Published online Nov 28, 2022. doi: 10.3748/wjg.v28.i44.6230
Peer-review started: September 11, 2022
First decision: September 29, 2022
Revised: October 7, 2022
Accepted: November 16, 2022
Article in press: November 16, 2022
Published online: November 28, 2022
The liver is a key organ involved in a wide range of functions, whose damage can lead to chronic liver disease (CLD). CLD accounts for more than two million deaths worldwide, becoming a social and economic burden for most countries. Among the different factors that can cause CLD, alcohol abuse, viruses, drug treatments, and unhealthy dietary patterns top the list. These conditions prompt and perpetuate an inflammatory environment and oxidative stress imbalance that favor the development of hepatic fibrogenesis. High stages of fibrosis can eventually lead to cirrhosis or hepatocellular carcinoma (HCC). Despite the advances achieved in this field, new approaches are needed for the prevention, diagnosis, treatment, and prognosis of CLD. In this context, the scientific com-munity is using machine learning (ML) algorithms to integrate and process vast amounts of data with unprecedented performance. ML techniques allow the integration of anthropometric, genetic, clinical, biochemical, dietary, lifestyle and omics data, giving new insights to tackle CLD and bringing personalized medicine a step closer. This review summarizes the investigations where ML techniques have been applied to study new approaches that could be used in inflammatory-related, hepatitis viruses-induced, and coronavirus disease 2019-induced liver damage and enlighten the factors involved in CLD development.
Core Tip: Chronic liver disease has become a global burden, and new approaches need to be explored to tackle this disease. In this context, machine learning techniques bring a whole new set of opportunities to study novel approaches and biomarkers for prevention, diagnosis, treatment, and prognosis of inflammatory and virus-related liver diseases. The application of machine learning algorithms constitutes a pivotal piece of personalized medicine, allowing the integration of different phenotypical and genotypical data for a precision outcome concerning inflammatory liver comorbidities in non-communicable and viral diseases.
- Citation: Martínez JA, Alonso-Bernáldez M, Martínez-Urbistondo D, Vargas-Nuñez JA, Ramírez de Molina A, Dávalos A, Ramos-Lopez O. Machine learning insights concerning inflammatory and liver-related risk comorbidities in non-communicable and viral diseases. World J Gastroenterol 2022; 28(44): 6230-6248
- URL: https://www.wjgnet.com/1007-9327/full/v28/i44/6230.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i44.6230
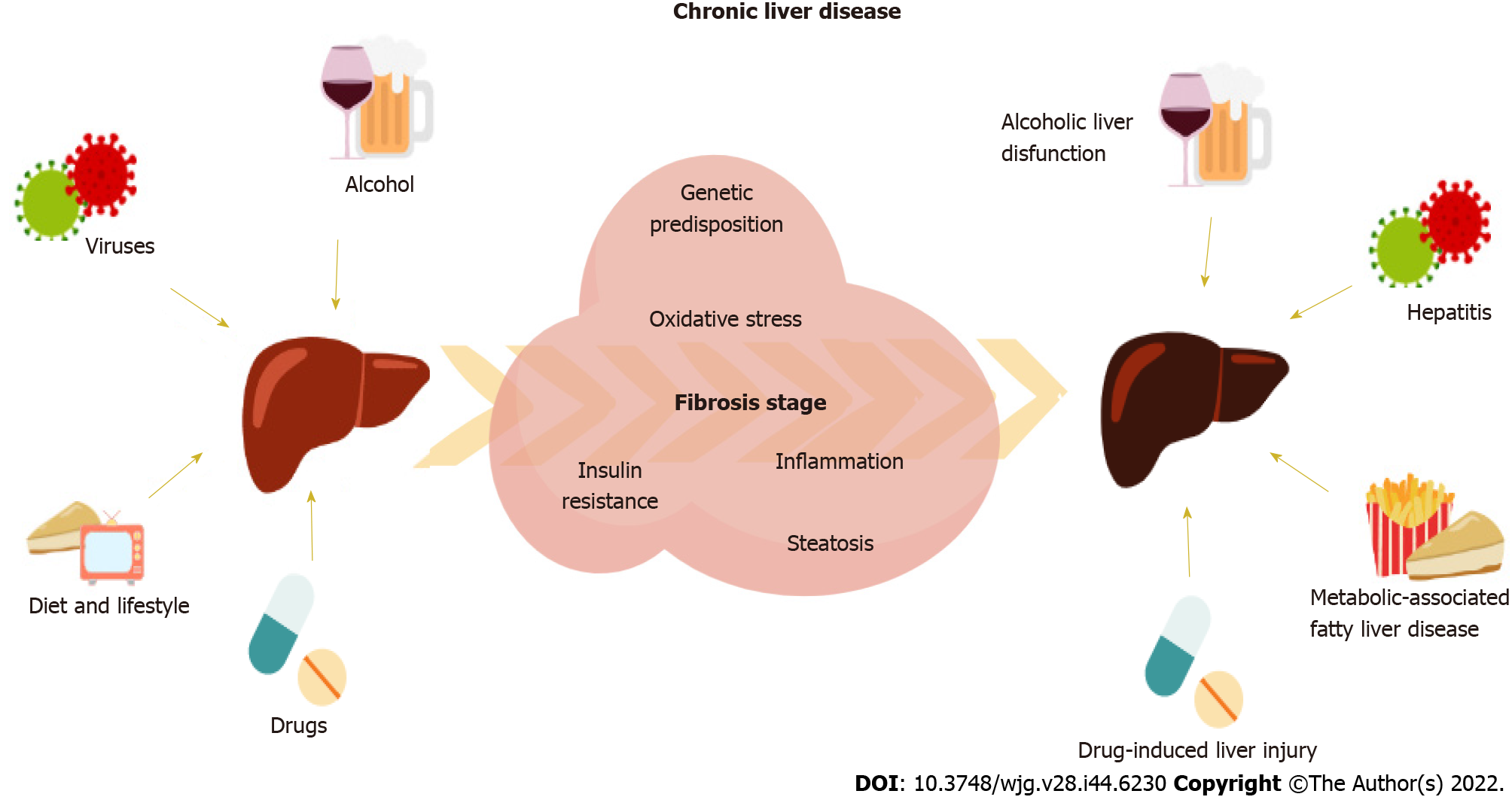
The liver is a key organ involved in relevant homeostatic metabolic and detoxifying human functions[1]. Thus, the liver is the epicenter of an organ-organ network weaving a series of complex interactions in the organism, which makes liver damage an underlying adverse condition in a whole set of diseases. Chronic liver disease (CLD) can be caused mainly by alcoholic liver-related dysfunctions, hepatitis B virus (HBV), hepatitis C virus (HCV), drug treatments, or non-alcoholic fatty liver disease (NAFLD), as recently updated to the term metabolic-associated FLD (Figure 1)[2,3]. Patients with liver-related diseases need frequent follow-ups and careful monitoring since CLD can eventually lead to cirrhosis or hepatocellular carcinoma (HCC) if not diagnosed on time for treatment or surgery. These CLD-related conditions have become a global burden, whose mortality associated rates have increased over the years reaching more than 2 million deaths worldwide[4].
CLD is usually accompanied by an unhealthy inflammatory environment[5]. The immune response is a fundamental process to maintain homeostasis within the organism defense machinery and is characterized by the secretion of proinflammatory cytokines, like interleukin (IL)-1, tumor necrosis factor-α (TNF-α), and prostaglandin E2, in an acute manner in order to resolve sudden damage[5]. However, if sustained over time, these abnormal levels of inflammatory cytokines cause low-grade inflammation (LGI). LGI is a silent condition that predisposes to the development of metabolic and infectious diseases that has become a worldwide health issue[6]. Patients with CLD, such as non-alcoholic steatohepatitis (NASH), present impaired immune function, dysbiosis, insulin resistance (IR) and LGI, all of which can aggravate infectious disease progression and perpetuate excess of adipose tissue, are characterized by overstimulation of the production of adipose-derived inflammatory molecules[5,7-9].
The liver also secretes important hepatokines that act as signaling proteins modulating functions in other organs and are involved in a wide range of conditions, such as IR and adipogenesis[1]. For instance, fibroblast growth factor-21 (FGF-21) is a mediator participating in glucose metabolism mainly secreted by the liver that modulates adipogenesis, while fetuins, liver-derived plasma proteins, are participating in metabolic impairment and inflammation[1]. A dysregulation in systemic cytokines prompts fat accumulation in hepatocytes, which in turn promotes local secretion of proinflammatory hepatokines, leading to liver steatosis and IR. In addition, immune cells also find difficulty in this inflammatory environment to exert their role appropriately. Persistent inflammatory signals over time also abnormally activate immune cells, impairing the body’s ability to fight infection, repair tissue damage, or recover from possible poisoning. Inflammation comes hand in hand with an increase in oxidative stress, a state characterized by an imbalance in favoring the accumulation of higher reactive oxygen (ROS) and nitrogen species. These molecules in unusual concentrations damage the cell and environmental milieu by promoting the expression of proinflammatory genes, resulting in a vicious cycle. Thus, CLD presents an oxidative atmosphere, probably linked to the proinflammatory state[10,11]. This environment is the perfect setting for the fibrogenic process to unfold, an underlying condition of CLD that is characterized by progressive accumulation of fibrillar extracellular matrix in the liver[12]. The stage of hepatic fibrosis has been associated with the risk of mortality and liver-related morbidity in patients with NAFLD[13], virus-induced hepatitis[14,15], and alcoholic-derived liver disease[16], eventually leading to HCC.
In this context, infection by human hepatitis viruses (HHVs) is the most common cause of hepatitis, leading to the activation of the immune system, and the subsequent inflammatory response[17]. HBV and HCV acute infections can be resolved with antiviral and immune therapy. However, in a significant percentage they can progress to chronic hepatitis. This persistent infection can lead to comorbidities outside the liver, like arthritis, vasculitis, myalgia, and peripheral neuropathies[18]. Moreover, another new infectious disease appeared in late 2019 that can cause liver damage: Coronavirus disease 2019 (COVID-19). COVID-19 is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, and it has become a global health issue since its outbreak in 2020 was declared a pandemic. Beyond lung function, COVID-19 can affect a wide variety of tissues, like the gastrointestinal tract, kidneys, and liver, with an underlying adverse inflammatory environment[19]. This inflammatory-related condition has been strongly associated to metabolic status and worsening diseases like obesity, diabetes, and hypertension[7,20-22]. For instance, COVID-19 can increase hepatic lipid accumulation by mitochondrial and endoplasmic reticulum (ER) dysfunction or worsen NAFLD if it was already present. A recent systematic review depicted that the parameters normally used for liver impairment screening were significantly increased in COVID-19 patients[23], placing CLD as a risk factor for progressive and severe COVID-19[24,25].
CLD is a global health problem, and new methods are needed to tackle this life-threatening condition. In this line, this review aims to explore machine learning (ML)-based approaches to manage CLD and develop biomarkers for diagnosis and prognosis. Its goal is to shed light on the factors involved in CLD to help health professionals in clinical management with the support of ML and identify new targets that can define therapeutic care lines in viral infections and non-communicable diseases (NCD), with an impact on liver functions with an inflammatory component. This includes the new disease, COVID-19.
The incidence of NCD, such as cardiovascular diseases and diabetes, has skyrocketed in the last decades, pressing authorities to establish developmental goals to achieve in the near future in terms of decreasing NCD-caused mortality[26]. Some of the risk factors that contribute to the development of NCD are excess of adipose tissue and high levels of glycemia. In this context, adipose tissue plays a key role in the development of FLD by secreting adipokines and other molecules, like free fatty acids (FFA)[8].
An energy excess prompts fat accumulation in the organism and the subsequent dysregulation of this tissue. This is of relevance since an inflamed adipose tissue results in increased levels of FFA and pro-inflammatory cytokines, IR, and infiltration of macrophages in the liver by the activation of Th1 and Th17 cells[8]. FFA enter the liver through the portal vein and trigger a series of reactions. For instance, they serve as ligands to toll-like receptor-4 complex, stimulating the production of TNF-α through the activation of nuclear factor-kappa B, favoring an inflammatory environment. Moreover, the excess of fat drives the polarization state of this increased number of macrophages from anti-inflammatory M2 to proinflammatory M1 macrophages and prompts fat accumulation in the liver and IR[8]. Adipose-derived macrophages also secrete inflammatory molecules, like TNF-α and IL-6, and adipokines, such as visfatin [also named nicotinamide phosphoribosyl transferase (NAMPT)]. NAMPT has gained relevance as a pivotal molecule linking adipose tissue and FLD. NAMPT is a pleiotropic molecule that can be found in an extracellular (eNAMPT) or an intracellular (iNAMP) form. Studies indicate that eNAMPT has enzyme and cytokine-like activity, stimulating the release of proinflammatory cytokines. Meanwhile, iNAMPT catalyzes the rate-limiting step in nicotinamide adenine dinucleotide (NAD+) formation. Because of this NAD+ boosting property, levels of iNAMPT have been proposed as beneficial for the homeostasis of the cell due to influencing the activity of NAD-dependent enzymes, such as sirtuins (SIRT). Remarkably, SIRT1 plays a key role in the liver by modulating the acetylation status of target molecules in lipid metabolism[27].
Furthermore, IR is characterized by hyperglycemia and the subsequent hyperinsulinemia to counteract high glucose levels, being a risk factor for NCDs, particularly type 2 diabetes, where it has been closely linked to oxidative stress[28]. A normal insulin signaling pathway starts with the activation of the insulin receptor so that it can bind to phosphoinositide 3-kinase to ultimately activate protein kinase B (Akt). Activated Akt drives glucose entry into the cell by promoting GLUT4 expression and glycogen synthesis[29]. Oxidative stress impairs this signal transduction through many different mechanisms, like inhibiting the transcription factors insulin promoter factor 1 and peroxisome proliferator-activated receptor gamma, which mediate insulin and GLUT-4 expression, respectively. Moreover, under hyperglycemic conditions, fetuin A hepatokine inhibits the insulin receptor and promotes inflammation, while FGF-21 inhibits lipid accumulation and increases insulin sensitivity. Dysregulation of this hormones, together with oxidative stress imbalance, lead to impaired insulin signaling[30].
The metabolic conditions underlying the development of NCD are complex, and they often reinforce each other, perpetuating an inflammatory environment and oxidative stress imbalance. As the orchestrating organ, these processes converge in the liver, affecting metabolic functions and setting the basis for the onset of the fibrogenic process characteristic of CLD.
Persistent virus-associated liver damage can progress to CLD, which pressures health systems with a big social and economic burden. Although lots of resources have been invested to study the molecular mechanisms that mediate this process, results are diverse and still under investigation by the scientific community. HHVs directly infect hepatocytes, and the internalization into the cell is believed to happen by endocytosis, requiring the interaction with several host cell factors[17]. However, viral entry of HBV and HCV within hepatocytes is unclear, and further research is needed to elucidate this question. Sodium taurocholate co-transporting polypeptide was recently identified as an HBV receptor that would mediate HBV cell entry[31]. In the case of HCV, specific intercellular adhesion molecules appear key to cell adhesion and subsequent internalization[32].
Regarding HBV and HCV replication, it has been found that liver X receptor-α (LXR-α) plays a key role. LXR-α is a transcription factor whose activation triggers the expression of different genes that directly or indirectly modulate these viruses’ replication as well as the lipid and inflammatory alterations associated to CLD[33]. This inflammation is also mediated by the nucleotide-binding oligomerization domain-like receptor protein 3, which is activated by the abnormal production of ROS after a viral infection occurs in the liver. This ROS increase is associated with a decreased expression of nuclear factor-e2-related factor-2, a transcription factor that regulates ROS/recepteur d’origine nantais balance by maintaining redox homeostasis. These alterations compromise the normal state of the cell, laying the foundations on which the fibrotic process of CLD begins[11].
In the case of COVID-19, the mechanisms by which liver damage can occur are more unclear, but it is widely accepted that inflammation plays a huge role. This infection can trigger an exaggerated immune response leading to an uncontrolled cytokine release, also known as a “cytokine storm”. It is characterized by abnormal levels of IL-6, IL-1, C-C motif chemokine ligand (CCL)-5, chemokine (C-X-C motif) ligand (CXCL)-8, CXCL-1, and TNF-α among others[19]. This inflammatory cascade affects bile duct function since cytokines like TNF-α, IL-1, and IL-6, can induce hepatocellular cholestasis by downregulating hepatobiliary uptake and excretory systems[34].
Furthermore, the presence of this inflammatory environment can upregulate the expression of angiotensin converting enzyme 2 (ACE2) receptor in different tissues, like the adipose tissue and the liver[35-39]. This is of relevance since ACE2 receptors are the main cell entrance of the SARS-CoV-2 virus, and they are present in different tissues. Particularly in the liver, the cholangiocytes (characteristic cells of the bile duct)[40], as well as liver vascular endothelial cells[41], express ACE2 receptors. Hepatocytes and cholangiocytes are permissive to the SARS-CoV-2 virus, mediating subsequent entrance into the liver[42]. Several studies have found that ACE2 expression in hepatocytes is increased under hypoxia[43], a frequent condition in COVID-19 patients, and fibrotic conditions[44]. Besides ACE2 receptors, transmembrane serine protease 2 (TMPRSS2) and paired basic amino acid cleaving enzyme (FURIN) have been noted as significant for infection in the liver[45,46]. In this context, ACE2 expression is increased in patients in HCV-related cirrhosis[44], whereas TMPRSS2 and FURIN expression are upregulated in patients with obesity and NAFLD[47]. Moreover, infection by SARS-CoV-2 increases glucose-regulated protein 78 and 94, two biomarkers of ER stress[48,49], and impairs mitochondrial function[50]. This process is of interest since this state has been associated with de novo lipogenesis in hepatocytes[51], which could eventually lead to steatosis in these patients.
The use of therapeutic drugs can be another underlying cause of liver damage[3]. Because of detoxifying functions, the liver is subject to drug-induced damage coming from a wide range of approved drugs. Oncology drugs account for most hepatotoxicity cases, followed by those used for infectious diseases[3]. Since the beginning of the COVID-19 pandemic, a wide range of different treatments (antivirals, antibiotics, antimalaria, or corticosteroids) have been used in the absence of an efficient drug to treat severe infections. This pharmacological administration could explain that drug-induced liver injury appears in nearly 25% of COVID-19 patients[23], a consequence to consider when addressing liver damage in this disease.
Despite all the advances in the mechanisms driving the onset of these diseases, new techniques to detect innovative biomarkers for diagnosis and prognosis as well as to discover novel drugs are needed, for example artificial intelligence (AI). AI seeks to mimic human behavior, and within this science, ML is the most common approach[52]. The advances in computational science in the last decades have permitted the development of powerful algorithms based on this science. ML algorithms are particularly relevant for biological research because they allow the processing and integration of the huge amount of data that the latest advances in this field have brought by applying statistical methods to enable machines to improve experiences. This methodological approach can be categorized into two big groups: Supervised and unsupervised learning. In supervised algorithms, data is tagged in order to train the algorithm and fit it appropriately, whereas if it is unsupervised, the algorithm learns patterns from unlabeled data[53]. ML algorithms are generally assessed by simple methodologies like sensitivity, specificity, and accuracy. While sensitivity evaluates the proportion of true positives correctly identified, specificity evaluates the proportion of true negatives. Meanwhile, the accuracy value indicates the number of times the model is correct[54].
Supervised algorithms can be divided into two categories depending on the purpose: Prediction, in which the algorithm is fed and trained predictive models to data; or classification, which consists in clustering data within explanatory groups[55,56]. Predictive algorithms are based on regression models, and the most used are linear and logistic regression (LR), support vector machine (SVM), support vector regression (SVR), extra tree regression (ETR), artificial neural networks (ANN), and decision trees (DT). Regression models analyze the influence of one or multiple variables on a nominal or ordinal categorical outcome. ANN are more complex mathematical models (deep learning algorithms) that mimic the brain neural network, like the convolutional neural network (CNN), in which an input is fed through a hidden layer of many different well connected and structured nodes to produce a final output. In deep neuronal network (DNN) models, a great number of hidden successive layers use the output from the previous layer as input in a more complex algorithm. DT can also classify data, like random forest (RF) or gradient boosting (GB) models. Instead of minimizing error, these models determine thresholds derived from input data, assigning weight values to variables. Other models of classification are the Ada-Boost, Bayesian network (BN), Naïve Bayes (NB), K-Nearest Neighbors (KNN), and linear discriminant analysis (LDA) that group data into clusters[55,56]. All these models can shed light into biological questions and are normally used indistinctively to obtain the best performance with the same dataset. For instance, Mijwil and Aggarwal[57] analyzed and compared 7 ML algorithms to predict appendix illness in the same dataset, revealing that certain models performed better than others, allowing for higher accuracy and results.
In FLD, the common techniques used in diagnostics are based on techniques like ultrasonography and magnetic resonance imagining (MRI). These methods are subjective, and the informed outcome mainly relies on the interpretation of the professional carrying out the procedure. Several investigations have studied the implementation of ML in order to classify FLD and other liver diseases by using images from ultrasounds, computed tomography (CT), and MRI[58,59]. However, the downside of this approach is that the quality of the images differs from one another because of several factors, such as equipment precision and interpersonal differences, for instance. Therefore, there is a need for ML approaches to help in image segmentation, and some authors have already implemented this technique to improve clinical practice[60,61].
Moreover, ML can help with the integration of more complex information beyond imaging to study and diagnose liver diseases since patients with CLD in the developmental phase require frequent follow-ups to check the progress of the disease and early detection changes in the diagnosis[58]. For example, patients with HHV-induced CLD are normally on antivirals. However, there is no consensus or guidelines about when to stop antiviral therapy or even if quitting these drugs will increase HCC risk. Therefore, new approaches need to be established to classify and prevent the development of more severe illnesses, like cirrhosis or cancer. In this line, ML approaches can be used to measure liver fibrosis, optimize diagnosis, and predict disease progression of CLD[62]. Table 1 summarizes selected studies that have used ML for these purposes, which have been collected for this review, and Table 2 summarizes the most repeated inputs from all compiled ML models along with the most repeated predictive results for the main four inflammation-related liver conditions.
| Ref. | Objective | Subjects | Variables | ML model | Performance | Observations/remarks |
| Fialoke et al[63] | To predict NASH in NAFLD patients | n = 108139, NASH and healthy (non-NASH) populations | Demographic data, type 2 diabetes status, and blood biomarkers | RF, XGBoosting, DT, LR | AUROC of 88% by XGBoosting | The average and maximum value of ALT appeared was the most important variable |
| Ma et al[64] | To predict NAFLD in the general population | n = 10508, Subjects who attended a health examination | Age, blood biomarkers, and anthropometric data | LR, RF, SVM, baggin, DT, LR, KNN, BN, hidden NB, AdaBoosting, AODE | 83% accuracy, 0.878 specificity, 0.675 sensitivity, and 0.655 F-measure score by BN | BMI, TG, GGT, ALT and uric acid were the top five predictors |
| Yip et al[65] | To detect NAFLD for the general population | n = 500, involving NAFLD patients and healthy subjects | Demographic, clinical data and blood biomarkers | LR, RIDGE regression, AdaBoosting, DT | AUROC of 90% by AdaBoosting | ALT, HDL-c, TG, HbA1c and white blood cells to predictors |
| Pei et al[66] | To identify FLD in general patients | n = 3419, patients of which 845 had FLD | Age, anthropometric, and blood biomarkers | RF, ANN, KNN, XGBoosting, LDA | 0.9415 accuracy, 0.9306 AUC, and 0.9091 sensitivity by XGBoosting | Uric acid, BMI, and TG were the top three risk factors |
| Choi et al[67] | To stage liver fibrosis | n = 7461, patients with pathologically confirmed liver fibrosis | Age, sex, clinical data, CT images, and liver fibrosis stage | CNN | Overall staging accuracy of 79.4% and an AUROC of 0.96, 0.97, and 0.95 for diagnosing significant and advanced fibrosis, and cirrhosis, respectively | The model outperformed the radiologist’s interpretation, APRI, and FIB-4 index |
| Chen et al[68] | To stage liver fibrosis in patients with CHB | n = 513, patients with confirmed liver fibrosis | Age, sex, CT liver images | RF, KNN, SVM, NB | 0.8118-0.9125 accuracy by RF for all stages | The adopted classifiers significantly outperformed the liver fibrosis index method |
| Jeong et al[69] | To classify susceptible individuals for adjuvant treatment in patients with ICC after resection | n = 1421, ICC patients | Age, sex, clinical data, and blood biomarkers | DNN | AUC of 0.78 | The model was found to be more accurate than the traditional AJCC stage classifier |
| Wübbolding et al[70] | To identify immune profiles for the prediction of early virological relapse | n = 284, patients with CHB and treated with NA antivirals | Age, sex, and analytical and blood biomarkers | KNN, RF, LR | AUC of 0.89 | The combination of IL-2, MIG/CCL9, RANTES/CCL5, SCF, and TRAIL was reliable in predicting viral relapse |
| Hong et al[71] | To predict esophageal varices in patients with HBV related cirrhosis | n = 197, patients with HBV-related cirrhosis | PLT count, spleen width, and portal vein diameter | ANN | Sensitivity of 96.5%, specificity of 60.4%, accuracy of 86.8% | The model obtained a positive predictive value of 90.00%; and a negative predictive value of 80.85% |
| Zhong et al[72] | To compare the prognostic performance of ALBI and CTP grades for HCC treated with TACE combined with sorafenib as an initial treatment | n = 504, HCC patients | ALBI and CTP grades BCLC stage, clinical data and plasma α-fetoprotein | ANN | - | The ALBI grade had higher importance in survival prediction compared to the CTP one |
| Shi et al[73] | To predict in-hospital mortality after primary liver cancer surgery | n = 22926, HCC surgery patients | Age, sex, clinical, and hospital data | ANN, LR | 97.28% of accuracy and 84.67 % of AUROC by ANN | ANN model had higher overall performance indices and accurately predicted in-hospital mortality |
| Shi et al[74] | To predict 5-yr mortality after surgery for HCC | n = 22926, HCC surgery patients | Age, sex, clinical, and hospital data | ANN, LR | 96.57 % of accuracy and 88.51 % of AUROC by ANN | Surgeon volume was the top predictor parameter |
| Patnaik et al[75] | To predict liver function-related scores (MELD, APRI, CTP) using breath biomarkers | n = 28, healthy patients compared to n = 17, liver patients | Age, anthropometric data, blood biomarkers, breath analysis | LR, RF, SVR, ETR | R2 values of 0.78, 0.82, and 0.85 for CTP score, APRI score, and MELD, respectively, by ETR | Isoprene, limonene and dimethyl sulfide can be potential biomarkers for liver disease |
| Butt et al[85] | To diagnose the stage of hepatitis C | n = 968, patients with HCV | Age, anthropometric data, blood biomarkers, and histological staging | ANN, RF, SVM, XGBoosting | 98.89% precision by ANN | The model performed better than previously presented models by other authors |
| Wei et al[87] | To predict HBV and HCV-related hepatic fibrosis | n = 490, HBV patients; n = 254, and 230 HCV patients | Age, BMI, analytical data (FIB-4 score), and liver biopsy | GB, DT, RF | AUROC of 0.918 by GB | GB outperformed the FIB-4 predictive score |
| Barakat et al[89] | To predict and stage hepatic fibrosis in children with HCV | n = 166, children with CHC | Analytical data (APRI and FIB-4 scores) | RF | AUCs of 0.903 for any type of fibrosis | RF outperformed FIB-4 and APRI predictive score |
| Konerman et al[88] | To predict progression of HCV | n = 72683, veterans with CHC | Age, BMI, demographic, and blood biomarkers (APRI score) | CS and LGT Cox and boosting | AUROC of 0.830 and 0.77 sensitivity by LGT boosting model for 1 yr follow-up | APRI and PLT count were top predictors in the LGT boosting model |
| Wong et al[86] | To predict HCC in patients with CVH | n = 86804, CHV patients, of which 6821 with HCC | Age, sex, clinical data, and blood biomarkers | LR, RIDGE regression, AdaBoosting, RF, DT | AUROC of 0.992 and 0.837 by RF in training and validation cohort, respectively | ML models obtained better AUROCs than HCC traditional risk scores |
| Feldman et al[91] | To predict DAA therapy duration in hepatitis C | n = 3943, HCV patients with sofosbuvir/ledipasvir as the first course of DAA, of which n = 240, received the prolonged DAA treatment | Age, sex, and clinical data (including hepatitis C record data) | XGBoosting, RF, SVM | AUC of 0.745 by XGBoosting | Results showed age, comorbidity burden, and type 2 diabetes status as new predictors for DAA therapy duration |
| Kamboj et al[92] | To predict repurposed drugs for HCV | n = 17968, HCV molecular fingerprints | Experimentally validated small molecules from the ChEMBL database with bioactivity against HCV NS3, NS3/A4, NS5A and NS5B proteins | SVM, ANN, KNN, RF | R2 value of 0.92 by SVM | Results identified more than 8 repurposed treatments anti-HCV |
| Tian et al[93] | To predict HBsAg seroclearance | n = 2235, patients with CHB, of which 106 achieved HBsAg seroclearance | Age, BMI, demographic and clinical data, and blood biomarkers | LR, RF, DT, XGBoosting | AUC of 0.891 by XGBoosting | Level of HBsAg followed by age and HBV DNA were the top predictors |
| Chen et al[94] | To predict HBV-induced HCC using quasispecies patterns of HBV | n = 307, CHB patients; n = 237, HBV-related HCC patients | rt nucleic acid and rt/s amino acid sequences | SVM, RF, KNN, LR | AUC of 0.96, and accuracy of 0.90 by RF | HBV rt gene features can efficiently discriminate HCC from CHB |
| Mueller-Breckenridge et al[95] | To classify HBeAg status in HBV patients using virus full-length genome quasispecies | n = 352, CHB untreated patients | Matrix of allele frequencies (0.1-0.99) and the associated HBeAg status | RF | Range balanced accuracy of 0.8-1 | n1896GA, n1934AT, n1753TC mutants were the highest-ranking variables |
| Kayvanjoo et al[96] | To predict HCV interferon/ribavirin therapy outcome based on viral nucleotide attributes | n = 76, gene attributes | HCV nucleotide attributes | DT, SVM, NB, DNN | Accuracy of 84.17% by SVM in responder vs relapser of subtype 1b sequences | Dinucleotides UA and UU were top predictors in the combination treatment outcome |
| Li et al[98] | To distinguish influenza from COVID-19 patients | n = 398, COVID-19 and influenza cases | Age, sex, blood biomarkers, clinical data, and CT and X-ray scans | XGBoosting, RF, and LASSO and RIDGE regression models | AUC of 0.990, sensitivity of 92.5% and a specificity of 97.9% by XGBoosting | Age, CT scan result, and temperature were top three predictors |
| Bhargava et al[99] | To detect novel COVID-19 and discriminate between pneumonia | n = 31454, images acquired from nine distinct datasets of COVID-19 patients | CT or X-ray scans | KNN, SRC, ANN, SVM | 99.14 of accuracy by SVM | SVM model classified with the highest recognition rate the images as normal, pneumonia, and COVID-19 positive |
| Bennett et al[97] | To predict early severity and clinically characterize COVID-19 patients | n = 174568, patients with a positive lab test for COVID-19 | Age, sex, demographic, anthropometric and clinical data, and blood biomarkers | RF, LR, XGBoosting | AUROC of 0.87 by XGBoosting | Age, oxygen respiratory rate, and blood urea nitrogen were ranked as top predictor for severity outcome |
| Günster et al[100] | To identify independent risk factors for 180-d all-cause mortality in COVID-19 patients | n = 8679, hospitalized COVID-19 patients | Age, sex, BMI, and clinical data | LR | AUC of 0.81 | A high BMI and age were strong risk factors for 180-d all-cause mortality, while female sex was protective |
| Deng et al[101] | To identify clinical indicators for COVID-19 | n = 379, patients, 62 with COVID-19 and 317 with pneumonia | Age, sex, demographic and clinical data, and blood biomarkers | EBM | AUC of 0.948 | Variables grouped under liver function was top the predictor category for COVID-19 prediction |
| Lipták et al[102] | To identify gastrointestinal predictors for the risk of COVID-19-related hospitalization | n = 680, patients | Age, sex, clinical data, and blood biomarkers | RF | AUC of 0.799 | AST was top predictor for hospitalization |
| Elemam et al[103] | To identify immunological and clinical predictors of COVID-19 severity and sequelae | n = 37, COVID-19 patients; n = 40, controls | Age, sex, BMI, clinical data, and blood biomarkers | Stepwise linear regression | AUC of 0.93 for cytokines as predictors. AUC of 0.98 for biochemical markers as predictors | IL-6 and granzyme B were top potential predictors of liver injury in COVID-19 patients |
| Mashraqi et al[104] | To predict adverse effects on liver functions of COVID-19 ICU patients | n = 140, COVID-19 patients admitted to ICU | Blood biomarkers and existence of liver damage | SVM, KNN, ANN, NB, DT | AUC of 0.857 and precision of 0.95 by SVM | AST and ALT were top predictors of liver damage in these patients |
| Soltan et al[106] | To evaluate a laboratory-free COVID-19 triage for emergency care | n = 114957, emergency presentations prior to the global COVID-19 pandemic and n = 437, COVID-19 positive | Blood biomarkers, blood gas, and vital signs | LR, XGBoosting, RF | AUROC range of 0.9-0.94 by XGBoosting for datasets | The model could effectively triage patients presenting to hospital for COVID-19 without lab results |
| Gao et al[111] | To predict mortality in patients with alcoholic hepatitis | n = 210, alcoholic hepatitis patients | Age, clinical data, blood biomarkers, and omics data sets (metagenomics, lipidomics, and metabolomics) | GB, LR, SVM, RF | AUC of 0.87 by GB for 30-d mortality prediction using the dataset combining clinical data, bacteria and MetaCyc pathways and for and 90-d mortality prediction using the fungi dataset | The model performed better than the currently used MELD score |
| Inflammatory-related liver condition | Inputs | Most repeated predictors |
| FLD | Age, sex, blood biomarkers, and demographic, anthropometric, and clinical data | BMI, uric acid, TG, and ALT levels |
| Liver fibrosis | Age, sex, and CT images | Better diagnosis compared to classical methods like APRI and FIB-4 indexes |
| Virus-induced hepatitis | Age, sex, blood biomarkers, and demographic, anthropometric, and clinical data | AST, PLT levels, APRI index, and age |
| COVID-19 | Age, sex, blood biomarkers, CT images, and demographic, anthropometric, and clinical data | Age, BMI, CT images, oxygen rate, AST, and ALT levels |
In recent years, promising results have been found when applying ML approaches in CLD. Regarding prevention, Fialoke et al[63] screened 108139 patients to identify those diagnosed with benign steatosis and NASH, a type of NAFLD, train ML classifiers for NASH and healthy (non-NASH) populations, and predict NASH disease status on patients diagnosed with NAFLD according to aspartate transaminase (AST), alanine transaminase (ALT), and platelet (PLT) levels. In this line, another study detected body mass index (BMI), triglycerides (TG), gamma-glutamyl transpeptidase (GGT), ALT, and uric acid as the top 5 features contributing to NAFLD, with the BN model performing the best[64]. Accordingly, Yip et al[65] selected TG, ALT, white blood cell count, high-density lipoprotein cholesterol (HDL-c), glycated hemoglobin A1c (HbA1c), and the presence of hypertension as the six variables to build ML models, of which Ada-Boost outperformed the others individually and described the NAFLD status in 922 subjects.
More recently, Pei et al[66] designed a ML model that integrated medical records as a clinical variable to classify FLD. Concretely, they selected the variables of age, height, BMI, hemoglobin, AST, glucose, uric acid, low-density lipoprotein cholesterol (LDL-c), alpha-fetoprotein, TG, HLD-c, and carcinoembryonic antigen. They tested six different ML models in 3419 participants, of which 845 were diagnosed with FLD: LR, RF, ANN, KNN, extreme gradient boosting (XGBoost) (a type of GB model), and LDA. Results from these authors showed that the XGBoost model had the highest performance, followed by LR and ANN, to predict the risk of FLD. BMI, uric acid, and TG levels were the top three variables associated to FLD risk across the six analyzed models.
When it comes to diagnosis and treatment, several ML models have been tested for different purposes obtaining good specificity, sensitivity, and accuracy values[62]. For example, to determine the stage of liver fibrosis, some authors have used CT images processed by segmentation algorithms. Choi et al[67] used CNN upon CT images, whereas Chen et al[68] employed RF, KNN, SVM, and the NB classifiers with real-time tissue elastography imaging, age, and sex as feeding variables. In both cases, the ML approach outperformed the classical methods. Regarding treatment, different ML models have been used to define the best therapy for liver diseases such as carcinomas and virus-induced hepatitis. Jeong et al[69] used DNN to classify intrahepatic cholangiocarcinoma susceptible to adjuvant therapy following resection according to laboratory and clinicopathological markers and found it more accurate than the commonly used staging system.
Wübbolding et al[70] studied the prediction of early virological relapse analyzing soluble immune markers using supervised ML approaches like KNN, RF, and LR. This study showed that IL-2, monokine induced by interferon γ/CCL9, RANTES/CCL5, stem cell factor, and TNF-related apoptosis-inducing ligand in combination were more reliable in predicting virological relapse than viral antigens. In the same way, researchers have used ML classifiers to explore new methods able to better predict prognosis of liver diseases[71-74]. The weighted variables are usually CT images and/or biochemical parameters that involved invasive and costly methods. However, researchers have recently proposed volatile organic compounds as new biomarkers for progression and prognosis of liver disease. These researchers monitored isoprene, limonene, and dimethyl sulfide concentrations from a breath sample in liver patients compared to healthy subjects. They used regression ML models (LR, ETR, SVR, and RF) to demonstrate that these approaches together with breath profile data can predict clinical scores of liver disease[75]. These findings are promising and open the way for new, safe, and non-invasive approaches to study liver function and for diagnosis purposes.
ML methods have been employed when studying the comorbidities of liver-related diseases, like obesity, diabetes, and cardiovascular diseases[53,55,76]. For example, ML algorithms have been built to study the risk factors associated to overweight and obesity development, showing that BMI, age, dietary pattern, blood test results, socioeconomic status, and sedentarism were key factors when studying excess of adipose tissue[77]. In this line, further research has revealed by ML techniques that the minutes devoted to physical activity in one week[78], as well as specific species of gut microbiota[79], are also crucial for obesity prediction. ML algorithms have also elucidated the risk factors of childhood obesity, of which parental BMI and the upbringing environment play a huge role[80-82]. Furthermore, researchers have observed by training a multivariate LR model with a dataset of 3634 children and adolescents’ vitamin intake that vitamins A, D, B1, B2, and B12 were associated in a negative manner with obesity in this cohort[83]. These results are of interest since new insights are needed to discover novel targets to tackle comorbidities that affect liver function.
HBV and HCV infections can dangerously become chronic if not treated early and with the right treatment[84]. While scientists are still relentlessly working on an effective vaccine against HCV, a good and efficient diagnosis is key to prevent chronic HCV infection (CHC), and ML algorithms have been elucidated for this purpose. Thus, Butt et al[85] designed an ANN model and trained it with a dataset of 19 variables, among which age, sex, BMI, transaminase, and PLT count levels were included. The algorithm was able to better identify the stage of hepatitis C compared to other XGBoost, RF, and SVM models tested by other researchers with a higher precision rate and a decreased miss rate.
ML algorithms have been applied and compared to traditional methods used to follow HHV-induced advanced liver disease[86-88]. For instance, Wei et al[87] used a GB model trained with the same variables that the formula fibrosis-4 (FIB-4) uses, which are age, AST, ALT and PLT levels in a cohort of 490 HBV patients, and two cohorts of HCV patients (n = 240 each). The GB model outperformed FIB-4 score in classifying hepatic fibrosis and the existence of cirrhosis. Barakat et al[89] designed an RF model that also outperformed the FIB-4 score, as well as the AST/PLT ratio index (APRI), for prediction and staging of fibrosis in children with hepatitis C. In this line, data of 72683 veterans with CHC were used to predict the progression of the disease. GB models were used and compared with cross-sectional or linear models fed with variables like transaminases levels, alkaline phosphatase (ALP), PLT, AST, APRI, albumin, bilirubin, glucose, white blood cells, and BMI were included in the dataset. Results showed that APRI, PLT, AST, albumin, and AST/ALT ratio were the best predictors for featuring CHC progression[88].
Regarding therapy, CHC can be effectively treated with direct-acting antiviral (DAA) therapy, a novel treatment that targets viral non-structural proteins. Although it has null side effects compared to standard treatment, it has some downsides. Treatment failure in a low percentage of the cases, a very high cost, and no treatment duration established[90]. New methods to define this therapy duration are needed to optimize adherence and success. Feldman et al[91] studied the prediction of DAA treatment duration in hepatitis C patients using XGBoost, RF, and SVM models. They used the dataset of 240 patients with prolonged first course of DAA against another dataset of 3478 patients on standard duration. Age, sex, comorbidities, and previous hepatitis C treatment record were considered. The predictive model constructed with XGBoost obtained the best performance in predicting prolonged DAA treatment, in which the presence of cirrhosis, type 2 diabetes, age, HCC, and previous standard treatment were the most determining variables. Meanwhile, Kamboj et al[92] used ML approaches in the search of repurposed drugs that could target non-structural proteins, developing regression-based algorithms able to identify inhibitors of these proteins, and proposing new drugs to test in CHC.
A huge milestone when treating chronic HBV infection (CHB) is seroclearance of HBV surface antigen (HBsAg)[84]. It has been demonstrated that seroclearance of HBsAg is associated to a better prognosis in CHB. Some authors used ML models to predict HBsAg seroclearance in a cohort of 2235 patients, of which 106 achieved it. They used XGBoost, RF, and LR, among other models, and tested a total of 30 categorical and continuous variables, including sex, drinking history, initial diagnosis and treatment, age, BMI, and serum and radiological indicators. Results revealed that the XGBoost model showed the best predictive performance, indicating that HBsAg levels were the best predictor for HbsAg seroclearance, followed by age, and the DNA level of HBV[93].
Interestingly, ML has also contributed to personalized medicine in this field. HHVs evolve and adapt to different cellular environments in order to escape immune responses and drugs to survive. These adaptations rely on high mutagenetic activity, especially within the target genes of antivirals. Regarding HBV, Chen et al[94] used ML to identify patients with HCC or CHB based solely on genetic differences and found that the RF model impressively discriminated both cases based on the rt gene sequence of HBV. Moreover, Mueller-Breckenridge et al[95] ultra-deep sequenced 400 HBV samples and used an RF model to classify the status of a particular HBsAg according to the novel viral variants encountered. Results showed five genotypes that could benefit from personalized healthcare. In the case of HCV, Kayvanjoo et al[96] built several ML algorithms and trained them with two datasets of responders vs non-responders of antiviral therapy in HCV infection caused by two different strains. These investigations reported novel genetic markers that could predict therapy response with high accuracy. These results are very promising since they contribute to bringing personalized medicine to the public system.
A recent systematic review depicted that the parameters normally used for liver impairment screening were significantly increased in COVID-19 patients[23]. Particularly, several studies showed that levels of AST and/or ALT can increase in these patients up to 20%, bilirubin up to 14%, ALP up to 6%, and GGT levels up to 21%. Prothrombin is a protein synthesized in the liver that results in thrombin, a protein with a critical role in coagulation function. Prolonged prothrombin is a symptom of decreased production of coagulation factors, characteristic of liver disease. For this reason, the prolonged prothrombin time (PT) is another parameter usually checked when screening for liver injury, and it has been described that COVID-19 patients present nearly a 10% increase in PT[23]. Besides biochemical alterations, COVID-19 illness can lead to hypoxemia, impaired cardiac function, and secondary damage due to multiple organ dysfunction, which can result in liver injury in patients with or without prior liver disease. Therefore, new insights of the relationship between this recent infectious illness and liver disease are expected.
The use of ML approaches has been encouraged by the National COVID Cohort Collaborative Consortium for early detection, prediction, and follow-up of severe COVID-19 cases since the pandemic started[97]. For instance, some researchers used the XGBoost approach and found that age, CT scan result, body temperature, lymphocyte levels, fever, and coughing can classify influenza patients from COVID-19 patients[98]. Bhargava et al[99] tried different ML approaches to detect novel COVID-19 and discriminate between pneumonia using CT and X-ray scans as inputs. These authors pre-processed the images by normalization and then segmented them by fuzzy c-means clustering. Results showed that the SVM model was the one that better classified patients in COVID-19 positive, pneumonia, and healthy groups, obtaining a very high accuracy.
In this same line, obesity and liver disease were identified as risk factors for higher clinical severity in a cohort of 174568 adults with severe acute respiratory syndrome associated with SARS-CoV-2 infection by a multivariable LR model[97]. Interestingly, a German study of 8679 patients used an LR model and identified liver disease and BMI as determinant risk factors for 180-d all-cause mortality in hospitalized COVID-19 patients[100]. A case-control study with COVID-19 patients compared to patients with community-acquired pneumonia showed how, by applying a GB model, the category of liver function appeared as one of the top systematic predictors for COVID-19 risk factors, with albumin, total bilirubin, and ALT among the most important input variables[101]. Furthermore, a study with 710 enrolled patients diagnosed with COVID-19 identified AST levels as the top predictor for COVID-19-related hospitalization based on an RF algorithm, followed by age and diabetes mellitus[102].
A stepwise linear regression model identified IL-6 and granzyme B as potential predictors of liver dysfunction, characterized by an elevation in the levels of ALT and/or AST[103]. Other authors designed a model for detecting liver damage testing different ML approaches with laboratory parameters as the input variables. SVM was the model with the best accuracy, and AST and ALT levels were the variables with the best predictive scores[104]. In this context, the newest version of the CURIAL model was developed to identify COVID-19 patients using vital signs, blood gas, and laboratory blood tests. It showed greater sensitivity, making this model a potential emergency workflow[105,106]. All these ML-based methods would dramatically improve the time to diagnosis, free hospital laboratories and rooms of potential positive subjects, and reduce costs if implemented in the public health system.
AI has also been employed to discover potential efficient new drugs to tackle SARS-CoV-2 infection[107]. Baricitinib is a drug initially approved for rheumatoid arthritis that was selected by ML as a potential drug to treat COVID-19. Researchers proved the anti-inflammatory and antiviral properties of this drug in human liver spheroids infected with live SARS-CoV-2 to check any potential drug-induced liver injury[107]. Due to the good results, researchers moved on to a clinical trial where they tested baricitinib in a few COVID-19 patients. Levels of liver enzymes were not altered, except for a transient increase in liver aminotransferases in all patients that remitted in the following 72 h without interrupting treatment. The authors stated that this might be reflective of disease severity rather than a drug-induced injury, showing overall good tolerance and results in this pilot study[108]. In summary, ML approaches support liver biochemistry as a prognostic tool in COVID-19 disease.
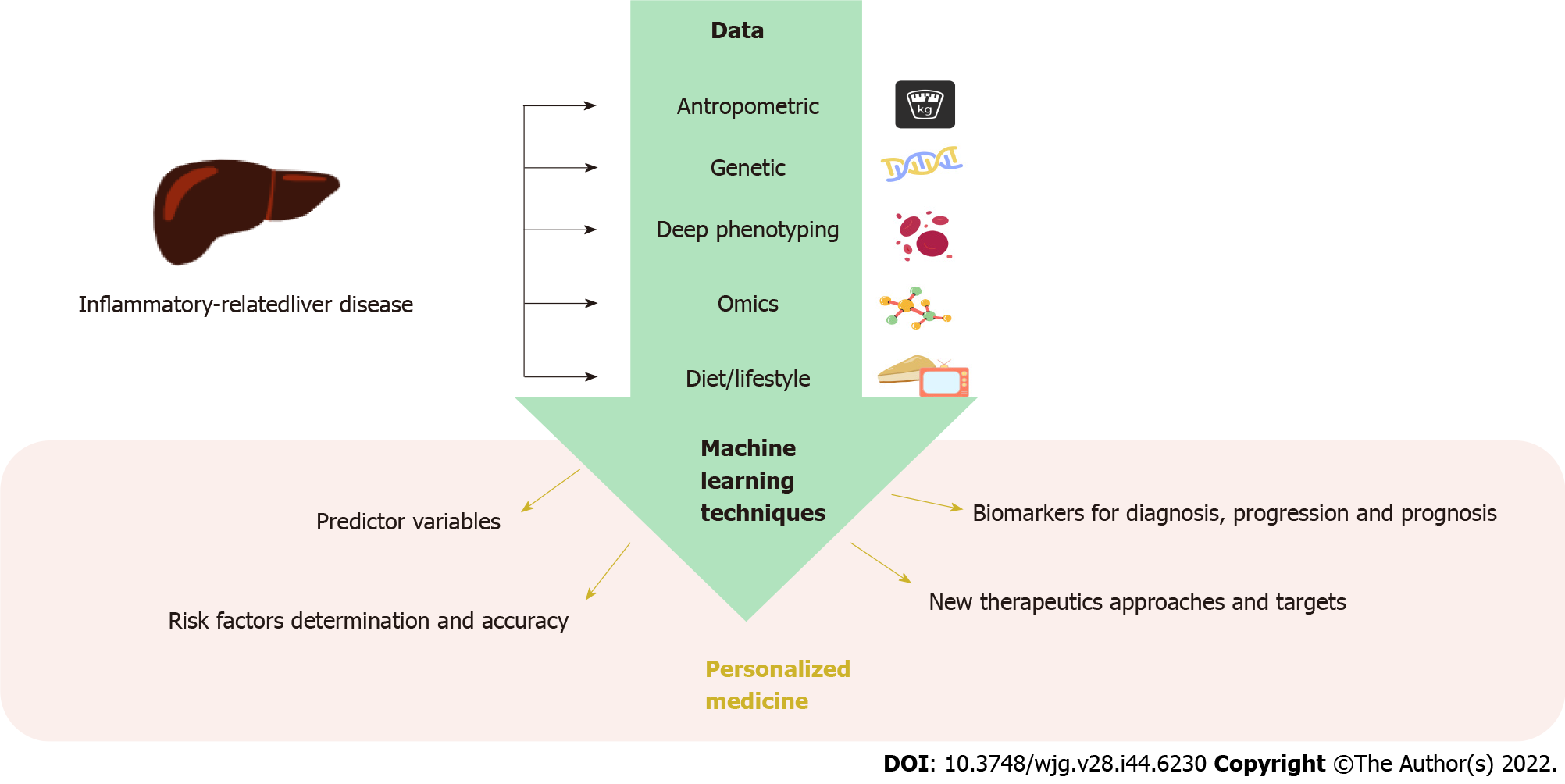
In the early 21st century, the Human Genome Project started the genomic era in which new disciplines like precision medicine appeared. Precision medicine aims to deliver targeted treatments based on a group of individual factors that greatly influence the onset and progression of a disease, like omics sciences. This approach covers a great number of patients, overcoming potential adverse effects and ensuring effectiveness of the treatment. In this context, computational advances have greatly contributed to the escalation of this science by lowering the costs of omics analysis and allowing the processing and integration of an enormous amount of data based on ML algorithms (Figure 2).
ML has permitted the development of diagnostics and therapeutics based on the integration of omics data (genomics, epigenomic, transcriptomics, proteomics, metabolomics, and metagenomics) with clinical data. The ultimate goal is to bridge these omics data with the phenotype to bring molecular accuracy to the diagnosis, treatment, prognosis, and recurrence process of a pathological condition. This methodology has been used in a wide range of diseases in the search for more efficient and effective approaches, like heart and liver diseases[109,110]. For example, ML algorithms fed with omics data have been able to predict mortality in patients with alcoholic hepatitis. In this study, routine clinical variables of 210 patients with this disease were used to build six different datasets to assess mortality at 30 d and 90 d. Five different ML models were tested, obtaining the best performance in predicting 30-d mortality with a GB model using bacteria, MetaCyc pathways, and clinical data, as well as LR using viral and clinical data[111].
In hepatitis B, it has been found that ML algorithms can be very useful in assessing HBV-associated HCC progression. Ye et al[112] analyzed 67 HBV-positive HCC samples with or without intrahepatic metastases and discovered key genes for metastatic progression and survival training ML models. The majority of them were inflammatory or related to the inflammation process, like IL-2 receptor and osteopontin, which encodes an extracellular cytokine ligand whose overexpression favors metastasis. These authors were able for the first time to draw a molecular signature useful to classify metastatic HBV-HCC patients, opening the way for early detection and new treatments to increase patient survival. In hepatitis C, the CC and CT genotypes of the rs12979860 polymorphism in the IL28B gene have been associated with liver fibrosis progression, being able to predict antiviral treatment effectiveness[113].
Moreover, ML algorithms have allowed the diagnosis of advanced liver fibrosis according to the rs12979860 genotype with higher performance compared to APRI and FIB-4 scores[114]. In this study, patients were divided into two groups according to HCV-related liver fibrosis stage: None to moderate fibrosis (n = 204); or with advanced fibrosis (n = 223). ML algorithms revealed the IL28B genotype as the first predictor, while the second predictor depended on the mentioned genotype. For instance, in CT patients, PLT, albumin, and age were the determining variables, while for patients with the TT genotype, white blood cell count was the decisive feature to assess advanced fibrosis probability.
ML approaches have also helped to categorize obesity in different subtypes based on metabolic status[115-117]. For example, Masi et al[115] studied a cohort of 2567 subjects suffering from obesity and made clusters of metabolically healthy or metabolically unhealthy patients based on clinical and biochemical variables using two ML models. The first model showed that IR, body fat, HbA1c, red blood cells, age, ALT, uric acid, white blood cells, insulin growth factor-1, and GGT were the top predictors of a metabolically healthy obesity, revealing the importance of liver function.
Other authors have also used ML models to classify 882 obese patients in subtypes of obesity according to glucose, insulin, and uric acid levels[116]. Results showed four stable metabolic clusters in this cohort, which were characterized by a healthy metabolic status, or by hyperuricemia, hyperinsulinemia, and hyperglycemia, respectively. Furthermore, Lee et al[117] explored three-way interactions between genome, epigenome, and dietary/lifestyle factors using GB and RF models in a subset (n = 394) of the exam 8 of the Framingham Offspring Study cohort. Interestingly, GB obtained the best performance, revealing 21 single nucleotide polymorphisms, 230 methylation sites in relevant genes (like CPT1A, ABCG1, and SREBF1), and 26 dietary factors as top predictors for obesity. Intake of processed meat, artificially sweetened beverages, French fries, and alcohol intake, among other dietary factors, were highly associated with overweight/obesity.
Personalized and precision medicine aims to harmonize the greatest number of factors so that diagnosis, prognosis, and treatment are based on the greatest number of decision elements. Much remains to be investigated to establish guidelines in the context of personalized medicine. However, it is safe to say that precision medicine will drive modern medicine, combining the most classic variables with the newest digital variables. Health professionals must be prepared to understand and implement these new technologies in the near future.
In summary, ML science can process and integrate a vast amount of different data with unprecedented outstanding performance. The objective of this article was to collect the information derived from ML techniques in liver damage induced by inflammatory conditions, including the new disease COVID-19. The main role of ML in liver pathologies is to help identify high risk patients for referral to specialized centers. Results show that the use of ML models have brought new insights into biology and medicine questions that can be very useful in determining the next directions for research in diagnosis, prognosis, and treatment of inflammatory and virus-related liver diseases, leading the way to personalized medicine. Also inflammation/IR biomarkers related to liver disease can be boosted by ML strategies. This review clarified and compiled the importance of the different factors involved in CLD and analyzed by ML algorithms, which can be useful information for clinicians, like endocrinologists and gastroenterologists, and other healthcare professionals with a focus on hepatology and bioinformatics.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Mexico
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu XQ, China; Mijwil MM, Iraq; Özlem Ş, Turkey S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ
| 1. | Wang F, So KF, Xiao J, Wang H. Organ-organ communication: The liver's perspective. Theranostics. 2021;11:3317-3330. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 2. | Eslam M, Sanyal AJ, George J; International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology. 2020;158:1999-2014.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1188] [Cited by in F6Publishing: 1576] [Article Influence: 394.0] [Reference Citation Analysis (1)] |
| 3. | Clinton JW, Kiparizoska S, Aggarwal S, Woo S, Davis W, Lewis JH. Drug-Induced Liver Injury: Highlights and Controversies in the Recent Literature. Drug Saf. 2021;44:1125-1149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 30] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 4. | Paik JM, Golabi P, Younossi Y, Mishra A, Younossi ZM. Changes in the Global Burden of Chronic Liver Diseases From 2012 to 2017: The Growing Impact of NAFLD. Hepatology. 2020;72:1605-1616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 336] [Cited by in F6Publishing: 361] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 5. | Tanwar S, Rhodes F, Srivastava A, Trembling PM, Rosenberg WM. Inflammation and fibrosis in chronic liver diseases including non-alcoholic fatty liver disease and hepatitis C. World J Gastroenterol. 2020;26:109-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 78] [Cited by in F6Publishing: 93] [Article Influence: 23.3] [Reference Citation Analysis (2)] |
| 6. | Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, Ferrucci L, Gilroy DW, Fasano A, Miller GW, Miller AH, Mantovani A, Weyand CM, Barzilai N, Goronzy JJ, Rando TA, Effros RB, Lucia A, Kleinstreuer N, Slavich GM. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25:1822-1832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1084] [Cited by in F6Publishing: 1867] [Article Influence: 373.4] [Reference Citation Analysis (0)] |
| 7. | Ramos-Lopez O, San-Cristobal R, Martinez-Urbistondo D, Micó V, Colmenarejo G, Villares-Fernandez P, Daimiel L, Martinez JA. Proinflammatory and Hepatic Features Related to Morbidity and Fatal Outcomes in COVID-19 Patients. J Clin Med. 2021;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Luo Y, Lin H. Inflammation initiates a vicious cycle between obesity and nonalcoholic fatty liver disease. Immun Inflamm Dis. 2021;9:59-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 9. | Wu H, Ballantyne CM. Metabolic Inflammation and Insulin Resistance in Obesity. Circ Res. 2020;126:1549-1564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 380] [Article Influence: 95.0] [Reference Citation Analysis (0)] |
| 10. | Monserrat-Mesquida M, Quetglas-Llabrés M, Abbate M, Montemayor S, Mascaró CM, Casares M, Tejada S, Abete I, Zulet MA, Tur JA, Martínez JA, Sureda A. Oxidative Stress and Pro-Inflammatory Status in Patients with Non-Alcoholic Fatty Liver Disease. Antioxidants (Basel). 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 41] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 11. | Ramos-Tovar E, Muriel P. Molecular Mechanisms That Link Oxidative Stress, Inflammation, and Fibrosis in the Liver. Antioxidants (Basel). 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 118] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 12. | Böttcher K, Pinzani M. Pathophysiology of liver fibrosis and the methodological barriers to the development of anti-fibrogenic agents. Adv Drug Deliv Rev. 2017;121:3-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 13. | Taylor RS, Taylor RJ, Bayliss S, Hagström H, Nasr P, Schattenberg JM, Ishigami M, Toyoda H, Wai-Sun Wong V, Peleg N, Shlomai A, Sebastiani G, Seko Y, Bhala N, Younossi ZM, Anstee QM, McPherson S, Newsome PN. Association Between Fibrosis Stage and Outcomes of Patients With Nonalcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Gastroenterology. 2020;158:1611-1625.e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 537] [Article Influence: 134.3] [Reference Citation Analysis (0)] |
| 14. | Chinnaratha MA, Jeffrey GP, MacQuillan G, Rossi E, de Boer BW, Speers DJ, Adams LA. Prediction of morbidity and mortality in patients with chronic hepatitis C by non-invasive liver fibrosis models. Liver Int. 2014;34:720-727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Sohn W, Chang Y, Cho YK, Hong YS, Shin H, Ryu S. Liver fibrosis scores and risk of liver-related mortality in young adults with chronic hepatitis B: A cohort study. J Viral Hepat. 2022;29:69-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Chrostek L, Panasiuk A. Liver fibrosis markers in alcoholic liver disease. World J Gastroenterol. 2014;20:8018-8023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 47] [Cited by in F6Publishing: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Rasche A, Sander AL, Corman VM, Drexler JF. Evolutionary biology of human hepatitis viruses. J Hepatol. 2019;70:501-520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 18. | Wang CR, Tsai HW. Human hepatitis viruses-associated cutaneous and systemic vasculitis. World J Gastroenterol. 2021;27:19-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 21] [Cited by in F6Publishing: 13] [Article Influence: 4.3] [Reference Citation Analysis (2)] |
| 19. | Premkumar M, Kedarisetty CK. Cytokine Storm of COVID-19 and Its Impact on Patients with and without Chronic Liver Disease. J Clin Transl Hepatol. 2021;9:256-264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 20. | Martínez-Urbistondo M, Moreno-Torres V, Mora-Vargas A, Expósito-Palomo E, Castejón-Díaz R, Daimiel L, Ramos-Lopez O, San-Cristóbal R, Vargas JA, Martínez JA. Interaction of ACEI antihypertensive agent's administration with the inflammatory status at admission concerning COVID-19 clinical stay outcomes. Vascul Pharmacol. 2022;143:106955. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Martinez-Urbistondo M, Mora-Vargas A, Expósito-Palomo E, Castejón R, Citores MJ, Rosado S, de Mendoza C, Baños I, Fernández-Cruz A, Daimiel L, San-Cristóbal R, Vargas JA, Martinez JA. Inflammatory-Related Clinical and Metabolic Outcomes in COVID-19 Patients. Mediators Inflamm. 2020;2020:2914275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 22. | McGurnaghan SJ, Weir A, Bishop J, Kennedy S, Blackbourn LAK, McAllister DA, Hutchinson S, Caparrotta TM, Mellor J, Jeyam A, O'Reilly JE, Wild SH, Hatam S, Höhn A, Colombo M, Robertson C, Lone N, Murray J, Butterly E, Petrie J, Kennon B, McCrimmon R, Lindsay R, Pearson E, Sattar N, McKnight J, Philip S, Collier A, McMenamin J, Smith-Palmer A, Goldberg D, McKeigue PM, Colhoun HM; Public Health Scotland COVID-19 Health Protection Study Group; Scottish Diabetes Research Network Epidemiology Group. Risks of and risk factors for COVID-19 disease in people with diabetes: a cohort study of the total population of Scotland. Lancet Diabetes Endocrinol. 2021;9:82-93. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 203] [Article Influence: 67.7] [Reference Citation Analysis (0)] |
| 23. | Kulkarni AV, Kumar P, Tevethia HV, Premkumar M, Arab JP, Candia R, Talukdar R, Sharma M, Qi X, Rao PN, Reddy DN. Systematic review with meta-analysis: liver manifestations and outcomes in COVID-19. Aliment Pharmacol Ther. 2020;52:584-599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 155] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 24. | Hegyi PJ, Váncsa S, Ocskay K, Dembrovszky F, Kiss S, Farkas N, Erőss B, Szakács Z, Hegyi P, Pár G. Metabolic Associated Fatty Liver Disease Is Associated With an Increased Risk of Severe COVID-19: A Systematic Review With Meta-Analysis. Front Med (Lausanne). 2021;8:626425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 25. | Marjot T, Moon AM, Cook JA, Abd-Elsalam S, Aloman C, Armstrong MJ, Pose E, Brenner EJ, Cargill T, Catana MA, Dhanasekaran R, Eshraghian A, García-Juárez I, Gill US, Jones PD, Kennedy J, Marshall A, Matthews C, Mells G, Mercer C, Perumalswami PV, Avitabile E, Qi X, Su F, Ufere NN, Wong YJ, Zheng MH, Barnes E, Barritt AS 4th, Webb GJ. Outcomes following SARS-CoV-2 infection in patients with chronic liver disease: An international registry study. J Hepatol. 2021;74:567-577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 348] [Cited by in F6Publishing: 330] [Article Influence: 110.0] [Reference Citation Analysis (0)] |
| 26. | NCD Countdown 2030 collaborators. NCD Countdown 2030: worldwide trends in non-communicable disease mortality and progress towards Sustainable Development Goal target 3.4. Lancet. 2018;392:1072-1088. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 540] [Cited by in F6Publishing: 578] [Article Influence: 96.3] [Reference Citation Analysis (1)] |
| 27. | Cordeiro A, Costa R, Andrade N, Silva C, Canabrava N, Pena MJ, Rodrigues I, Andrade S, Ramalho A. Does adipose tissue inflammation drive the development of non-alcoholic fatty liver disease in obesity? Clin Res Hepatol Gastroenterol. 2020;44:394-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Yaribeygi H, Farrokhi FR, Butler AE, Sahebkar A. Insulin resistance: Review of the underlying molecular mechanisms. J Cell Physiol. 2019;234:8152-8161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 409] [Article Influence: 68.2] [Reference Citation Analysis (0)] |
| 29. | Yaribeygi H, Sathyapalan T, Atkin SL, Sahebkar A. Molecular Mechanisms Linking Oxidative Stress and Diabetes Mellitus. Oxid Med Cell Longev. 2020;2020:8609213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 263] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 30. | Mohamed J, Nazratun Nafizah AH, Zariyantey AH, Budin SB. Mechanisms of Diabetes-Induced Liver Damage: The role of oxidative stress and inflammation. Sultan Qaboos Univ Med J. 2016;16:e132-e141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 227] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 31. | Ni Y, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, Fälth M, Stindt J, Königer C, Nassal M, Kubitz R, Sültmann H, Urban S. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070-1083. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 540] [Cited by in F6Publishing: 548] [Article Influence: 54.8] [Reference Citation Analysis (0)] |
| 32. | Zhu YZ, Qian XJ, Zhao P, Qi ZT. How hepatitis C virus invades hepatocytes: the mystery of viral entry. World J Gastroenterol. 2014;20:3457-3467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 43] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 33. | Zhao L, Lei W, Deng C, Wu Z, Sun M, Jin Z, Song Y, Yang Z, Jiang S, Shen M, Yang Y. The roles of liver X receptor α in inflammation and inflammation-associated diseases. J Cell Physiol. 2021;236:4807-4828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 34. | Horvatits T, Drolz A, Trauner M, Fuhrmann V. Liver Injury and Failure in Critical Illness. Hepatology. 2019;70:2204-2215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 35. | Gkogkou E, Barnasas G, Vougas K, Trougakos IP. Expression profiling meta-analysis of ACE2 and TMPRSS2, the putative anti-inflammatory receptor and priming protease of SARS-CoV-2 in human cells, and identification of putative modulators. Redox Biol. 2020;36:101615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 36. | Suárez-Fariñas M, Tokuyama M, Wei G, Huang R, Livanos A, Jha D, Levescot A, Irizar H, Kosoy R, Cording S, Wang W, Losic B, Ungaro RC, Di'Narzo A, Martinez-Delgado G, Suprun M, Corley MJ, Stojmirovic A, Houten SM, Peters L, Curran M, Brodmerkel C, Perrigoue J, Friedman JR, Hao K, Schadt EE, Zhu J, Ko HM, Cho J, Dubinsky MC, Sands BE, Ndhlovu L, Cerf-Bensusan N, Kasarskis A, Colombel JF, Harpaz N, Argmann C, Mehandru S. Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps With the Pathogenesis of SARS-CoV-2-related Disease. Gastroenterology. 2021;160:287-301.e20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 75] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 37. | Bonyek-Silva I, Machado AFA, Cerqueira-Silva T, Nunes S, Silva Cruz MR, Silva J, Santos RL, Barral A, Oliveira PRS, Khouri R, Serezani CH, Brodskyn C, Caldas JR, Barral-Netto M, Boaventura V, Tavares NM. LTB4-Driven Inflammation and Increased Expression of ALOX5/ACE2 During Severe COVID-19 in Individuals With Diabetes. Diabetes. 2021;70:2120-2130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 38. | Iwasaki M, Saito J, Zhao H, Sakamoto A, Hirota K, Ma D. Inflammation Triggered by SARS-CoV-2 and ACE2 Augment Drives Multiple Organ Failure of Severe COVID-19: Molecular Mechanisms and Implications. Inflammation. 2021;44:13-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 119] [Article Influence: 39.7] [Reference Citation Analysis (0)] |
| 39. | Herman-Edelstein M, Guetta T, Barnea A, Waldman M, Ben-Dor N, Barac YD, Kornowski R, Arad M, Hochhauser E, Aravot D. Expression of the SARS-CoV-2 receptorACE2 in human heart is associated with uncontrolled diabetes, obesity, and activation of the renin angiotensin system. Cardiovasc Diabetol. 2021;20:90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 40. | Chai X, Hu L, Zhang Y, Han W, Lu Z, Ke A, Zhou J, Shi G, Fang N, Fan J, Cai J, Lan F. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. 2020 Preprint. Available from: bioRxiv 2020.02.03.931766v1. [DOI] [Cited in This Article: ] |
| 41. | Liu Y, Wu Q, Wan D, He H, Lin H, Wang K, Que G, Wang Y, Chen Y, Tang X, Wu L, Yang X. Expression and Possible Significance of ACE2 in the Human Liver, Esophagus, Stomach, and Colon. Evid Based Complement Alternat Med. 2021;2021:6949902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Yang L, Han Y, Nilsson-Payant BE, Gupta V, Wang P, Duan X, Tang X, Zhu J, Zhao Z, Jaffré F, Zhang T, Kim TW, Harschnitz O, Redmond D, Houghton S, Liu C, Naji A, Ciceri G, Guttikonda S, Bram Y, Nguyen DT, Cioffi M, Chandar V, Hoagland DA, Huang Y, Xiang J, Wang H, Lyden D, Borczuk A, Chen HJ, Studer L, Pan FC, Ho DD, tenOever BR, Evans T, Schwartz RE, Chen S. A Human Pluripotent Stem Cell-based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell. 2020;27:125-136.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 466] [Article Influence: 116.5] [Reference Citation Analysis (0)] |
| 43. | Huang Q, Xie Q, Shi CC, Xiang XG, Lin LY, Gong BD, Zhao GD, Wang H, Jia NN. Expression of angiotensin-converting enzyme 2 in CCL4-induced rat liver fibrosis. Int J Mol Med. 2009;23:717-723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 44. | Paizis G, Tikellis C, Cooper ME, Schembri JM, Lew RA, Smith AI, Shaw T, Warner FJ, Zuilli A, Burrell LM, Angus PW. Chronic liver injury in rats and humans upregulates the novel enzyme angiotensin converting enzyme 2. Gut. 2005;54:1790-1796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 249] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 45. | Pirola CJ, Sookoian S. SARS-CoV-2 virus and liver expression of host receptors: Putative mechanisms of liver involvement in COVID-19. Liver Int. 2020;40:2038-2040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 89] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 46. | Zhao B, Ni C, Gao R, Wang Y, Yang L, Wei J, Lv T, Liang J, Zhang Q, Xu W, Xie Y, Wang X, Yuan Z, Zhang R, Lin X. Recapitulation of SARS-CoV-2 infection and cholangiocyte damage with human liver ductal organoids. Protein Cell. 2020;11:771-775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 278] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 47. | Fondevila MF, Mercado-Gómez M, Rodríguez A, Gonzalez-Rellan MJ, Iruzubieta P, Valentí V, Escalada J, Schwaninger M, Prevot V, Dieguez C, Crespo J, Frühbeck G, Martinez-Chantar ML, Nogueiras R. Obese patients with NASH have increased hepatic expression of SARS-CoV-2 critical entry points. J Hepatol. 2021;74:469-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 48. | Versteeg GA, van de Nes PS, Bredenbeek PJ, Spaan WJ. The coronavirus spike protein induces endoplasmic reticulum stress and upregulation of intracellular chemokine mRNA concentrations. J Virol. 2007;81:10981-10990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 89] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 49. | Chan CP, Siu KL, Chin KT, Yuen KY, Zheng B, Jin DY. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2006;80:9279-9287. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 177] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 50. | Wang Y, Liu S, Liu H, Li W, Lin F, Jiang L, Li X, Xu P, Zhang L, Zhao L, Cao Y, Kang J, Yang J, Li L, Liu X, Li Y, Nie R, Mu J, Lu F, Zhao S, Lu J, Zhao J. SARS-CoV-2 infection of the liver directly contributes to hepatic impairment in patients with COVID-19. J Hepatol. 2020;73:807-816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 397] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 51. | Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J, Katze MG, Hussain MM, Song B, Swathirajan J, Wang J, Yau GD, Kaufman RJ. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829-840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 436] [Cited by in F6Publishing: 451] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 52. | Aggarwal K, Mijwil MM, Garg S, Al-Mistarehi A-H, Alomari S, Gök M, Zein Alaabdin AM, Abdulrhman SH. Has the Future Started? IJCSM. 2022;. [DOI] [Cited in This Article: ] |
| 53. | Quer G, Arnaout R, Henne M. Machine Learning and the Future of Cardiovascular Care: JACC State-of-the-Art Review. J Am Coll Cardiol. 2021;77:300-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 141] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 54. | Sidey-Gibbons JAM, Sidey-Gibbons CJ. Machine learning in medicine: a practical introduction. BMC Med Res Methodol. 2019;19:64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 614] [Cited by in F6Publishing: 415] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 55. | DeGregory KW, Kuiper P, DeSilvio T, Pleuss JD, Miller R, Roginski JW, Fisher CB, Harness D, Viswanath S, Heymsfield SB, Dungan I, Thomas DM. A review of machine learning in obesity. Obes Rev. 2018;19:668-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 56. | Greener JG, Kandathil SM, Moffat L, Jones DT. A guide to machine learning for biologists. Nat Rev Mol Cell Biol. 2022;23:40-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 406] [Article Influence: 203.0] [Reference Citation Analysis (0)] |
| 57. | Mijwil MM, Aggarwal K. A diagnostic testing for people with appendicitis using machine learning techniques. Multimed Tools Appl. 2022;81:7011-7023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 58. | Lee HW, Sung JJY, Ahn SH. Artificial intelligence in liver disease. J Gastroenterol Hepatol. 2021;36:539-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 59. | Lysdahlgaard S. Comparing Radiomics features of tumour and healthy liver tissue in a limited CT dataset: A machine learning study. Radiography (Lond). 2022;28:718-724. [PubMed] [DOI] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 60. | Furtado P. Testing Segmentation Popular Loss and Variations in Three Multiclass Medical Imaging Problems. J Imaging. 2021;7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 61. | Jiřík M, Hácha F, Gruber I, Pálek R, Mírka H, Zelezny M, Liška V. Why Use Position Features in Liver Segmentation Performed by Convolutional Neural Network. Front Physiol. 2021;12:734217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 62. | Christou CD, Tsoulfas G. Challenges and opportunities in the application of artificial intelligence in gastroenterology and hepatology. World J Gastroenterol. 2021;27:6191-6223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 17] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (7)] |
| 63. | Fialoke S, Malarstig A, Miller MR, Dumitriu A. Application of Machine Learning Methods to Predict Non-Alcoholic Steatohepatitis (NASH) in Non-Alcoholic Fatty Liver (NAFL) Patients. AMIA Annu Symp Proc. 2018;2018:430-439. [PubMed] [Cited in This Article: ] |
| 64. | Ma H, Xu CF, Shen Z, Yu CH, Li YM. Application of Machine Learning Techniques for Clinical Predictive Modeling: A Cross-Sectional Study on Nonalcoholic Fatty Liver Disease in China. Biomed Res Int. 2018;2018:4304376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 65. | Yip TC, Ma AJ, Wong VW, Tse YK, Chan HL, Yuen PC, Wong GL. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment Pharmacol Ther. 2017;46:447-456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 66. | Pei X, Deng Q, Liu Z, Yan X, Sun W. Machine Learning Algorithms for Predicting Fatty Liver Disease. Ann Nutr Metab. 2021;77:38-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (1)] |
| 67. | Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, Yun J, Choi JY, Lee Y, Kang BK, Kim JH, Kim SY, Yu ES. Development and Validation of a Deep Learning System for Staging Liver Fibrosis by Using Contrast Agent-enhanced CT Images in the Liver. Radiology. 2018;289:688-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 120] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 68. | Chen Y, Luo Y, Huang W, Hu D, Zheng RQ, Cong SZ, Meng FK, Yang H, Lin HJ, Sun Y, Wang XY, Wu T, Ren J, Pei SF, Zheng Y, He Y, Hu Y, Yang N, Yan H. Machine-learning-based classification of real-time tissue elastography for hepatic fibrosis in patients with chronic hepatitis B. Comput Biol Med. 2017;89:18-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 69. | Jeong S, Ge Y, Chen J, Gao Q, Luo G, Zheng B, Sha M, Shen F, Cheng Q, Sui C, Liu J, Wang H, Xia Q, Chen L. Latent Risk Intrahepatic Cholangiocarcinoma Susceptible to Adjuvant Treatment After Resection: A Clinical Deep Learning Approach. Front Oncol. 2020;10:143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 70. | Wübbolding M, Lopez Alfonso JC, Lin CY, Binder S, Falk C, Debarry J, Gineste P, Kraft ARM, Chien RN, Maasoumy B, Wedemeyer H, Jeng WJ, Meyer Hermann M, Cornberg M, Höner Zu Siederdissen C. Pilot Study Using Machine Learning to Identify Immune Profiles for the Prediction of Early Virological Relapse After Stopping Nucleos(t)ide Analogues in HBeAg-Negative CHB. Hepatol Commun. 2021;5:97-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 71. | Hong WD, Ji YF, Wang D, Chen TZ, Zhu QH. Use of artificial neural network to predict esophageal varices in patients with HBV related cirrhosis. Hepat Mon. 2011;11:544-547. [PubMed] [Cited in This Article: ] |
| 72. | Zhong BY, Yan ZP, Sun JH, Zhang L, Hou ZH, Yang MJ, Zhou GH, Wang WS, Li Z, Huang P, Zhang S, Zhu XL, Ni CF. Prognostic Performance of Albumin-Bilirubin Grade With Artificial Intelligence for Hepatocellular Carcinoma Treated With Transarterial Chemoembolization Combined With Sorafenib. Front Oncol. 2020;10:525461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Shi HY, Lee KT, Lee HH, Ho WH, Sun DP, Wang JJ, Chiu CC. Comparison of artificial neural network and logistic regression models for predicting in-hospital mortality after primary liver cancer surgery. PLoS One. 2012;7:e35781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 74. | Shi HY, Lee KT, Wang JJ, Sun DP, Lee HH, Chiu CC. Artificial neural network model for predicting 5-year mortality after surgery for hepatocellular carcinoma: a nationwide study. J Gastrointest Surg. 2012;16:2126-2131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 75. | Patnaik RK, Lin YC, Agarwal A, Ho MC, Yeh JA. A pilot study for the prediction of liver function related scores using breath biomarkers and machine learning. Sci Rep. 2022;12:2032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 76. | Tuppad A, Patil SD. Machine learning for diabetes clinical decision support: a review. Adv Comput Intell. 2022;2:22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 77. | Chatterjee A, Gerdes MW, Martinez SG. Identification of Risk Factors Associated with Obesity and Overweight-A Machine Learning Overview. Sensors (Basel). 2020;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 78. | Cheng X, Lin SY, Liu J, Liu S, Zhang J, Nie P, Fuemmeler BF, Wang Y, Xue H. Does Physical Activity Predict Obesity-A Machine Learning and Statistical Method-Based Analysis. Int J Environ Res Public Health. 2021;18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 79. | Liu W, Fang X, Zhou Y, Dou L, Dou T. Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 2022;24:104892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 80. | Marcos-Pasero H, Colmenarejo G, Aguilar-Aguilar E, Ramírez de Molina A, Reglero G, Loria-Kohen V. Ranking of a wide multidomain set of predictor variables of children obesity by machine learning variable importance techniques. Sci Rep. 2021;11:1910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 81. | Fu Y, Gou W, Hu W, Mao Y, Tian Y, Liang X, Guan Y, Huang T, Li K, Guo X, Liu H, Li D, Zheng JS. Integration of an interpretable machine learning algorithm to identify early life risk factors of childhood obesity among preterm infants: a prospective birth cohort. BMC Med. 2020;18:184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 82. | LeCroy MN, Kim RS, Stevens J, Hanna DB, Isasi CR. Identifying Key Determinants of Childhood Obesity: A Narrative Review of Machine Learning Studies. Child Obes. 2021;17:153-159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 83. | Tang W, Zhan W, Wei M, Chen Q. Associations Between Different Dietary Vitamins and the Risk of Obesity in Children and Adolescents: A Machine Learning Approach. Front Endocrinol (Lausanne). 2021;12:816975. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 84. | Torre P, Aglitti A, Masarone M, Persico M. Viral hepatitis: Milestones, unresolved issues, and future goals. World J Gastroenterol. 2021;27:4603-4638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 20] [Cited by in F6Publishing: 15] [Article Influence: 5.0] [Reference Citation Analysis (5)] |
| 85. | Butt MB, Alfayad M, Saqib S, Khan MA, Ahmad M, Elmitwally NS. Diagnosing the Stage of Hepatitis C Using Machine Learning. J Healthc Eng. 2021;2021:8062410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 86. | Wong GL, Hui VW, Tan Q, Xu J, Lee HW, Yip TC, Yang B, Tse YK, Yin C, Lyu F, Lai JC, Lui GC, Chan HL, Yuen PC, Wong VW. Novel machine learning models outperform risk scores in predicting hepatocellular carcinoma in patients with chronic viral hepatitis. JHEP Rep. 2022;4:100441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 87. | Wei R, Wang J, Wang X, Xie G, Wang Y, Zhang H, Peng CY, Rajani C, Kwee S, Liu P, Jia W. Clinical prediction of HBV and HCV related hepatic fibrosis using machine learning. EBioMedicine. 2018;35:124-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 88. | Konerman MA, Beste LA, Van T, Liu B, Zhang X, Zhu J, Saini SD, Su GL, Nallamothu BK, Ioannou GN, Waljee AK. Machine learning models to predict disease progression among veterans with hepatitis C virus. PLoS One. 2019;14:e0208141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 89. | Barakat NH, Barakat SH, Ahmed N. Prediction and Staging of Hepatic Fibrosis in Children with Hepatitis C Virus: A Machine Learning Approach. Healthc Inform Res. 2019;25:173-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 90. | Pawlotsky JM. Treatment failure and resistance with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742-1751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 263] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 91. | Feldman TC, Dienstag JL, Mandl KD, Tseng YJ. Machine-learning-based predictions of direct-acting antiviral therapy duration for patients with hepatitis C. Int J Med Inform. 2021;154:104562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 92. | Kamboj S, Rajput A, Rastogi A, Thakur A, Kumar M. Targeting non-structural proteins of Hepatitis C virus for predicting repurposed drugs using QSAR and machine learning approaches. Comput Struct Biotechnol J. 2022;20:3422-3438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 93. | Tian X, Chong Y, Huang Y, Guo P, Li M, Zhang W, Du Z, Li X, Hao Y. Using Machine Learning Algorithms to Predict Hepatitis B Surface Antigen Seroclearance. Comput Math Methods Med. 2019;2019:6915850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 94. | Chen S, Zhang Z, Wang Y, Fang M, Zhou J, Li Y, Dai E, Feng Z, Wang H, Yang Z, Huang X, Jia J, Li S, Huang C, Tong L, Xiao X, He Y, Duan Y, Zhu S, Gao C. Using Quasispecies Patterns of Hepatitis B Virus to Predict Hepatocellular Carcinoma With Deep Sequencing and Machine Learning. J Infect Dis. 2021;223:1887-1896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 95. | Mueller-Breckenridge AJ, Garcia-Alcalde F, Wildum S, Smits SL, de Man RA, van Campenhout MJH, Brouwer WP, Niu J, Young JAT, Najera I, Zhu L, Wu D, Racek T, Hundie GB, Lin Y, Boucher CA, van de Vijver D, Haagmans BL. Machine-learning based patient classification using Hepatitis B virus full-length genome quasispecies from Asian and European cohorts. Sci Rep. 2019;9:18892. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 96. | KayvanJoo AH, Ebrahimi M, Haqshenas G. Prediction of hepatitis C virus interferon/ribavirin therapy outcome based on viral nucleotide attributes using machine learning algorithms. BMC Res Notes. 2014;7:565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 97. | Bennett TD, Moffitt RA, Hajagos JG, Amor B, Anand A, Bissell MM, Bradwell KR, Bremer C, Byrd JB, Denham A, DeWitt PE, Gabriel D, Garibaldi BT, Girvin AT, Guinney J, Hill EL, Hong SS, Jimenez H, Kavuluru R, Kostka K, Lehmann HP, Levitt E, Mallipattu SK, Manna A, McMurry JA, Morris M, Muschelli J, Neumann AJ, Palchuk MB, Pfaff ER, Qian Z, Qureshi N, Russell S, Spratt H, Walden A, Williams AE, Wooldridge JT, Yoo YJ, Zhang XT, Zhu RL, Austin CP, Saltz JH, Gersing KR, Haendel MA, Chute CG. The National COVID Cohort Collaborative: Clinical Characterization and Early Severity Prediction. medRxiv. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 98. | Li WT, Ma J, Shende N, Castaneda G, Chakladar J, Tsai JC, Apostol L, Honda CO, Xu J, Wong LM, Zhang T, Lee A, Gnanasekar A, Honda TK, Kuo SZ, Yu MA, Chang EY, Rajasekaran MR, Ongkeko WM. Using machine learning of clinical data to diagnose COVID-19: a systematic review and meta-analysis. BMC Med Inform Decis Mak. 2020;20:247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 59] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 99. | Bhargava A, Bansal A, Goyal V. Machine learning-based automatic detection of novel coronavirus (COVID-19) disease. Multimed Tools Appl. 2022;81:13731-13750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 100. | Günster C, Busse R, Spoden M, Rombey T, Schillinger G, Hoffmann W, Weber-Carstens S, Schuppert A, Karagiannidis C. 6-month mortality and readmissions of hospitalized COVID-19 patients: A nationwide cohort study of 8,679 patients in Germany. PLoS One. 2021;16:e0255427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 51] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 101. | Deng X, Li H, Liao X, Qin Z, Xu F, Friedman S, Ma G, Ye K, Lin S. Building a predictive model to identify clinical indicators for COVID-19 using machine learning method. Med Biol Eng Comput. 2022;60:1763-1774. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 5] [Reference Citation Analysis (0)] |
| 102. | Lipták P, Banovcin P, Rosoľanka R, Prokopič M, Kocan I, Žiačiková I, Uhrik P, Grendar M, Hyrdel R. A machine learning approach for identification of gastrointestinal predictors for the risk of COVID-19 related hospitalization. PeerJ. 2022;10:e13124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 103. | Elemam NM, Hammoudeh S, Salameh L, Mahboub B, Alsafar H, Talaat IM, Habib P, Siddiqui M, Hassan KO, Al-Assaf OY, Taneera J, Sulaiman N, Hamoudi R, Maghazachi AA, Hamid Q, Saber-Ayad M. Identifying Immunological and Clinical Predictors of COVID-19 Severity and Sequelae by Mathematical Modeling. Front Immunol. 2022;13:865845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 104. | Mashraqi A, Halawani H, Alelyani T, Mashraqi M, Makkawi M, Alasmari S, Shaikh A, Alshehri A. Prediction Model of Adverse Effects on Liver Functions of COVID-19 ICU Patients. J Healthc Eng. 2022;2022:4584965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 105. | Soltan AAS, Yang J, Pattanshetty R, Novak A, Yang Y, Rohanian O, Beer S, Soltan MA, Thickett DR, Fairhead R, Zhu T, Eyre DW, Clifton DA; CURIAL Translational Collaborative. Real-world evaluation of rapid and laboratory-free COVID-19 triage for emergency care: external validation and pilot deployment of artificial intelligence driven screening. Lancet Digit Health. 2022;4:e266-e278. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 13] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 106. | Soltan AAS, Kouchaki S, Zhu T, Kiyasseh D, Taylor T, Hussain ZB, Peto T, Brent AJ, Eyre DW, Clifton DA. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: development and prospective validation of an artificial intelligence screening test. Lancet Digit Health. 2021;3:e78-e87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 60] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 107. | Schultz MB, Vera D, Sinclair DA. Can artificial intelligence identify effective COVID-19 therapies? EMBO Mol Med. 2020;12:e12817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 108. | Stebbing J, Krishnan V, de Bono S, Ottaviani S, Casalini G, Richardson PJ, Monteil V, Lauschke VM, Mirazimi A, Youhanna S, Tan YJ, Baldanti F, Sarasini A, Terres JAR, Nickoloff BJ, Higgs RE, Rocha G, Byers NL, Schlichting DE, Nirula A, Cardoso A, Corbellino M; Sacco Baricitinib Study Group. Mechanism of baricitinib supports artificial intelligence-predicted testing in COVID-19 patients. EMBO Mol Med. 2020;12:e12697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in F6Publishing: 194] [Article Influence: 48.5] [Reference Citation Analysis (0)] |
| 109. | Mijwil MM, Salman Shukur B. A Scoping Review of Machine Learning Techniques and Their Utilisation in Predicting Heart Diseases. Ibn AL-Haitham J Pure Appl Sci. 2022;35:175-189. [DOI] [Cited in This Article: ] |
| 110. | Spann A, Yasodhara A, Kang J, Watt K, Wang B, Goldenberg A, Bhat M. Applying Machine Learning in Liver Disease and Transplantation: A Comprehensive Review. Hepatology. 2020;71:1093-1105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 78] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 111. | Gao B, Wu TC, Lang S, Jiang L, Duan Y, Fouts DE, Zhang X, Tu XM, Schnabl B. Machine Learning Applied to Omics Datasets Predicts Mortality in Patients with Alcoholic Hepatitis. Metabolites. 2022;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 112. | Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZQ, Ye SL, Liu YK, Tang ZY, Wang XW. Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning. Nat Med. 2003;9:416-423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 642] [Cited by in F6Publishing: 687] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 113. | Petta S, Cabibbo G, Enea M, Macaluso FS, Plaia A, Bruno R, Gasbarrini A, Craxì A, Cammà C; WEF Study Group. Cost-effectiveness of sofosbuvir-based triple therapy for untreated patients with genotype 1 chronic hepatitis C. Hepatology. 2014;59:1692-1705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 114. | Shousha HI, Awad AH, Omran DA, Elnegouly MM, Mabrouk M. Data Mining and Machine Learning Algorithms Using IL28B Genotype and Biochemical Markers Best Predicted Advanced Liver Fibrosis in Chronic Hepatitis C. Jpn J Infect Dis. 2018;71:51-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 115. | Masi D, Risi R, Biagi F, Vasquez Barahona D, Watanabe M, Zilich R, Gabrielli G, Santin P, Mariani S, Lubrano C, Gnessi L. Application of a Machine Learning Technology in the Definition of Metabolically Healthy and Unhealthy Status: A Retrospective Study of 2567 Subjects Suffering from Obesity with or without Metabolic Syndrome. Nutrients. 2022;14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 116. | Lin Z, Feng W, Liu Y, Ma C, Arefan D, Zhou D, Cheng X, Yu J, Gao L, Du L, You H, Zhu J, Zhu D, Wu S, Qu S. Machine Learning to Identify Metabolic Subtypes of Obesity: A Multi-Center Study. Front Endocrinol (Lausanne). 2021;12:713592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 117. | Lee YC, Christensen JJ, Parnell LD, Smith CE, Shao J, McKeown NM, Ordovás JM, Lai CQ. Using Machine Learning to Predict Obesity Based on Genome-Wide and Epigenome-Wide Gene-Gene and Gene-Diet Interactions. Front Genet. 2021;12:783845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |