QIAGEN Company), and subsequently concentrated to 6.7 mL by ethanol precipitation method.
Based on the instructions of Clontech PCR-SelectTM cDNA Subtraction Kit, sample I and sample II were used as test 1 and driver 1, correspondingly sample II and sample I were used as test 2 and driver 2. Mixture of sample III and ω× 174/Hae III DNA was treated as test 3, correspondingly sample III alone was performed as driver 3. Each sample test was divided into two parts, and each part was ligated separately with Adaptor 1 and Adaptor 2, and then hybridized with the corresponding sample driver. The mixture of two parts was hybridized with corresponding sample driver again. The fragments with both Adaptor 1 and Adaptor 2, namely specially expressed cDNA fragments in sample test rather than in sample driver, were amplified by nested PCR. Adeptors possessed outside primer and inside primer. So forward and reverse subtraction fragments were obtained. Subtraction efficiency was checked by G3PDH, a housekeeping gene, according to PCR cycles needed in subtracted sample and unsubtracted sample, with which the gene could be observed on agarose/EtBr gel.
QIAquick PCR Purification Kit was utilized to purify the subtraction fragments.
1 μL, 2 μL, 3 μL PCR fragments of subtraction and non-subtraction and 2 μL control DNA were respectively taken to ligate with 1 μL pGEM-T easy Vector. 10-μL ligation reaction solutions were transformed into 150 μL competent cells JM109. Background control was set up by pGEM-T easy Vector without any insert, and transformation control was also set up by an uncut plasmid pGEM@-5Zf (+) Vector. The transformation culture was plated on petri dishes containing LB/ampicillin/IPTG/X-Gal, and then screened white colony for inset fragment.
Select 100 white colonies separately from forward and reverse library and replant 5 mL LB/Amp solution. Then it was shaken at 37 °C overnight. Take 1 μg culture solution as model and Nested Primer 1 and Nested Primer 2R to amplify the insert and test it by electrophoresis.
Select white colonies separately from forward and reverse library and inoculate 5 mL LB/Amp solution. Then it was shaken at 37 °C overnight. Add 700 μL culture solution into 1.5 mL EP tube containing 50% glycerin and keep it at -80 °C.
cDNA library of stomach cancer tissue, normal stomach tissue and vascular endothelial cell were analyzed using GLS, cDNA xProfiler, DDD, DGED and Library Finder in Cancer Genome Anatomy Project (CGAP). dbEST was categorized using the GLS tool of CGAP. We considered precancer libraries as cancer libraries. All of the cDNA libraries were categorized according to tissue type (tissue origin), tissue histology (cancerous, normal, or fetal), and the library preparation method (microdissected, bulk, cell line, or flow cytometric sorted).
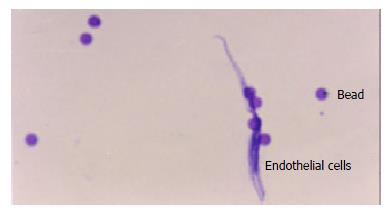
Through Dynabeads® CD31, about 8 × 106 endothelial cells were obtained with almost 100% purity. Slides were made and stained with H&E (Figure 1).
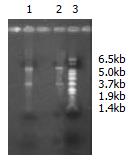
The amount of RNA extracted from stomach cancer and normal stomach tissue was respectively 1.0 μg and 0.85 μg with OD260/OD280 ratio 2.10 and 1.96. By formaldehyde agarose gel electrophoresis, the integrity and size were analyzed and clear bands of 18 s and 28 s were seen (Figure 2).
The first strands of cDNA have been amplified by LD-PCR with different cycles: 15, 18, 21, 24, and 27. The products could be viewed on 2% agarose gel electrophoresis (Figure 3). For both normal tissue and cancer tissue, a total of 27 cycles were performed, respectively, and for placental tissue, totally 18 cycles were performed.
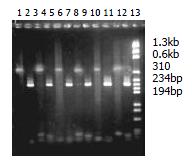
cDNA, before digestion with RsaI, appeared as a smear of 0.5-10 kb on 1% agarose gel electrophoresis, and after digestion the average cDNA size was smaller (0.1-2 kb) (Figure 4).
