Published online Nov 14, 2021. doi: 10.3748/wjg.v27.i42.7210
Peer-review started: March 21, 2021
First decision: April 30, 2021
Revised: May 12, 2021
Accepted: October 25, 2021
Article in press: October 25, 2021
Published online: November 14, 2021
Clostridioides difficile (C. difficile) is a gram-positive, anaerobic spore-forming bacterium and a major cause of antibiotic-associated diarrhea. Humans are naturally resistant to C. difficile infection (CDI) owing to the protection provided by healthy gut microbiota. When the gut microbiota is disturbed, C. difficile can colonize, produce toxins, and manifest clinical symptoms, ranging from asymptomatic diarrhea and colitis to death. Despite the steady-if not rising-prevalence of CDI, it will certainly become more problematic in a world of antibiotic overuse and the post-antibiotic era. C. difficile is naturally resistant to most of the currently used antibiotics as it uses multiple resistance mechanisms. Therefore, current CDI treatment regimens are extremely limited to only a few antibiotics, which include vancomycin, fidaxomicin, and metronidazole. Therefore, one of the main challenges experienced by the scientific community is the development of alternative approaches to control and treat CDI. In this Frontier article, we collectively summarize recent advances in alternative treatment approaches for CDI. Over the past few years, several studies have reported on natural product-derived compounds, drug repurposing, high-throughput library screening, phage therapy, and fecal microbiota tran
Core Tip: Clostridioides difficile is considered a threat to public health owing to increases in treatment failure over the past few years. Current antibiotic treatment options are highly limited. Therefore, alternative strategies are critical. Herein, we review recent advances in alternative therapeutics, including the development of new chemical entities, fecal microbiota transplantation, pre- and pro-biotic, antitoxin antibodies, use of bacteriophages, and vaccines. We also highlight the concerns, limitations, and directions for each of these developments.
- Citation: Phanchana M, Harnvoravongchai P, Wongkuna S, Phetruen T, Phothichaisri W, Panturat S, Pipatthana M, Charoensutthivarakul S, Chankhamhaengdecha S, Janvilisri T. Frontiers in antibiotic alternatives for Clostridioides difficile infection. World J Gastroenterol 2021; 27(42): 7210-7232
- URL: https://www.wjgnet.com/1007-9327/full/v27/i42/7210.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i42.7210
Clostridioides difficile (C. difficile) is a gram-positive, spore-forming, toxin-producing, rod-shaped anaerobic bacterium. C. difficile infection (CDI) is a leading cause of nosocomial infections in several countries. The source of C. difficile still remains debatable. Studies have proposed that C. difficile was part of the human gut commensal bacterial community, as the bacterium could often be isolated from neonates[1,2]. The pathogenicity of C. difficile was believed to be attributed to its toxin-production properties; however, recent developments have suggested that outgrowth and colonization are also a pivotal feature of pathogenicity. Under normal conditions, when the gut microbiota is in balance, in a stage called “eubiosis”, C. difficile can neither multiply nor colonize the gut, thus preventing it from causing disease. On the other hand, when the composition of the gut microbiota is altered from its normal state, so-called “dysbiosis” occurs, which allows C. difficile to multiply and colonize[3,4]. Once colonized, C. difficile can produce up to 3 toxins, i.e., toxin A (TcdA), toxin B (TcdB), and binary toxin (CDT). The first two are prominent virulent factors, whilst the latter is controversial[5-7]. The binary toxin is believed to enhance the toxicity and pathogenicity of the primary toxins; however, only a few reports have shown that PCR-negative, but CDT-positive, TcdA and TcdB can cause CDI[8,9]. Details of the pathogenesis of C. difficile can be found in reviews elsewhere[5,7,10,11].
CDI is one of the most prominent causes of healthcare-associated infections (HAI). In the United States and European countries, C. difficile has been ranked among the top 10 causes of HAI[12]. The challenges of CDI control and prevention are intrinsic drug resistance and environmental resistant spore of C. difficile. To date, CDI treatment with most antibiotics has generally resulted in failure. Use of certain antibiotics, such as clindamycin, cephalosporins, quinolones, and penicillins reportedly increased the risk of CDI[13]. Since these antibiotics are broad-spectrum and deplete other gut microbiota that inhibit C. difficile multiplication[13], the use of such antibiotics increases CDI risk. To the best of our knowledge, some classes of antibiotics do not increase the risk of CDI, including tetracyclines and aminoglycosides[14]. The antibiotics currently used to treat CDI are vancomycin and fidaxomicin, whereas metronidazole is the antibiotic of choice only when the first two are not available[15]. Along with treatment, prevention is a good control strategy for CDI in most settings. Multiple strategies are being implemented to reduce the rate of CDI, e.g., case management, infrastructure, and antibiotic stewardship, in hospitals[16]. To make all these control measures meaningful, monitoring of C. difficile situations is necessary to ensure that C. difficile does not develop resistance to current antibiotics, which would threaten the control and treatment of CDI. There has been an increase in the number of fatal cases of CDI in the past decade[17]. In most countries, a national surveillance system for CDI is still lacking, resulting in under-represented cases of CDI.
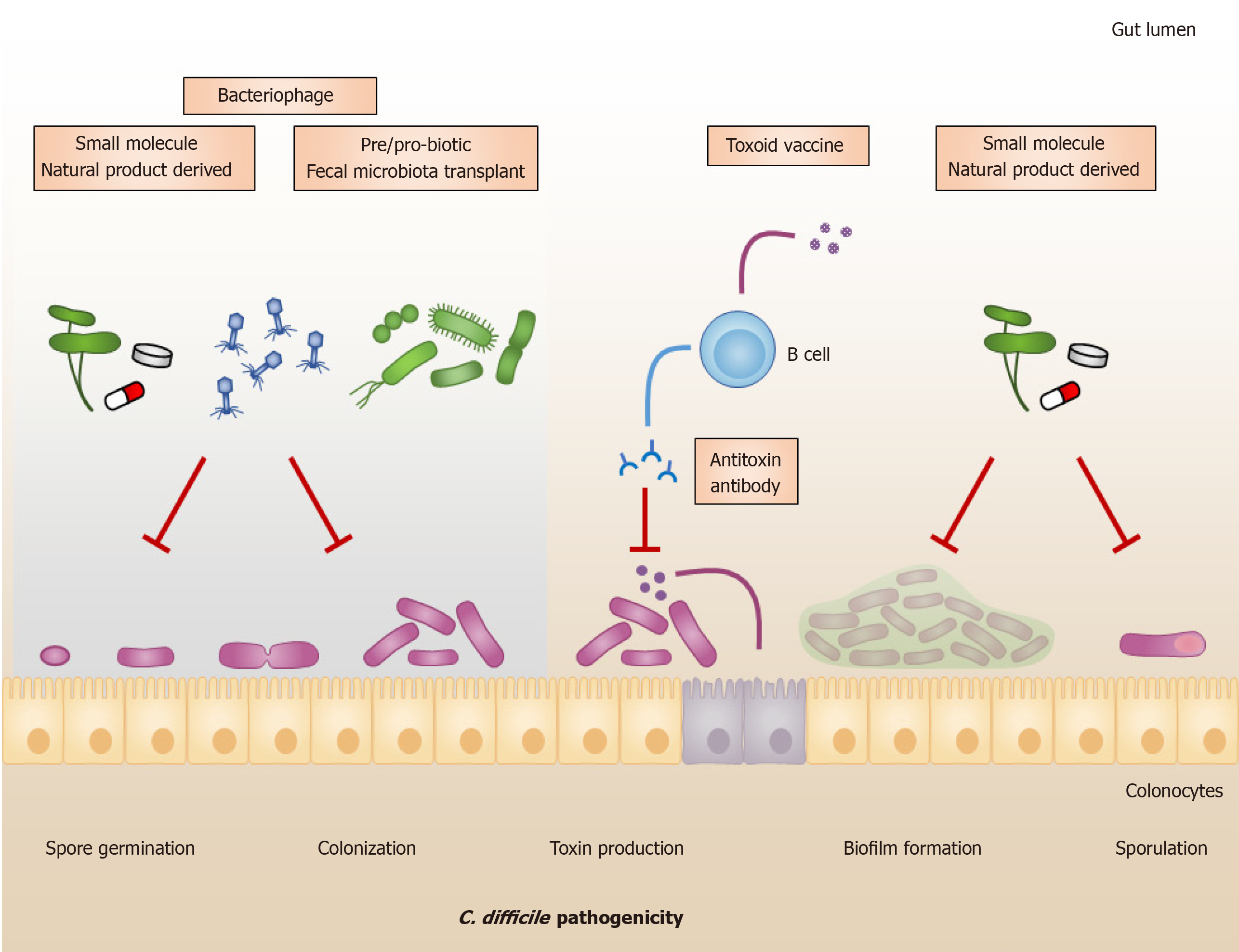
Current antibiotic options seem to have reduced efficacy. Recent CDI treatment guidelines have moved away from metronidazole because its efficacy has reduced from 95% pre-2000 to 75% since[15]. A similar reduction has been observed for vancomycin, from 98% to 85%[15]. The increases in recurrent episodes of CDI have reflected the failure of current antibiotics to sustain positive treatment outcomes[18]. Therefore, there is a critical need to develop alternative treatment approaches for CDI (Figure 1). In fact, several alternative approaches are under development, and some are more advanced than others. The development of new antibiotics is the most active research field. Fecal microbiota transplantation (FMT), including pre/probiotic approaches, has also gained extensive research interest in the past few years, as evidenced by several clinical trial registrations. Phage therapy, antitoxin antibody, and vaccines are trying to catch up with others in the league. Notably, there is one antitoxin antibody, bezlotoxumab, approved for preventing the recurrence of C. difficile by the United States Food and Drug Administration (FDA), in 2016.
We explored the developments that are currently in the pipeline for alternative therapies by examining data from the ClinicalTrials website. Several trials are investigating current antibiotics for CDI but in various aspects, most commonly in conjunction with FMT or pre/probiotic or antitoxin antibodies. Several molecular entities, such as nitazoxanide, are under investigation. Based on this insight, we extrapolated that the developing treatment options aim to treat/prevent recurrent CDI rather than the initial episode of CDI. Interestingly, currently no trials have been reported on bacteriophage therapy, despite this technology having been active since the early 1990s[19], and its therapeutic application having been demonstrated in other pathogenic bacteria[20]. This indicates that some complexities underline the technology.
In the realm of drug discovery and development, small molecules dominate the pharmacy shelf, irrespective of the sources of the small molecules, e.g., natural product-derived/inspired or (semi-)synthetics. According to the United States FDA database, small molecules account for most approved drugs. Regarding the nature and pathogenesis of C. difficile, the drugs used to treat CDI have a specific set of requirements, although no target product profiles have been proposed. We believe that the community consensus for the expected properties of the CDI drugs is that they could effectively kill vegetative and spore stages with low systemic absorption (high colonic concentration)[21,22]. Currently in clinical trials, several candidate compounds are being investigated for the development of CDI drugs. Petrosillo et al[23] have published a comprehensive review on the development of small molecules for CDI drugs.
Cadazolid is a synthetic oxazolidinone, a derivative of linezolid, which was developed for CDI and also exhibited potency against other enterococci[24]. Cadazolid exhibits good clearance of C. difficile with sub-mM MIC and limited effects on other gut microorganisms[25]. Its mechanism of action is protein synthesis inhibition by binding to peptidyl transferase center (PTC). Although cadazolid shares similar mechanism to linezolid, cadazolid is active against linezolid-resistant C. difficile[24]. The results from phase 3 clinical trials of cadazolid demonstrated a comparable clinical cure to vancomycin, good safety profile and well tolerated. However, it did not show non-inferiority to vancomycin, which subsequently resulted in the discontinuation of the development by the developer.
Nitazoxanide is an FDA-approved drug with an indication for treatments of cryptosporidiosis and giardiasis. At the millimolar range, this drug inhibited multiple gram-negative and gram-positive anaerobic bacteria, e.g., Bacteroides spp., Prevotella spp., C. difficile, C. perfringens and Mycobacterium tuberculosis[26]. The mechanism of action of nitazoxanide has been suggested to involve the inhibition of pyruvate ferredoxin oxidoreductase (PFOR) in anaerobic energy metabolism[27] and the disruption of membrane potential in aerobic bacteria[28]. In addition, studies have shown that nitazoxanide is effective against various RNA and DNA viruses including coronavirus[29] and SARS-CoV-2[30]. The result of an early phase-3 trial have suggested that the effectiveness of nitazoxanide is comparable to that of vancomycin, although noninferiority to vancomycin is inconclusive owing to small sample size and early termination[31]. Hence, the mechanism of action of nitazoxanide compared with current CDI treatments is anticipated from other ongoing trials because of the high expectations for nitazoxanide in the treatment of recurrent CDI[32].
Ridinilazole (SMT19969) is an antibiotic developed specifically for CDI. It exhibits good anticlostridial activity, with different magnitude, against multiple Clostridium spp. and also C. difficile with minimal effect to other gut microbiome[33,34] and bile acid profiles[35]. The mechanism of action of ridinilazole is believed to be unique from that of other drugs. It does not interfere with cell division by inhibiting cell wall synthesis; it does so by decreasing septum formation[36]. Ridinilazole has a low systemic absorption and, therefore, has a high colonic concentration, making it a good agent for CDI treatment[34]. The results obtained from a phase-2 clinical trial demonstrated sustained clinical response with ridinilazole treatment, noninferiority over vancomycin, and good tolerance[37]. These results altogether support further development of ridinilazole for clinical use. Phase-3 clinical trials are ongoing.
Advancements in technology and compound libraries has helped considerably in screening thousands of compounds, and has been made possible within a much shorter time. In the past few decades, several FDA-approved drugs were initially screened from large compound libraries through phenotypic screening[38]. A recent analysis showed that more than 16 million compounds can be purchased from various chemical suppliers[39]. This extends our opportunity to discover even more hits for drug discovery and development.
Apart from a rationally designed drug approach or large library screening, one recent popular drug development approach is drug repurposing or repositioning. The approach is regarded as a shortcut for a lengthy conventional drug development pipeline, as safety and pharmacokinetic data are readily available, which allows the compounds to proceed directly into clinical trials. Many works have demonstrated approved drugs can be repurposed for C. difficile[40,41]. The screening of both random compound libraries and approved drugs has become a very intriguing approach for drug development in the past years. Several studies conducted using this approach have been published[30,40,41]. However, if the route of administration is different from the approved indication, preclinical stages are necessary to evaluate the safety profile[42].
We examined the molecular similarity among the FDA-approved drugs from screening campaigns mentioned in this paper[40,43]. Molecular clustering of these drugs will help us predict a key biomolecular pathway or target the hits interact with or act upon. Molecular analysis was performed using the KNIME Analytics Platform, free and open-source software, aiming to solve large-data analysis problems[44]. KNIME offers an extensive toolset for data pre-processing and transformation as well as visualization and it can be equipped with extensions and nodes that are suitable for pharmaceutical research. We employed KNIME 4.1.2 equipped with various extensions, including RDKit KNIME integration, KNIME Distance Matrix Extension, and KNIME-CDK to create our workflow[45,46]. The workflow of the molecular analysis of the hits is shown in Scheme 1 of the supplement file.
The results shown in Table 1 and Supplementary material revealed that those hits can be categorized structurally into two large clusters and six smaller clusters based on molecular clustering analysis (> 3 hits per cluster). The largest group of clusters belongs to the β-lactam antibiotics (clusters 88–97, Supplementary material in SI) and the second largest is the tetracycline and its derivatives (clusters 98–99, Supplementary material in SI). Although these drugs are well-known antibiotics and are expected to exert some biological activity against C. difficile, the clinical use of these materials might not be of best interest since they are broad-spectrum antibiotics that could cause concern in gut microbiota dysbiosis and may drive an undesired side-effect of antibiotic resistance elsewhere in the body[47]. Other known antibiotics, such as aminoglycosides (cluster 123) and benzalkoniums cationic surfactant (cluster 61) which could be used topically as antiseptic were also identified.
| Cluster No. | Compound class |
| 21 | Antifungal imidazoles |
| 42 | Metronidazole and its derivatives |
| 61 | Benzalkonium cationic surfactants |
| 88–97 | β-lactams |
| 98–99 | Tetracycline and its derivatives |
| 123 | Aminoglycosides |
Interestingly, metronidazole, one of the current treatments for C. difficile, and its derivatives (cluster 42) also show up as one cluster. Further exploration in this chemical moiety seems a way forward to avoid cross-resistance with current antibiotics as the drugs within this class exert their biological activity through the formation of nitroso radical. The mechanism that causes microbial DNA damage is not related to a certain enzyme and, therefore, makes it more challenging for the bacteria to develop resistance. Antifungal imidazoles were identified in another interesting cluster (cluster 21). These drugs act as an inhibitor of lanosterol 14α-demethylase, which catalyzes the formation of ergosterol, an important sterol found in eukaryotic cell membranes[48]. Although the mechanism of this class of drugs in C. difficile is not known, the result may prompt scientists to investigate the potential of these materials further, since these azoles are not currently in use as antibiotics, and it could avoid unwanted antibiotic cross-resistance with other current drugs[40,43]. Furthermore, most of the drugs identified from the FDA-approved panels resulted in drugs with anthelmintic (parasitic) indication[40]. It is possible that these parasites share a similar anaerobic/microaerophilic environment, and to some extent anaerobic metabolism, to that of C. difficile, thereby allowing those drugs to be active against C. difficile. It would be very interesting to observe whether this hypothesis remains valid when more screenings are performed. Although several drugs and lead compounds have been identified in the literature, only a few have been put forward into the development pipeline. This is largely due to the lack of academic and industrial partnerships. We have seen what the scientific community has achieved, in an unprecedented time scale, with the development of COVID-19 vaccines with the leading role of big pharmaceutical companies and their academic partners. This emphasizes the importance of academic-industrial partnerships in drug and vaccine developments, as this field of research is resource-intensive and requires considerable funding and workforce. Nevertheless, non-profit organizations can play an important role in drug discovery and development as well evidenced by the approval of anti-tubercular pretomanid, which was led by the TB Alliance[49].
Humans have been using natural products as traditional medicine for treating several illnesses for centuries. It has been estimated that approximately 500000 plant species exist on the planet, but only 1% of them have been explored for bioactive compounds[50]. A detailed analysis of FDA-approved drugs from 1931 to 2013 revealed that natural products and their derivatives represented over 30% of new drugs[51]. Due to the increasing prevalence of antibiotic resistance in several pathogenic bacteria, the prescribed antibiotics may no longer be effective. Therefore, exploration of plant-derived compounds could provide us a tremendous opportunity to discover novel bioactive agents.
Several plant extracts and plant-derived compounds possess antibacterial activity against C. difficile and their action has been investigated previously. Roshan et al[52] reported the anti-C. difficile activities of natural products that are commercially available as both processed and unprocessed products. Some processed products such as aloe vera gel, peppermint oil, artichoke capsules, and garlic tablets demonstrated antimicrobial activity against C. difficile with MICs of 16% (v/v), 8% (v/v), 75–150 mg/mL, and 37.5–75 mg/mL, respectively[52]. Regarding the unprocessed products, allicin (derived from garlic) and cinnamon powder demonstrated antimicrobial activity against C. difficile with MICs of 2.3–4.7 and 75 mg/mL, respectively. Zingerone (derived from ginger) and menthol (derived from peppermint) inhibited C. difficile at the same inhibition concentration of 9.4 mg/mL, whereas trans-cinnamaldehyde, an active constituent found in cinnamon bark, exhibited strong inhibitory activity with an MIC of 0.2 mg/mL[52]. δ-3-Carene, a monoterpene derived from the root of Asarum heterotropoides, exhibited anticlostridial activity with an MIC of 0.7 mg/mL with a less potential of suppressing beneficial intestinal bacteria[53]. Moreover, an essential oil extract that contains 16.5% of δ-3-carene exhibited antimicrobial activity against C. difficile at a concentration of 0.25% (v/v)[54]. Xanthohumol, derived from Humulus lupulus L., exhibited anti-C. difficile activity with MICs of 0.032–0.107 mg/mL against 28 different C. difficile isolates[55]. Crude methanol extract of the bark of Mammea africana demonstrated anti-C. difficile activity with an MIC of 2 µg/mL. The identified active flavonic compound in M. africana, mammeisin or mammea A/AA, exhibited strong inhibitory activity with an MIC of 0.25 µg/mL[56]. Curcumin (phenol), an active agent derived from Curcuma longa, is reportedly active against 27 C. difficile strains, with MICs ranging between 16 and 32 µg/mL. The inhibition was specific to C. difficile as it did not affect beneficial gut microbiota[57]. Curcumin was also able to reduce sporulation and toxin production in C. difficile[57].
Several studies have reported that the anticlostridial activity of crude plant extracts contain unidentified active compounds. For instance, pomegranate extract was shown to exhibit specific inhibitory activity against 23 tested isolates of C. difficile with MICs of 12.5–25 µg/mL, but no inhibitory effect was observed on the tested normal intestinal bacteria (MIC > 400 µg/mL)[58]. Investigation of a commercial animal supplement, BIOCITRO, a citrus fruit extract, revealed an inhibitory effect on C. difficile with MICs of 16–32 µg/mL with a possible mode of action of disrupting polysaccharides and carbohydrates of the cell wall[59]. In addition, methanolic extract of the leaf and rhizomes of Aristolochia paucinervis Pomel demonstrated an inhibitory effect on C. difficile, with concentrations ranging from 8 to 64 µg/mL[60].
Several compounds are known to kill C. difficile through changes in cell permeability. The ability of membrane disruption was suggested for zingerone, menthol, and trans-cinnamaldehyde[61]. Asiatic acid, an active triterpenoid derived from Centella asiatica, exhibited significant antimicrobial activity against C. difficile strains isolated from different sources by disrupting the cytoplasmic membrane with MICs of 10–20 µg/mL[62]. Lauric acid, an active component found in virgin coconut oil, exhibited anticlostridial activity at a 250-µmol/L concentration through the membrane disruption mechanism[63]. Cannabidiol, an active ingredient derived from cannabis, could inhibit C. difficile at concentrations of 2–4 µg/mL, possibly by disrupting the bacterial membrane[64]. Another study of cannabidiol in caco-2 cells infected with C. difficile reported its potential to inhibit toxin A-induced cytotoxicity[65]. Interestingly, data analysis of hospitalization from the Healthcare Cost and Utilization Project in 2014 suggested that patients associated with cannabis usage had potentially lower risk for CDI by 28%[66].
The dormant spores of C. difficile contribute to transmission and link with the pathogenesis of CDI. C. difficile spores can persist in harsh conditions. They can then translocate to the intestinal tract, germinate in response to specific bile salts, and initiate infection. Most antibiotics are inactive against C. difficile spores because of the spores’ intrinsic durability. In addition to vegetative cell activity, natural products are active against several spore stages of C. difficile. Peppermint oil and trans-cinnamaldehyde have been shown to exhibit sporicidal activity, which reduces the number of spores by up to 200 times when spores were exposed for 7 d[67]. Several compounds such as allicin and carvacrol, essential oils found in oregano, and fresh onion bulb extracts could inhibit spore outgrowth[67,68]. As C. difficile spores play an essential role in CDI, the inhibition of sporulation is an attractive strategy to reduce infection. A study conducted by Roshan et al[67] showed that the subinhibitory concentration of coconut oil, fresh onion bulb, and fresh ginger can reduce the number of spores production by 90%. Another study conducted on Manuka honey or Leptospermum honey also reported the inhibition of sporulation in C. difficile[69]. Baicalin, a flavonoid derivative present in the root of Scutellaria baicalensis, could inhibit both sporulation and outgrowth of C. difficile at a concentration of 1.6 mmol/L. It also reduced toxin production by downregulating tcdA and tcdB gene expression[70]. Toxin A and toxin B produced by C. difficile are cytotoxic and cause colitis. Leptospermum honey, fresh onion bulb, and trans-cinnamaldehyde were able to reduce the cytotoxicity of C. difficile toxins in Vero cells by 70% and toxin production by 40%[71].
In an in vivo mouse model, it was observed that berberine, a compound derived from the genus Berberis, has the potential to prevent recurrent CDI and restore gut community, either alone or in combination with vancomycin[72]. It is noteworthy that successful therapy by endoscopic lavage with Manuka honey was reported in the patient with vancomycin treatment failure[73].
Toxin production has long been associated with CDI pathogenicity. Classical antibiotic treatments effectively eliminate the pathogen, although they inevitably affect the normal gut microbiota to some extent. Antitoxin antibody treatment aims to neutralize the toxicity of the toxin, rather than interfere directly with the bacterium. Therefore, it can reduce clinical severity, in conjunction with antibiotic treatment, and reduce recurrent CDI. To date, the only FDA-approved antitoxin antibody for CDI treatment is bezlotoxumab, a human monoclonal antibody against toxin B. A detailed study demonstrated that bezlotoxumab binds to carbohydrate-binding pockets on toxin B, which directly prevents the interaction between the toxin and host cells[74]. This hypothesis was supported by a mutant antibody that does not bind to Fc receptors of host immune cells, which provides similar toxin neutralization and protection effects as wild-type antibody[75]. Its toxin A counterpart, actoxumab, did not demonstrate improvement in clinical efficacy, and therefore it was discontinued during the phase 3 trial[76]. These results support the hypothesis that TcdB is a more prominent virulent factor than other toxins and that the interplay between these toxins is a complex process.
IM-01 is an experimental polyclonal antibody for inflammatory bowel disease, including Crohn’s disease, and CDI. IM-01 is produced using a chicken egg technique by immunizing hens with toxin A, toxin B, and C. difficile spores. Currently, detailed information on this intervention is lacking, but its phase 2 clinical trial is being recruited. The results from patent filing showed good antibody production, toxin neutralization, reduction of spore burden, and inhibition of vegetative cell growth[77]. These results indicate that IM-01 is a promising development for CDI treatment; however, it is too early to state whether it would work effectively independently or in combination with antibiotics.
Colostrum is a fluid produced by the mammary glands immediately after the birth of a newborn. It has been demonstrated that cows persistently injected with antigens will produce antibodies in the colostrum, which are collectively referred to as hyperimmune bovine colostrum (HBC). A preclinical piglet model study showed that HBC can reduce the severity of CDI[78]. A limitation of using colostrum is the scale of production, which is limited to a few hours after birth. Therefore, some studies have examined the use of milk or whey protein isolate (WPI) instead of colostrum for CDI treatment. WPI exhibited superior and sustained treatment outcome compared with vancomycin in a hamster model[79]. Considering its clinical use, we found only one terminated phase 2 trial, which was completed, but there were no results, of the whey protein concentrate MucoMilk.
Altogether, antitoxin antibodies in any form appear to be a significant candidate for alternative treatment of CDI, since preclinical studies have demonstrated good inhibition of vegetative cells and spores and neutralization effect of toxins in initial and recurrent episodes of CDI. Nonetheless, it will take some time to observe this group of treatments for clinical use, as production is more complicated and potentially expensive.
Although antibiotic administration is the first-line treatment for CDI, severe outbreaks and recurrent episodes remain excessive due to a significant increase in resistant strains[80]. Treatment options for CDI, especially recurrent CDI, become limited, due to which the development of alternative therapeutics is highly required. In particular, CDI develops when the indigenous microbiota is dysbiotic, typically via exposure to antibiotics, thereby allowing C. difficile to acquire nutrient niches in the gut and cause disease[13,81]. Based on this concept, restoration of the healthy microbial community, eubiosis, can help eliminate and prevent this growing problem. To date, researchers have expressed interest in biotherapeutic strategies that confer curative effects on recurrences. A burgeoning alternative treatment approach for recurrent CDI is FMT, which refers to the administration of feces collected from a healthy individual to the intestinal tract of a patient[82,83]. In general, FMT is applied to a patient with refractory CDI who had failed standard antibiotic treatments in an initial episode of CDI[84]. This approach emerged due to its unique feature, restoring the balance of the gut microbiota, which is unlikely to be achieved by other approaches. FMT provides multiple mechanisms of colonization resistance, such as competition for nutrients, production of antimicrobial peptides, and production of short-chain fatty acids (SCFAs) to inhibit vegetative growth and spore germination of C. difficile[85,86].
The first clinical trial of FMT, whose efficacy was significantly greater than the use of antibiotics for recurrent CDI treatment, was performed by van Nood et al[87] and published in 2013. Among the 16 patients, 13 with FMT recovered from CDI after the first infusion and the symptoms of 2 patients resolved after a second infusion, whereas only 3 of 13 patients resolved with vancomycin treatment alone. Other clinical studies have demonstrated that FMT provides better benefits than standard antibiotics for treating recurrent CDI by replenishing the balance of the gut microbiota. Currently, clinical studies are ongoing with the recruitment of patients with relapsing CDI to evaluate the safety and efficacy of FMT (Table 2). The upper and lower route of administration has been used in delivering the transplant. Several methods, including nasoduodenal, enema, upper endoscopy, colonoscopy, and oral capsules, are commonly used for administering FMT, with diverse outcomes[88]. A study using colonoscopy demonstrated the resolution of CDI in 18 of 20 patients treated with FMT compared with 5 of 19 patients treated with vancomycin[89]. Conversely, FMT administered by enema was found to have a low success rate in treating acute episodes of recurrent CDI[90]. To simplify administration, oral capsules of fecal filtrates have been developed and widely used. Patients with relapsing CDI had a decent response to FMT administered by oral capsules, wherein the resolution rate of recurrent CDI was similar to that observed with colonoscopy[91]. These findings demonstrated no definite preference of the administration route for FMT.
| Current phase | Title | ClinicalTrials.gov Identifier | First posted |
| NA | Fecal microbiota transplantation for C. difficile infection | NCT01905709 | July 23, 2013 |
| Immune response to FMT for C. difficile | NCT02797288 | June 13, 2016 | |
| Outcomes and data collection for fecal microbiota transplantation for the treatment of recurrent C. difficile | NCT03562741 | June 19, 2018 | |
| Fecal microbiota transplantation (FMT) for C. difficile (CEFTA) | NCT03712722 | October 1, 2018 | |
| Rescue fecal microbiota transplantation for national refractory intestinal infection | NCT03895593 | March 29, 2019 | |
| Safety and efficacy of fecal microbiota transplantation | NCT04014413 | July 10, 2019 | |
| 1 | Fecal transplant for pediatric patients who have recurrent C. difficile infection (FMT) | NCT02134392 | May 9, 2014 |
| 2 | Stool transplants to treat refractory C. difficile | NCT02127398 | April 30, 2014 |
| FMT versus antimicrobials for initial treatment of recurrent CDI | NCT02255305 | October 2, 2014 | |
| Fecal microbiota therapy for recurrent C. difficile infection | NCT02686645 | February 19, 2016 | |
| Phase II trial of fecal microbiota transplant (FMT) for VRE and CRE patients | NCT03643887 | August 23, 2018 | |
| Fecal microbiota transplantation (FMT) plus fidaxomicin for severe of fulminant C. difficile | NCT03760484 | November 30, 2018 | |
| Multicentre blinded comparison of lyophilized sterile fecal filtrate to lyophilized fecal microbiota transplant in recurrent C. difficile infection | NCT03806803 | January 16, 2019 | |
| FMT and bezltoxumab compared to FMT and placebo for patients with IBD and CDI (ICON-2) | NCT03829475 | February 4, 2019 | |
| PMT for severe-CDI | NCT03970200 | May 31, 2019 | |
| Penn microbiome therapy (PMT) for recurrent C. difficile infection | NCT03973697 | June 4, 2019 | |
| 3 | Fecal transplantation for primary C. difficile infection (COLONIZE) | NCT03796650 | January 8, 2019 |
| Microbiota restoration therapy for recurrent C. difficile infection (PUNCH CD3-OLS) (CD3-OLS) | NCT03931941 | April 30, 2019 | |
| Fecal microbiota transplantation for primary C. difficile diarrhea | NCT02801656 | June 16, 2016 |
Another factor questioned in FMT is the preparation of fecal material. Initially, fresh stools from donors were used for infusion. As the process of collecting fresh stools is difficult, frozen and lyophilized stools were then considered for transplant delivery. Several clinical trials claimed that frozen stool was comparable to fresh stool with no loss of effectiveness[92,93]. In contrast, lyophilization reduced the efficacy of the transplant material compared with fresh and frozen materials[94]. In particular, frozen fecal material is becoming available for commercial purchase. In addition to its use for treating multiple episodes of recurrent CDI, FMT has been proposed as a promising treatment approach for severe and complicated CDI. Evidence obtained from clinical trials has shown that FMT improved survival in severe cases, including immunocompromised patients[95,96].
Although FMT is a favorable therapeutic option for recurrent CDI, a major concern is that there is no universal or industrial standard such as a defined bacterial formula; moreover, there is no information on the mechanism associated with FMT-screening methods for adverse transmission events and the period of treatment[97,98]. Such limitations of FMT led to the discovery of beneficial microorganisms in fecal materials that activate colonization resistance and the determination of their roles in inhibiting C. difficile. Correspondingly, the refined stool-derived microbial suspension RBX2660 has been successfully used to treat relapsing CDI, and is currently under a phase 2b study[99]; furthermore, SER-109 and SER-262, defined microbial preparations containing spore-forming bacteria purified from human feces and formulated as a capsule, are currently being investigated in a phase 3 and 1b trial[100,101]. These data conclude that FMT continues to be a recommended therapy for recurrent CDI, and in the meantime, a number of studies continue identifying essential bacteria species from the feces of healthy donors to formulate standard microbial preparations approved by the FDA.
Another point of concern for FMT is the unintentional transfer of drug-resistant bacteria, which could possibly lead to complicated infection and death. There have been multiple bacteremia incidents in patients who received FMT[102], some of which were severe and life-threatening. This indicates the need for a standardized protocol for stool preparation, to minimize potential drug-resistant bacterial infection from FMT, including, but not limited to, extended-spectrum beta-lactamase (EBSL)-producing E. coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, etc.[103]. Recently, a caution has been issued regarding the transmission of SARS-CoV-2 in FMT[104].
Probiotics are live microorganisms that confer health benefits on the host when administered in adequate amounts[105,106]. They can improve host immunity by producing beneficial metabolites, such as SCFAs, as well as preventing enteric infections via colonization resistance mechanisms, such as competition for nutrients, inhibition of bile acid conversion, and production of antimicrobial peptides[107,108]. Numerous probiotic strains and probiotic mixtures have been evaluated to combat CDI in in vitro studies; however, there are limitations due to the lack of evidence from human clinical trials.
Lactobacillus rhamnosus GG and Saccharomyces boulardii, the most common probiotics, have provided promising applications against CDI and have been widely investigated in clinical trials. The efficacy of S. boulardii in CDI management was first examined by McFarland et al[109], who reported a significant reduction in recurrence rates after the administration of S. boulardii twice a day for 4 wk during and after antibiotic treatment. Their study demonstrated a significant reduction in the CDI recurrence rate in the S. boulardii treatment group compared with the placebo group. Moreover, patients with relapsing CDI had a statistically significant response to S. boulardii compared with placebo. The probiotic effect of L. rhamnosus GG in a clinical study was first verified by Gorbach et al[110], in 1987. They successfully used the organism to treat 5 patients with multiple recurrent CDI episodes. In addition to its beneficial effects in treating refractory CDI, L. rhamnosus GG displayed a protective potential in healthy individuals[111]. Further meta-analytical studies have been performed to confirm the usefulness of S. boulardii and L. rhamnosus GG in the prevention of CDI[112,113]. In addition to single-probiotic strains, probiotic mixtures have been developed for quite some time. The probiotic mixture BioK+ containing L. acidophilus CL1285, L. rhamnosus CL2, and L. casei LBC80R was able successfully to reduce the CDI rate from 18.0 to 2.3 cases per 10000 patients. Furthermore, new mixtures have been developed and are in the process of clinical trials[114]. The probiotic mixture of L. casei DN-114 001, S. thermophilus, and L. bulgaricus was randomly administered to hospital inpatients twice-daily during, and for 1 wk after, antibiotic treatment[115]. This probiotic mixture showed a positive result, wherein no individual in the probiotic group developed CDI compared with 9 of 53 individuals who contracted CDI in the placebo group. Another study evaluated the efficacy of the probiotic mixture of L. acidophilus and L. casei[116]. The probiotic doses were varied and administered within 1.5 d of initial antibiotic therapy and then continued for 5 d after the final antibiotic dose. Both higher dose and lower dose probiotic groups showed significantly reduced CDI incidence rates, at 1.2% and 9.4%, respectively, compared with 23.8% in the placebo group. The most recent clinical trial of a multi-strain probiotic consisting of L. acidophilus NCFM, ATCC700396, L. paracasei Lcp-37, ATCC SD5275, Bifidobacterium lactis Bi-07, ATCC SC5220, and B. lactis B1-04, ATCC SD5219 has been conducted[117]. A phase 2 study evaluated the potential benefits of this probiotic mixture by administration daily for 4 wk. The results revealed a shorter duration of diarrhea in patients with an initial episode of mild-to-moderate CDI compared with the placebo group. Based on the available evidence, probiotics are strongly advocated as an alternative for preventing and treating CDI. However, the high heterogeneity in existing studies indicates that the beneficial effects of probiotics are rather subjective[118]. Therefore, discovering novel probiotics and understanding their functions and interactions have been continuing to improve the probiotic effects in a clinical setting.
It is known that diet has a key influence on the composition and functions of the gut microbiota[119]. Several types of fiber, particularly, galacto-oligosaccharides (GOS) and fructo-oligosaccharides (FOS), have been found to increase the abundance of common probiotic bacteria, such as Bifidobacterium and Lactobacillus species[120]. Hence, plant-based foods containing dietary fiber are generally accepted as favorable for gut health. Certain fiber types, including FOS, GOS, and inulin, are considered to be prebiotics, which are defined as substrates that are selectively used by host microorganisms conferring a health benefit[121]. The combination of probiotics and prebiotics has been proposed as an alternative, known as synbiotics, to prevent and treat refractory CDI. A recent study investigated the effects of a synbiotic, L. plantarum DSM 21379 and xylitol, on the germination of C. difficile spores[122]. In vitro experiments demonstrated that the synbiotic completely inhibited the germination of C. difficile spores. Moreover, the administration of this synbiotic for 5–6 d before ampicillin and C. difficile challenge in mice reduced the CDI incidence from 44% to 22% mortality. Another in vitro study examined the inhibitory capability of four different Bifidobacterium sp. strains combined with various prebiotics against C. difficile growth. Using oligo-fructosaccharides as a carbon source, it was observed that B. longum and B. breve rescued the survival of a cell line exposed to C. difficile cell-free supernatant. These findings indicate that a probiotic strain requires a specific prebiotic substrate. To produce more effective synbiotics to control CDI, it is necessary to determine the optimal prebiotic for each probiotic.
As bacteria-infecting viruses, bacteriophages or phages have received attention for potential use as an alternative treatment for several bacterial infections. The high specificity to their bacterial hosts and the self-replicating mechanism of phages are claimed to have advantages over other approaches. Although C. difficile phages have been discovered and investigated since 1983[123], the use of phages for human infection is restricted due to some limitations. The key bacteriophage studies of C. difficile are summarized in Table 3.
| Phage | Experiment | Finding | Ref. |
| phiCD140 | A single dose of phage treatment for C. difficile infection in hamsters | Surviving of phage treated hamster | [135] |
| phiCD27 | Phage treatment of CDI in an in vitro batch fermentation and human colon model | (1) Reduction of both vegetative cell and toxin A and toxin B productions from C. difficile; and (2) No impact on others gut microbes | [135] |
| phiCDHM1 to phiCDHM6, and phiCDHS1 | (1) Investigation for an effective phage combination; and (2) Phage delivered orally in hamster model every 8 h after C. difficile challenge | (1) Discovery of phage-resistant colonies after a single phage treatment; and (2) Reduction of C. difficile amount and colonization using phage combination in vivo | [124] |
| phiCDHM1, 2, 5, and 6 | (1) Phage treatment before and after the biofilm formation; (2) First time using Galleria mellonella (wax moth) model for C. difficile phage; and (3) Using phage in combination with antibiotics (vancomycin) | (1) Reduction and prevention of the biofilm establishment in vitro; and (2) Disease prevention in the prophylaxis group and increasing the wax moth survival rates | [130] |
| phiCDHM1, 2, 5, and 6 | (1) Optimized temperate phage cocktail to treat in batch fermentation model; and (2) First metagenomic analysis of phage treatment on gut microbiome | (1) C. difficile elimination after 24 h in prophylactic condition while maintain other microbiota components; and (2) No significant impact on other bacterial groups in human gut | [127] |
| phiCDHS1 | Measurement of planktonic and adhered C. difficile cells and free phage to human colon tumorigenic cell line HT-29 | (1) Reduction of planktonic and adhered C. difficile; and (2) No cytotoxicity to human cells | [129] |
| phiCD24-2 | (1) Using engineered phage delivered Type 1-B CRISPR system as antimicrobial agent in vitro and in vivo; and (2) Mutation of phage lysogenic gene by the cI repressor and integrase gene deletion | (1) C. difficile eradication effectively in engineered phage comparing with wild-type phage; and (2) Detection of lysogen due to potentially functional complements from C. difficile prophage | [125] |
C. difficile phages were first described for bacterial typing purposes[123]. Morphological analysis showed that most of them are either Myoviridae or Siphoviridae, belonging to the order Caudovirales. They possess dsDNA as their genetic materials[124,125]. Currently, all 26 complete genomes of C. difficile phages are characterized as temperate; they can alternate their life cycles between lytic and lysogenic cycles[126,127]. This is a major constraint, as most therapeutic phage applications require a virulent phage, i.e., a phage with a strictly lytic life cycle. Therefore, current research has been focusing on the use of temperate phage and phage-derived proteins to combat CDI[128-131]. At the very beginning, phage therapy was performed as a single-phage treatment to ensure its capacity. It has been demonstrated that phiCD27 significantly reduced the growth of C. difficile cells, as well as the production of toxin A and toxin B in an in vitro batch fermentation and artificial gut model. Furthermore, phiCD27 treatment did not affect commensal microbiota in both models, suggesting high specificity on the bacterial target[132]. However, using only one phage has some limitations due to narrow host range and lysogenic capacity. Therefore, a combination of different phages or phage cocktails has become more intriguing[133-135].
A phage cocktail was successfully developed both in vitro and in vivo, as observed in the study conducted by Nale et al[134]. Optimized cocktails of phiCDHM1 to phiCDHM6 and phiCDHS1 were tested against C. difficile ribotypes 076, 014/020, and 027 strains. The best combination included phiCDHM 1, 2, 4, and 6, which could completely kill C. difficile without regrowth. The CDI hamster model showed a significantly lower number of spores in the cecum and colon with the combination of phiCDHM 1, 2, 5, and 6. The same phage mixture was used in the wax moth larva Galleria mellonella. The efficiency of using phage combined with antibiotics, including vancomycin and clindamycin, was evaluated. The results suggested that phage could be used as a supplement to antibiotic treatment and prevent the onset of CDI. Prophylaxis was the most effective therapy with 100% protection, and efficiency was reduced when used as a remedial treatment. Moreover, the authors found that the phage could penetrate and prevent biofilm formation in the C. difficile ribotype 014/020[136]. Phage treatment in an artificial gut model was further investigated using these sets of phage combinations. A six-log reduction in C. difficile growth was observed after 5 h in the prophylaxis group, and the vegetative cells were completely removed within 24 h[136]. Furthermore, metagenomic analysis was conducted using fecal samples of volunteers to observe the impact of phage therapy on the total gut microbiome. Another in vitro study conducted on the human colonic cell line HT-29, which is the CDI site, reported that phiCDHS1 preferentially adsorbed onto HT-29 cells, thereby promoting the interaction between the phage and bacterial cells[137]. It has also been shown that either bacterial lysis by phage or the phage itself was nontoxic to the colonic cells.
Recently, phiCD24-2 has been engineered to contain a genome targeting CRISPR-Cas3, which is commonly found in the genome of C. difficile[135]. The major characteristics of the engineered phage or CRISPR-enhanced phage (crPhage) have been investigated. There was no difference in phage morphology or host range compared with the wild-type phage (wtPhage). The efficiency of phage treatment for C. difficile was determined both in vitro and in vivo. crPhage demonstrated a higher efficiency to reduce the growth of vegetative cells than wtPhage in both models. However, the bacterial number rebounded by 24 h, suggesting lysogination into the host genome rather than bacterial lysis. The mouse model exhibited a significant difference in the number of C. difficile cells recovered from mouse feces between wtPhage and crPhage treatments, indicating the superiority of crPhage treatment in vivo. The CRISPR approach enhances phage efficiency during the lytic cycle as bacterial lysis can occur via two independent mechanisms, including genome damage and phage lytic activity via endolysin and holin expression. The challenge of engineered phages remains due to lysogenic conversion. Therefore, the removal of cI repressor and integrase genes was performed to generate lysogenic phage mutants[135]. Although lysogen from in vitro culture was not detected, it was detectable in mouse feces. These findings suggested the functional complement in the C. difficile genome for those removed genes.
An alternative to using temperate phages for therapeutic purposes is to utilize their products. Endolysin, a peptidoglycan hydrolase enzyme, is encoded by the phage genome. Endolysin is required to disrupt the bacterial cell wall to release phage progeny at the final step of viral infection. The endolysin CD27L is derived from the phage CD27, which is the first phage endolysin characterized in C. difficile. The specificity test demonstrated that CD27L was active against a panel of 30 C. difficile strains, including a hypervirulent ribotype 027[125]. The N-terminal truncated CD27L, CD27L1–179, improved the lytic activity when tested against C. difficile. CD27L1–179 exhibited a slightly broader lytic range than the full-length phage. It was also active against Listeria spp. However, both CD27L and CD27L1–179 did not harm the selected gut commensal bacteria[130]. Another endolysin retrieved from C. difficile 630 prophage has been described. The lytic activity of full-length PlyCD, and that of a truncated N-terminal containing the catalytic domain PlyCD1–174, were evaluated. Similar to CD27L, the truncated PlyCD1–174 exhibited greater lytic activity than its full-length counterpart and also displayed a broader activity range against C. difficile strains than the full-length PlyCD. The bactericidal assay demonstrated that PlyCD1–174 reduced more than 4-log growth of the C. difficile hypervirulent MLST2 strains 217B, 615H, and UK1 (027). This result highlighted the potency of PlyCD1–174 against the crucial clinical strains. Interestingly, PlyCD1–174 exhibited a synergistic effect with vancomycin pretreatment. The combination treatment between vancomycin and PlyCD1–174 in vivo demonstrated significant inhibition of C. difficile growth (> 2-log). Unfortunately, the in vivo study of PlyCD1–174 was unsuccessful due to inconsistent results. Ex vivo experiments were conducted using infected mouse cecum and anus as an infecting area. PlyCD1–174 exhibited 2-log reduction of C. difficile cell growth, indicating the activity of PlyCD1–174 in the gastrointestinal environment[131]. The mechanism by which truncated endolysins exhibit higher efficiency than the wild-type counterpart remains to be explored.
Phage-derived proteins with high specificity to C. difficile strains could be developed for targeted therapy. Diffiocin, a contractile R-type phage tail-like bacteriocin, was originally derived from the C. difficile strain CD4 (Diffiocin-4). This protein exhibits a higher efficiency when fused with the receptor-binding protein (RBP) of prophage phi027 in the genome of strain R20291 (Av-CD291.1 and Av-CD291.2). Interestingly, avidocin is stably active throughout the mouse gastrointestinal tract when supplied in drinking water containing 4% sucrose and 1% sodium bicarbonate. The specificity of avidocin-CDs was evaluated across C. difficile strains, based on which the relationship between RBPs in avidocin-CDs construct and slpA allele was concluded[129]. Despite the advances in the knowledge of bacteriophage biology, the use of phage for therapeutic purposes remains much of a challenge. The first challenge is lysogenic conversion, which results in ineffective treatment outcomes. Therefore, the remaining challenge is to identify or engineer the phage to obtain a strictly lytic phage. The subsequent concern is the battle between the host and the phage that occurs during phage infection. Some bacteria naturally mutate and become resistant to phage infection using different mechanisms, e.g., extracellular modification or intracellular modification, as mentioned in a review elsewhere[138]. Still, there is room for lytic phage hunters and phage modification, which are desperately needed. However, to apply this technology effectively in clinical practice, further research is warranted to overcome these limitations.
At the other end of the spectrum, one alternative treatment is the removal of part of the gut, i.e., colectomy, ileostomy, loop ileostomy, and colonic lavage. This treatment approach is largely reserved for severe and complicated (fulminant) CDI as it is the most invasive treatment. Earlier, colectomy was opted, but it did not improve the clinical outcome, resulting in some mortality[139]. Diverting loop ileostomy and colonic lavage were developed at the Pittsburgh School of Medicine and therefore referred to as the Pittsburgh protocol. This protocol exhibited benefits over conventional colectomy and end ileostomy, which decreased the mortality rate and preserved the colon[140]. We found 2 clinical trials that were terminated due to slow recruitment of participants; hence, there is limited insight into how this method confers benefit to patients with CDI. This indicates to some degree that the current treatment regimen is sufficient to save patients’ lives without involving this invasive intervention.
Current treatment approaches for CDI concentrate primarily on antibiotics. Although antibiotic administration commonly serves as the first-line therapy, prolonged disruption of the normal microbiota can result in recurrent CDI, with incidences of up to 45% after antibiotic therapy[141]. The incidence of healthcare-associated CDI has reduced over time, although there have been recent reports on the growing number of cases of community-associated CDI[142]. Therefore, there exists an urgent need to control and prevent CDI transmission. To date, several preventive remedies for recurrent CDI have been proposed, including antibiotics and FMT. Although their efficacy has been proven, none are recommended by the Infectious Disease Society of America (IDSA), and the protracted efficacy is still questionable. Therefore, vaccines could be a valuable option to serve as long-term prophylaxis for CDI due to their memory function.
Several vaccine candidates have been developed and launched to animal and clinical trials. Currently, a total of 21 clinical trials on C. difficile vaccines have been reported, of which 3 were terminated. Most of the vaccine studies have been developed based on 3 formulations, viz., toxoid, recombinant peptide, and surface-associated antigen. In this section, we shall briefly describe each vaccine formulation regarding its development and efficacy in preventing CDI.
Because toxin A and toxin B are key virulence determinants of CDI and associated with the severity of colon damage, they become a remarkable target for vaccine development. The alteration of toxin structure by chemical treatment, thereby inactivating toxicity while preserving its immunogenicity—termed as toxoid—has been developed for over 3 decades. Toxoid-based vaccine use in animal models has demonstrated a satisfactory production level of serum antibody responses against both toxins and a preventive efficacy against a lethal outcome of CDI[143]. The first clinical trial on toxoid-based vaccine in healthy individuals was introduced 20 years ago. It was observed that the majority of study subjects developed a massive level of specific antibodies for both toxins. Although some adverse reactions were documented, the vaccine was demonstrated to be safe and immunogenic in healthy adults[144]. This information paves the way for further development of the next generation of formulated vaccine candidates that are currently under investigation.
The use of a recombinant protein-based design is another approach for vaccine development to avoid residual toxins, which are not completely inactivated by chemical, and also maximizes the protective effect using only the toxin domain responsible for immunogenicity. Furthermore, this genetic modification allows uncomplicated manufacture of vaccine preparations compared with toxoid production. Three functional domains of both toxins, including glucosyl-transferase (GT) and cysteine protease (CP), central translocation (T), and C-terminal receptor-binding domain (RBD), were tested for immunogenic validation. The first study conducted by Lyerly et al[145] on the use of the RBD of toxin A reported partial protection against CDI and death in a hamster model. The genetic modifications of toxin A RBD were widely investigated by several research groups. These modifications included the use of RBD subdomain, recombinant fusion with other immunogenic proteins, and combination with a mucosal adjuvant[146]. These constructions exhibited strong action by evoking a complete seroconversion for toxin A and preventing fluid secretion and histological changes. Although seroconversion against toxin A was found to be greater than that against toxin B, an optimal vaccine needs to include both toxin A and toxin B fragments to achieve maximum protective efficacy. Therefore, later developments focused on the combination of recombinant peptides with/without immune adjuvants. The fusion peptide, containing fragments of toxin A and toxin B, generates an immune response to both toxins. Complete protection against toxin A was observed at all doses, whereas less immunogenicity toward toxin B was noticed[147]. Co-administration of the combined recombinant protein with the adjuvant also demonstrated satisfactory positive protection, hence this vaccine formulation is currently in phase 1 clinical trial.
To overcome the limitations of previous vaccine candidates related to the prevention of CDI, surface protein antigens have emerged as an alternative target for vaccine development. Several studies have confirmed the induction of immune response during infection against surface layer protein (SLP), including flagella components, adhesin, fibronectin-binding protein, and cysteine protease[148,149]. However, none of the vaccinations provided significant protection in animal models. Furthermore, glycans, the polysaccharide coat on the surface of the bacterium, serve as another target for eliciting specific antibodies. It has been documented that all 3 glycan structures, PSI, PSII and PSIII, conjugated with other immune inducers stimulated specific antibodies, including IgA and IgG, in animal models[150,151]. It was also shown that immune cells can recognize both native and synthesized glycans, supporting the importance of this molecule to be developed as a vaccine and/or a vaccine additive.
There have been several attempts to develop vaccines for CDI. Unfortunately, there is still no approved vaccine to prevent initial and/or recurrent CDI. Nevertheless, there are 2 potential vaccine candidates reaching the final stage of development[152-154]. VLA84, a chimeric protein containing truncated toxin A and toxin B, developed by Valneva Austria GmbH, demonstrated good efficacy in a phase 1 trial[155,156]. It is currently under phase 2 clinical trial (NCT02316470). This clinical phase is a randomized placebo-controlled study of a total of 500 healthy subjects aged > 50 years. The subjects were separated into 4 groups, which individually received different vaccine doses, including VLA84 75 µg without alum, VLA84 200 µg with and without alum, and phosphate-buffered saline as the placebo group. All subjects received 3 doses of vaccination on days 0, 7, and 28. The immunogenicity and safety of VLA84 were evaluated after the last vaccination for up to 6 mo. Based on the primary outcome, it was reported that seroconversion for IgG was ≥ 4-fold increase for toxin A and toxin B on day 56. However, efficacy and data analysis are yet to be reported.
Pfizer’s vaccine has been developed using genetically modified full-length toxin A and toxin B, using a novel detoxification process that preserves the critical epitopes responsible for immunogenicity for maximizing the production of neutralizing antibodies[157,158]. This vaccine entered clinical trials in 2012 and received Fast Track designation from the United States FDA in 2014. The phase 2 clinical trial was completed on patients aged 50–85 years (NCT02117570). The vaccine induced robust immune responses and exerted protective effects in preclinical models. The Pfizer vaccine is currently undergoing phase 3 clinical trials with ≥ 17000 subjects in 23 countries. Subjects aged ≥ 50 years will receive 3-dose vaccinations at months 0, 1, and 6. Volunteers will be followed up for 3 years after the last vaccination. As the trial is ongoing, the results of data analysis from this vaccine are expected in the near future.
Although several vaccines have been developed for decades, aiming to serve as a prophylaxis for CDI, there are concerns and limitations for rapid, long-lasting, and protective immunity. Further efforts are still required to identify optimal dose, dosing schedule, and vaccine formulation, and also to determine potential application for high-risk healthy populations and immunocompromised individuals. Regarding the promising efficacy of a CDI vaccine, if approved, it will provide the primary prevention and reduction of CDI cases worldwide.
CDI is a serious healthcare concern as most countries move towards aging societies[159]; C. difficile can have maximum impact on this age group. CDI treatment is also threatened by treatment failures, especially recurrent CDI. Therefore, an urgent need exists to develop alternative treatment approaches. Small molecules and natural products have been subjected to the most advanced progress compared with other approaches, followed by an erupting trend of FMT. Both single and combinational therapies appear to be the way forward, such as antibiotic/FMT, antibiotic/antitoxin antibody, and antibiotic/synbiotic. Vaccine development is highly anticipated as the results are extremely promising and would provide a significant tool for CDI prevention and control in community and healthcare settings. Bacteriophage therapy has to overcome the grand challenge before it can be used in clinical practice. These developments are the future of CDI treatment; they require a huge amount of effort and capital; meanwhile, the management of antibiotic use, hygiene precautions and education, and monitoring systems, must be implemented to reduce the incidence of CDI.
Authors would like to thank Paul Adams for kindly proofreading the final manuscript and the Faculty of Tropical Medicine for subsidizing the English editing service on the first manuscript.
Manuscript source: Invited manuscript
Specialty type: Microbiology
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ong LT, Sales-Campos H S-Editor: Ma YJ L-Editor: A P-Editor: Ma YJ
| 1. | Kuiper GA, van Prehn J, Ang W, Kneepkens F, van der Schoor S, de Meij T. Clostridium difficile infections in young infants: Case presentations and literature review. IDCases. 2017;10:7-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 2. | Tullus K, Aronsson B, Marcus S, Möllby R. Intestinal colonization with Clostridium difficile in infants up to 18 months of age. Eur J Clin Microbiol Infect Dis. 1989;8:390-393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 82] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 3. | Hooks KB, O'Malley MA. Dysbiosis and Its Discontents. mBio. 2017;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 180] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 4. | Seekatz AM, Young VB. Clostridium difficile and the microbiota. J Clin Invest. 2014;124:4182-4189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 5. | Chandrasekaran R, Lacy DB. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev. 2017;41:723-750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 185] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 6. | Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 164] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 7. | Smits WK, Lyras D, Lacy DB, Wilcox MH, Kuijper EJ. Clostridium difficile infection. Nat Rev Dis Primers. 2016;2:16020. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 439] [Cited by in F6Publishing: 472] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 8. | Androga GO, Hart J, Foster NF, Charles A, Forbes D, Riley TV. Infection with Toxin A-Negative, Toxin B-Negative, Binary Toxin-Positive Clostridium difficile in a Young Patient with Ulcerative Colitis. J Clin Microbiol. 2015;53:3702-3704. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 9. | Eckert C, Emirian A, Le Monnier A, Cathala L, De Montclos H, Goret J, Berger P, Petit A, De Chevigny A, Jean-Pierre H, Nebbad B, Camiade S, Meckenstock R, Lalande V, Marchandin H, Barbut F. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect. 2015;3:12-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 88] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 10. | Abt MC, McKenney PT, Pamer EG. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol. 2016;14:609-620. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 320] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 11. | Poxton IR, McCoubrey J, Blair G. The pathogenicity of Clostridium difficile. Clin Microbiol Infect. 2001;7:421-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 103] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, Jans B, Hopkins S, Hansen S, Lyytikäinen O, Reilly J, Deptula A, Zingg W, Plachouras D, Monnet DL; The Healthcare-Associated Infections Prevalence Study Group. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Euro Surveill. 2018;23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 375] [Cited by in F6Publishing: 337] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 13. | Vardakas KZ, Trigkidis KK, Boukouvala E, Falagas ME. Clostridium difficile infection following systemic antibiotic administration in randomised controlled trials: a systematic review and meta-analysis. Int J Antimicrob Agents. 2016;48:1-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 14. | O'Grady K, Knight DR, Riley TV. Antimicrobial resistance in Clostridioides difficile. Eur J Clin Microbiol Infect Dis. 2021;. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis. 2018;66:e1-e48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 995] [Cited by in F6Publishing: 1196] [Article Influence: 239.2] [Reference Citation Analysis (0)] |
| 16. | Baur D, Gladstone BP, Burkert F, Carrara E, Foschi F, Döbele S, Tacconelli E. Effect of antibiotic stewardship on the incidence of infection and colonisation with antibiotic-resistant bacteria and Clostridium difficile infection: a systematic review and meta-analysis. Lancet Infect Dis. 2017;17:990-1001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 381] [Cited by in F6Publishing: 442] [Article Influence: 63.1] [Reference Citation Analysis (0)] |
| 17. | Negrut N, Nistor-Cseppento DC, Khan SA, Pantis C, Maghiar TA, Maghiar O, Aleya S, Rus M, Tit DM, Aleya L, Rahdar A, Bungau S. Clostridium difficile infection epidemiology over a period of 8 years—a single centre study. Sustainability. 2020;12:4439. [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Marra AR, Perencevich EN, Nelson RE, Samore M, Khader K, Chiang HY, Chorazy ML, Herwaldt LA, Diekema DJ, Kuxhausen MF, Blevins A, Ward MA, McDanel JS, Nair R, Balkenende E, Schweizer ML. Incidence and Outcomes Associated With Clostridium difficile Infections: A Systematic Review and Meta-analysis. JAMA Netw Open. 2020;3:e1917597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 19. | Principi N, Silvestri E, Esposito S. Advantages and Limitations of Bacteriophages for the Treatment of Bacterial Infections. Front Pharmacol. 2019;10:513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 209] [Article Influence: 41.8] [Reference Citation Analysis (0)] |
| 20. | Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med. 2019;25:730-733. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 779] [Cited by in F6Publishing: 710] [Article Influence: 142.0] [Reference Citation Analysis (0)] |
| 21. | Amidon S, Brown JE, Dave VS. Colon-targeted oral drug delivery systems: design trends and approaches. AAPS PharmSciTech. 2015;16:731-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 218] [Cited by in F6Publishing: 239] [Article Influence: 26.6] [Reference Citation Analysis (0)] |
| 22. | Jarrad AM, Karoli T, Blaskovich MA, Lyras D, Cooper MA. Clostridium difficile drug pipeline: challenges in discovery and development of new agents. J Med Chem. 2015;58:5164-5185. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 23. | Petrosillo N, Granata G, Cataldo MA. Novel Antimicrobials for the Treatment of Clostridium difficile Infection. Front Med (Lausanne). 2018;5:96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 24. | Mendes RE, Rhomberg PR, Locher HH, Jones RN. Activity of cadazolid against Gram-positive clinical isolates, including linezolid-resistant subsets with defined resistance mechanisms. ICACC; 2013; North Liberty, IA, USA. Available from: https://www.jmilabs.com/data/posters/ICAAC2013/E-144.PDF. [Cited in This Article: ] |
| 25. | Chilton CH, Crowther GS, Baines SD, Todhunter SL, Freeman J, Locher HH, Athanasiou A, Wilcox MH. In vitro activity of cadazolid against clinically relevant Clostridium difficile isolates and in an in vitro gut model of C. difficile infection. J Antimicrob Chemother. 2014;69:697-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Odingo J, Bailey MA, Files M, Early JV, Alling T, Dennison D, Bowman J, Dalai S, Kumar N, Cramer J, Masquelin T, Hipskind PA, Parish T. In Vitro Evaluation of Novel Nitazoxanide Derivatives against Mycobacterium tuberculosis. ACS Omega. 2017;2:5873-5890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 27. | Hoffman PS, Sisson G, Croxen MA, Welch K, Harman WD, Cremades N, Morash MG. Antiparasitic drug nitazoxanide inhibits the pyruvate oxidoreductases of Helicobacter pylori, selected anaerobic bacteria and parasites, and Campylobacter jejuni. Antimicrob Agents Chemother. 2007;51:868-876. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 170] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 28. | de Carvalho LP, Darby CM, Rhee KY, Nathan C. Nitazoxanide Disrupts Membrane Potential and Intrabacterial pH Homeostasis of Mycobacterium tuberculosis. ACS Med Chem Lett. 2011;2:849-854. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 29. | Rossignol JF. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res. 2014;110:94-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 315] [Cited by in F6Publishing: 341] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 30. | Stachulski AV, Taujanskas J, Pate SL, Rajoli RKR, Aljayyoussi G, Pennington SH, Ward SA, Hong WD, Biagini GA, Owen A, Nixon GL, Leung SC, O'Neill PM. Therapeutic Potential of Nitazoxanide: An Appropriate Choice for Repurposing versus SARS-CoV-2? ACS Infect Dis. 2021;7:1317-1331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Musher DM, Logan N, Bressler AM, Johnson DP, Rossignol JF. Nitazoxanide versus vancomycin in Clostridium difficile infection: a randomized, double-blind study. Clin Infect Dis. 2009;48:e41-e46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 149] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 32. | Rafiullah F, Kanwal S, Majeed UM, Korsten MA, Cheema FH, Luthra M, Sohail MR. Successful use of nitazoxanide in the treatment of recurrent Clostridium difficile infection. BMJ Case Rep. 2011;2011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 33. | Brook I, Wexler HM, Goldstein EJ. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev. 2013;26:526-546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 193] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 34. | Vickers R, Robinson N, Best E, Echols R, Tillotson G, Wilcox M. A randomised phase 1 study to investigate safety, pharmacokinetics and impact on gut microbiota following single and multiple oral doses in healthy male subjects of SMT19969, a novel agent for Clostridium difficile infections. BMC Infect Dis. 2015;15:91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 35. | Qian X, Yanagi K, Kane AV, Alden N, Lei M, Snydman DR, Vickers RJ, Lee K, Thorpe CM. Ridinilazole, a narrow spectrum antibiotic for treatment of Clostridioides difficile infection, enhances preservation of microbiota-dependent bile acids. Am J Physiol Gastrointest Liver Physiol. 2020;319:G227-G237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 36. | Bassères E, Endres BT, Khaleduzzaman M, Miraftabi F, Alam MJ, Vickers RJ, Garey KW. Impact on toxin production and cell morphology in Clostridium difficile by ridinilazole (SMT19969), a novel treatment for C. difficile infection. J Antimicrob Chemother. 2016;71:1245-1251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 37. | Vickers RJ, Tillotson GS, Nathan R, Hazan S, Pullman J, Lucasti C, Deck K, Yacyshyn B, Maliakkal B, Pesant Y, Tejura B, Roblin D, Gerding DN, Wilcox MH; CoDIFy study group. Efficacy and safety of ridinilazole compared with vancomycin for the treatment of Clostridium difficile infection: a phase 2, randomised, double-blind, active-controlled, non-inferiority study. Lancet Infect Dis. 2017;17:735-744. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 84] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 38. | Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1274] [Cited by in F6Publishing: 1179] [Article Influence: 90.7] [Reference Citation Analysis (0)] |
| 39. | Volochnyuk DM, Ryabukhin SV, Moroz YS, Savych O, Chuprina A, Horvath D, Zabolotna Y, Varnek A, Judd DB. Evolution of commercially available compounds for HTS. Drug Discov Today. 2019;24:390-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 40. | AbdelKhalek A, Mohammad H, Mayhoub AS, Seleem MN. Screening for potent and selective anticlostridial leads among FDA-approved drugs. J Antibiot (Tokyo). 2020;73:392-409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | AbdelKhalek A, Seleem MN. Repurposing the Veterinary Antiprotozoal Drug Ronidazole for the Treatment of Clostridioides difficile Infection. Int J Antimicrob Agents. 2020;56:106188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Pushpakom S, Iorio F, Eyers PA, Escott KJ, Hopper S, Wells A, Doig A, Guilliams T, Latimer J, McNamee C, Norris A, Sanseau P, Cavalla D, Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov. 2019;18:41-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1664] [Cited by in F6Publishing: 1726] [Article Influence: 287.7] [Reference Citation Analysis (0)] |
| 43. | Pal R, Seleem MN. Screening of Natural Products and Approved Oncology Drug Libraries for Activity against Clostridioides difficile. Sci Rep. 2020;10:5966. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Fillbrunn A, Dietz C, Pfeuffer J, Rahn R, Landrum GA, Berthold MR. KNIME for reproducible cross-domain analysis of life science data. J Biotechnol. 2017;261:149-156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 45. | Beisken S, Meinl T, Wiswedel B, de Figueiredo LF, Berthold M, Steinbeck C. KNIME-CDK: Workflow-driven cheminformatics. BMC Bioinformatics. 2013;14:257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 95] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 46. | Mazanetz MP, Marmon RJ, Reisser CB, Morao I. Drug discovery applications for KNIME: an open source data mining platform. Curr Top Med Chem. 2012;12:1965-1979. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 47. | Kong KF, Schneper L, Mathee K. Beta-lactam antibiotics: from antibiosis to resistance and bacteriology. APMIS. 2010;118:1-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 48. | Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12:40-79. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 825] [Cited by in F6Publishing: 721] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 49. | de la Torre BG, Albericio F. The Pharmaceutical Industry in 2019. An Analysis of FDA Drug Approvals from the Perspective of Molecules. Molecules. 2020;25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 98] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 50. | Palombo EA. Phytochemicals from traditional medicinal plants used in the treatment of diarrhoea: modes of action and effects on intestinal function. Phytother Res. 2006;20:717-724. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 153] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 51. | Patridge E, Gareiss P, Kinch MS, Hoyer D. An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discov Today. 2016;21:204-207. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 421] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 52. | Roshan N, Riley TV, Hammer KA. Antimicrobial activity of natural products against Clostridium difficile in vitro. J Appl Microbiol. 2017;123:92-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 53. | Perumalsamy H, Jung MY, Hong SM, Ahn YJ. Growth-Inhibiting and morphostructural effects of constituents identified in Asarum heterotropoides root on human intestinal bacteria. BMC Complement Altern Med. 2013;13:245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 54. | Fraternale D, Flamini G, Ricci D. Essential oil composition and antimicrobial activity of Angelica archangelica L. (Apiaceae) roots. J Med Food. 2014;17:1043-1047. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 55. | Cermak P, Olsovska J, Mikyska A, Dusek M, Kadleckova Z, Vanicek J, Nyc O, Sigler K, Bostikova V, Bostik P. Strong antimicrobial activity of xanthohumol and other derivatives from hops (Humulus lupulus L.) on gut anaerobic bacteria. APMIS. 2017;125:1033-1038. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 48] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 56. | Canning C, Sun S, Ji X, Gupta S, Zhou K. Antibacterial and cytotoxic activity of isoprenylated coumarin mammea A/AA isolated from Mammea africana. J Ethnopharmacol. 2013;147:259-262. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 57. | Mody D, Athamneh AIM, Seleem MN. Curcumin: A natural derivative with antibacterial activity against Clostridium difficile. J Glob Antimicrob Resist. 2020;21:154-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 58. | Finegold SM, Summanen PH, Corbett K, Downes J, Henning SM, Li Z. Pomegranate extract exhibits in vitro activity against Clostridium difficile. Nutrition. 2014;30:1210-1212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 59. | de Nova PJG, Carvajal A, Prieto M, Rubio P. In vitro Susceptibility and Evaluation of Techniques for Understanding the Mode of Action of a Promising Non-antibiotic Citrus Fruit Extract Against Several Pathogens. Front Microbiol. 2019;10:884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 60. | Gadhi CA, Weber M, Mory F, Benharref A, Lion C, Jana M, Lozniewski A. Antibacterial activity of Aristolochia paucinervis Pomel. J Ethnopharmacol. 1999;67:87-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 61. | Roshan N, Riley TV, Knight DR, Steer JH, Hammer KA. Natural products show diverse mechanisms of action against Clostridium difficile. J Appl Microbiol. 2019;126:468-479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Harnvoravongchai P, Chankhamhaengdecha S, Ounjai P, Singhakaew S, Boonthaworn K, Janvilisri T. Antimicrobial Effect of Asiatic Acid Against Clostridium difficile Is Associated With Disruption of Membrane Permeability. Front Microbiol. 2018;9:2125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 63. | Shilling M, Matt L, Rubin E, Visitacion MP, Haller NA, Grey SF, Woolverton CJ. Antimicrobial effects of virgin coconut oil and its medium-chain fatty acids on Clostridium difficile. J Med Food. 2013;16:1079-1085. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 64. | Blaskovich MAT, Kavanagh AM, Elliott AG, Zhang B, Ramu S, Amado M, Lowe GJ, Hinton AO, Pham DMT, Zuegg J, Beare N, Quach D, Sharp MD, Pogliano J, Rogers AP, Lyras D, Tan L, West NP, Crawford DW, Peterson ML, Callahan M, Thurn M. The antimicrobial potential of cannabidiol. Commun Biol. 2021;4:7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 92] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 65. | Gigli S, Seguella L, Pesce M, Bruzzese E, D'Alessandro A, Cuomo R, Steardo L, Sarnelli G, Esposito G. Cannabidiol restores intestinal barrier dysfunction and inhibits the apoptotic process induced by Clostridium difficile toxin A in Caco-2 cells. United European Gastroenterol J. 2017;5:1108-1115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 66. | Adejumo AC, Bukong TN. Cannabis use and risk of Clostridioides difficile infection: Analysis of 59,824 hospitalizations. Anaerobe. 2020;61:102095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | Roshan N, Riley TV, Hammer KA. Effects of natural products on several stages of the spore cycle of Clostridium difficile in vitro. J Appl Microbiol. 2018;125:710-723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 68. | Mooyottu S, Flock G, Venkitanarayanan K. Carvacrol reduces Clostridium difficile sporulation and spore outgrowth in vitro. J Med Microbiol. 2017;66:1229-1234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 69. | Yu L, Palafox-Rosas R, Luna B, She RC. The Bactericidal Activity and Spore Inhibition Effect of Manuka Honey against Clostridioides Difficile. Antibiotics (Basel). 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 70. | Pellissery AJ, Vinayamohan PG, Venkitanarayanan K. In vitro antivirulence activity of baicalin against Clostridioides difficile. J Med Microbiol. 2020;69:631-639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 71. | Roshan N, Riley TV, Knight DR, Hammer KA. Effect of natural products on the production and activity of Clostridium difficile toxins in vitro. Sci Rep. 2018;8:15735. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 72. | Wultańska D, Piotrowski M, Pituch H. The effect of berberine chloride and/or its combination with vancomycin on the growth, biofilm formation, and motility of Clostridioides difficile. Eur J Clin Microbiol Infect Dis. 2020;39:1391-1399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 73. | Giles SL, Laheij RJF. Successful treatment of persistent Clostridium difficile infection with manuka honey. Int J Antimicrob Agents. 2017;49:522-523. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 74. | Orth P, Xiao L, Hernandez LD, Reichert P, Sheth PR, Beaumont M, Yang X, Murgolo N, Ermakov G, DiNunzio E, Racine F, Karczewski J, Secore S, Ingram RN, Mayhood T, Strickland C, Therien AG. Mechanism of action and epitopes of Clostridium difficile toxin B-neutralizing antibody bezlotoxumab revealed by X-ray crystallography. J Biol Chem. 2014;289:18008-18021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 75. | Yang Z, Ramsey J, Hamza T, Zhang Y, Li S, Yfantis HG, Lee D, Hernandez LD, Seghezzi W, Furneisen JM, Davis NM, Therien AG, Feng H. Mechanisms of protection against Clostridium difficile infection by the monoclonal antitoxin antibodies actoxumab and bezlotoxumab. Infect Immun. 2015;83:822-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 76. | Wilcox MH, Gerding DN, Poxton IR, Kelly C, Nathan R, Birch T, Cornely OA, Rahav G, Bouza E, Lee C, Jenkin G, Jensen W, Kim YS, Yoshida J, Gabryelski L, Pedley A, Eves K, Tipping R, Guris D, Kartsonis N, Dorr MB; MODIFY I and MODIFY II Investigators. Bezlotoxumab for Prevention of Recurrent Clostridium difficile Infection. N Engl J Med. 2017;376:305-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 521] [Article Influence: 74.4] [Reference Citation Analysis (0)] |
| 77. | Maiti PK, inventor Immunimed Inc. , assignee. Use of polyclonal antibodies against Clostridium Difficile for treatment of inflammatory bowel disease. US patent US20180362618A1. 2017. Available from: https://patents.google.com/patent/US10513552B2/en. [Cited in This Article: ] |
| 78. | Sponseller JK, Steele JA, Schmidt DJ, Kim HB, Beamer G, Sun X, Tzipori S. Hyperimmune bovine colostrum as a novel therapy to combat Clostridium difficile infection. J Infect Dis. 2015;211:1334-1341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 79. | Heidebrecht HJ, Weiss WJ, Pulse M, Lange A, Gisch K, Kliem H, Mann S, Pfaffl MW, Kulozik U, von Eichel-Streiber C. Treatment and Prevention of Recurrent Clostridium difficile Infection with Functionalized Bovine Antibody-Enriched Whey in a Hamster Primary Infection Model. Toxins (Basel). 2019;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 80. | Sholeh M, Krutova M, Forouzesh M, Mironov S, Sadeghifard N, Molaeipour L, Maleki A, Kouhsari E. Antimicrobial resistance in Clostridioides (Clostridium) difficile derived from humans: a systematic review and meta-analysis. Antimicrob Resist Infect Control. 2020;9:158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 81. | Reeves AE, Theriot CM, Bergin IL, Huffnagle GB, Schloss PD, Young VB. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile Infection. Gut Microbes. 2011;2:145-158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 207] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 82. | Gupta S, Allen-Vercoe E, Petrof EO. Fecal microbiota transplantation: in perspective. Therap Adv Gastroenterol. 2016;9:229-239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 240] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 83. | Sbahi H, Di Palma JA. Faecal microbiota transplantation: applications and limitations in treating gastrointestinal disorders. BMJ Open Gastroenterol. 2016;3:e000087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 84. | Mullish BH, Quraishi MN, Segal JP, McCune VL, Baxter M, Marsden GL, Moore DJ, Colville A, Bhala N, Iqbal TH, Settle C, Kontkowski G, Hart AL, Hawkey PM, Goldenberg SD, Williams HRT. The use of faecal microbiota transplant as treatment for recurrent or refractory Clostridium difficile infection and other potential indications: joint British Society of Gastroenterology (BSG) and Healthcare Infection Society (HIS) guidelines. Gut. 2018;67:1920-1941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 202] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 85. | Khoruts A, Staley C, Sadowsky MJ. Faecal microbiota transplantation for Clostridioides difficile: mechanisms and pharmacology. Nat Rev Gastroenterol Hepatol. 2021;18:67-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 70] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 86. | Segal JP, Mullish BH, Quraishi MN, Iqbal T, Marchesi JR, Sokol H. Mechanisms underpinning the efficacy of faecal microbiota transplantation in treating gastrointestinal disease. Therap Adv Gastroenterol. 2020;13:1756284820946904. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 87. | van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2582] [Cited by in F6Publishing: 2489] [Article Influence: 226.3] [Reference Citation Analysis (0)] |
| 88. | Kim KO, Gluck M. Fecal Microbiota Transplantation: An Update on Clinical Practice. Clin Endosc. 2019;52:137-143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 104] [Article Influence: 20.8] [Reference Citation Analysis (1)] |
| 89. | Cammarota G, Masucci L, Ianiro G, Bibbò S, Dinoi G, Costamagna G, Sanguinetti M, Gasbarrini A. Randomised clinical trial: faecal microbiota transplantation by colonoscopy vs. vancomycin for the treatment of recurrent Clostridium difficile infection. Aliment Pharmacol Ther. 2015;41:835-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 382] [Cited by in F6Publishing: 400] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 90. | Hui W, Li T, Liu W, Zhou C, Gao F. Fecal microbiota transplantation for treatment of recurrent C. difficile infection: An updated randomized controlled trial meta-analysis. PLoS One. 2019;14:e0210016. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 85] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 91. | Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, Chang HJ, Coward S, Goodman KJ, Xu H, Madsen K, Mason A, Wong GK, Jovel J, Patterson J, Louie T. Effect of Oral Capsule- vs Colonoscopy-Delivered Fecal Microbiota Transplantation on Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA. 2017;318:1985-1993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 380] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 92. | Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ. High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes. 2013;4:125-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 237] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 93. | Lee CH, Steiner T, Petrof EO, Smieja M, Roscoe D, Nematallah A, Weese JS, Collins S, Moayyedi P, Crowther M, Ropeleski MJ, Jayaratne P, Higgins D, Li Y, Rau NV, Kim PT. Frozen vs Fresh Fecal Microbiota Transplantation and Clinical Resolution of Diarrhea in Patients With Recurrent Clostridium difficile Infection: A Randomized Clinical Trial. JAMA. 2016;315:142-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 436] [Cited by in F6Publishing: 430] [Article Influence: 53.8] [Reference Citation Analysis (0)] |
| 94. | Jiang ZD, Ajami NJ, Petrosino JF, Jun G, Hanis CL, Shah M, Hochman L, Ankoma-Sey V, DuPont AW, Wong MC, Alexander A, Ke S, DuPont HL. Randomised clinical trial: faecal microbiota transplantation for recurrent Clostridum difficile infection - fresh, or frozen, or lyophilised microbiota from a small pool of healthy donors delivered by colonoscopy. Aliment Pharmacol Ther. 2017;45:899-908. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 95. | Agrawal M, Aroniadis OC, Brandt LJ, Kelly C, Freeman S, Surawicz C, Broussard E, Stollman N, Giovanelli A, Smith B, Yen E, Trivedi A, Hubble L, Kao D, Borody T, Finlayson S, Ray A, Smith R. The Long-term Efficacy and Safety of Fecal Microbiota Transplant for Recurrent, Severe, and Complicated Clostridium difficile Infection in 146 Elderly Individuals. J Clin Gastroenterol. 2016;50:403-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 96. | Fischer M, Sipe B, Cheng YW, Phelps E, Rogers N, Sagi S, Bohm M, Xu H, Kassam Z. Fecal microbiota transplant in severe and severe-complicated Clostridium difficile: A promising treatment approach. Gut Microbes. 2017;8:289-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 97. | DeFilipp Z, Bloom PP, Torres Soto M, Mansour MK, Sater MRA, Huntley MH, Turbett S, Chung RT, Chen YB, Hohmann EL. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N Engl J Med. 2019;381:2043-2050. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 575] [Cited by in F6Publishing: 666] [Article Influence: 133.2] [Reference Citation Analysis (0)] |
| 98. | Wilcox MH, McGovern BH, Hecht GA. The Efficacy and Safety of Fecal Microbiota Transplant for Recurrent Clostridium difficile Infection: Current Understanding and Gap Analysis. Open Forum Infect Dis. 2020;7:ofaa114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 99. | Kwak S, Choi J, Hink T, Reske KA, Blount K, Jones C, Bost MH, Sun X, Burnham CD, Dubberke ER, Dantas G; CDC Prevention Epicenter Program. Impact of investigational microbiota therapeutic RBX2660 on the gut microbiome and resistome revealed by a placebo-controlled clinical trial. Microbiome. 2020;8:125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 100. | Ford C, Litcofsky K, McGovern B, Pardi D, Nathan R, Hansen V, Brennan R, Pullman J, Bernardo P, Tomlinson A, Horgan K, Bryant J, Walsh E, Rodriguez M, Rogalin H, Wang E, Henn M. 1503. Engraftment of investigational microbiome drug, SER-262, in subjects receiving vancomycin is associated with reduced rates of recurrence after primary Clostridium infection (CDI). Open Forum Infect Dis. 2019;6:S547-S548. [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 101. | McGovern BH, Ford CB, Henn MR, Pardi DS, Khanna S, Hohmann EL, O'Brien EJ, Desjardins CA, Bernardo P, Wortman JR, Lombardo MJ, Litcofsky KD, Winkler JA, McChalicher CWJ, Li SS, Tomlinson AD, Nandakumar M, Cook DN, Pomerantz RJ, Auninš JG, Trucksis M. SER-109, an Investigational Microbiome Drug to Reduce Recurrence After Clostridioides difficile Infection: Lessons Learned From a Phase 2 Trial. Clin Infect Dis. 2021;72:2132-2140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 87] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 102. | Quera R, Espinoza R, Estay C, Rivera D. Bacteremia as an adverse event of fecal microbiota transplantation in a patient with Crohn's disease and recurrent Clostridium difficile infection. J Crohns Colitis. 2014;8:252-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 113] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 103. | Vendrik KEW, Terveer EM, Kuijper EJ, Nooij S, Boeije-Koppenol E, Sanders IMJG, van Lingen E, Verspaget HW, Berssenbrugge EKL, Keller JJ, van Prehn J; Netherlands Donor Faeces Bank Study Group. Periodic screening of donor faeces with a quarantine period to prevent transmission of multidrug-resistant organisms during faecal microbiota transplantation: a retrospective cohort study. Lancet Infect Dis. 2021;21:711-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 104. | US Food and Drug Administration. Safety alert regarding use of fecal microbiota for transplantation and additional safety protections pertaining to SARS-CoV-2 and COVID-19. 2020. Available from: https://www.fda.gov/vaccines-blood-biologics/safety-availability-biologics/safety-alert-regarding-use-fecal-microbiota-transplantation-and-additional-safety-protections. [Cited in This Article: ] |
| 105. | Joint FAO/WHO. Guidelines for the evaluation of probiotics in food. 2002. Available from: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf. [Cited in This Article: ] |
| 106. | Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, Calder PC, Sanders ME. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4055] [Cited by in F6Publishing: 4408] [Article Influence: 440.8] [Reference Citation Analysis (2)] |
| 107. | Hudson LE, Anderson SE, Corbett AH, Lamb TJ. Gleaning Insights from Fecal Microbiota Transplantation and Probiotic Studies for the Rational Design of Combination Microbial Therapies. Clin Microbiol Rev. 2017;30:191-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 108. | Mills JP, Rao K, Young VB. Probiotics for prevention of Clostridium difficile infection. Curr Opin Gastroenterol. 2018;34:3-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 93] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 109. | McFarland LV, Surawicz CM, Greenberg RN, Fekety R, Elmer GW, Moyer KA, Melcher SA, Bowen KE, Cox JL, Noorani Z. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA. 1994;271:1913-1918. [PubMed] [Cited in This Article: ] |
| 110. | Gorbach SL, Chang TW, Goldin B. Successful treatment of relapsing Clostridium difficile colitis with Lactobacillus GG. Lancet. 1987;2:1519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 295] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 111. | Pace F, Pace M, Quartarone G. Probiotics in digestive diseases: focus on Lactobacillus GG. Minerva Gastroenterol Dietol. 2015;61:273-292. [PubMed] [Cited in This Article: ] |
| 112. | Cameron D, Hock QS, Kadim M, Mohan N, Ryoo E, Sandhu B, Yamashiro Y, Jie C, Hoekstra H, Guarino A. Probiotics for gastrointestinal disorders: Proposed recommendations for children of the Asia-Pacific region. World J Gastroenterol. 2017;23:7952-7964. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 50] [Cited by in F6Publishing: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 113. | Valdés-Varela L, Gueimonde M, Ruas-Madiedo P. Probiotics for Prevention and Treatment of Clostridium difficile Infection. Adv Exp Med Biol. 2018;1050:161-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 114. | McFarland LV, Ship N, Auclair J, Millette M. Primary prevention of Clostridium difficile infections with a specific probiotic combining Lactobacillus acidophilus, L. casei, and L. rhamnosus strains: assessing the evidence. J Hosp Infect. 2018;99:443-452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 115. | Hickson M, D'Souza AL, Muthu N, Rogers TR, Want S, Rajkumar C, Bulpitt CJ. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: randomised double blind placebo controlled trial. BMJ. 2007;335:80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 423] [Cited by in F6Publishing: 343] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 116. | Gao XW, Mubasher M, Fang CY, Reifer C, Miller LE. Dose-response efficacy of a proprietary probiotic formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea prophylaxis in adult patients. Am J Gastroenterol. 2010;105:1636-1641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 117. | Barker AK, Duster M, Valentine S, Hess T, Archbald-Pannone L, Guerrant R, Safdar N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J Antimicrob Chemother. 2017;72:3177-3180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 118. | Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, Federici S, Cohen Y, Linevsky R, Rothschild D, Moor AE, Ben-Moshe S, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Shapiro H, Pevsner-Fischer M, Sharon I, Halpern Z, Segal E, Elinav E. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388-1405.e21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 702] [Cited by in F6Publishing: 725] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 119. | Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1342] [Cited by in F6Publishing: 1527] [Article Influence: 254.5] [Reference Citation Analysis (0)] |
| 120. | So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT, Shanahan ER, Staudacher HM, Campbell KL. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. 2018;107:965-983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 306] [Cited by in F6Publishing: 345] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 121. | Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol. 2017;14:491-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2256] [Cited by in F6Publishing: 2491] [Article Influence: 355.9] [Reference Citation Analysis (0)] |
| 122. | Rätsep M, Kõljalg S, Sepp E, Smidt I, Truusalu K, Songisepp E, Stsepetova J, Naaber P, Mikelsaar RH, Mikelsaar M. A combination of the probiotic and prebiotic product can prevent the germination of Clostridium difficile spores and infection. Anaerobe. 2017;47:94-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 123. | Sell TL, Schaberg DR, Fekety FR. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J Clin Microbiol. 1983;17:1148-1152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 92] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 124. | Fortier LC, Moineau S. Morphological and genetic diversity of temperate phages in Clostridium difficile. Appl Environ Microbiol. 2007;73:7358-7366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 125. | Mayer MJ, Narbad A, Gasson MJ. Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol. 2008;190:6734-6740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 91] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 126. | Fortier LC. Bacteriophages Contribute to Shaping Clostridioides (Clostridium) difficile Species. Front Microbiol. 2018;9:2033. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 34] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 127. | Li T, Zhang Y, Dong K, Kuo CJ, Li C, Zhu YQ, Qin J, Li QT, Chang YF, Guo X, Zhu Y. Isolation and Characterization of the Novel Phage JD032 and Global Transcriptomic Response during JD032 Infection of Clostridioides difficile Ribotype 078. mSystems. 2020;5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 128. | Drulis-Kawa Z, Majkowska-Skrobek G, Maciejewska B. Bacteriophages and phage-derived proteins--application approaches. Curr Med Chem. 2015;22:1757-1773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 123] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 129. | Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio. 2015;6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 81] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 130. | Mayer MJ, Garefalaki V, Spoerl R, Narbad A, Meijers R. Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range. J Bacteriol. 2011;193:5477-5486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 131. | Wang Q, Euler CW, Delaune A, Fischetti VA. Using a Novel Lysin To Help Control Clostridium difficile Infections. Antimicrob Agents Chemother. 2015;59:7447-7457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 132. | Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A. Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe. 2013;22:25-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 133. | Loc-Carrillo C, Abedon ST. Pros and cons of phage therapy. Bacteriophage. 2011;1:111-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 467] [Cited by in F6Publishing: 522] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 134. | Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepiński P, Douce GR, Clokie MR. Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob Agents Chemother. 2016;60:968-981. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 149] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 135. | Selle K, Fletcher JR, Tuson H, Schmitt DS, McMillan L, Vridhambal GS, Rivera AJ, Montgomery SA, Fortier LC, Barrangou R, Theriot CM, Ousterout DG. In Vivo Targeting of Clostridioides difficile Using Phage-Delivered CRISPR-Cas3 Antimicrobials. mBio. 2020;11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 102] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 136. | Nale JY, Redgwell TA, Millard A, Clokie MRJ. Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiotics (Basel). 2018;7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 137. | Shan J, Ramachandran A, Thanki AM, Vukusic FBI, Barylski J, Clokie MRJ. Bacteriophages are more virulent to bacteria with human cells than they are in bacterial culture; insights from HT-29 cells. Sci Rep. 2018;8:5091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 138. | Caflisch KM, Suh GA, Patel R. Biological challenges of phage therapy and proposed solutions: a literature review. Expert Rev Anti Infect Ther. 2019;17:1011-1041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 139. | Hall JF, Berger D. Outcome of colectomy for Clostridium difficile colitis: a plea for early surgical management. Am J Surg. 2008;196:384-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 104] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 140. | Neal MD, Alverdy JC, Hall DE, Simmons RL, Zuckerbraun BS. Diverting loop ileostomy and colonic lavage: an alternative to total abdominal colectomy for the treatment of severe, complicated Clostridium difficile associated disease. Ann Surg. 2011;254:423-7; discussion 427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 272] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 141. | McFarland LV, Elmer GW, Surawicz CM. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol. 2002;97:1769-1775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 523] [Cited by in F6Publishing: 516] [Article Influence: 23.5] [Reference Citation Analysis (0)] |
| 142. | Popovich KJ, Calfee DP, Patel PK, Lassiter S, Rolle AJ, Hung L, Saint S, Chopra V. The Centers for Disease Control and Prevention STRIVE Initiative: Construction of a National Program to Reduce Health Care-Associated Infections at the Local Level. Ann Intern Med. 2019;171:S2-S6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 143. | Anosova NG, Brown AM, Li L, Liu N, Cole LE, Zhang J, Mehta H, Kleanthous H. Systemic antibody responses induced by a two-component Clostridium difficile toxoid vaccine protect against C. difficile-associated disease in hamsters. J Med Microbiol. 2013;62:1394-1404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 144. | Sehgal K, Khanna S. Immune response against Clostridioides difficile and translation to therapy. Therap Adv Gastroenterol. 2021;14:17562848211014817. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 145. | Lyerly DM, Johnson JL, Frey SM, Wilkins TD. Vaccination against lethal Clostridium difficile enterocolitis with a nontoxic recombinant peptide of toxin A. Curr Microbiol. 1990;21:29-32. [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 16] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 146. | Ward SJ, Douce G, Dougan G, Wren BW. Local and systemic neutralizing antibody responses induced by intranasal immunization with the nontoxic binding domain of toxin A from Clostridium difficile. Infect Immun. 1999;67:5124-5132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 147. | Ghose C, Verhagen JM, Chen X, Yu J, Huang Y, Chenesseau O, Kelly CP, Ho DD. Toll-like receptor 5-dependent immunogenicity and protective efficacy of a recombinant fusion protein vaccine containing the nontoxic domains of Clostridium difficile toxins A and B and Salmonella enterica serovar typhimurium flagellin in a mouse model of Clostridium difficile disease. Infect Immun. 2013;81:2190-2196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 148. | Péchiné S, Janoir C, Collignon A. Variability of Clostridium difficile surface proteins and specific serum antibody response in patients with Clostridium difficile-associated disease. J Clin Microbiol. 2005;43:5018-5025. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 149. | Wright A, Drudy D, Kyne L, Brown K, Fairweather NF. Immunoreactive cell wall proteins of Clostridium difficile identified by human sera. J Med Microbiol. 2008;57:750-756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 150. | Adamo R, Romano MR, Berti F, Leuzzi R, Tontini M, Danieli E, Cappelletti E, Cakici OS, Swennen E, Pinto V, Brogioni B, Proietti D, Galeotti CL, Lay L, Monteiro MA, Scarselli M, Costantino P. Phosphorylation of the synthetic hexasaccharide repeating unit is essential for the induction of antibodies to Clostridium difficile PSII cell wall polysaccharide. ACS Chem Biol. 2012;7:1420-1428. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 151. | Monteiro MA, Ma Z, Bertolo L, Jiao Y, Arroyo L, Hodgins D, Mallozzi M, Vedantam G, Sagermann M, Sundsmo J, Chow H. Carbohydrate-based Clostridium difficile vaccines. Expert Rev Vaccines. 2013;12:421-431. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 152. | Legenza LM, Barnett SG, Rose WE. Vaccines in development for the primary prevention of Clostridium difficile infection. J Am Pharm Assoc (2003). 2017;57:547-549. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 153. | Leuzzi R, Adamo R, Scarselli M. Vaccines against Clostridium difficile. Hum Vaccin Immunother. 2014;10:1466-1477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 154. | Riley TV, Lyras D, Douce GR. Status of vaccine research and development for Clostridium difficile. Vaccine. 2019;37:7300-7306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 155. | Bézay N, Ayad A, Dubischar K, Firbas C, Hochreiter R, Kiermayr S, Kiss I, Pinl F, Jilma B, Westritschnig K. Safety, immunogenicity and dose response of VLA84, a new vaccine candidate against Clostridium difficile, in healthy volunteers. Vaccine. 2016;34:2585-2592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 156. | Tian JH, Glenn G, Flyer D, Zhou B, Liu Y, Sullivan E, Wu H, Cummings JF, Elllingsworth L, Smith G. Clostridium difficile chimeric toxin receptor binding domain vaccine induced protection against different strains in active and passive challenge models. Vaccine. 2017;35:4079-4087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 157. | Donald RGK, Flint M, Kalyan N, Johnson E, Witko SE, Kotash C, Zhao P, Megati S, Yurgelonis I, Lee PK, Matsuka YV, Severina E, Deatly A, Sidhu M, Jansen KU, Minton NP, Anderson AS. A novel approach to generate a recombinant toxoid vaccine against Clostridium difficile. Microbiology (Reading). 2013;159:1254-1266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 158. | Vidunas E, Mathews A, Weaver M, Cai P, Koh EH, Patel-Brown S, Yuan H, Zheng ZR, Carriere M, Johnson JE, Lotvin J, Moran J. Production and Characterization of Chemically Inactivated Genetically Engineered Clostridium difficile Toxoids. J Pharm Sci. 2016;105:2032-2041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 159. | United Nations, Department of Economic and Social Affairs, Population Division. World population aging 2019: Highlight. 2019. Available from: https://www.un.org/en/development/desa/population/publications/pdf/ageing/WorldPopulationAgeing2019-Report.pdf. [Cited in This Article: ] |