Published online Dec 28, 2021. doi: 10.3748/wjg.v27.i48.8283
Peer-review started: June 18, 2021
First decision: August 19, 2021
Revised: September 9, 2021
Accepted: December 2, 2021
Article in press: December 7, 2021
Published online: December 28, 2021
A symbiotic relationship has set up between the gut microbiota and its host in the course of evolution, forming an interkingdom consortium. The gut offers a favorable ecological niche for microbial communities, with the whole body and external factors (e.g., diet or medications) contributing to modulating this microenvironment. Reciprocally, the gut microbiota is important for maintaining health by acting not only on the gut mucosa but also on other organs. However, failure in one or another of these two partners can lead to the breakdown in their symbiotic equilibrium and contribute to disease onset and/or progression. Several microbial and host processes are devoted to facing up the stress that could alter the symbiosis, ensuring the resilience of the ecosystem. Among these processes, autophagy is a host catabolic process integrating a wide range of stress in order to maintain cell survival and homeostasis. This cytoprotective mechanism, which is ubiquitous and operates at basal level in all tissues, can be rapidly down- or up-regulated at the transcriptional, post-transcriptional, or post-translational levels, to respond to various stress conditions. Because of its sensitivity to all, metabolic-, immune-, and microbial-derived stimuli, autophagy is at the crossroad of the dialogue between changes occurring in the gut microbiota and the host responses. In this review, we first delineate the modulation of host autophagy by the gut microbiota locally in the gut and in peripheral organs. Then, we describe the autophagy-related mechanisms affecting the gut microbiota. We conclude this review with the current challenges and an outlook toward the future interventions aiming at modulating host autophagy by targeting the gut microbiota.
Core Tip: We are now aware that maintaining a fine equilibrium between the host and its gut microbiota is a prerequisite to maintain host homeostasis and promote long-term health. Several host and microbial processes interact dynamically to respond to external stresses. Among these processes, host autophagy acts as a cytoprotective mechanism responsive to a wide range of stress conditions, including metabolic, immune, and microbial stimuli. Autophagy was initially described as a degradative process active upon nutrient starvation. However, this process fulfils a wide range of other functions that are essential to host homeostasis. We discuss herein reciprocal interactions of autophagy with the gut microbiota in health and disease conditions.
- Citation: Lapaquette P, Bizeau JB, Acar N, Bringer MA. Reciprocal interactions between gut microbiota and autophagy. World J Gastroenterol 2021; 27(48): 8283-8301
- URL: https://www.wjgnet.com/1007-9327/full/v27/i48/8283.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i48.8283
The commensal microbiota living in the human gut is a unique ecosystem that has co-evolved with human to establish a symbiotic relationship. This microbial community is estimated to encompass about 1014 resident microorganisms, dominated by bacteria, but containing also populations of archaea, fungi, protozoa, and viruses[1]. The host provides nutrients and a favorable environment (i.e., ecological niches) for its microbial inhabitants. In return, the gut microbiota plays multiple roles that contribute to the host whole-body homeostasis, in particular by metabolizing dietary nutrients, by preventing colonization by enteric pathogens, and by regulating the host immune system and metabolism. The gut microbiota is, for instance, essential for the synthesis of vitamins (e.g., K and B-group vitamins) and the fermentation of dietary fibers and carbohydrates, which generate short-chain fatty acids (SCFAs). These fermentation products are used as energy source by organs and are also involved in the regulation of various cellular processes (e.g., intestinal barrier integrity, mucus production, and inflammation)[2,3].
Through their interactions with the host, gut microbes and their derived products are involved not only in the physiological regulation of the gut mucosa but also in that of organs located at distance from the gut mucosa, as illustrated by the studies detailing molecular features of the gut-microbiota-brain axis[4-6]. Keeping the mutualistic relationship between the gut microbiota and the host throughout host’s life is thus essential to maintain the health status of the host[7]. Deleterious shifts in the composition of the gut microbiota, called dysbiosis, can unbalance its functions, leading to the disruption of host homeostasis. This is particularly well illustrated by the ability of fecal microbiota transplantation (FMT) to transmit detrimental metabolic and/or pro-inflammatory traits from a sick donor to healthy recipient mice[8-10]. In addition to environmental stresses, the symbiotic equilibrium of the gut microbiota and the host can also be broken by dysfunctions/alterations in the host metabolism and immune system, which are conditions that can contribute to dysbiosis[8,11,12]. In this context, the roles of autophagy in strengthening the intestinal barrier and in maintaining host metabolic and inflammatory balance position it as the cornerstone of the symbiotic relationship between the gut microbiota and the host[4,13].
Macroautophagy/autophagy is an intracellular and multistep process starting with the formation of a membranous cup-shaped structure, called phagophore, which engulfs portions of the cytoplasm. The phagophore elongates and finally closes to form a sealed double-membraned vacuole, called autophagosome, whose maturation ends by its fusion with lysosomes[14-16]. Autophagy was initially described as a lysosomal catabolic process occurring under starvation that degrades and recycles cytoplasmic macromolecules (e.g., proteins, lipids, and carbohydrates) for the biosynthesis of essential cellular components and to restore energy balance[17]. Nowadays, autophagy process and autophagy-related proteins are recognized as key cellular components whose roles are not restricted to the regulation of energy balance[18,19]. These roles include, but are not limited to, the regulation of the inflammatory response, the cytoprotection by preventing the accumulation of intracellular waste (e.g., damaged organelles and misfolded or aggregated proteins), the protection against intracellular pathogens (e.g., bacteria, fungi, or viruses), the membrane dynamic (e.g., transport or secretion), and the regulation of cell differentiation and survival. Autophagy also regulates specific functions related to the features of organs. For example, at the gut mucosa - the first tissue at the interface between the gut microbiota and the host - autophagy is involved in the regulation of the functions of the secretory cells and of the intestinal stem cell[4]. In the central nervous system, autophagy plays roles in neuronal development and survival and other various functions[20]. The central role of autophagy in maintaining homeostasis, and thus the health status, is supported by the observed embryonic or neonatal lethality of mice deficient for most autophagy-related (Atg) core genes (Becn1, Vps34, Atg9a, Ulk1/2, Atg3, Atg5, Atg7, and Atg16l1) as well as association of numerous diseases and disorders with autophagy defects[19,21].
Of note, a growing number of recent studies highlight that most of the proteins of the autophagy machinery also mediate autophagy-independent functions, including phagocytosis, exocytosis, cytokinesis, DNA repair, or innate and adaptive immune signaling[22]. To exert their numerous functions, the machineries involving autophagy proteins are intricated with molecular sensors specialized in the detection of various stimuli such as microbial sensors [e.g., Toll-like receptors (TLR) and Nod-like receptors (NLR)], stress sensors (e.g., HMGB1, Sestrins, ER-stress sensor proteins, P2XR, and cGAS-STING pathway), or energy status sensors (e.g., AMPK and mTOR pathways)[23-29].
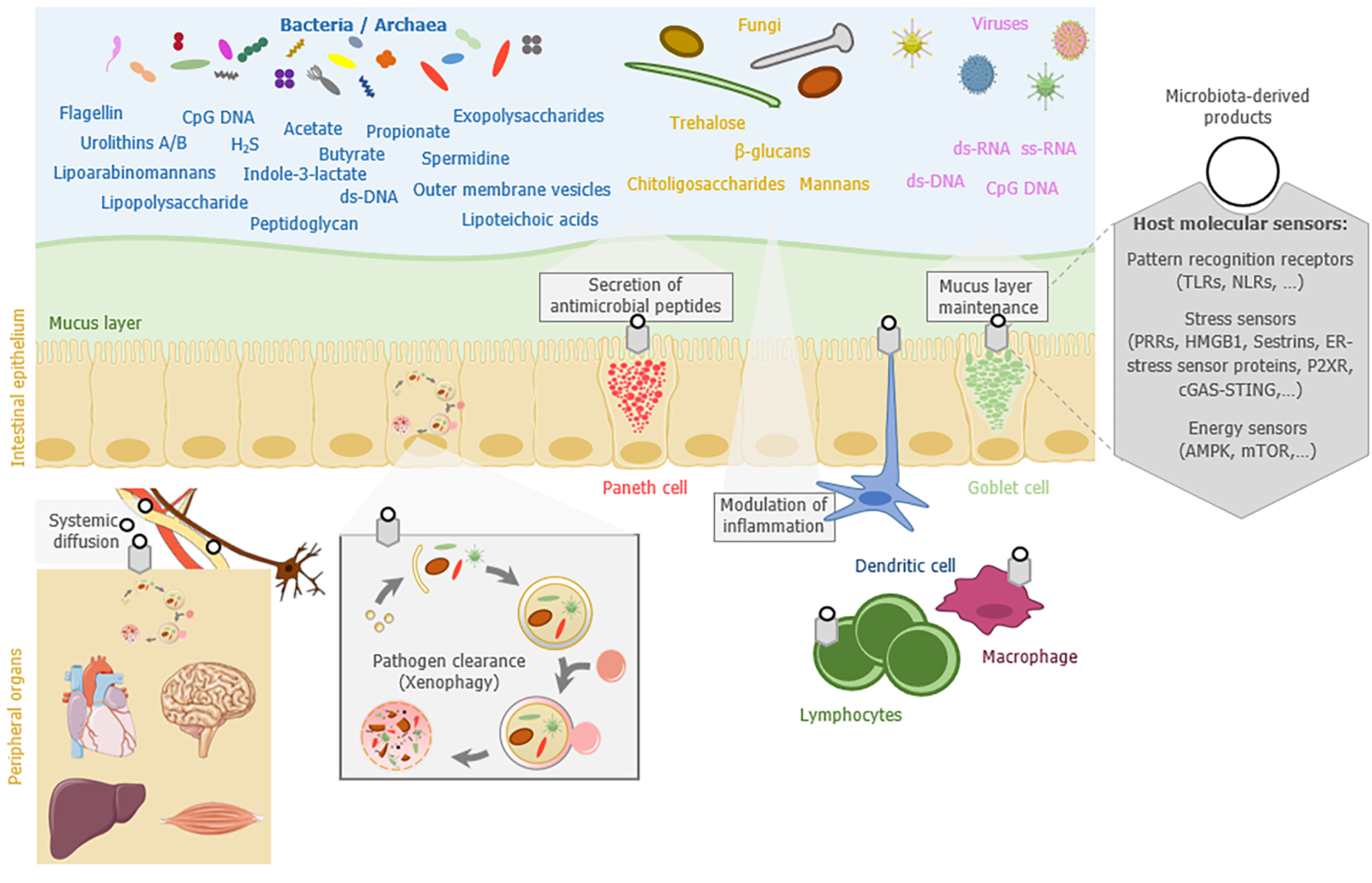
In this review, we summarize the current knowledge on how the gut microbiota influences host autophagy locally in the gut mucosa or remotely in peripheral organs (brain, heart, liver, or muscles), and how autophagy or autophagy-related proteins can reciprocally shape the gut microbiota composition and modify its functions (Figure 1). We finally discuss the potential of targeting the gut microbiota as a strategy to modulate autophagy or restore its functionality in pathological context.
A first clue that points out a direct implication of the gut microbiota in the regulation of host autophagy has been provided by analyzing autophagy in germ-free mice (i.e., mice lacking microorganisms and bred in isolators without any microbial exposure). Basal autophagy was decreased in the colonic epithelium of germ-free mice compared to conventionally raised mice, suggesting that the gut microbiota influences intestinal autophagy in physiological condition[30]. The increase in basal activity of autophagy in germ-free mice was attributed to an energy-deprived status of colonocytes. Treatment of these cells with butyrate, a SCFA generated by some gut bacteria and serving as main energy source for colonocytes, was sufficient to reverse the phenotype. In vivo, colonization of germ-free mice with the butyrate-producing bacterial strain Butyrivibrio fibrisolvens was sufficient to restore autophagy steady state. In addition to butyrate, other bacteria-derived metabolites may have the ability to reduce basal autophagy in the colon. They include indole-3-lactate, which is a tryptophan metabolite produced notably by the bacteria belonging to the Lacticaseibacillus, Lactobacillus, Bifidobacterium, Megamonas, Roseburia, or Ruminococcus genus[31,32].
Pathogen-associated molecular patterns (PAMPs), which are conserved microbial molecules, are also able to modulate autophagy usually by stimulating the process[23]. These effects have been particularly well described for pathogens. PAMPs mainly act by interacting with specific host cell receptors that belong to the TLR and NLR families. This has been illustrated by the ability of the lipopolysaccharide (LPS) from Gram negative bacteria to stimulate autophagy through its binding to TLR4[33], or the peptidoglycan (PGN) from Gram positive bacteria through NOD1-, NOD2-, and TLR2-associated signaling[34,35]. Besides those of bacteria, fungal PAMPs can also mobilize components of the autophagy machinery. This is true for β-glucans that are found in fungal cell walls and stimulate autophagy-related processes though their binding to the host receptor Dectin-1[36,37]. Trehalose, a non-reducing disaccharide produced by bacteria and fungi, is also a potent autophagy inducer, for which the ability to stimulate colonic autophagy during colitis in mice has been described[38,39]. In addition, in-depth studies of the infectious cycle of some pathogenic bacteria have shed the light on the existence of secreted bacterial effectors able to activate (e.g., Ats-1 protein from Anaplasma phagocytophilum) or inhibit (e.g., RavZ protein from Legionella pneumophila) autophagy at various stages of the process[40,41]. It is not excluded that some commensal microorganisms in the gut express such proteins that influence host autophagy.
Given the influence of gut microbiota-related factors on autophagy, one could expect that alterations in the composition of the gut microbiota would affect autophagy in the gut mucosa. Indeed, an increase in the expression of some autophagy-related proteins (FoxO1, FoxO3, GABARAP, and ATG7) and LC3-II/LC3-I ratio and a decrease in AKT activation have been reported in newborn piglets receiving FMT[42]. In addition, alteration of the gut microbiota resulting from the administration of a cocktail of broad-spectrum antibiotics increased the basal activity of autophagy as well as the expression of some autophagy-related proteins (ATG16L1, ATG5, and IRGM1) in the ileal mucosa of mice[43,44]. Interestingly, oral administration of a single bacterial species (e.g., Desulfovibrio spp., Fusobacterium nucleatum, or Escherichia coli) in conventional mice can also be sufficient to modulate gut autophagy[42,44,45]. Altogether, these studies suggest that autophagy regulatory network is sensitive to changes in the gut microbiota.
Microbial-derived metabolites (e.g., PAMPs), compounds that are issued from the gut microbiota metabolism (e.g., neuroactive compounds and SCFAs) and host bioactive molecules that are produced in response to its interaction with the gut microbiota (e.g., cytokines), can have large systemic effects and modulate the physiology of organs that are distant from the gut. Influence of the gut microbiota on the brain is a well-documented example of such effects[6]. Several communication routes (immune system, autonomic nervous system, neuroendocrine system, hypothalamic – pituitary – adrenal axis, and other metabolic pathways) between the microbiota and the brain have been identified[6]. It is very likely that similar pathways and microbiota-derived players, or at least some of them, modulate as well the physiology of other organs in the body. Evidence is accumulating on the modulation of autophagy by the gut microbiota in distant organs and several of these are presented below (Table 1).
| Ref. | Impact on autophagy | |||
| Brain | Liver | Muscles | ||
| [49,74-76] | Diet-induced changes in the gut microbiota | Feeding of mother mice with an HSHF diet: Changes in the expression levels of LC3A-I/LC3A-II/ LC3B-I/LC3B-II in the offspring. | Feeding mice or rats with an HF diet: Changes in the expression levels of LC3, p62, mTOR, and p-AKT and modulation of the LC3-II amount. | |
| [55,56,59,70] | Mice with specific gut microbiota | AD mice1: Modulation of the lysosomal activity (Cathepsin L) and SIRT1 activity and changes in the expression levels of Beclin-1, p62, and SIRT1. | ASF colonized mice: Changes in the expression of a set of genes related to autophagy/membrane trafficking (Uvrag, Atg14, Becn1, Bcl2l1, and Pik3c3) and lysosomal functions (Chmp4c and Chmp2a) compared to germ-free mice. | |
| FMT from patients with AIS to mice: Changes in the expression levels of Becn1, ATG12, and LC3 expression and in the amount of LC3-II. | ||||
| [71,79] | Germ free or antibiotic-treated animals | Antibiotic treatment of mice fed a normal diet: Alteration of the basal expression of LC3 compared to controls. | Germ free piglets: Changes in the expression levels of LC3A, LC3B, and Becn1 and of mTOR, p-mTOR, AKT, and p-AKT levels compared to normal and/or FMT piglets. | |
| [55,56,75,76,78] | Probiotics | SLAB512: Modulation of SIRT1 activity and changes in the expression levels of Beclin-1, p62, and SIRT-1 as well as in the LC3-II amount in AD mice1. | Limosilactobacillus reuteri: Modulation of the expression levels of mTOR and p-AKT in HFD-fed rats. | Lacticaseibacillus rhamnosus, Pediococcus acidilactici, Bifidobacterium adolescentis: Changes in the expression levels of LC3 and ATG7 in rats fed a high-calorie diet. |
| [52,71,74,77,80] | Gut microbiota-derived products | UA: Modulation of LC3-II/LC3-I and p-mTOR/mTOR ratio and changes in the expression levels of ATG7 and p62 in mouse models of aging3. | SCFAs: Activation of the PPARγ-UCP2-AMPK pathway, and induction of autophagy flux and lysosomal activity in mouse hepatocyte AML-12 cells. | UA: Induction of mitophagy in Caenorhabditis elegans and in rodents. |
| FXR and TGR54: Involved in autophagy modulation. | UB: Modulation of LC3-II/LC3-I, p-mTOR/mTOR and p-ULK1/ULK1 ratio and change in the expression level of p62 in a rat model of ischemia/reperfusion injury. | |||
Although few studies are available on this emerging topic, they suggest that the gut microbiota could influence autophagy in the brain throughout life in both physiological and pathological conditions.
Diet is a key environmental factor that drives the composition and metabolic functions of the gut microbiota[46,47]. In particular, maternal diet can influence post-natal gut microbiota and neurological development of the offspring[48]. In a recent study, Wang and colleagues reported that feeding mothers with a high sugar and high fat (HSHF) diet, a condition that modifies the gut microbiota of the offspring, modulates also the expression of neuronal and autophagy markers in the brain during early life stage[49]. Particularly, they observed that the LC3A and LC3B levels were modified in the brain of the offspring in the HSHF group compared to controls before 28 d of age, and then decreased, meaning that autophagy may be differentially regulated in HSHF offspring[49].
Aging is associated with a decline of host autophagy including in the brain[50]. Influence of the gut microbiota on brain autophagy in aging has been evidenced in in vivo models. Alteration of autophagy has been reported in the brain of D-gal-treated mice, a model of accelerated aging[51,52]. These alterations were characterized by decreases in the LC3-II/LC3-I ratio and in the expression of ATG7 and SIRT1, as well as by increased phosphorylation of the master negative regulator of autophagy mTOR (S2448) and expression of p62 in the hippocampus tissue of D-gal-induced aging mice[52]. Interestingly, the administration of urolithin A (UA), a bioactive metabolite generated by the gut microbiota, was efficient in rescuing these autophagy-related defects. To note, UA administration also allowed to reverse increases in the LC3-II/LC3-I ratio, the expression of p62, and the phosphorylation of mTOR (S2448), as well as the decreased expression of Sirt-1 and ATG7 observed in the hippocampus of 12-mo-old mice[52].
Autophagy defect is thought to play a role in neurodegenerative processes associated with numerous diseases, including Alzheimer’s disease (AD)[53]. Interestingly, although a causal relationship remains to be demonstrated, a few studies suggest that dysbiosis associated with AD could influence brain autophagy[54]. Decreased Beclin-1 expression and increased expression of p62 have been observed in the brain of old 3xTg-AD mice (a transgenic mouse model of AD) compared to young control mice, indicating alterations in autophagy[55]. Interestingly, in addition to modifying the composition and predicted function of the gut microbiota, oral supplementation of old 3xTg-AD mice with a combination of nine probiotic strains (Streptococcus thermophilus, Bifidobacterium longum, B. breve, B. infantis, Lactobacillus acidophilus, Lactiplantibacillus plantarum, Lacticaseibacillus paracasei, Lactobacillus delbrueckii subsp. bulgaricus, and Levilactobacillus brevis; SLAB51 formulation) also partially restored defects in autophagy[55]. Moreover, SLAB51 was also effective in restoring the impaired expression level and activity of SIRT1, a positive regulator of autophagy, in the brain of 3xTg-AD mice[56,57].
In another context, changes in the composition of the fecal microbiota have been reported in patients with acute ischemic stroke (AIS), a common cerebrovascular disease caused by sudden loss of blood circulation in a specific brain area[58,59]. Interestingly, anal administration of the fecal supernatant obtained from an AIS patient to antibiotics-treated mice resulted in increased expression of genes encoding Beclin-1, ATG12, and LC3 as well as increased expression of Beclin-1 at the protein level and an increased level of LC3-II in brain tissue compared to antibiotics-treated mice that received the fecal supernatant of healthy controls[59].
The retina, which is the light sensitive neural tissue that lines the back of the eyes, displays numerous similarities with the brain either anatomically or functionally[60]. Neurodegenerative conditions that affect the brain seem to compromise the retina, and vice versa[60-62]. Similarly to the brain, the retina is also highly sensitive to nutritional variations[63]. Retina autophagy[64,65] as well as modifications in the gut microbiota[66-69] is suspected to contribute to retinal diseases such as diabetic retinopathy, age-related macular degeneration, and glaucoma. Although no causal relationship has been yet established, one can assume that, as in the brain, the gut microbiota might influence retinal autophagy and that changes in its composition might alter retinal autophagy and contribute to the development of retinopathies.
Evidence of the influence of the gut microbiota on liver autophagy came from studies in gut microbiota-deprived mouse models. Comparison of germ-free mice and altered Schaedler’s flora (a community of eight bacterial species) colonized mice revealed that absence of the gut microbiota altered hepatic expression of genes involved in autophagy and lysosomal functions[70]. In another study, a decrease in the expression of LC3 at the protein level has been reported in the liver of mice deprived from gut microbiota as a consequence of chronic treatment with antibiotics (ampicillin and neomycin) compared to control mice[71]. In addition, those authors showed that microbial-derived SCFAs (propionate and butyrate) activated autophagy, induced lysosomal activity, and increased autophagy flux in vitro in mouse hepatocyte AML-12 cells[71]. The mechanism involves the activation of the PPARγ-UCP2-AMPK pathway[71].
Primary bile acids are synthesized from cholesterol in the liver and are converted into secondary bile acids by the gut microbiota[72]. Bile acids are signaling molecules that can activate nuclear hormone receptors including FXR and TGR5 (also known as GPBAR1), which is a cell-surface receptor of the G protein-coupled receptor family[73]. These two bile acid receptors have been described to modulate autophagy in the liver and adipose tissue in fed and fasted states[74].
Several alterations of autophagy, including a decreased amount of LC3 mRNA and LC3-II and an increased amount of p62, have been observed in the liver of mice fed a high-fat diet (HFD), a potent inducer of dysbiosis[74]. Chronic exposure of rats to an HFD can lead to NASH (non-alcoholic fatty steatohepatitis). Development of this liver disease has been associated with dysbiosis and alterations in autophagy, particularly increased expression of hepatic mTOR and p-AKT[75,76]. Interestingly, supplementation of an HFD with a probiotic strain (Limosilactobacillus reuteri) and/or treatment of NASH mice with antibiotics (metronidazole) tended to normalize the hepatic content of these two autophagy-related proteins, as well as SCFAs and Firmicutes and Bacteroidetes fecal contents, thus suggesting a role of the gut microbiota in the modulation of hepatic autophagy[75,76]. To note, some data suggest a role for TGR5 in the regulation of autophagy in response to HFD[74].
An induction of autophagy, characterized by decreased phosphorylation of mTOR (S2448) and ULK1 (S757), an increased amount of LC3-II, and decreased expression of p62, has been reported in a rat model of ischemia/reperfusion injury[77]. Interestingly, intraperitoneal injection of urolithin B (UB), a gut microbiota-derived metabolite, was able to reverse this phenotype[77]. The inhibitory effect of UB on autophagy is thought to activate the Nrf2-related antioxidant response by increasing p62 accumulation and favoring p62-Keap1interaction[77]. Another argument that suggests the influence of the gut microbiota on heart autophagy has been provided by changes in the expression levels of LC3 and ATG7 observed in heart tissue of rats fed a high-calorie diet supplemented with probiotics (Lacticaseibacillus rhamnosus, Pediococcus acidilactici, and Bifidobacterium adolescentis)[78].
In addition to the heart, autophagy might be regulated by the gut microbiota in other muscles. Recently, high-throughput RNA-seq analysis revealed that the expression levels of autophagy-related genes (LC3A, LC3B, and Beclin-1) were modulated in the skeletal muscles of germ-free piglets compared to control piglets[79]. Moreover, germ-free piglets harbored decreased expression of mTOR and AKT and their phosphorylated forms, phospho-mTOR (S2448) and phospho-AKT (S473), respectively, compared to control piglets[79]. FMT of germ-free piglets with stools collected on healthy donors pigs was effective in restoring the amounts of phospho-AKT and mTOR to a level similar to that of controls[79]. Some microbial-derived metabolites able to influence the muscle autophagy have been identified. For example, a role of UA as a mitophagy (selective degradation of mitochondria by autophagy) inducer in the muscle tissue has been described in the model organism Caenorhabditis elegans and in rodents[80].
As developed in the first part of this review, the gut microbiota is able to influence host autophagy by several pathways and through complex regulatory networks governing the autophagy machinery. Reciprocally, autophagy and autophagy-related proteins can shape the gut microbiota (Figure 1). This is particularly well illustrated by changes in the gut microbiota composition observed in mice conditionally deficient for autophagy (Atg5-/-, Atg7-/-, and ATG16L1 T300A knock-in) in the gut[81-83]. Interestingly, alterations of autophagy in peripheral organs such as the liver have been shown to influence the composition of the gut microbiota[84].
A first overall reason that would explain why autophagy activity in the gut mucosa can modulate the abundance of gut microorganisms is that this process is essential to maintain homeostasis of their ecological niche. Indeed, basal autophagy is crucial to maintain the integrity of Lgr5-positive intestinal stem cells that give rise to all differentiated lineages of the intestinal epithelium throughout life[85]. In addition, autophagy contributes to the maintaining of intestinal barrier integrity, particularly by regulating proteins involved in tight junctions (e.g., Claudin-2 and Occludin) on the apical side of intestinal epithelial cells and by promoting cell survival upon various stress (e.g., bacterial or viral infection, inflammation, or chemical stress)[4,86-88].
The main cellular mechanisms by which host autophagy shapes the gut microbiota (including pathosymbionts) are described below.
Autophagy mediates the bulk or selective lysosomal degradation of cellular components. In selective autophagy, selective autophagy receptors (SARs) recognize and bind specific cargoes to promote phagophore formation around them, ultimately leading to their degradation into a mature autolysosome. These specific cargoes can be for instance mitochondria (mitophagy), lipid droplets (lipophagy), protein aggregates (aggrephagy), or peroxysomes (pexophagy)[89]. A selective form of autophagy termed xenophagy is dedicated to the elimination of intracellular pathogens (e.g., bacteria, viruses, fungi, or protozoa) and is supported by the expression of several SARs including NDP52, Optineurin, p62, TAX1BP1, Galectin 8, and TECPR1[90]. Xenophagy has been shown to restrict or avoid the intracellular persistence and the replication of various human pathogenic or pathosymbiotic bacteria, residing either in damaged vacuoles [e.g., Salmonella Typhimurium or adherent-invasive Escherichia coli (AIEC)] or free in the host cytosol (Group A Streptococcus)[91-93]. Thus, by limiting the dissemination of invasive pathogens from the gut lumen to extra-intestinal sites, autophagy also restrains their persistency in the gut microbiota[94,95]. Defects in xenophagy are thought to contribute to the etiology of Crohn’s disease (CD) an inflammatory bowel disease (IBD) characterized by chronic and severe intestinal inflammation associated with dysbiosis[96]. In particular, a coding polymorphism (Thr300Ala) in the autophagy-related gene ATG16L1 that confers an increased risk for the development of CD has been shown in vitro and in vivo to alter the xenophagy process, thus favoring persistency of the CD-associated AIEC bacteria[92,97,98]. CD risk polymorphisms have also been identified in other autophagy-related genes, including core autophagy genes (IRGM, ULK1, ATG4a, and ATG4d) and genes involved more specifically in xenophagy (NOD2 and NDP52)[99-101].
One important point is that, besides xenophagy, non-canonical autophagy such as LC3-associated phagocytosis (LAP) can also contribute to the clearance of intracellular pathogens. This specific form of phagocytosis requires an important set of core autophagy proteins (UVRAG, BECN1, VPS34, LC3, ATG3, ATG4, ATG5, ATG7, ATG12, and ATG16L1), but some other proteins involved in canonical autophagy remain dispensable (ATG14, ULK1, FIP200, and AMBRA1). LAP also distinguishes from canonical autophagy by the formation of single-membrane vacuoles called LAPosomes[102]. Efficiency of LAP to increase clearance of pathogens such as Listeria monocytogenes or Aspergillus fumigatus has been shown[103,104].
A mucus layer composed of highly glycosylated proteins (mucins) overlays the gut epithelium and represents an important physical barrier limiting the contact of luminal microbes with the epithelium, thus avoiding their potential translocation into underlying tissues[105]. The mucus layer differs between the small and large intestine in terms of physicochemical properties (e.g., thickness, density, and composition) and it is under the influence of numerous factors, including the gut microbiota and the diet[106-108]. Whereas in the small intestine the mucus is non-attached and constitutes a discontinuous layer, it is organized in two layers - the inner and outer mucus layers - in the large intestine. Compared to the intestinal lumen, only few bacterial species are able to live and to persist in the mucus layer. This is partly due to the important amount of various antimicrobial compounds (e.g., IgA, lysozyme, defensins, REG3γ, and phospholipase A2-IIA) found in the mucus layer, particularly in the small intestine. However, some commensal bacteria are molecularly equipped to bind, degrade the mucus glycans, and/or harvest the oligosaccharides, giving them a selective advantage in colonizing this particular ecological niche[109]. Among others, mucin-degrading specialists include species belonging to the genera Bacteroides (e.g., B. thetaiotaomicron and B. fragilis), Ruminococcus (e.g., R. gnavus and R. torques), and Akkermansia (e.g., A. muciniphila). Interestingly, A. muciniphila, a bacterial species belonging to the phylum Verrucomicrobia, is considered as a healthy marker of the intestine since its presence in high abundance is associated with a healthy mucosa whereas reduction of its abundance is associated with intestinal disorders (e.g., obesity and IBD)[110,111]. Studies suggest that the composition of mucus-associated microbiota differs depending on the intestinal segment or the mucus layer (outer or inner layer) that is considered[105]. Bacteria belonging to the phylum Firmicutes have been found in higher abundance in the mucus layer than Bacteroidetes, both in humans and in rodents[105].
Mucus plays a critical role in the maintenance of the symbiotic relationship between the host and the gut microbiota[112]. Deletion of the Muc2 gene in mice results in changes in the gut microbiota composition characterized in particular by an increase in the abundance of potential pathobionts (e.g., Desulfovibrio, Escherichia, and Erysipelotrichaceae), and the reduction of beneficial bacteria (e.g., Lactobacilli) and Lachnospiraceae[112]. In addition to ensuring an habitat and energy sources for a specific part of the gut microbiota, the mucus constitutes a protective layer against pathogen invasion and infection, although some pathogenic bacteria have developed efficient strategies to colonize this special environment and reach the intestinal epithelium (e.g., Shigella flexneri and AIEC)[113,114]. Thus, modifications in mucus layer structure or composition by genetic and environmental factors, such as diet, can modify the gut microbiota[105]. These changes can be beneficial when they strengthen the mucus barrier properties, but they can also be deleterious by favoring emergence of pathobionts, by bringing harmful bacteria closer to the epithelial barrier and by destabilizing the symbiotic relationship between the gut microbiota and the host, at the gut mucosa as well as at systemic levels[107].
Mucus secretion into the gut lumen is achieved by specialized secretory cells, the goblet cells. Mucins, the proteins forming the mucus, are packed into secretory granules that are localized on the apical side of the goblet cells and constitutively secreted by fusion of the granules with the plasma membrane. Proteins belonging to the core autophagy machinery (ATG5, ATG7, and LC3B) are critical in mice for the release of these secretory granules by supporting the generation of reactive oxygen species[115].
The NLRP6 inflammasome has been identified, among others roles, as a key factor involved in autophagy-induced regulation of goblet cell secretory functions[116,117]. NLRP6-deficient mice exhibit defective autophagy in intestinal cells including in goblet cells, a phenotype that is associated with impaired mucus layer formation. This mucus alteration may contribute, together with the other NLRP6-related defects, to modulating the composition of the gut microbiota and abnormally bring microbes closer to the epithelial barrier in NLRP6-deficient mice. Analyses of the gut microbiota in NLRP6-deficient mice revealed an abnormal representation of the bacterial phyla Bacteroidetes (Prevotellaceae) and Saccharibacteria (formerly known as TM7)[116]. In addition, alteration of the mucus layer in NLRP6-deficient mice enables Citrobacter rodentium, a mouse-specific pathogen, to penetrate deeper into the crypts and be more invasive[117]. The role of autophagy in shaping the gut microbiota through the regulation of mucus layer maintenance is also supported by observations made in Atg7-deficient mice. Secretion of mucins from goblet cells was diminished in colonic-epithelial cell-specific Atg7 knock-out mice[82]. This phenotype was associated with an abnormal composition of the gut microbiota characterized in particular by an increased abundance of Clostridia and Prevotellaceae in Atg7-deficient mice. In addition, those authors observed an increased bacterial burden in the colon, a phenotype that could contribute to the exacerbated sensitivity to experimental colitis observed in Atg7 knock-out mice. Interestingly, stimulation of the autophagy-related process, either by a beneficial bacterial strain (Bifidobacterium dentium) or by a polyphenol (oxyresveratrol), has been shown to enhance mucin production by goblet cells in in vivo and in vitro models[118,119].
Autophagy and autophagy-related proteins can also affect the composition of the gut microbiota by regulating the secretion of some antimicrobial compounds released into the gut lumen by enterocytes, Paneth cells, or immune cells. Among them, immunoglobulins of the A class (IgAs) are daily released in huge amount (several grams per day) into the gut lumen and shape the composition of the gut microbiota. Alterations of the gut microbial ecosystem have been reported in the absence of hypermutated intestinal IgA in mice with deficiency of activation-induced cytidine deaminase[120-122]. Changes in the gut microbiota were particularly characterized by expansion of anaerobic bacteria in the small intestine, with a domination by segmented filamentous bacteria[121]. Several other studies in mouse models support the role of IgAs in regulating the diversity and composition of microbiota[123,124]. Data obtained in humans showed that selective IgA-deficiency (sIgAd) is associated with a mild intestinal dysbiosis, characterized by expansion of pro-inflammatory bacteria (e.g., E. coli, Prevotella), reduction of anti-inflammatory commensals (e.g., Faecalibacterium), and perturbation of bacterial dependency association network[125]. In addition, Catanzaro and colleagues reported also a trend toward a decreased alpha diversity and shifts in the relative abundance of some taxa (e.g., increase in Eubacterium dolichum and Ruminococcus bromii and decrease in Paraprevotellaceae) in human sIgAd subjects compared to controls[126]. IgAs are produced by gut-resident antibody-secreting plasma cells (PCs) that display important metabolic adaptations and endoplasmic reticulum expansion to cope with the stress of producing very large amounts of IgAs[127]. Some studies suggest that autophagy is required for sustainable production of immunoglobulins by PCs since mice with conditional deficiency of Atg5 in B cells had defective antibody responses, with an increased sensitivity of PCs to cell death[128]. In addition, mice deficient for Atg5 in B cells harbored a decreased number of IgA-secreting PCs isolated from the gut-associated lamina propria, Peyer’s patches, and mesenteric lymph nodes in comparison to control mice[129].
Another important antimicrobial compound to which commensal bacteria are directly exposed in the gut lumen is the lysozyme secreted by Paneth cells, which are secretory epithelial cells located at the bottom of the crypts in the small intestine. This antimicrobial protein is also produced by macrophages and neutrophils in the lamina propria. Three types of lysozyme have been described so far across the animal kingdom[130]. Lysozyme causes bacterial lysis by hydrolyzing bacterial cell wall PGN, but it can also induce cationic killing of bacteria by inserting into and forming pores into the lipid bilayer of the bacterial cell membrane. This is the case with c-type lysozyme expressed in human[130]. Not all bacteria are equally sensitive to lysozyme and some pathogenic bacteria have developed strategies to escape its antimicrobial activity[130]. The contribution of lysozyme in shaping the gut microbiota is illustrated by the dysbiosis observed in lysozyme-deficient mice (Lyz1−/− mice) that is characterized by the expansion of some mucolytic bacteria such as Blautia gnavus (formerly known as Ruminococcus gnavus)[130,131]. No change in luminal bacterial load and alpha-diversity was observed in the cecum- and mucosal-associated bacteria in the ileum and the colon of Lyz1−/−mice[131]. However, changes occurred in the composition of the fecal microbiota (expansion of Dorea formicigenerans and reduction of Candidatus Arthromitus) as well as the ileal microbiota (expansion of B. gnavus and D. formicigenerans and reduction of C. Arthromitus) in Lyz1−/−mice[131].
Alpha-defensins (also called crypt defensins or cryptdins) are another example of antimicrobial factors that are produced by Paneth cells, whose roles in host defense against enteric pathogens and regulation of the composition of the gut indigenous microbiota have been described[132]. Interestingly, abnormal packaging and secretion of antimicrobial compounds by Paneth cells have been reported in mice harboring Paneth cells deficient for the autophagy-related genes Atg5, Atg7, and Atg16l1 and in patients with CD-associated NOD2 and ATG16L1 variants[133-135]. Of note, this defect in lysozyme packaging in autophagy-deficient mice required an infectious (viral or bacterial) trigger[136,137].
Even if canonical autophagy is considered as a degradative process, some infectious agents such as Salmonella Typhimurium can trigger a secretory autophagy resulting in the formation of LC3-positive, double-membraned lysozyme granules[136]. These autophagosome-like vacuoles are not directed for the fusion with the lysosomes but instead reach the plasma membrane for the release of their content into the gut lumen. Thus, the autophagy machinery participates in the unconventional protein secretion of lysozyme, thereby affecting the composition of the gut microbiota by counter-selecting the lysozyme-sensitive bacteria. In this context, it has been suggested that vitamin D, via binding to the vitamin D receptor expressed by Paneth cells, can sustain autophagy activities in these cells[138]. To note, several studies suggest that expression and secretion of other antimicrobial peptides than lyzozyme, such as the defensins and cathelicidins, would be regulated by autophagy. However, the exact molecular mechanisms remain to be determined[82,139].
Cell stimulation by microorganisms (e.g., invasive pathogens) or danger signals (e.g., extracellular ATP, uric acid, or HMGB1) are usually associated with the triggering of inflammatory processes through the release of cytokines and chemokines. Inflammation is a protective response that results in tissue repair. However, this response needs to be tightly regulated in order to avoid excessive and/or chronic inflammation that could be detrimental for host tissues. In the gut mucosa, immune tolerance toward the resident gut microbiota should be maintained to avoid chronic gut inflammation and sustain homeostasis[140]. Unbalanced inflammatory responses can also alter the gut microbiota as shown in mouse models of colitis that mimic human IBD, in which inflammation induces microbial dysbiosis[141,142]. Chronic inflammatory state was also suggested to contribute to dysbiosis in IBD patients[143]. This inflammation-driven bacterial dysbiosis is commonly characterized by an overall decrease in bacterial diversity, especially in Firmicutes (Clostridium groups) and an overgrowth of species belonging to Enterobacteriaceae[143,144].
Autophagy machinery and autophagy-related proteins are key contributors to the regulation of the inflammatory processes. Thus, one could assume that modulation of inflammation by autophagy could influence the composition of the gut microbiota. Autophagy is usually considered as an anti-inflammatory process, particularly since it controls activation of inflammasomes that are multimeric protein complexes involved in the maturation of pro-inflammatory cytokines[145]. Mice deficient for Atg16l1 in haematopoietic cells have been shown to be highly sensitive to chemically-induced colitis and produce increased levels of IL-1β and IL-18, two cytokines processed by inflammasomes[146]. Atg16l1-deficient macrophages that were stimulated by LPS also produced higher amounts of these cytokines compared to wild-type macrophages. Autophagy can alleviate activation of inflammasomes, at least by removing stimuli that induced them (e.g., intracellular infectious agents) and by degrading some inflammasome components (e.g., NLRP1, NLRP3, AIM2, or pro-CASP1)[147]. Interestingly, alterations of the gut microbiota (e.g., increased abundance of Bacteroidetes) as well as enhancement of the local Th1 and Th17 immune responses have been reported in mice with dextran sodium sulfate (DSS) colitis that express the CD risk allele ATG16L1 T300A - a genetic context known to impair some autophagy-related functions - compared to DSS-treated wild-type mice[81]. Similar observations have been made in gnotobiotic mice expressing the CD risk allele ATG16L1 T300A and inoculated with human stools from active CD patients[81]. These data illustrated how a subtle polymorphism in an autophagy-related gene could deeply impact the equilibrium between immune responses and the gut microbiota.
Autophagy is also able to modulate signaling of interferons, notably by degrading key players of type-I interferon responses (e.g., RIG-I, STING, MDA5, IRF3, MAVS, and cGAS)[148]. Abnormal regulation of interferon signaling can lead to alterations of the gut microbiota as described in knock-out mice and viral infection models[149]. Interestingly, the gut microbiota has been described to stimulate intestinal autophagy via the induction of the type-II interferon, and this microbiota-mediated activation of autophagy has been shown to protect the host against infection by the protozoan parasite Toxoplasma gondii by limiting the deleterious production of the pro-inflammatory cytokine TNF-α[150]. Autophagy has also been described to limit the production and the secretion of various cytokines including TNF-α, IL-1β, IL-23, IL-6, TGF-β, and MIF[151,152]. However, the molecular mechanisms by which autophagy regulates their expression remain elusive. In many cases, autophagy reduces secretion of cytokines by simply alleviating cellular stress that triggers the inflammatory responses.
Given its crucial role in regulating homeostasis at both cell and tissue levels, it is not surprising that alterations of autophagy are connected to a large number of disorders (e.g., IBD, cancers, and neurodegenerative diseases). To assume its various functions, autophagy activation is tightly regulated and the gut microbiota has recently emerged as a contributor in its regulatory networks in both the gut mucosa and other tissues. This advance in the understanding of the molecular mechanisms supporting this highly integrated cellular process that tip the balance between health and disease offers new opportunities to develop preventive or therapeutic tools. Indeed, the gut microbiota appears as a promising target to restore functional autophagy or to prevent its alterations in various disease conditions. The growing interest that was aroused from the discovery of such a hub position occupied by the gut microbiota in maintaining physical and mental health status has led to the conceptualization, development, and/or examination of various tools to manipulate the gut microbiota (probiotics, prebiotics, synbiotics, postbiotics, FMT, Crispr/Cas9, diet…). In the era of personalized medicine, such a toolbox could constitute a key element that could be integrated in the therapeutic strategies. However, further explorations of the interplay between the gut microbiota and autophagy are needed. Important advances have been made in understanding the local dialogue between the gut microbiota and autophagy at the level of the gut mucosa, but less is known about how and in which extent they communicate at the systemic level. Bi-directionality of the interactions between the gut microbiota and the autophagy network, plasticity and complexity of the gut microbiota and its multiple effects on host, as well as pleiotropy of the functions of autophagy are all factors that increase the level of complexity of the system. Better characterization of the cellular and molecular actors from both sides - the gut microbiota and autophagy - that contribute and regulate the framework of their interactions to maintain homeostasis constitutes a prerequisite to propose new preventive and therapeutic tools in pathological conditions associated with dysbiosis and/or autophagy dysfunction.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Microbiology
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Shi JL, Zhai Q S-Editor: Liu M L-Editor: Wang TQ P-Editor: Liu M
| 1. | Matijašić M, Meštrović T, Paljetak HČ, Perić M, Barešić A, Verbanac D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int J Mol Sci. 2020;21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 104] [Article Influence: 26.0] [Reference Citation Analysis (0)] |
| 2. | Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, Nauta A, Scott K, Stahl B, van Harsselaar J, van Tol R, Vaughan EE, Verbeke K. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11:411-455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 193] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 3. | LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160-168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 790] [Cited by in F6Publishing: 825] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 4. | Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. How autophagy controls the intestinal epithelial barrier. Autophagy. 2021;1-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 115] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 5. | Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22:1079-1089. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 665] [Cited by in F6Publishing: 695] [Article Influence: 86.9] [Reference Citation Analysis (0)] |
| 6. | Morais LH, Schreiber HL 4th, Mazmanian SK. The gut microbiota-brain axis in behaviour and brain disorders. Nat Rev Microbiol. 2021;19:241-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 727] [Article Influence: 181.8] [Reference Citation Analysis (0)] |
| 7. | Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369-2379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1826] [Cited by in F6Publishing: 1887] [Article Influence: 235.9] [Reference Citation Analysis (0)] |
| 8. | Schaubeck M, Clavel T, Calasan J, Lagkouvardos I, Haange SB, Jehmlich N, Basic M, Dupont A, Hornef M, von Bergen M, Bleich A, Haller D. Dysbiotic gut microbiota causes transmissible Crohn's disease-like ileitis independent of failure in antimicrobial defence. Gut. 2016;65:225-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 255] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 9. | Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027-1031. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7820] [Cited by in F6Publishing: 8095] [Article Influence: 476.2] [Reference Citation Analysis (1)] |
| 10. | Zenewicz LA, Yin X, Wang G, Elinav E, Hao L, Zhao L, Flavell RA. IL-22 deficiency alters colonic microbiota to be transmissible and colitogenic. J Immunol. 2013;190:5306-5312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 187] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 11. | Al Nabhani Z, Lepage P, Mauny P, Montcuquet N, Roy M, Le Roux K, Dussaillant M, Berrebi D, Hugot JP, Barreau F. Nod2 Deficiency Leads to a Specific and Transmissible Mucosa-associated Microbial Dysbiosis Which Is Independent of the Mucosal Barrier Defect. J Crohns Colitis. 2016;10:1428-1436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Yang M, Liu Y, Xie H, Wen Z, Zhang Y, Wu C, Huang L, Wu J, Xie C, Wang T, Peng W, Liu S, Chen L, Liu X. Gut Microbiota Composition and Structure of the Ob/Ob and Db/Db Mice. Int J Endocrinol. 2019;2019:1394097. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Lahiri V, Hawkins WD, Klionsky DJ. Watch What You (Self-) Eat: Autophagic Mechanisms that Modulate Metabolism. Cell Metab. 2019;29:803-826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 177] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 14. | Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, Choi AM, Chu CT, Codogno P, Colombo MI, Cuervo AM, Debnath J, Deretic V, Dikic I, Eskelinen EL, Fimia GM, Fulda S, Gewirtz DA, Green DR, Hansen M, Harper JW, Jäättelä M, Johansen T, Juhasz G, Kimmelman AC, Kraft C, Ktistakis NT, Kumar S, Levine B, Lopez-Otin C, Madeo F, Martens S, Martinez J, Melendez A, Mizushima N, Münz C, Murphy LO, Penninger JM, Piacentini M, Reggiori F, Rubinsztein DC, Ryan KM, Santambrogio L, Scorrano L, Simon AK, Simon HU, Simonsen A, Tavernarakis N, Tooze SA, Yoshimori T, Yuan J, Yue Z, Zhong Q, Kroemer G. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811-1836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1125] [Cited by in F6Publishing: 1076] [Article Influence: 153.7] [Reference Citation Analysis (0)] |
| 15. | Zhao YG, Codogno P, Zhang H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat Rev Mol Cell Biol. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 199] [Article Influence: 66.3] [Reference Citation Analysis (0)] |
| 16. | Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 767] [Cited by in F6Publishing: 884] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 17. | Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24:9-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 700] [Article Influence: 63.6] [Reference Citation Analysis (0)] |
| 18. | Khandia R, Dadar M, Munjal A, Dhama K, Karthik K, Tiwari R, Yatoo MI, Iqbal HMN, Singh KP, Joshi SK, Chaicumpa W. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 19. | Levine B, Kroemer G. Biological Functions of Autophagy Genes: A Disease Perspective. Cell. 2019;176:11-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in F6Publishing: 1551] [Article Influence: 310.2] [Reference Citation Analysis (0)] |
| 20. | Nikoletopoulou V, Papandreou ME, Tavernarakis N. Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ. 2015;22:398-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 150] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 21. | Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619-1628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 22. | Galluzzi L, Green DR. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019;177:1682-1699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 339] [Cited by in F6Publishing: 513] [Article Influence: 128.3] [Reference Citation Analysis (0)] |
| 23. | Oh JE, Lee HK. Pattern recognition receptors and autophagy. Front Immunol. 2014;5:300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Rashid HO, Yadav RK, Kim HR, Chae HJ. ER stress: Autophagy induction, inhibition and selection. Autophagy. 2015;11:1956-1977. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 437] [Cited by in F6Publishing: 524] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 25. | Ro SH, Fay J, Cyuzuzo CI, Jang Y, Lee N, Song HS, Harris EN. SESTRINs: Emerging Dynamic Stress-Sensors in Metabolic and Environmental Health. Front Cell Dev Biol. 2020;8:603421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Kang R, Livesey KM, Zeh HJ 3rd, Lotze MT, Tang D. HMGB1 as an autophagy sensor in oxidative stress. Autophagy. 2011;7:904-906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2401] [Cited by in F6Publishing: 2580] [Article Influence: 184.3] [Reference Citation Analysis (0)] |
| 28. | Young CN, Sinadinos A, Lefebvre A, Chan P, Arkle S, Vaudry D, Gorecki DC. A novel mechanism of autophagic cell death in dystrophic muscle regulated by P2RX7 receptor large-pore formation and HSP90. Autophagy. 2015;11:113-130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Zierhut C, Funabiki H. Regulation and Consequences of cGAS Activation by Self-DNA. Trends Cell Biol. 2020;30:594-605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517-526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1156] [Cited by in F6Publishing: 1153] [Article Influence: 88.7] [Reference Citation Analysis (0)] |
| 31. | Fan Q, Guan X, Hou Y, Liu Y, Wei W, Cai X, Zhang Y, Wang G, Zheng X, Hao H. Paeoniflorin modulates gut microbial production of indole-3-lactate and epithelial autophagy to alleviate colitis in mice. Phytomedicine. 2020;79:153345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 32. | Meng D, Sommella E, Salviati E, Campiglia P, Ganguli K, Djebali K, Zhu W, Walker WA. Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr Res. 2020;88:209-217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 127] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 33. | Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135-144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 714] [Cited by in F6Publishing: 688] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 34. | Arroyo DS, Soria JA, Gaviglio EA, Garcia-Keller C, Cancela LM, Rodriguez-Galan MC, Wang JM, Iribarren P. Toll-like receptor 2 Ligands promote microglial cell death by inducing autophagy. FASEB J. 2013;27:299-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 35. | Travassos LH, Carneiro LA, Ramjeet M, Hussey S, Kim YG, Magalhães JG, Yuan L, Soares F, Chea E, Le Bourhis L, Boneca IG, Allaoui A, Jones NL, Nuñez G, Girardin SE, Philpott DJ. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55-62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 916] [Cited by in F6Publishing: 967] [Article Influence: 64.5] [Reference Citation Analysis (0)] |
| 36. | Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012;287:34149-34156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 161] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 37. | Öhman T, Teirilä L, Lahesmaa-Korpinen AM, Cypryk W, Veckman V, Saijo S, Wolff H, Hautaniemi S, Nyman TA, Matikainen S. Dectin-1 pathway activates robust autophagy-dependent unconventional protein secretion in human macrophages. J Immunol. 2014;192:5952-5962. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 38. | Macias-Ceja DC, Cosín-Roger J, Ortiz-Masiá D, Salvador P, Hernández C, Esplugues JV, Calatayud S, Barrachina MD. Stimulation of autophagy prevents intestinal mucosal inflammation and ameliorates murine colitis. Br J Pharmacol. 2017;174:2501-2511. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 39. | Sarkar S, Davies JE, Huang Z, Tunnacliffe A, Rubinsztein DC. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem. 2007;282:5641-5652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 802] [Cited by in F6Publishing: 827] [Article Influence: 45.9] [Reference Citation Analysis (0)] |
| 40. | Choy A, Dancourt J, Mugo B, O'Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012;338:1072-1076. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 377] [Cited by in F6Publishing: 342] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 41. | Niu H, Xiong Q, Yamamoto A, Hayashi-Nishino M, Rikihisa Y. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A. 2012;109:20800-20807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 42. | Cheng S, Ma X, Geng S, Jiang X, Li Y, Hu L, Li J, Wang Y, Han X. Fecal Microbiota Transplantation Beneficially Regulates Intestinal Mucosal Autophagy and Alleviates Gut Barrier Injury. mSystems. 2018;3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 43. | Feng Y, Huang Y, Wang Y, Wang P, Song H, Wang F. Antibiotics induced intestinal tight junction barrier dysfunction is associated with microbiota dysbiosis, activated NLRP3 inflammasome and autophagy. PLoS One. 2019;14:e0218384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 114] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 44. | Singh SB, Wilson M, Ritz N, Lin HC. Autophagy Genes of Host Responds to Disruption of Gut Microbial Community by Antibiotics. Dig Dis Sci. 2017;62:1486-1497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 45. | Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, Chen Y, Chen H, Hong J, Zou W, Fang JY. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170:548-563.e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 814] [Cited by in F6Publishing: 1154] [Article Influence: 164.9] [Reference Citation Analysis (0)] |
| 46. | Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. Influence of diet on the gut microbiome and implications for human health. J Transl Med. 2017;15:73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1089] [Cited by in F6Publishing: 1317] [Article Influence: 188.1] [Reference Citation Analysis (0)] |
| 47. | Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1187] [Cited by in F6Publishing: 1288] [Article Influence: 161.0] [Reference Citation Analysis (0)] |
| 48. | Al Rubaye H, Adamson CC, Jadavji NM. The role of maternal diet on offspring gut microbiota development: A review. J Neurosci Res. 2021;99:284-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 49. | Wang D, Zhang H, Zeng M, Tang X, Zhu X, Guo Y, Qi L, Xie Y, Zhang M, Chen D. Maternal high sugar and fat diet benefits offspring brain function via targeting on the gut-brain axis. Aging (Albany NY). 2021;13:10240-10274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 50. | Barbosa MC, Grosso RA, Fader CM. Hallmarks of Aging: An Autophagic Perspective. Front Endocrinol (Lausanne). 2018;9:790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 147] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 51. | Azman KF, Zakaria R. D-Galactose-induced accelerated aging model: an overview. Biogerontology. 2019;20:763-782. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 189] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 52. | Chen P, Chen F, Lei J, Li Q, Zhou B. Activation of the miR-34a-Mediated SIRT1/mTOR Signaling Pathway by Urolithin A Attenuates D-Galactose-Induced Brain Aging in Mice. Neurotherapeutics. 2019;16:1269-1282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 53. | Chen S, Zhou Q, Ni Y, Le W. Autophagy and Alzheimer's Disease. Adv Exp Med Biol. 2020;1207:3-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 54. | Bostanciklioğlu M. The role of gut microbiota in pathogenesis of Alzheimer's disease. J Appl Microbiol. 2019;127:954-967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 55. | Bonfili L, Cecarini V, Berardi S, Scarpona S, Suchodolski JS, Nasuti C, Fiorini D, Boarelli MC, Rossi G, Eleuteri AM. Microbiota modulation counteracts Alzheimer's disease progression influencing neuronal proteolysis and gut hormones plasma levels. Sci Rep. 2017;7:2426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 266] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 56. | Bonfili L, Cecarini V, Cuccioloni M, Angeletti M, Berardi S, Scarpona S, Rossi G, Eleuteri AM. SLAB51 Probiotic Formulation Activates SIRT1 Pathway Promoting Antioxidant and Neuroprotective Effects in an AD Mouse Model. Mol Neurobiol. 2018;55:7987-8000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 146] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 57. | Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008;105:3374-3379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1134] [Cited by in F6Publishing: 1112] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 58. | Battaglini D, Pimentel-Coelho PM, Robba C, Dos Santos CC, Cruz FF, Pelosi P, Rocco PRM. Gut Microbiota in Acute Ischemic Stroke: From Pathophysiology to Therapeutic Implications. Front Neurol. 2020;11:598. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 59. | Xu N, Kan P, Yao X, Yang P, Wang J, Xiang L, Zhu Y. Astragaloside IV reversed the autophagy and oxidative stress induced by the intestinal microbiota of AIS in mice. J Microbiol. 2018;56:838-846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 60. | London A, Benhar I, Schwartz M. The retina as a window to the brain-from eye research to CNS disorders. Nat Rev Neurol. 2013;9:44-53. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 586] [Cited by in F6Publishing: 610] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 61. | Byun MS, Park SW, Lee JH, Yi D, Jeon SY, Choi HJ, Joung H, Ghim UH, Park UC, Kim YK, Shin SA, Yu HG, Lee DY; KBASE Research Group. Association of Retinal Changes With Alzheimer Disease Neuroimaging Biomarkers in Cognitively Normal Individuals. JAMA Ophthalmol. 2021;139:548-556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 24] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 62. | Nucci C, Martucci A, Cesareo M, Garaci F, Morrone LA, Russo R, Corasaniti MT, Bagetta G, Mancino R. Links among glaucoma, neurodegenerative, and vascular diseases of the central nervous system. Prog Brain Res. 2015;221:49-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 63. | Lien EL, Hammond BR. Nutritional influences on visual development and function. Prog Retin Eye Res. 2011;30:188-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 64. | Boya P, Esteban-Martínez L, Serrano-Puebla A, Gómez-Sintes R, Villarejo-Zori B. Autophagy in the eye: Development, degeneration, and aging. Prog Retin Eye Res. 2016;55:206-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 65. | Rosa MD, Distefano G, Gagliano C, Rusciano D, Malaguarnera L. Autophagy in Diabetic Retinopathy. Curr Neuropharmacol. 2016;14:810-825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 88] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 66. | Das T, Jayasudha R, Chakravarthy S, Prashanthi GS, Bhargava A, Tyagi M, Rani PK, Pappuru RR, Sharma S, Shivaji S. Alterations in the gut bacterial microbiome in people with type 2 diabetes mellitus and diabetic retinopathy. Sci Rep. 2021;11:2738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 74] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 67. | Gong H, Zhang S, Li Q, Zuo C, Gao X, Zheng B, Lin M. Gut microbiota compositional profile and serum metabolic phenotype in patients with primary open-angle glaucoma. Exp Eye Res. 2020;191:107921. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 68. | Huang Y, Wang Z, Ma H, Ji S, Chen Z, Cui Z, Chen J, Tang S. Dysbiosis and Implication of the Gut Microbiota in Diabetic Retinopathy. Front Cell Infect Microbiol. 2021;11:646348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 58] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 69. | Lin P, McClintic SM, Nadeem U, Skondra D. A Review of the Role of the Intestinal Microbiota in Age-Related Macular Degeneration. J Clin Med. 2021;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 70. | Juanola O, Hassan M, Kumar P, Yilmaz B, Keller I, Simillion C, Engelmann C, Tacke F, Dufour JF, De Gottardi A, Moghadamrad S. Intestinal microbiota drives cholestasis-induced specific hepatic gene expression patterns. Gut Microbes. 2021;13:1-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 71. | Iannucci LF, Sun J, Singh BK, Zhou J, Kaddai VA, Lanni A, Yen PM, Sinha RA. Short chain fatty acids induce UCP2-mediated autophagy in hepatic cells. Biochem Biophys Res Commun. 2016;480:461-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Dawson PA, Karpen SJ. Intestinal transport and metabolism of bile acids. J Lipid Res. 2015;56:1085-1099. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 285] [Cited by in F6Publishing: 329] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 73. | Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 474] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 74. | Carino A, Marchianò S, Biagioli M, Scarpelli P, Bordoni M, Di Giorgio C, Roselli R, Fiorucci C, Monti MC, Distrutti E, Zampella A, Fiorucci S. The bile acid activated receptors GPBAR1 and FXR exert antagonistic effects on autophagy. FASEB J. 2021;35:e21271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 75. | Ahmed LA, Salem MB, Seif El-Din SH, El-Lakkany NM, Ahmed HO, Nasr SM, Hammam OA, Botros SS, Saleh S. Gut microbiota modulation as a promising therapy with metformin in rats with non-alcoholic steatohepatitis: Role of LPS/TLR4 and autophagy pathways. Eur J Pharmacol. 2020;887:173461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 76. | Seif El-Din SH, Salem MB, El-Lakkany NM, Hammam OA, Nasr SM, Okasha H, Ahmed LA, Saleh S, Botros SS. Early intervention with probiotics and metformin alleviates liver injury in NAFLD rats via targeting gut microbiota dysbiosis and p-AKT/mTOR/LC-3II pathways. Hum Exp Toxicol. 2021;40:1496-1509. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 77. | Zheng D, Liu Z, Zhou Y, Hou N, Yan W, Qin Y, Ye Q, Cheng X, Xiao Q, Bao Y, Luo J, Wu X. Urolithin B, a gut microbiota metabolite, protects against myocardial ischemia/reperfusion injury via p62/Keap1/Nrf2 signaling pathway. Pharmacol Res. 2020;153:104655. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 73] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 78. | Lai CH, Tsai CC, Kuo WW, Ho TJ, Day CH, Pai PY, Chung LC, Huang CC, Wang HF, Liao PH, Huang CY. Multi-Strain Probiotics Inhibit Cardiac Myopathies and Autophagy to Prevent Heart Injury in High-Fat Diet-Fed Rats. Int J Med Sci. 2016;13:277-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 79. | Qi R, Sun J, Qiu X, Zhang Y, Wang J, Wang Q, Huang J, Ge L, Liu Z. The intestinal microbiota contributes to the growth and physiological state of muscle tissue in piglets. Sci Rep. 2021;11:11237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 80. | Ryu D, Mouchiroud L, Andreux PA, Katsyuba E, Moullan N, Nicolet-Dit-Félix AA, Williams EG, Jha P, Lo Sasso G, Huzard D, Aebischer P, Sandi C, Rinsch C, Auwerx J. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22:879-888. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 481] [Cited by in F6Publishing: 584] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 81. | Lavoie S, Conway KL, Lassen KG, Jijon HB, Pan H, Chun E, Michaud M, Lang JK, Gallini Comeau CA, Dreyfuss JM, Glickman JN, Vlamakis H, Ananthakrishnan A, Kostic A, Garrett WS, Xavier RJ. The Crohn's disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 70] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 82. | Tsuboi K, Nishitani M, Takakura A, Imai Y, Komatsu M, Kawashima H. Autophagy Protects against Colitis by the Maintenance of Normal Gut Microflora and Secretion of Mucus. J Biol Chem. 2015;290:20511-20526. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 75] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 83. | Yang L, Liu C, Zhao W, He C, Ding J, Dai R, Xu K, Xiao L, Luo L, Liu S, Li W, Meng H. Impaired Autophagy in Intestinal Epithelial Cells Alters Gut Microbiota and Host Immune Responses. Appl Environ Microbiol. 2018;84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 84. | Yan S, Khambu B, Chen X, Dong Z, Guo G, Yin XM. Hepatic Autophagy Deficiency Remodels Gut Microbiota for Adaptive Protection via FGF15-FGFR4 Signaling. Cell Mol Gastroenterol Hepatol. 2021;11:973-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 85. | Trentesaux C, Fraudeau M, Pitasi CL, Lemarchand J, Jacques S, Duche A, Letourneur F, Naser E, Bailly K, Schmitt A, Perret C, Romagnolo B. Essential role for autophagy protein ATG7 in the maintenance of intestinal stem cell integrity. Proc Natl Acad Sci U S A. 2020;117:11136-11146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 86. | Chang SY, Lee SN, Yang JY, Kim DW, Yoon JH, Ko HJ, Ogawa M, Sasakawa C, Kweon MN. Autophagy controls an intrinsic host defense to bacteria by promoting epithelial cell survival: a murine model. PLoS One. 2013;8:e81095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 87. | Liu TC, Kern JT, VanDussen KL, Xiong S, Kaiko GE, Wilen CB, Rajala MW, Caruso R, Holtzman MJ, Gao F, McGovern DP, Nunez G, Head RD, Stappenbeck TS. Interaction between smoking and ATG16L1T300A triggers Paneth cell defects in Crohn's disease. J Clin Invest. 2018;128:5110-5122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 46] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 88. | Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, Hubbard-Lucey VM, Cammer M, Neil J, Dewan MZ, Lieberman SR, Lazrak A, Marinis JM, Beal A, Harris PA, Bertin J, Liu C, Ding Y, van den Brink MRM, Cadwell K. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med. 2017;214:3687-3705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 204] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 89. | Gubas A, Dikic I. A guide to the regulation of selective autophagy receptors. FEBS J. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 80] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 90. | Viret C, Duclaux-Loras R, Nancey S, Rozières A, Faure M. Selective Autophagy Receptors in Antiviral Defense. Trends Microbiol. 2021;29:798-810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 91. | Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374-11383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 492] [Cited by in F6Publishing: 491] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 92. | Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 93. | Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037-1040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 831] [Cited by in F6Publishing: 869] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 94. | Benjamin JL, Sumpter R Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 217] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 95. | Bretin A, Carrière J, Dalmasso G, Bergougnoux A, B'chir W, Maurin AC, Müller S, Seibold F, Barnich N, Bruhat A, Darfeuille-Michaud A, Nguyen HT. Activation of the EIF2AK4-EIF2A/eIF2α-ATF4 pathway triggers autophagy response to Crohn disease-associated adherent-invasive Escherichia coli infection. Autophagy. 2016;12:770-783. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 96. | Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy. 2020;16:38-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 362] [Article Influence: 90.5] [Reference Citation Analysis (0)] |
| 97. | Lassen KG, Kuballa P, Conway KL, Patel KK, Becker CE, Peloquin JM, Villablanca EJ, Norman JM, Liu TC, Heath RJ, Becker ML, Fagbami L, Horn H, Mercer J, Yilmaz OH, Jaffe JD, Shamji AF, Bhan AK, Carr SA, Daly MJ, Virgin HW, Schreiber SL, Stappenbeck TS, Xavier RJ. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014;111:7741-7746. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 263] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 98. | Sadaghian Sadabad M, Regeling A, de Goffau MC, Blokzijl T, Weersma RK, Penders J, Faber KN, Harmsen HJ, Dijkstra G. The ATG16L1-T300A allele impairs clearance of pathosymbionts in the inflamed ileal mucosa of Crohn's disease patients. Gut. 2015;64:1546-1552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 99. | Ellinghaus D, Zhang H, Zeissig S, Lipinski S, Till A, Jiang T, Stade B, Bromberg Y, Ellinghaus E, Keller A, Rivas MA, Skieceviciene J, Doncheva NT, Liu X, Liu Q, Jiang F, Forster M, Mayr G, Albrecht M, Häsler R, Boehm BO, Goodall J, Berzuini CR, Lee J, Andersen V, Vogel U, Kupcinskas L, Kayser M, Krawczak M, Nikolaus S, Weersma RK, Ponsioen CY, Sans M, Wijmenga C, Strachan DP, McArdle WL, Vermeire S, Rutgeerts P, Sanderson JD, Mathew CG, Vatn MH, Wang J, Nöthen MM, Duerr RH, Büning C, Brand S, Glas J, Winkelmann J, Illig T, Latiano A, Annese V, Halfvarson J, D'Amato M, Daly MJ, Nothnagel M, Karlsen TH, Subramani S, Rosenstiel P, Schreiber S, Parkes M, Franke A. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology. 2013;145:339-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 100. | Girardelli M, Basaldella F, Paolera SD, Vuch J, Tommasini A, Martelossi S, Crovella S, Bianco AM. Genetic profile of patients with early onset inflammatory bowel disease. Gene. 2018;645:18-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 101. | Hoefkens E, Nys K, John JM, Van Steen K, Arijs I, Van der Goten J, Van Assche G, Agostinis P, Rutgeerts P, Vermeire S, Cleynen I. Genetic association and functional role of Crohn disease risk alleles involved in microbial sensing, autophagy, and endoplasmic reticulum (ER) stress. Autophagy. 2013;9:2046-2055. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 102. | Birgisdottir ÅB, Johansen T. Autophagy and endocytosis - interconnections and interdependencies. J Cell Sci. 2020;133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 103. | Gluschko A, Herb M, Wiegmann K, Krut O, Neiss WF, Utermöhlen O, Krönke M, Schramm M. The β2 Integrin Mac-1 Induces Protective LC3-Associated Phagocytosis of Listeria monocytogenes. Cell Host Microbe. 2018;23:324-337.e5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 76] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 104. | Sprenkeler EG, Gresnigt MS, van de Veerdonk FL. LC3-associated phagocytosis: a crucial mechanism for antifungal host defence against Aspergillus fumigatus. Cell Microbiol. 2016;18:1208-1216. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 105. | Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: the expected slimy partners? Gut. 2020;69:2232-2243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 693] [Cited by in F6Publishing: 603] [Article Influence: 150.8] [Reference Citation Analysis (0)] |
| 106. | Arike L, Holmén-Larsson J, Hansson GC. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. 2017;27:318-328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 62] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 107. | Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Núñez G, Martens EC. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339-1353.e21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1285] [Cited by in F6Publishing: 1557] [Article Influence: 222.4] [Reference Citation Analysis (0)] |
| 108. | Jakobsson HE, Rodríguez-Piñeiro AM, Schütte A, Ermund A, Boysen P, Bemark M, Sommer F, Bäckhed F, Hansson GC, Johansson ME. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015;16:164-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 388] [Cited by in F6Publishing: 441] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 109. | Schroeder BO. Fight them or feed them: how the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (Oxf). 2019;7:3-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 250] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 110. | Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, Moayyedi P. Differences in Gut Microbiota in Patients With vs Without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology. 2020;158:930-946.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in F6Publishing: 276] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 111. | Xu Y, Wang N, Tan HY, Li S, Zhang C, Feng Y. Function of Akkermansia muciniphila in Obesity: Interactions With Lipid Metabolism, Immune Response and Gut Systems. Front Microbiol. 2020;11:219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 148] [Cited by in F6Publishing: 223] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 112. | Wu M, Wu Y, Li J, Bao Y, Guo Y, Yang W. The Dynamic Changes of Gut Microbiota in Muc2 Deficient Mice. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 69] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 113. | Gibold L, Garenaux E, Dalmasso G, Gallucci C, Cia D, Mottet-Auselo B, Faïs T, Darfeuille-Michaud A, Nguyen HT, Barnich N, Bonnet R, Delmas J. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn's disease-associated Escherichia coli. Cell Microbiol. 2016;18:617-631. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 114. | Sperandio B, Fischer N, Joncquel Chevalier-Curt M, Rossez Y, Roux P, Robbe Masselot C, Sansonetti PJ. Virulent Shigella flexneri affects secretion, expression, and glycosylation of gel-forming mucins in mucus-producing cells. Infect Immun. 2013;81:3632-3643. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 115. | Patel KK, Miyoshi H, Beatty WL, Head RD, Malvin NP, Cadwell K, Guan JL, Saitoh T, Akira S, Seglen PO, Dinauer MC, Virgin HW, Stappenbeck TS. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32:3130-3144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 116. | Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, Peaper DR, Bertin J, Eisenbarth SC, Gordon JI, Flavell RA. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745-757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1392] [Cited by in F6Publishing: 1476] [Article Influence: 113.5] [Reference Citation Analysis (0)] |
| 117. | Wlodarska M, Thaiss CA, Nowarski R, Henao-Mejia J, Zhang JP, Brown EM, Frankel G, Levy M, Katz MN, Philbrick WM, Elinav E, Finlay BB, Flavell RA. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156:1045-1059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 453] [Cited by in F6Publishing: 512] [Article Influence: 51.2] [Reference Citation Analysis (0)] |
| 118. | Engevik MA, Luk B, Chang-Graham AL, Hall A, Herrmann B, Ruan W, Endres BT, Shi Z, Garey KW, Hyser JM, Versalovic J. Bifidobacterium dentium Fortifies the Intestinal Mucus Layer via Autophagy and Calcium Signaling Pathways. mBio. 2019;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 133] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 119. | Yeom J, Ma S, Lim YH. Oxyresveratrol Induces Autophagy via the ER Stress Signaling Pathway, and Oxyresveratrol-Induced Autophagy Stimulates MUC2 Synthesis in Human Goblet Cells. Antioxidants (Basel). 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 120. | Fagarasan S, Muramatsu M, Suzuki K, Nagaoka H, Hiai H, Honjo T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 2002;298:1424-1427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 470] [Cited by in F6Publishing: 451] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 121. | Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, Fagarasan S. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981-1986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 521] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 122. | Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 182] [Cited by in F6Publishing: 197] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 123. | Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, Hattori M, Fagarasan S. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 344] [Cited by in F6Publishing: 283] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 124. | Kawamoto S, Tran TH, Maruya M, Suzuki K, Doi Y, Tsutsui Y, Kato LM, Fagarasan S. The inhibitory receptor PD-1 regulates IgA selection and bacterial composition in the gut. Science. 2012;336:485-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 332] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 125. | Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, Pons N, Le Chatelier E, Levenez F, Kennedy S, Galleron N, de Barros JP, Malphettes M, Galicier L, Boutboul D, Mathian A, Miyara M, Oksenhendler E, Amoura Z, Doré J, Fieschi C, Ehrlich SD, Larsen M, Gorochov G. Microbial ecology perturbation in human IgA deficiency. Sci Transl Med. 2018;10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 173] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 126. | Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, Schofield WB, Palm NW. IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep. 2019;9:13574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 111] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 127. | Ma Y, Hendershot LM. The stressful road to antibody secretion. Nat Immunol. 2003;4:310-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 128. | Pengo N, Scolari M, Oliva L, Milan E, Mainoldi F, Raimondi A, Fagioli C, Merlini A, Mariani E, Pasqualetto E, Orfanelli U, Ponzoni M, Sitia R, Casola S, Cenci S. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298-305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 306] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 129. | Conway KL, Kuballa P, Khor B, Zhang M, Shi HN, Virgin HW, Xavier RJ. ATG5 regulates plasma cell differentiation. Autophagy. 2013;9:528-537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 120] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 130. | Ragland SA, Criss AK. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017;13:e1006512. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 324] [Cited by in F6Publishing: 403] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 131. | Yu S, Balasubramanian I, Laubitz D, Tong K, Bandyopadhyay S, Lin X, Flores J, Singh R, Liu Y, Macazana C, Zhao Y, Béguet-Crespel F, Patil K, Midura-Kiela MT, Wang D, Yap GS, Ferraris RP, Wei Z, Bonder EM, Häggblom MM, Zhang L, Douard V, Verzi MP, Cadwell K, Kiela PR, Gao N. Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine. Immunity. 2020;53:398-416.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 132. | Sankaran-Walters S, Hart R, Dills C. Guardians of the Gut: Enteric Defensins. Front Microbiol. 2017;8:647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 133. | Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW 4th. A key role for autophagy and the autophagy gene Atg16 L1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1084] [Cited by in F6Publishing: 1171] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 134. | Cadwell K, Patel KK, Komatsu M, Virgin HW 4th, Stappenbeck TS. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250-252. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 135. | VanDussen KL, Liu TC, Li D, Towfic F, Modiano N, Winter R, Haritunians T, Taylor KD, Dhall D, Targan SR, Xavier RJ, McGovern DP, Stappenbeck TS. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn's disease. Gastroenterology. 2014;146:200-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 134] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 136. | Bel S, Pendse M, Wang Y, Li Y, Ruhn KA, Hassell B, Leal T, Winter SE, Xavier RJ, Hooper LV. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047-1052. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 228] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 137. | Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135-1145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 688] [Cited by in F6Publishing: 677] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 138. | Lu R, Zhang YG, Xia Y, Zhang J, Kaser A, Blumberg R, Sun J. Paneth Cell Alertness to Pathogens Maintained by Vitamin D Receptors. Gastroenterology. 2021;160:1269-1283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 139. | Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol. 2012;3:310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 140. | Sun M, He C, Cong Y, Liu Z. Regulatory immune cells in regulation of intestinal inflammatory response to microbiota. Mucosal Immunol. 2015;8:969-978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 169] [Cited by in F6Publishing: 179] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 141. | Kozik AJ, Nakatsu CH, Chun H, Jones-Hall YL. Comparison of the fecal, cecal, and mucus microbiome in male and female mice after TNBS-induced colitis. PLoS One. 2019;14:e0225079. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 142. | Park H, Yeo S, Kang S, Huh CS. Longitudinal Microbiome Analysis in a Dextran Sulfate Sodium-Induced Colitis Mouse Model. Microorganisms. 2021;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 143. | Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017;10:18-26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 444] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 144. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J; MetaHIT Consortium, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7052] [Cited by in F6Publishing: 7123] [Article Influence: 508.8] [Reference Citation Analysis (3)] |
| 145. | Malik A, Kanneganti TD. Inflammasome activation and assembly at a glance. J Cell Sci. 2017;130:3955-3963. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 279] [Cited by in F6Publishing: 307] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 146. | Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264-268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1523] [Cited by in F6Publishing: 1563] [Article Influence: 97.7] [Reference Citation Analysis (0)] |
| 147. | Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 353] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 148. | Hou P, Yang K, Jia P, Liu L, Lin Y, Li Z, Li J, Chen S, Guo S, Pan J, Wu J, Peng H, Zeng W, Li C, Liu Y, Guo D. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31:62-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 48] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 149. | Pott J, Stockinger S. Type I and III Interferon in the Gut: Tight Balance between Host Protection and Immunopathology. Front Immunol. 2017;8:258. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 150. | Burger E, Araujo A, López-Yglesias A, Rajala MW, Geng L, Levine B, Hooper LV, Burstein E, Yarovinsky F. Loss of Paneth Cell Autophagy Causes Acute Susceptibility to Toxoplasma gondii-Mediated Inflammation. Cell Host Microbe. 2018;23:177-190.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 151. | Bussi C, Peralta Ramos JM, Arroyo DS, Gaviglio EA, Gallea JI, Wang JM, Celej MS, Iribarren P. Autophagy down regulates pro-inflammatory mediators in BV2 microglial cells and rescues both LPS and alpha-synuclein induced neuronal cell death. Sci Rep. 2017;7:43153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 71] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 152. | Cadwell K. Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis. Nat Rev Immunol. 2016;16:661-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 267] [Cited by in F6Publishing: 308] [Article Influence: 38.5] [Reference Citation Analysis (0)] |