That the intensity ratio of PCR products determined by G3PD primer 3’ and Adaptor primer 1 to PCR products determined by G3PD primer 3’ and 5’ was over 1:4 on 2.0% agarose/EB gel showed that ligation efficiency was above 25% (Figure 5).
The second hybridization products were amplified by outside primers. The products presented unclear smear on 2% agarose/EB gel. After second PCR with inside primers, several bands could be seen clearly among the smears. Based on the manual, the experiment was successful (Figure 6).
After PCR amplification, the housekeeping gene G3PDHs appeared at 18 cycles in unsubtraction samples and at 33 cycles in subtraction samples. This result indicated that G3PDHs expressed in both parts have been greatly decreased through the subtraction method. If five cycles corresponded roughly to 20-fold cDNA enrichment, G3PDHs would have been decreased almost 300 times. It implied that other genes expressed in both tissues have been reduced the same fold and the specially expressed genes in the test sample were selected (Figure 7).
720 white colonies were observed on two petri dishes of positive control with 7% presence ratio of blue colony. Two white colonies were observed on one petri dish of background control with 15 blue colonies. On one petri dish of transformation control, 160 white colonies without blue colony were observed. 992 white colonies appeared on 5 petri dishes of forward subtraction, and 890 white colonies appeared on 5 petri dishes of reverse subtraction. 60 white colonies were selected from forward and reverse subtractions, and amplified with nested Primer 1 and nested Primer 2R. 54 and 50 fragments ranged from 100 bp to 1000 bp were selected from the corresponding forward and reverse subtraction libraries.
dbEST was categorized using the GLS tool of the CGAP. All of the cDNA libraries were categorized according to tissue type (tissue origin), tissue histology (cancerous, normal, or fetal), and the library preparation method (microdissected, bulk, cell line, or flow cytometric sorted). Among 319 stomach cDNA libraries, all 73 libraries of normal stomach were from bulk tissues instead of cells and none was established by SSH method except those without label. Among the 245-cDNA libraries of stomach cancer, 28 originated from cell lines with 9 libraries built by SSH, and none of the other 217 libraries originating from tissue was built by SSH. All 15 vascular cDNA libraries came from normal tissue or cell line or cultured cells, and SSH method was not used. And appropriate VECs related to tumor have not been built into cDNA library. Among the libraries derived from cell lines, two derived from umbilical vein endothelium, one derived from aortic endothelium, another one came from endothelial cells of foreskin through primary culture of dermal microvascular endothelial cells. Among the 11 non-cell line libraries, 6 were from aorta, 2 umbilical veins, 1 unlabeled vein; another two were from choroidal plexus and basilar artery.
Bio-behavior such as growth and metastasis of cancer is closely related with proliferation of microvessel. Newborn capillaries of cancer differ from normal ones in growth process or distribution. For instance, VECs of breast cancer can grow 50 times faster than VEC in normal tissue[19] and vascular endothelial growth factors (VEGFs) play an important role in tumor angiogenesis[20,21], and their over- expression is closely related to clinical staging, lymph node metastasis and recurrence of gastric carcinoma[22]. So increasing attention has been paid to VECs[23].
Differentially expressed genes between the corresponding normal and cancer tissue can help us understand the molecular basis of malignancy and potentially serve as biomarkers or prognostic markers of malignancy. The identification and characterization of human genes expressed exclusively or preferentially in microvascular system of tumor will hopefully shed light on the mechanisms of tumor development and provide useful genetic markers for screening, diagnosis, prognosis, therapeutic monitoring and development of therapeutic vaccines. There are many techniques that aim at producing an inventory of differential transcripts between two populations of mRNAs. High-throughput gene expression techniques (microarrays, genechips) to identify cancer-specific genes are becoming available[24-26], however, the technology is not cost effective for average laboratories. SSH method allows identifying overexpressed genes (designated forward +SSH) but also underexpressed genes (designated reverse -SSH) by exchanging the driver and tester populations during the procedure (Clontech, Palo Alto, USA). Since this technique was established by Diatchenko, many new genes have been separated from almost all tissues, such as renal cell cancer, lung cancer, liver cancer[27-30], etc.
The CGAP database of the National Cancer Institute has thousands of expressed sequences, both known and novel, in the form of expressed sequence tags (ESTs). These ESTs derive from diverse normal and tumor cDNA libraries. In CGAP database, there are 8221 libraries from various tissues. Among these libraries, 54 libraries are based on SSH method, and 69 material samples are prepared through microdissection. Among those 54 libraries with SSH method, none of the material sample has been prepared through microdissection.
CGAP also offers different data-mining tools: tools of the GLS, the cDNA xProfiler, the DDD, and the DGED. With these tools and database in CGAP, differently expressed genes can be predicted too[31-33]. Using DGED tool to compare normal stomach libraries and cancer libraries, 117 differently expressed genes can be found. But endothelial cells related to cancer have no appropriate library that can be matched and compared. So cDNA library of endothelial cells related to cancer needs to be built, and the more the better, just like prostate libraries.
TO separate VECs from microvessel, tissues were usually treated with collagenase at 37 °C for 30 min according to present common techniques, and magnetic beads coupled with monoclonal antibody CD31 (Dynabeads CD31) have also been used[34,35]. VECs separated by monoclonal antibody have been identified without change[36]. Instead of using collagenase and cell culture, we separated VECs with Dynalbeads CD31 after mechanically grinding tissue and filtering through a sterile 80-μL-nylon filter. In this way, we obtained about 106 VECs, from which 1 μg total RNA was harvested. Although the amount of the cells was limited and even some cells were connected by fibers, they were relatively pure. The process of separating VECs from tissue with Danalbeads CD31 lasted almost 1 h, as a result RNAlater, solution-inhibiting degradation of RNA, has been used to substitute PBS required by the Danalbeads CD31 Kit. Because the amount was far from 1-2 mg total RNA required by SSH method, Smart cDNA Synthesis Kit was introduced to synthesize and amplify cDNA from the relatively few RNA, which requires only 50 ng total RNA. In the process of amplification, after the first 15 cycles, for each three more cycles, a little sample was taken and tested in order to get suitable copies of cDNA, as more copies would add burden to later screening work.
In performing SSH, each step was operated exactly according to the manual of the kit and the results were verified correct before each following step. G3PDH was used to identify the forward and reverse subtractions. On agarose/EB gel, appearance of G3PDH band was 15 PCR cycles later in subtraction sample than in unsubtraction sample. It implied the amount of G3PDH decreased 300 times by subtraction technique. Finally, with T/A technique, subtracted PCR products were ligated to T vector and transformed into bacteria JM109. So, both the forward subtraction cDNA library containing cDNA fragments only expressed in VECs of stomach cancer but not normal stomach tissue, and the reverse subtraction cDNA library containing cDNA fragments not expressed in VECs of stomach cancer tissue but normal stomach tissue were built up successfully, which was a good beginning for researching into new genes of VECs related to gastrocarcinoma and gene therapy of gastrocarcinoma.
We thank Dr. Xu ZF, Dr. Pan Wi, Dr. Wang XL, from the Life Science Academy of Zhongshan University; and Mr. Zhang QX, Mr. Ding Y, Mr. Jin H, from the Histological Department of Medical College of Zhengzhou University.
Edited by Zhu LH
| 1. | Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025-6030. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2262] [Cited by in F6Publishing: 1997] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 2. | Diatchenko L, Lukyanov S, Lau YF, Siebert PD. Suppression subtractive hybridization: A versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 283] [Cited by in F6Publishing: 302] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 3. | Ji W, Wright MB, Cai L, Flament A, Lindpaintner K. Efficacy of SSH PCR in isolating differentially expressed genes. BMC Genomics. 2002;3:12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 70] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 4. | Wang X, Feuerstein GZ. Suppression subtractive hybridisation: Application in the discovery of novel pharmacological targets. Pharmacogenomics. 2000;1:101-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 5. | Villalva C, Trempat P, Zenou RC, Delsol G, Brousset P. [Gene expression profiling by suppression subtractive hybridization (SSH): A example for its application to the study of lymphomas]. Bull Cancer. 2001;88:315-319. [PubMed] [Cited in This Article: ] |
| 6. | Li YJ, Tian F, Chen ZC, Guan YJ, He CM, Yang XM, Xie DH. Isolation and Identification of cDNA Sequences Differentially Expressed in Laryngeal Carcinoma. Shengwu Huaxue Yu Shengwu Wuli Xuebao (. Shanghai). 2000;32:153-157. [PubMed] [Cited in This Article: ] |
| 7. | Eleveld-Trancíková D, Kúdela P, Majerciak V, Regendová M, Zelník V, Pastorek J, Pastoreková S, Bízik J. Suppression subtractive hybridisation to isolate differentially expressed genes involved in invasiveness of melanoma cell line cultured under different conditions. Int J Oncol. 2002;20:501-508. [PubMed] [Cited in This Article: ] |
| 8. | Langley RR, Ramirez KM, Tsan RZ, Van Arsdall M, Nilsson MB, Fidler IJ. Tissue-specific microvascular endothelial cell lines from H-2K (b)-tsA58 mice for studies of angiogenesis and metastasis. Cancer Res. 2003;63:2971-2976. [PubMed] [Cited in This Article: ] |
| 9. | Fend F, Emmert-Buck MR, Chuaqui R, Cole K, Lee J, Liotta LA, Raffeld M. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 290] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 10. | Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 134] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Pedersen TX, Leethanakul C, Patel V, Mitola D, Lund LR, Danø K, Johnsen M, Gutkind JS, Bugge TH. Laser capture microdissection-based in vivo genomic profiling of wound keratinocytes identifies similarities and differences to squamous cell carcinoma. Oncogene. 2003;22:3964-3976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 12. | Yim SH, Ward JM, Dragan Y, Yamada A, Scacheri PC, Kimura S, Gonzalez FJ. Microarray analysis using amplified mRNA from laser capture microdissection of microscopic hepatocellular precancerous lesions and frozen hepatocellular carcinomas reveals unique and consistent gene expression profiles. Toxicol Pathol. 2003;31:295-303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | Mori M, Mimori K, Yoshikawa Y, Shibuta K, Utsunomiya T, Sadanaga N, Tanaka F, Matsuyama A, Inoue H, Sugimachi K. Analysis of the gene-expression profile regarding the progression of human gastric carcinoma. Surgery. 2002;131:S39-S47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 14. | Jung MH, Kim SC, Jeon GA, Kim SH, Kim Y, Choi KS, Park SI, Joe MK, Kimm K. Identification of differentially expressed genes in normal and tumor human gastric tissue. Genomics. 2000;69:281-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 68] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Yoshikawa Y, Mukai H, Hino F, Asada K, Kato I. Isolation of two novel genes, down-regulated in gastric cancer. Jpn J Cancer Res. 2000;91:459-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 99] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 16. | Su N, Yan H, Li YP. Isolation, purification, identification and gene transfer of microvascular endothelial cells. Xibao Shengwuxue Zazhi. 2000;22:94-98. [Cited in This Article: ] |
| 17. | Muczynski KA, Ekle DM, Coder DM, Anderson SK. Normal human kidney HLA-DR-expressing renal microvascular endothelial cells: characterization, isolation, and regulation of MHC class II expression. J Am Soc Nephrol. 2003;14:1336-1348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 18. | Stier S, Totzke G, Grünewald E, Neuhaus T, Fronhoffs S, Sachinidis A, Vetter H, Schulze-Osthoff K, Ko Y. Identification of syntenin and other TNF-inducible genes in human umbilical arterial endothelial cells by suppression subtractive hybridization. FEBS Lett. 2000;467:299-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Jin HM, Li XT. Neovascularization and disease. Zhongguo Weixunhuan. 2001;5:173-177. [Cited in This Article: ] |
| 20. | Liu DH, Zhang XY, Fan DM, Huang YX, Zhang JS, Huang WQ, Zhang YQ, Huang QS, Ma WY, Chai YB. Expression of vascular endothelial growth factor and its role in oncogenesis of human gastric carcinoma. World J Gastroenterol. 2001;7:500-505. [PubMed] [Cited in This Article: ] |
| 21. | Tao HQ, Lin YZ, Wang RN. Significance of vascular endothelial growth factor messenger RNA expression in gastric cancer. World J Gastroenterol. 1998;4:10-13. [PubMed] [Cited in This Article: ] |
| 22. | Konno H, Baba M, Tanaka T, Kamiya K, Ota M, Oba K, Shoji A, Kaneko T, Nakamura S. Overexpression of vascular endothelial growth factor is responsible for the hematogenous recurrence of early-stage gastric carcinoma. Eur Surg Res. 2000;32:177-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Liu C, Zhang L, Shao ZM, Beatty P, Sartippour M, Lane TF, Barsky SH, Livingston E, Nguyen M. Identification of a novel endothelial-derived gene EG-1. Biochem Biophys Res Commun. 2002;290:602-612. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Khan J, Bittner ML, Saal LH, Teichmann U, Azorsa DO, Gooden GC, Pavan WJ, Trent JM, Meltzer PS. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc Natl Acad Sci USA. 1999;96:13264-13269. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 249] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 25. | Elek J, Park KH, Narayanan R. Microarray-based expression profiling in prostate tumors. In Vivo. 2000;14:173-182. [PubMed] [Cited in This Article: ] |
| 26. | Wikman H, Kettunen E, Seppänen JK, Karjalainen A, Hollmén J, Anttila S, Knuutila S. Identification of differentially expressed genes in pulmonary adenocarcinoma by using cDNA array. Oncogene. 2002;21:5804-5813. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 139] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 27. | Pitzer C, Stassar M, Zöller M. Identification of renal-cell-carcinoma-related cDNA clones by suppression subtractive hybridization. J Cancer Res Clin Oncol. 1999;125:487-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 28. | Ai JK, Huang X, Wang YJ, Bai Y, Lu YQ, Ye XJ, Xin DQ, Na YQ, Zhang ZW, Guo YL. [Screening of novel genes differentially expressed in human renal cell carcinoma by suppression subtractive hybridization]. Ai Zheng. 2002;21:1065-1069. [PubMed] [Cited in This Article: ] |
| 29. | Zhang L, Cilley RE, Chinoy MR. Suppression subtractive hybridization to identify gene expressions in variant and classic small cell lung cancer cell lines. J Surg Res. 2000;93:108-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Li J, Han BL, Huang GJ, Qian GS, Liang P, Yang TH, Chen J. [Screening and identification for cDNA of differentially expressed genes in human primary hepatocellular carcinoma]. Zhonghua Yixue Yichuanxue Zazhi. 2003;20:49-52. [PubMed] [Cited in This Article: ] |
| 31. | Schmitt AO, Specht T, Beckmann G, Dahl E, Pilarsky CP, Hinzmann B, Rosenthal A. Exhaustive mining of EST libraries for genes differentially expressed in normal and tumour tissues. Nucleic Acids Res. 1999;27:4251-4260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 88] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 32. | Scheurle D, DeYoung MP, Binninger DM, Page H, Jahanzeb M, Narayanan R. Cancer gene discovery using digital differential display. Cancer Res. 2000;60:4037-4043. [PubMed] [Cited in This Article: ] |
| 33. | Asmann YW, Kosari F, Wang K, Cheville JC, Vasmatzis G. Identification of differentially expressed genes in normal and malignant prostate by electronic profiling of expressed sequence tags. Cancer Res. 2002;62:3308-3314. [PubMed] [Cited in This Article: ] |
| 34. | Zhou H, Xu GW, Sheng ML, Tang J. Construction of human vascular endothelial cell database. Shengwu Huaxue Zazhi. 1993;9:507-509. [Cited in This Article: ] |
| 35. | van Leeuwen EB, Molema G, de Jong KP, van Luyn MJ, Dijk F, Slooff MJ, Ruiters MH, van der Meer J. One-step method for endothelial cell isolation from large human blood vessels using fibrin glue. Lab Invest. 2000;80:987-989. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |