Published online Dec 15, 2001. doi: 10.3748/wjg.v7.i6.821
Revised: August 19, 2001
Accepted: September 26, 2001
Published online: December 15, 2001
AIM: To study and clone a novel liver cancer related gene, and to explore the molecular basis of liver cancer genesis.
METHODS: Using mRNA differential display polymerase chain reaction (DDPCR), we investigated the difference of mRNA in human hepatocellular carcinoma (HCC) and paired surrounding liver tissues, and got a gene probe. By screening a human placenta cDNA library and genomic homologous extend, we obtained a full-length cDNA named HCCA3. We analyzed the expression of this novel gene in 42 pairs of HCC and the surrounding liver tissues, and distribution in human normal tissues by means of Northern blot assay.
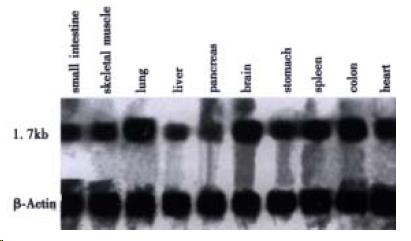
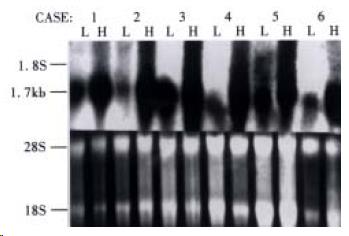
RESULTS: A full-length cDNA of liver cancer associated gene HCCA3 has been submitted to the GeneBank nucleotide sequence databases (Accession No. AF276707). The positive expression rate of this gene was 78.6% (33/42) in HCC tissues, and the clinical pathological data showed that the HCCA3 was closely associated with the invasion of tumor capsule (P = 0.023) and adjacant small metastasis satellite nodules lesions (P = 0.041). The HCCA3 was widely distributed in the human normal tissues, which was intensively expressed in lungs, brain and colon tissues, while lowly expressed in the liver tissues.
CONCLUSION: A novel full-length cDNA was cloned and differentiated, which was highly expressed in liver cancer tissues. The high expression was closely related to the tumor invasiveness and metastasis, that may be the late heredited change in HCC genesis.
- Citation: Wang ZX, Hu GF, Wang HY, Wu MC. Expression of liver cancer associated gene HCCA3. World J Gastroenterol 2001; 7(6): 821-825
- URL: https://www.wjgnet.com/1007-9327/full/v7/i6/821.htm
- DOI: https://dx.doi.org/10.3748/wjg.v7.i6.821
Primary hepatocellular carcinoma (HCC) is one of the most common fatal malignant tumors in China[1-26]. According to the statistics of our country, primary liver cancer claims 20.40 lives per 100000 people annually, with 19.98 per 100000 in cities and 23.59 per 100000 in rural areas, ranking as the 2nd and the 1st leading cause of cancer death, respectively. Of all the newly enrolled cases in the world each year, 45% are found in the mainland of China. In the southeast areas of high incidence, the situation is even worse with tumors tending to occur in a younger age group. The molecular events for HCC development are very complex, and HCC has proved to be genetically heterogenous neoplasm[27-30]. But to date, the identified genes have not yet fully disclosed the mechanisms of HCC[31-38]. In an attempt to identify HCC susceptible genes, differential display method was employed in this study. In the analysis of altered expression genes between HCC tissues and their nontumor counterparts, we isolated a novel gene named HCCA3 with a full length of cDNA.
PCR polymerase is a product of Promega (Madison, USA); isotope α-32P-dATP was purchased from Amersham Pharmacia Biotech (Arlington Heights, IL); Qiaex II gel extraction kit were from Qiagen (Hilden, Germany); pGEM-T vector were from Promega; human placental cDNA library were from Clontech (USA); nitrocellulose were from Amersham Pharmacia Biotech; Prime-a-Gene Labeling System were from Promega; 14-mer anchored oligo (dT) primer (dT12 CA) and an arbitrary 10-base olignucleotide A2 (5’-AATCGGGCTG-3’) were donated by Professor Pei.
Primary HCC and their surrounding liver tissues were obtained from 42 patients who received surgical resection at the Eastern Hepatobilliary Surgical Hospital of the Second Military Medical University, Shanghai, China. These included 41 male and 1 female patients with a median age of 49 years (range 24-72 years, mean age of 49.8 years). Thirty-five (83.3%) patients had serological evidence of hepatitis B virus infection. The serum AFP level was above 25 μg•L¯¹ in 23 cases (54.8%). The tumor size was smaller than 5 cm (small HCC) in 13 patients and larger than 5 cm in 29. Histologically, 40 patients (95.2%) were complicated with cirrhosis. There were 7 well differentiated (Edmondson’s grades I and II) and 35 poorly differentiated (Edmondson’s grades III and IV) HCCs. Macroscopic portal vein tumor spread was found in 3 patients, and microscopic surrounding liver vascular cancer thrombi were found in 26. Gross and microscopic intrahepatic adjacent small satellite nodules lesions were found in 28, and tumor capsule invasion of liver cancer in 32. Adult normal tissues were obtained from a healthy young man who died of a traffic accident.
RNA extraction and differential display Total cytoplasmatic RNA was extracted by the acid guanidinium thiocyanate-phenol-chloroform extraction methods[39,40]. The differential display method was performed as described previously[39,41,42]. Amplification consisted of initial den aturation at 94 °C for 4 min, followed by 40 reaction cycles (60 s at 93 °C, 2 min at 40 °C, and 90 s at 70 °C) and a final cycle at 72 °C for 10 min. PCR fragments were then reamplified by the same primer, separated on a 16 g•L¯¹ agarose gel, purified by Qiaex II gel extraction kit, and subcloned into the pGEM-T vector using standard molecular cloning techniques.
Library screening and DNA sequencing Fragment contained in the PCR clone (length in 350 base pairs) from DDPCR served as probe to screen placental cDNA library, using the standard filter hybridization techniques described[43,44]. At the end of the third screening, we got several plagues containing the target DNA sequence and sequenced them by DNA automated sequencing system. To obtain the cDNA in a full length, genomic homologous screening was used through comparing the cDNA sequence obtained by screening the library with the NCBI GeneBank EST database. We used the PCR assay and sequencing to confirm the correct ness of the cDNA sequence.
Northern blot assay For Northern blot analysis[37,40,44], 40 μg of total RNA was denatured, loaded on a 15 g•L¯¹ agarose gel and ran at 5 V·cm-1 for about 3 h. The collected gels were then transferred to nitrocellulose. Hybridization of the filters was performed using specific probe of HCCA3 cDNA fragment (length in 1125 bp) obtained from screening the library. The probe was labeled with 50 μCi α-32P-dATP using Prime-a-Gene labeling kit according to the given protocol. After prehybridization at 42 °C for 3 h, the membranes were hybridized in the same solution containing the labeled probe for 6 h at the same temperature, and exposed to X-ray film for 10 d at -70 °C. In order to calibrate relative quantities of loaded RNAs, the blot was rehybridized with a cDNA probe of the β-Actin gene.
χ² test or Fisher’s exact test was used to examine the differences and relationship among groups of patients classified by HCCA3 expression. Differences at P < 0.05 were judged to be statistically significant.
By DDPCR, we found a differentially expressed gene fragment that exclusively present in the liver cancer lane. This fragment (length in 350 bp) was then subcloned into pGEM-T vector and served as specific probe to screen human placental cDNA library. We have obtained the gene fragment of 1125 bp in length, which shortened nearly 600 bp according to the location of Northern hybridization by screening the library. We also obtained the full-length cDNA of 1706 bp, which was in good agreement with the size of the mRNA species observed by Northern blotting through genomic homologous extend, along with the EST sequence (GeneBank Accession No. AP001077, length in 197663 bp) of NCBI GeneBank EST databases. The sequence of 1706 bp in length was corrected by PCR assay and sequencing. It was named HCCA3 (HCC associated gene 3, also named STW-2) and submitted to EMBL/GeneBank/DDBJ nucleotide sequence databases (Accession No. AF27 6707).
HCCA3 contains a consensus initiation codon[45,46] at position 681 followed by a single open reading frame of 792 bp encoding 264 amino acids. The 3’ untranslated region of 230 bp had a consensus polyadenylation signal (AATAAA) beginning 16 bases upstream the poly (A) tail. Alignment at nucleotide and amino acid level showed no significant homologues with known genes. The deduced protein was estimated to be 29396 da lton and has a pI of 6.93. Amino acid sequence analysis by GCG sequence analysis software package (version 9.1, Genetics Computer Group, Madison, Wisconsin) and PC/GENE (Version 5.03, Geneva University, Switzerland) showed that there were several putative modification sites. These include two N-glycosylation sites at amino acid of 40 and 67, three N-myristoylation sites at amino acid of 183, 185 and 258, five phosphorylation sites for protein kinase: two protein kinase C (PKC) phosphorylation sites at 82 and 250, three cases in kinase II phosphorylation sites at 8, 189 and 209. No transmembrane domain and signal peptide was found. Further more, there was no nuclear targeting sequence. The schematic presentation of HCCA3 cDNA is shown in Figure 1.
Northern blot analysis showed that HCCA3 mRNA appears to be widely expressed in human normal tissues (Figure 2). The HCCA3 gene was particularly highly expressed in human lungs, brain and colon, moderately expressed in muscle, stomach, spleen and heart tissues, weakly expressed in small intestines, pancreas, and liver tissues. Among the tissues with positive signals, HCCA3 mRNA was observed in a transcript of approximately 1.7 kb which well corresponded to the size of the cloned cDNA.
HCCA3 mRNA expression was noticed in 78.6% (33/42) patients, which was intensively expressed in HCC tissues (Figure 3), while lowly expressed in the surrounding liver tissues. To investigate the potential biological role of HCCA3 in the development of HCC, we further investigated its expression in HCC with extensive samples and compared with the clinically pathological parameters. The mRNA expression level of HCCA3 was associated with the invasion of liver cancer capsule and the adjacant small satellite nodules lesions (P < 0.05), but not with tumor size, tumor differentiation, serum AFP, hepatitis B vrius (HBV) infection and microscopic vascular cancer cell thrombi (P > 0.05). The results are listed in Table 1.
| Clinicopathology | n | Positive n (%) |
| Tumor size (cm) | ||
| < 5 | 10 | 6 (60.0) |
| > 5 | 32 | 27 (84.4) |
| Tumor differentiation | ||
| Well | 7 | 4 (57.1) |
| Poor | 35 | 29 (82.9) |
| Serum AFP (μg•L¯¹) | ||
| 25 | > 19 | 15 (79.0) |
| > 25 | 23 | 18 (78.3) |
| HBV infection | ||
| Positive | 35 | 28 (80.0) |
| Negative | 7 | 5 (71.4) |
| Capsule invasion | ||
| Positive | 32 | 28 (87.5) |
| Negative | 10 | 5 (50.0)a |
| Cancer thrombi | ||
| Positive | 26 | 21 (76.9) |
| Negative | 16 | 12 (82.4) |
| Satellite lesions | ||
| Positive | 28 | 25 (89.3) |
| Negative | 14 | 8 (57.1)a |
Differential display polymerase chain reaction (DDPCR) method is a useful tool for detecting and characterizing altered gene expression in eukaryotic cells[39,41,47,48]. Using this technique, we have successfully isolated a gene named HCCA3 with a transcript of 1706 bp. A consensus initiation coden is at 681 bp and the franking sequence of the predicted initiating methionine coincides with Kozak[45,46,49] criterion because the nucleotides at -3, +4 site of start codon are purine, and there is one stop codon at the upstream sequence. The length of HCCA3 cDNA agrees with the size of mRNA by Northern blot analysis. No significant homologues with known genes at nucleotide and amino acid levels were found[47-50]. No signal peptides searched by GCG and PC/GENE software were found, suggesting that HCCA3 was not a secretive protein[51,52].This protein also has no transmembrane domain and nuclear targeting sequence indicating that it may not be located on cell member or within nucleus. The putative protein of HCCA3 revealed several phosphorylation sites for protein kinase C and casein kinase II. It is generally accepted that protein phosphorylation-dephosphorylation plays a role in the regulation of essentially all cellular functions, and there is evidence that deregulation of protein phosphorylation is involved in several human cancers[22,53,54]. HCCA2 as a possible oncoprotein may be functionally abnormal, and phosphorylative deregulation may be a mechanism. The function of HCCA3 protein may also largely depend on its phosphorylation status [22,54]. However, this needs further studies.
Studies on the expression of HCCA3 can reveal its potential biological significance[22,36,37, 55]. By Northern blot analysis, we noted that HCCA3 mRNA was expressed widely in normal human tissues, indicating that HCCA3 is a normal cellular gene which may be involved in the physical process of the distributed tissues. Although HCCA3 mRNA was lowly expressed in normal liver tissues, it was significantly expressed in HCC tissues, showing that the high expression of HCCA3 might participate in the process of liver on cogenesis[36,37]. Because HCCA3 is a normal cellular gene, increased expression of HCCA3 in HCC implys the genetic abnormality and acts as an oncogene in the development of HCC. In 42 patients with HCC, HCCA3 mRNA was detected in 33 (78.6%), which was highly expressed in HCC tissues, suggesting that HCCA3 is a very common molecular event involved in the pathogenesis of HCC. These findings indicate that HCCA3 mRNA is overexpressed preferentially in HCC and can serve as a tumor biomarker for HCC[7,18,23,36,37]. The three patients with macroscopic portal vein tumor thrombi all had significantly high expression of HCCA3 mRNA, implying that HCCA3 is a late incidence in HCC carcinogenesis. Compared with pathological features, HCCA3 mRNA expression was associated with the invasion of tumor capsule and the adjacant small satellite nodules lesions (P < 0.0 5), indicating that HCCA3 mRNA expression is a factor of HCC invasiveness and metastasis[37,38]. Although the function of HCCA3 is still unknown, our results suggest that the up-regulation of expression of HCCA3 mRNA may play an important ro le in the development and/or progression of hepatocellular carcinoma[36-38,42,45]. This finding demonstrated that it is possible to identify the previously unknown, differential gene expression from a small amount of clinical samples [39-42,56]. Information about such alteration of HCCA3 in gene expression could be useful in elucidating the genetic events in HCC pathogenesis, developing new diagnostic markers, or determining novel therapeutic targets.
Edited by Ma JY
| 1. | Tang ZY. Hepatocellular carcinoma--cause, treatment and metastasis. World J Gastroenterol. 2001;7:445-454. [PubMed] [Cited in This Article: ] |
| 2. | Zhou XD, Tang ZY, Yang BH, Lin ZY, Ma ZC, Ye SL, Wu ZQ, Fan J, Qin LX, Zheng BH. Experience of 1000 patients who underwent hepatectomy for small hepatocellular carcinoma. Cancer. 2001;91:1479-1486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 9] [Reference Citation Analysis (0)] |
| 3. | Wu M. Clinical advances in primary liver cancer in China. Hepatogastroenterology. 2001;48:29-32. [PubMed] [Cited in This Article: ] |
| 4. | Wang Z, He Z, Wu Z, Zhang W. [Induction of apoptosis in human hepatocellular carcinoma cells by adenoviral-mediated transfer of human multigenes]. Zhonghua Gan Zang Bing Za Zhi. 2000;8:224-226. [PubMed] [Cited in This Article: ] |
| 5. | Jiang YF, Yang ZH, Hu JQ. Recurrence or metastasis of HCC: predictors, early detection and experimental antiangiogenic therapy. World J Gastroenterol. 2000;6:61-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 57] [Cited by in F6Publishing: 62] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 6. | Ma XD, Sui YF, Wang WL. Expression of gap junction genes connexin 32, connexin 43 and their proteins in hepatocellular carcinoma and normal liver tissues. World J Gastroenterol. 2000;6:66-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.0] [Reference Citation Analysis (0)] |
| 7. | Kong XB, Yang ZK, Liang LJ, Huang JF, Lin HL. Overexpression of P-glycoprotein in hepatocellular carcinoma and its clinical implication. World J Gastroenterol. 2000;6:134-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Sun BH, Zhao XP, Wang BJ, Yang DL, Hao LJ. FADD and TRADD expression and apoptosis in primary hepatocellular carcinoma. World J Gastroenterol. 2000;6:223-227. [PubMed] [Cited in This Article: ] |
| 9. | Yang DH, Zhang MQ, Du J, Xu C, Liang QM, Mao JF, Qin HR, Fan ZR. Inhibitory effect of IGF- II antisense RNA on malignant phenotype of hepatocellular carcinoma. World J Gastroenterol. 2000;6:266-267. [PubMed] [Cited in This Article: ] |
| 10. | Sithinamsuwan P, Piratvisuth T, Tanomkiat W, Apakupakul N, Tongyoo S. Review of 336 patients with hepatocellular carcinoma at Songklanagarind Hospital. World J Gastroenterol. 2000;6:339-343. [PubMed] [Cited in This Article: ] |
| 11. | Sun BH, Zhang J, Wang BJ, Zhao XP, Wang YK, Yu ZQ, Yang DL, Hao LJ. Analysis of in vivo patterns of caspase 3 gene expression in primary hepatocellular carcinoma and its relationship to p21 (WAF1) expression and hepatic apoptosis. World J Gastroenterol. 2000;6:356-360. [PubMed] [Cited in This Article: ] |
| 12. | Wang Y, Liu H, Zhou Q, Li X. Analysis of point mutation in site 1896 of HBV precore and its detection in the tissues and serum of HCC patients. World J Gastroenterol. 2000;6:395-397. [PubMed] [Cited in This Article: ] |
| 13. | Zhu HZ, Ruan YB, Wu ZB, Zhang CM. Kupffer cell and apoptosis in experimental HCC. World J Gastroenterol. 2000;6:405-407. [PubMed] [Cited in This Article: ] |
| 14. | Mei MH, Xu J, Shi QF, Yang JH, Chen Q, Qin LL. Clinical significance of serum intercellular adhesion molecule-1 detection in patients with hepatocellular carcinoma. World J Gastroenterol. 2000;6:408-410. [PubMed] [Cited in This Article: ] |
| 15. | Qin Y, Li B, Tan YS, Sun ZL, Zuo FQ, Sun ZF. Polymorphism of p16INK4a gene and rare mutation of p15INK4b gene exon2 in primary hepatocarcinoma. World J Gastroenterol. 2000;6:411-414. [PubMed] [Cited in This Article: ] |
| 16. | Shen LJ, Zhang ZJ, Ou YM, Zhang HX, Huang R, He Y, Wang MJ, Xu GS. Computed morphometric analysis and expression of alpha fetoprotein in hepatocellular carcinoma and its related lesion. World J Gastroenterol. 2000;6:415-416. [PubMed] [Cited in This Article: ] |
| 17. | Li J, Yang XK, Yu XX, Ge ML, Wang WL, Zhang J, Hou YD. Overexpression of p27 (KIP1) induced cell cycle arrest in G (1) phase and subsequent apoptosis in HCC-9204 cell line. World J Gastroenterol. 2000;6:513-521. [PubMed] [Cited in This Article: ] |
| 18. | Niu Q, Tang ZY, Ma ZC, Qin LX, Zhang LH. Serum vascular endothelial growth factor is a potential biomarker of metastatic recurrence after curative resection of hepatocellular carcinoma. World J Gastroenterol. 2000;6:565-568. [PubMed] [Cited in This Article: ] |
| 19. | Huang XF, Wang CM, Dai XW, Li ZJ, Pan BR, Yu LB, Qian B, Fang L. Expressions of chromogranin A and cathepsin D in human primary hepatocellular carcinoma. World J Gastroenterol. 2000;6:693-698. [PubMed] [Cited in This Article: ] |
| 20. | Xu HY, Yang YL, Guan XL, Song G, Jiang AM, Shi LJ. Expression of regulating apoptosis gene and apoptosis index in primary liver cancer. World J Gastroenterol. 2000;6:721-724. [PubMed] [Cited in This Article: ] |
| 21. | Fan J, Wu ZQ, Tang ZY, Zhou J, Qiu SJ, Ma ZC, Zhou XD, Ye SL. Multimodality treatment in hepatocellular carcinoma patients with tumor thrombi in portal vein. World J Gastroenterol. 2001;7:28-32. [PubMed] [Cited in This Article: ] |
| 22. | Feng DY, Zheng H, Tan Y, Cheng RX. Effect of phosphorylation of MAPK and Stat3 and expression of c-fos and c-jun proteins on hepatocarcinogenesis and their clinical significance. World J Gastroenterol. 2001;7:33-36. [PubMed] [Cited in This Article: ] |
| 23. | Xu J, Mei MH, Zeng SE, Shi QF, Liu YM, Qin LL. Expressions of ICAM-1 and its mRNA in sera and tissues of patients with hepatocellular carcinoma. World J Gastroenterol. 2001;7:120-125. [PubMed] [Cited in This Article: ] |
| 24. | Cui J, Yang DH, Bi XJ, Fan ZR. Methylation status of c-fms oncogene in HCC and its relationship with clinical pathology. World J Gastroenterol. 2001;7:136-139. [PubMed] [Cited in This Article: ] |
| 25. | Fan ZR, Yang DH, Cui J, Qin HR, Huang CC. Expression of insulin like growth factor II and its receptor in hepatocellular carcinogenesis. World J Gastroenterol. 2001;7:285-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 26. | Wang Q, Lin ZY, Feng XL. Alterations in metastatic properties of hepatocellular carcinoma cell following H-ras oncogene transfection. World J Gastroenterol. 2001;7:335-339. [PubMed] [Cited in This Article: ] |
| 27. | Liu WW. Study of etiology on liv er neoplasms. Shijie Huaren Xiaohua Zazhi. 1999;7:93-95. [Cited in This Article: ] |
| 28. | Roberts LR, LaRusso NF. Potential roles of tumor suppressor genes and microsatellite instability in hepatocellular carcinogenesis in southern African blacks. World J Gastroenterol. 2000;6:37-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Bian JC, Shen FM, Shen L, Wang TR, Wang XH, Chen GC, Wang JB. Susceptibility to hepatocellular carcinoma associated with null genotypes of GSTM1 and GSTT1. World J Gastroenterol. 2000;6:228-230. [PubMed] [Cited in This Article: ] |
| 30. | Li J, Yang XK, Yu XX, Ge ML, Wang WL, Zhang J, Hou YD. Overexpression of p27 (KIP1) induced cell cycle arrest in G (1) phase and subsequent apoptosis in HCC-9204 cell line. World J Gastroenterol. 2000;6:513-521. [PubMed] [Cited in This Article: ] |
| 31. | Zheng JY, Li KZ, Dou KF and Li J. Studies on p53 and nm23-H1 mRNA expression in hepatocellular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:40-42. [Cited in This Article: ] |
| 32. | Wang HY, Yan RQ, Long JB and Wu QL. Cyclin D1 ampli fication is associated with HBV DNA integration and pathology in human hepatocel lular carcinoma. Shijie Huaren Xiaohua Zazhi. 1999;7:98-100. [Cited in This Article: ] |
| 33. | Lin GY, Chen ZL, Lu CM, Li Y, Ping XJ, Huang R. Immunohistochemical study on p53, H-rasp21, c-erbB-2 protein and PCNA expression in HCC tissues of Han and minority ethnic patients. World J Gastroenterol. 2000;6:234-238. [PubMed] [Cited in This Article: ] |
| 34. | Laurent-Puig P, Legoix P, Bluteau O, Belghiti J, Franco D, Binot F, Monges G, Thomas G, Bioulac-Sage P, Zucman-Rossi J. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763-1773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 457] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 35. | Ozturk M. Genetic aspects of hepatocellular carcinogenesis. Semin Liver Dis. 1999;19:235-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 133] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 36. | Martins C, Kedda MA, Kew MC. Characterization of six tumor suppressor genes and microsatellite instability in hepatocellular carcinoma in southern African blacks. World J Gastroenterol. 1999;5:470-476. [PubMed] [Cited in This Article: ] |
| 37. | Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990-4996. [PubMed] [Cited in This Article: ] |
| 38. | Schlott T, Ahrens K, Ruschenburg I, Reimer S, Hartmann H, Droese M. Different gene expression of MDM2, GAGE-1, -2 and FHIT in hepatocellular carcinoma and focal nodular hyperplasia. Br J Cancer. 1999;80:73-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Wang L, Lu W, Chen YG, Zhou XM, Gu JR. Comparison of gene expression between normal colon mucosa and colon carcinoma by means of messenger RNA differential display. World J Gastroenterol. 1999;5:533-534. [PubMed] [Cited in This Article: ] |
| 40. | Tackels-Horne D, Goodman MD, Williams AJ, Wilson DJ, Eskandari T, Vogt LM, Boland JF, Scherf U, Vockley JG. Identification of differentially expressed genes in hepatocellular carcinoma and metastatic liver tumors by oligonucleotide expression profiling. Cancer. 2001;92:395-405. [PubMed] [Cited in This Article: ] |
| 41. | Thai SF, Allen JW, DeAngelo AB, George MH, Fuscoe JC. Detection of early gene expression changes by differential display in the livers of mice exposed to dichloroacetic acid. Carcinogenesis. 2001;22:1317-1322. [PubMed] [Cited in This Article: ] |
| 42. | Graveel CR, Jatkoe T, Madore SJ, Holt AL, Farnham PJ. Expression profiling and identification of novel genes in hepatocellular carcinomas. Oncogene. 2001;20:2704-2712. [PubMed] [Cited in This Article: ] |
| 43. | Yamashita T, Kaneko S, Hashimoto S, Sato T, Nagai S, Toyoda N, Suzuki T, Kobayashi K, Matsushima K. Serial analysis of gene expression in chronic hepatitis C and hepatocellular carcinoma. Biochem Biophys Res Commun. 2001;282:647-654. [PubMed] [Cited in This Article: ] |
| 44. | Fuchs M, Wang H, Ciossek T, Chen Z, Ullrich A. Differential expression of MAM-subfamily protein tyrosine phosphatases during mouse development. Mech Dev. 1998;70:91-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 45. | Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y, Ji Y, Wu X, Han Z, Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176-3181. [PubMed] [Cited in This Article: ] |
| 46. | Harada H, Nagai H, Tsuneizumi M, Mikami I, Sugano S, Emi M. Identification of DMC1, a novel gene in the TOC region on 17q25.1 that shows loss of expression in multiple human cancers. J Hum Genet. 2001;46:90-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 47. | Robb CW, Orihuela CJ, Ekkelenkamp MB, Niesel DW. Identification and characterization of an in vivo regulated D15/Oma87 homologue in Shigella flexneri using differential display polymerase chain reaction. Gene. 2001;262:169-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 48. | Tran YK, Bögler O, Gorse KM, Wieland I, Green MR, Newsham IF. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999;59:35-43. [PubMed] [Cited in This Article: ] |
| 49. | Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA. Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res. 2001;61:5370-5373. [PubMed] [Cited in This Article: ] |
| 50. | Kang D, Jiang H, Wu Q, Pestka S, Fisher PB. Cloning and characterization of human ubiquitin-processing protease-43 from terminally differentiated human melanoma cells using a rapid subtraction hybridization protocol RaSH. Gene. 2001;267:233-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | Cheung WM, Chu AH, Chu PW, Ip NY. Cloning and expression of a novel nuclear matrix-associated protein that is regulated during the retinoic acid-induced neuronal differentiation. J Biol Chem. 2001;276:17083-17091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 52. | Tateno M, Fukunishi Y, Komatsu S, Okazaki Y, Kawai J, Shibata K, Itoh M, Muramatsu M, Held WA, Hayashizaki Y. Identification of a novel member of the snail/Gfi-1 repressor family, mlt 1, which is methylated and silenced in liver tumors of SV40 T antigen transgenic mice. Cancer Res. 2001;61:1144-1153. [PubMed] [Cited in This Article: ] |
| 53. | Graves JD, Krebs EG. Protein phosphorylation and signal transduction. Pharmacol Ther. 1999;82:111-121. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 313] [Cited by in F6Publishing: 321] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 54. | Barthel A, Schmoll D, Krüger KD, Bahrenberg G, Walther R, Roth RA, Joost HG. Differential regulation of endogenous glucose-6-phosphatase and phosphoenolpyruvate carboxykinase gene expression by the forkhead transcription factor FKHR in H4IIE-hepatoma cells. Biochem Biophys Res Commun. 2001;285:897-902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 55. | Shirota Y, Kaneko S, Honda M, Kawai HF, Kobayashi K. Identification of differentially expressed genes in hepatocellular carcinoma with cDNA microarrays. Hepatology. 2001;33:832-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 124] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 56. | Miyasaka Y, Enomoto N, Nagayama K, Izumi N, Marumo F, Watanabe M, Sato C. Analysis of differentially expressed genes in human hepatocellular carcinoma using suppression subtractive hybridization. Br J Cancer. 2001;85:228-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 70] [Article Influence: 3.0] [Reference Citation Analysis (0)] |