Published online Sep 7, 2021. doi: 10.3748/wjg.v27.i33.5488
Peer-review started: February 2, 2021
First decision: March 14, 2021
Revised: March 26, 2021
Accepted: July 21, 2021
Article in press: July 21, 2021
Published online: September 7, 2021
Changes in gut microbiota influence both the gut and liver, which are strictly connected by the so-called “gut–liver axis”. The gut microbiota acts as a major determinant of this relationship in the onset and clinical course of liver diseases. According to the results of several studies, gut dysbiosis is linked to viral he
Core Tip: Changes within the gut microbiota have an impact on the mutual crosstalk between intestinal microbiota and the liver. Gut dysbiosis is linked to viral hepatitis, mainly hepatitis C virus and hepatitis B virus infection. However, concrete mecha
- Citation: Milosevic I, Russo E, Vujovic A, Barac A, Stevanovic O, Gitto S, Amedei A. Microbiota and viral hepatitis: State of the art of a complex matter. World J Gastroenterol 2021; 27(33): 5488-5501
- URL: https://www.wjgnet.com/1007-9327/full/v27/i33/5488.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i33.5488
The gut contains a large, complex microbial community that has much more genetic material than the total human genome[1]. Gut microbiota (GM) acts as a major de
The term gut-liver axis refers to the anatomical and physiological connection between the gut and the liver[3]. The gut-liver axis is based on the very close ana
The GM represents one of the main factors influencing the gut-liver axis and its role in the alteration of liver function has recently received considerable attention[3].
Gut dysbiosis may specifically lead to an inflammatory response due to increased production of pro-inflammatory cytokines, which are recognized by receptors in the portal circulation[10,12,14]. Production of these pro-inflammatory cytokines largely depends on the response of the innate immune system to the presence of microbial products[12]. It was documented that the GM profile in patients with chronic hepatitis differs considerably from that observed in healthy patients[15,16]. Moreover, the degree of liver insufficiency is closely related to the severity of gut dysbiosis[10]. Recent studies support the fact that GM dysbiosis helps the advancement of viral hepatitis infection[17]. During chronic viral hepatitis, the intestinal microbiota has a marked impact on viral host cell interaction as well as on viral replication[18]. Esche
This paper provides an overview of the state of the art of this complex matter. We discuss recent updates on the interactions of GM, liver diseases and viral hepatitis and potential therapeutic interventions that target the gut–liver axis.
As previously mentioned, the liver and intestine extensively interact via the biliary tract, the portal vein and systemic mediators. The liver is a crucial immunological organ, mainly enriched with innate immune cells and persistently exposed to nu
A large number of preclinical and clinical studies from the last decade have investigated the role of the GM in NAFLD and NASH, showing different results. In general, NAFLD patients display a decreased GM diversity[25,26], with a decrease of Bac
Several studies have been conducted in mouse and human models based on how alcohol can induce dysbiosis associated with microbiota, which in turn can lead to ALD pathogenesis and development[39]. Regarding the GM composition, ALD pa
Patients with PBC show a significant general decrease in bacterial diversity[57]. In
Several reports investigating the role of GM in PSC show an overall reduction in GM diversity[61]. Alterations in biliary and fecal microbiome in PSC patients are characterized by higher abundance of Lactobacillus, Fusobacterium, and Enterococcus and low diversity[62,63]. In addition, PSC patients showed an overrepresentation of Entero
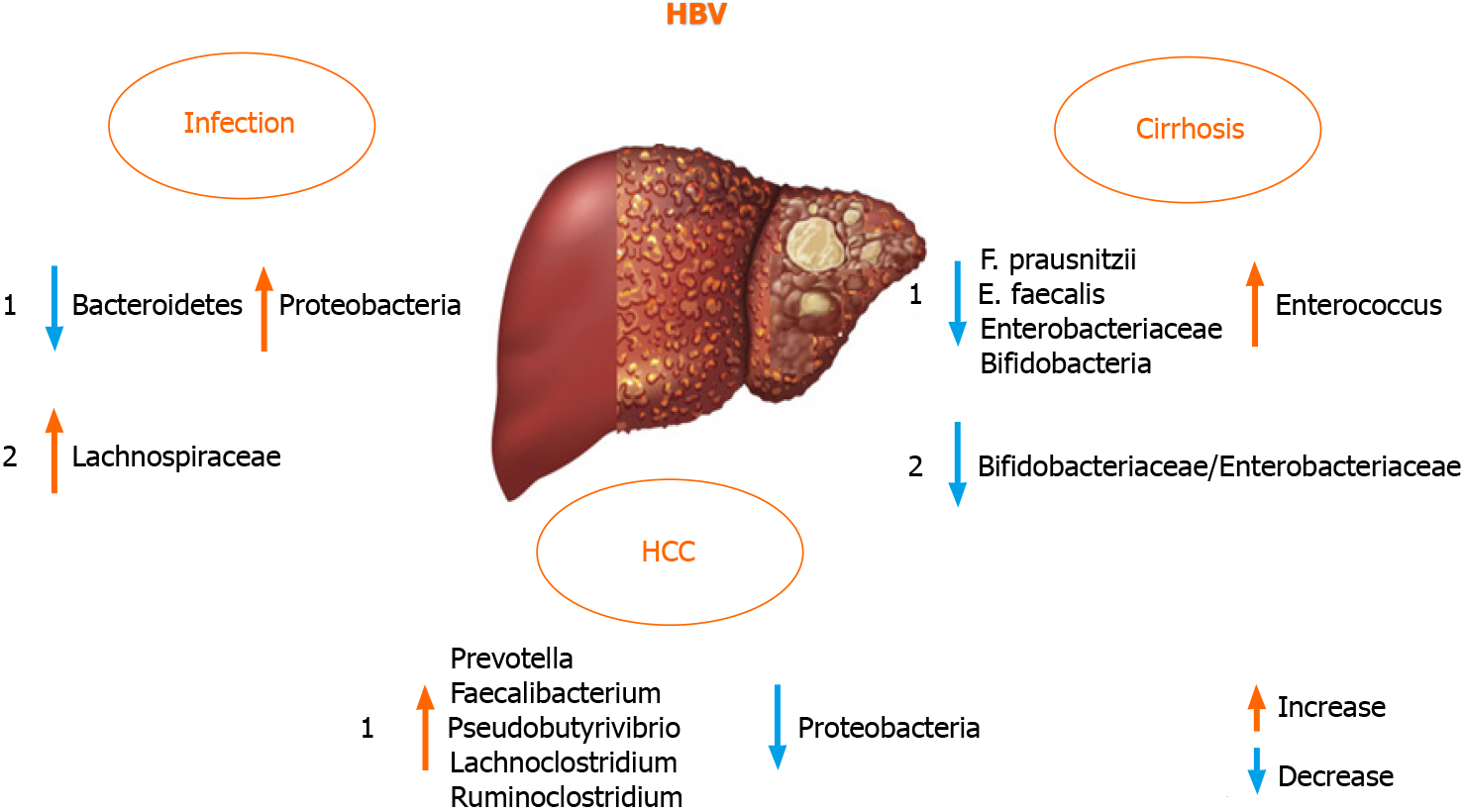
Liver failure and disease progression in patients with chronic HBV infection was found to be connected to GM dysbiosis in a large proportion[18,70]. There are di
On the contrary, HBV and HCV negative patients with HCC harbored fewer potential anti-inflammatory bacteria and more pro-inflammatory bacteria. Taken together, these data indicated that GM plays an important role in the progression of HBV or non-HBV non-HCV related HCC. These findings were different from previous reports on HBV-induced liver diseases[16,70,76]. The discrepancy in these findings was probably due to the progression of liver diseases and the group of patients in evaluated studies. To analyze the GM role in HBV positive patients, it is appropriate to use a defined cohort of patients suffering from HBV infection[77], excluding patients with different etiologies of liver cirrhosis (including alcohol abuse) that itself may change the composition of the intestinal microbial community and the same might be true for other liver diseases[70,78]. Therefore, the discrepancy of fecal microbiota between HBV positive and HBV negative patients in the study by Liu et al[70] is perhaps due to the HBV infection[70,77].
Not only gut dysbiosis, but also dysbiosis of the oral microbiota was observed in HBV patients, and yellow tongue coating is suggestive of a reduction in Bacteroidetes, but an increase in Proteobacteria. Zhao et al[79] also suggested a positive correlation between serum HBV-DNA and the number of Neisseriaceae in oral microbiota.
Furthermore, the GM composition differed according to the level of alanine ami
Recently, Wang et al[82] defined serum zonulin as an intestinal permeability marker and showed its association with AFP levels in HBV-associated liver cirrhosis and HCC, especially helpful in correlating it with advanced stages of these diseases[83]. It has been reported that the diagnostic model of one location may be not used in other locations, as the diagnostic efficiency declined when the geographic scale was in
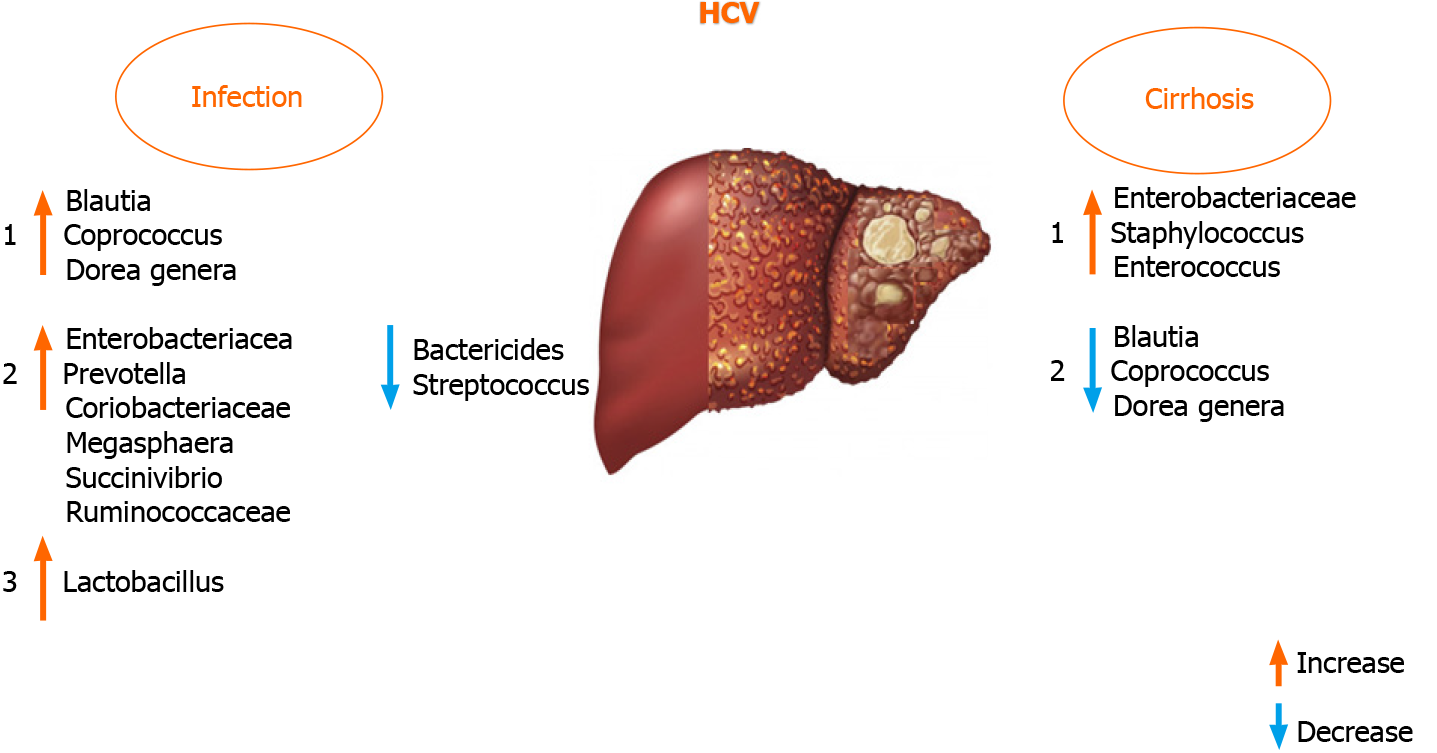
The liver-gut interaction controls GM homeostasis during HCV infection, which might be an ideal model to study the interaction between the GM and liver[18,80]. A review of previous reports provides the rationale for the hypothesis that GM dysbiosis could also be employed as a biomarker and a therapeutic target to mitigate disease pro
It was documented that antiviral HCV treatment with ribavirin + pegylated in
It was found that Blautia, Coprococcus, and Dorea genera were increased in HCV po
An increased presence of Lactobacillus in HCV-infected patients was found in very few studies, although the study cohorts were not uniform (treatment-naïve and pa
The GM varies in patients with different HCV genotypes[86]. Statistically significant differences in GM composition between patients with and without liver cirrhosis are only found in genotype non-1. In genotype 1, differences in the microbial composition are associated with the persistent HCV infection rather than with the fibrosis stage. Therefore, studies investigating patients with liver diseases compared to healthy controls might overlook the influence of the underlying disease on the intestinal microbial communities, and so other diseases and stages of diseases need to be investigated for potential associations[15,75].
A recent study has shown that the GM has a lower abundance of Bacteroidetes and higher levels of Prevotella, Enterococcus, Veillonella, Proteobacteria, Megasphaera, Burkhol
Alterations in the composition of the microbial profiles are suspected of having a role in carcinogenesis[1]. Indeed, recent studies suggested a correlation between particular bacterial profiles in HCC patients[74]. However, the contribution of GM to HCC pathogenesis is intricate: (1) A disturbed intestinal barrier brings a series of TLR ligands to the liver and activates the inflammatory response; (2) via downregulation of hepatocyte apoptosis and upregulation of hepatic stellar cell proliferation, the TLR signaling pathways mediate hepatocarcinogenesis[102]; and (3) Finally, impaired immunosurveillance is associated with abnormal GM in HCC. Moreover, microbiota dysbiosis can be related to HCC pathogenesis by increasing oxidative stress, steatosis, and the inflammatory response[103]. Additionally, the GM of HCC patients under
Different therapeutic approaches have been proposed to improve the health of patients with chronic viral hepatitis through the manipulation of GM composition, the mo
Ren et al[110] reported a clinical trial of FMT for the treatment of HBeAg positive patients with chronic HBV with ongoing entecavir and tenofovir therapy. The results demonstrated that FMT induced HBeAg clearance in a significant number of patients that had persistent positive HBeAg even after long-term antiviral treatment. This result was especially encouraging for HBeAg positive patients who otherwise could not stop oral antiviral treatment[110]. In addition, this trial provided evidence that FMT could be a beneficial treatment option for modulating GM in patients with chronic HBV. HBV carriers might be suitable donors for FMT as their GM composition seems to be more appropriate compared to the healthy population. The results of Yang et al[108] confirmed that HBV carriers have altered GM although they are asympto
The use of probiotics in HBV positive patients was shown to be beneficial and suggested that probiotic VSL#3 plays an important role in the management of HBV infection[111]. In addition, the use of probiotics in HCV-infected patients with cirr
This literature review provides evidence on the association between gut dysbiosis and the clinical course of both chronic HBV and HCV infection. The evidence in humans seems to confirm the potential role of intestinal overgrowth of pathogenic bacteria and the development of chronic viral hepatitis observed in animal-based studies. In ad
The authors thank Dr. Elisangela Miceli, PhD and Dr. Savannah Devente for English revisions.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Serbia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Zheng H S-Editor: Liu M L-Editor: Webster JR P-Editor: Yuan YY
| 1. | Russo E, Amedei A. The Role of the Microbiota in the Genesis of Gastrointestinal Cancers. In: Frontiers in Anti-Infective Drug Discovery. Bentham e Books: 1-44. [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Sultan S, El-Mowafy M, Elgaml A, El-Mesery M, El Shabrawi A, Elegezy M, Hammami R, Mottawea W. Alterations of the Treatment-Naive Gut Microbiome in Newly Diagnosed Hepatitis C Virus Infection. ACS Infect Dis. 2021;7:1059-1068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 3. | Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, Gmizic I, Stevanovic O, Djordjevic V, Lekic N, Russo E, Amedei A. Gut-Liver Axis, Gut Microbiota, and Its Modulation in the Management of Liver Diseases: A Review of the Literature. Int J Mol Sci. 2019;20:395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 255] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 4. | Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55-60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3971] [Cited by in F6Publishing: 4291] [Article Influence: 357.6] [Reference Citation Analysis (0)] |
| 5. | Mottawea W, Chiang CK, Mühlbauer M, Starr AE, Butcher J, Abujamel T, Deeke SA, Brandel A, Zhou H, Shokralla S, Hajibabaei M, Singleton R, Benchimol EI, Jobin C, Mack DR, Figeys D, Stintzi A. Altered intestinal microbiota-host mitochondria crosstalk in new onset Crohn's disease. Nat Commun. 2016;7:13419. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 251] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 6. | Distrutti E, Monaldi L, Ricci P, Fiorucci S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J Gastroenterol. 2016;22:2219-2241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 210] [Cited by in F6Publishing: 182] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 7. | Ray K. Gut microbiota: Adding weight to the microbiota's role in obesity--exposure to antibiotics early in life can lead to increased adiposity. Nat Rev Gastroenterol Hepatol. 2012;9:615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 8. | Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1374] [Cited by in F6Publishing: 1481] [Article Influence: 123.4] [Reference Citation Analysis (0)] |
| 9. | Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4:623-632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 750] [Cited by in F6Publishing: 784] [Article Influence: 156.8] [Reference Citation Analysis (0)] |
| 10. | Grąt M, Wronka KM, Krasnodębski M, Masior Ł, Lewandowski Z, Kosińska I, Grąt K, Stypułkowski J, Rejowski S, Wasilewicz M, Gałęcka M, Szachta P, Krawczyk M. Profile of Gut Microbiota Associated With the Presence of Hepatocellular Cancer in Patients With Liver Cirrhosis. Transplant Proc. 2016;48:1687-1691. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 11. | Brenner DA, Paik YH, Schnabl B. Role of Gut Microbiota in Liver Disease. J Clin Gastroenterol. 2015;49 Suppl 1:S25-S27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 12. | Giannelli V, Di Gregorio V, Iebba V, Giusto M, Schippa S, Merli M, Thalheimer U. Microbiota and the gut-liver axis: bacterial translocation, inflammation and infection in cirrhosis. World J Gastroenterol. 2014;20:16795-16810. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 156] [Cited by in F6Publishing: 149] [Article Influence: 14.9] [Reference Citation Analysis (1)] |
| 13. | Vassallo G, Mirijello A, Ferrulli A, Antonelli M, Landolfi R, Gasbarrini A, Addolorato G. Review article: Alcohol and gut microbiota - the possible role of gut microbiota modulation in the treatment of alcoholic liver disease. Aliment Pharmacol Ther. 2015;41:917-927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 14. | Chassaing B, Etienne-Mesmin L, Gewirtz AT. Microbiota-liver axis in hepatic disease. Hepatology. 2014;59:328-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 245] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 15. | Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1230] [Cited by in F6Publishing: 1325] [Article Influence: 132.5] [Reference Citation Analysis (2)] |
| 16. | Lu H, Wu Z, Xu W, Yang J, Chen Y, Li L. Intestinal microbiota was assessed in cirrhotic patients with hepatitis B virus infection. Intestinal microbiota of HBV cirrhotic patients. Microb Ecol. 2011;61:693-703. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 145] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 17. | Xu D, Huang Y, Wang J. Gut microbiota modulate the immune effect against hepatitis B virus infection. Eur J Clin Microbiol Infect Dis. 2015;34:2139-2147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Sehgal R, Bedi O, Trehanpati N. Role of Microbiota in Pathogenesis and Management of Viral Hepatitis. Front Cell Infect Microbiol. 2020;10:341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 4.3] [Reference Citation Analysis (3)] |
| 19. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 659] [Cited by in F6Publishing: 713] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 20. | Chen Y, Ji F, Guo J, Shi D, Fang D, Li L. Dysbiosis of small intestinal microbiota in liver cirrhosis and its association with etiology. Sci Rep. 2016;6:34055. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 129] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 21. | Mohamadkhani A. On the potential role of intestinal microbial community in hepatocarcinogenesis in chronic hepatitis B. Cancer Med. 2018;7:3095-3100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Cui L, Morris A, Ghedin E. The human mycobiome in health and disease. Genome Med. 2013;5:63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 220] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 23. | Hartmann P, Seebauer CT, Schnabl B. Alcoholic liver disease: the gut microbiome and liver cross talk. Alcohol Clin Exp Res. 2015;39:763-775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 175] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 24. | Wang R, Tang R, Li B, Ma X, Schnabl B, Tilg H. Gut microbiome, liver immunology, and liver diseases. Cell Mol Immunol. 2021;18:4-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 150] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 25. | Zhu L, Baker SS, Gill C, Liu W, Alkhouri R, Baker RD, Gill SR. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57:601-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1015] [Cited by in F6Publishing: 1101] [Article Influence: 100.1] [Reference Citation Analysis (1)] |
| 26. | Jiang W, Wu N, Wang X, Chi Y, Zhang Y, Qiu X, Hu Y, Li J, Liu Y. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 421] [Cited by in F6Publishing: 388] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 27. | Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer SE, Lou W, Allard JP. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep. 2018;8:1466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 160] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 28. | Alferink LJM, Radjabzadeh D, Erler NS, Vojinovic D, Medina-Gomez C, Uitterlinden AG, de Knegt RJ, Amin N, Ikram MA, Janssen HLA, Kiefte-de Jong JC, Metselaar HJ, van Duijn CM, Kraaij R, Darwish Murad S. Microbiomics, Metabolomics, Predicted Metagenomics, and Hepatic Steatosis in a Population-Based Study of 1,355 Adults. Hepatology. 2021;73:968-982. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 29. | Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, Hunault G, Oberti F, Calès P, Diehl AM. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764-775. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 763] [Cited by in F6Publishing: 862] [Article Influence: 107.8] [Reference Citation Analysis (0)] |
| 30. | Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, Greenwood R, Sikaroodi M, Lam V, Crotty P, Bailey J, Myers RP, Rioux KP. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11:868-75.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 470] [Article Influence: 42.7] [Reference Citation Analysis (0)] |
| 31. | Schwenger KJP, Chen L, Chelliah A, Da Silva HE, Teterina A, Comelli EM, Taibi A, Arendt BM, Fischer S, Allard JP. Markers of activated inflammatory cells are associated with disease severity and intestinal microbiota in adults with nonalcoholic fatty liver disease. Int J Mol Med. 2018;42:2229-2237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 32. | Loomba R, Seguritan V, Li W, Long T, Klitgord N, Bhatt A, Dulai PS, Caussy C, Bettencourt R, Highlander SK, Jones MB, Sirlin CB, Schnabl B, Brinkac L, Schork N, Chen CH, Brenner DA, Biggs W, Yooseph S, Venter JC, Nelson KE. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2019;30:607. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 33. | Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, Waget A, Delmée E, Cousin B, Sulpice T, Chamontin B, Ferrières J, Tanti JF, Gibson GR, Casteilla L, Delzenne NM, Alessi MC, Burcelin R. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761-1772. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4095] [Cited by in F6Publishing: 4129] [Article Influence: 242.9] [Reference Citation Analysis (0)] |
| 34. | Lang S, Demir M, Martin A, Jiang L, Zhang X, Duan Y, Gao B, Wisplinghoff H, Kasper P, Roderburg C, Tacke F, Steffen HM, Goeser T, Abraldes JG, Tu XM, Loomba R, Stärkel P, Pride D, Fouts DE, Schnabl B. Intestinal Virome Signature Associated With Severity of Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;159:1839-1852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 86] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 35. | Caussy C, Tripathi A, Humphrey G, Bassirian S, Singh S, Faulkner C, Bettencourt R, Rizo E, Richards L, Xu ZZ, Downes MR, Evans RM, Brenner DA, Sirlin CB, Knight R, Loomba R. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun. 2019;10:1406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 184] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 36. | Corbin KD, Zeisel SH. Choline metabolism provides novel insights into nonalcoholic fatty liver disease and its progression. Curr Opin Gastroenterol. 2012;28:159-165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 300] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 37. | Song M, Yuan F, Li X, Ma X, Yin X, Rouchka EC, Zhang X, Deng Z, Prough RA, McClain CJ. Analysis of sex differences in dietary copper-fructose interaction-induced alterations of gut microbial activity in relation to hepatic steatosis. Biol Sex Differ. 2021;12:3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 38. | Scorletti E, Afolabi PR, Miles EA, Smith DE, Almehmadi A, Alshathry A, Childs CE, Del Fabbro S, Bilson J, Moyses HE, Clough GF, Sethi JK, Patel J, Wright M, Breen DJ, Peebles C, Darekar A, Aspinall R, Fowell AJ, Dowman JK, Nobili V, Targher G, Delzenne NM, Bindels LB, Calder PC, Byrne CD. Synbiotics Alter Fecal Microbiomes, But Not Liver Fat or Fibrosis, in a Randomized Trial of Patients With Nonalcoholic Fatty Liver Disease. Gastroenterology. 2020;158:1597-1610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 116] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 39. | Li F, Duan K, Wang C, McClain C, Feng W. Probiotics and Alcoholic Liver Disease: Treatment and Potential Mechanisms. Gastroenterol Res Pract. 2016;2016:5491465. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 54] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 40. | Bull-Otterson L, Feng W, Kirpich I, Wang Y, Qin X, Liu Y, Gobejishvili L, Joshi-Barve S, Ayvaz T, Petrosino J, Kong M, Barker D, McClain C, Barve S. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PLoS One. 2013;8:e53028. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 382] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 41. | Tuomisto S, Pessi T, Collin P, Vuento R, Aittoniemi J, Karhunen PJ. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14:40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 42. | Bluemel S, Williams B, Knight R, Schnabl B. Precision medicine in alcoholic and nonalcoholic fatty liver disease via modulating the gut microbiota. Am J Physiol Gastrointest Liver Physiol. 2016;311:G1018-G1036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 57] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 43. | Davis BC, Bajaj JS. The Human Gut Microbiome in Liver Diseases. Semin Liver Dis. 2017;37:128-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Kakiyama G, Hylemon PB, Zhou H, Pandak WM, Heuman DM, Kang DJ, Takei H, Nittono H, Ridlon JM, Fuchs M, Gurley EC, Wang Y, Liu R, Sanyal AJ, Gillevet PM, Bajaj JS. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G929-G937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 137] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 45. | Kirpich IA, Solovieva NV, Leikhter SN, Shidakova NA, Lebedeva OV, Sidorov PI, Bazhukova TA, Soloviev AG, Barve SS, McClain CJ, Cave M. Probiotics restore bowel flora and improve liver enzymes in human alcohol-induced liver injury: a pilot study. Alcohol. 2008;42:675-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 340] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 46. | Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K, de Timary P, Delzenne NM. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci USA. 2014;111:E4485-E4493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 571] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 47. | Bode C, Kolepke R, Schäfer K, Bode JC. Breath hydrogen excretion in patients with alcoholic liver disease--evidence of small intestinal bacterial overgrowth. Z Gastroenterol. 1993;31:3-7. [PubMed] [Cited in This Article: ] |
| 48. | Hauge T, Persson J, Danielsson D. Mucosal bacterial growth in the upper gastrointestinal tract in alcoholics (heavy drinkers). Digestion. 1997;58:591-595. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 49. | Morencos FC, de las Heras Castaño G, Martín Ramos L, López Arias MJ, Ledesma F, Pons Romero F. Small bowel bacterial overgrowth in patients with alcoholic cirrhosis. Dig Dis Sci. 1995;40:1252-1256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 101] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 50. | Parlesak A, Schäfer C, Schütz T, Bode JC, Bode C. Increased intestinal permeability to macromolecules and endotoxemia in patients with chronic alcohol abuse in different stages of alcohol-induced liver disease. J Hepatol. 2000;32:742-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 461] [Cited by in F6Publishing: 445] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 51. | Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148:30-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 442] [Cited by in F6Publishing: 473] [Article Influence: 52.6] [Reference Citation Analysis (0)] |
| 52. | Mutlu EA, Gillevet PM, Rangwala H, Sikaroodi M, Naqvi A, Engen PA, Kwasny M, Lau CK, Keshavarzian A. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302:G966-G978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 479] [Cited by in F6Publishing: 493] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 53. | Yan AW, Fouts DE, Brandl J, Stärkel P, Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA, Schnabl B. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53:96-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 529] [Cited by in F6Publishing: 549] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 54. | Llopis M, Cassard AM, Wrzosek L, Boschat L, Bruneau A, Ferrere G, Puchois V, Martin JC, Lepage P, Le Roy T, Lefèvre L, Langelier B, Cailleux F, González-Castro AM, Rabot S, Gaudin F, Agostini H, Prévot S, Berrebi D, Ciocan D, Jousse C, Naveau S, Gérard P, Perlemuter G. Intestinal microbiota contributes to individual susceptibility to alcoholic liver disease. Gut. 2016;65:830-839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 326] [Cited by in F6Publishing: 346] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 55. | Chen P, Torralba M, Tan J, Embree M, Zengler K, Stärkel P, van Pijkeren JP, DePew J, Loomba R, Ho SB, Bajaj JS, Mutlu EA, Keshavarzian A, Tsukamoto H, Nelson KE, Fouts DE, Schnabl B. Supplementation of saturated long-chain fatty acids maintains intestinal eubiosis and reduces ethanol-induced liver injury in mice. Gastroenterology. 2015;148:203-214.e16. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 214] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 56. | Cresci GA, Glueck B, McMullen MR, Xin W, Allende D, Nagy LE. Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J Gastroenterol Hepatol. 2017;32:1587-1597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 57. | Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, Yang F, Miao Q, Xiao X, Zhang H, Lian M, Jiang X, Zhang J, Cao Q, Fan Z, Wu M, Qiu D, Fang JY, Ansari A, Gershwin ME, Ma X. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut. 2018;67:534-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 243] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 58. | Lv LX, Fang DQ, Shi D, Chen DY, Yan R, Zhu YX, Chen YF, Shao L, Guo FF, Wu WR, Li A, Shi HY, Jiang XW, Jiang HY, Xiao YH, Zheng SS, Li LJ. Alterations and correlations of the gut microbiome, metabolism and immunity in patients with primary biliary cirrhosis. Environ Microbiol. 2016;18:2272-2286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 153] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 59. | Chen W, Wei Y, Xiong A, Li Y, Guan H, Wang Q, Miao Q, Bian Z, Xiao X, Lian M, Zhang J, Li B, Cao Q, Fan Z, Zhang W, Qiu D, Fang J, Gershwin ME, Yang L, Tang R, Ma X. Comprehensive Analysis of Serum and Fecal Bile Acid Profiles and Interaction with Gut Microbiota in Primary Biliary Cholangitis. Clin Rev Allergy Immunol. 2020;58:25-38. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 76] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 60. | Furukawa M, Moriya K, Nakayama J, Inoue T, Momoda R, Kawaratani H, Namisaki T, Sato S, Douhara A, Kaji K, Kitade M, Shimozato N, Sawada Y, Saikawa S, Takaya H, Kitagawa K, Akahane T, Mitoro A, Yamao J, Tanaka Y, Yoshiji H. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol Res. 2020;50:840-852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 39] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 61. | Karlsen TH. Primary sclerosing cholangitis: 50 years of a gut-liver relationship and still no love? Gut. 2016;65:1579-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 62. | Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C; Saint-Antoine IBD Network, Chazouillères O, Housset C, Sokol H. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut. 2020;69:92-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 115] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 63. | Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, Trøseid M, Marschall HU, Schrumpf E, Moum B, Røsjø H, Aukrust P, Karlsen TH, Hov JR. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 257] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 64. | Bajer L, Kverka M, Kostovcik M, Macinga P, Dvorak J, Stehlikova Z, Brezina J, Wohl P, Spicak J, Drastich P. Distinct gut microbiota profiles in patients with primary sclerosing cholangitis and ulcerative colitis. World J Gastroenterol. 2017;23:4548-4558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 215] [Cited by in F6Publishing: 214] [Article Influence: 30.6] [Reference Citation Analysis (1)] |
| 65. | Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, Ferrante M, Van Assche G, Van der Merwe S, Vermeire S, Raes J. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut. 2016;65:1681-1689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 270] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 66. | Torres J, Bao X, Goel A, Colombel JF, Pekow J, Jabri B, Williams KM, Castillo A, Odin JA, Meckel K, Fasihuddin F, Peter I, Itzkowitz S, Hu J. The features of mucosa-associated microbiota in primary sclerosing cholangitis. Aliment Pharmacol Ther. 2016;43:790-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 67. | Rossen NG, Fuentes S, Boonstra K, D'Haens GR, Heilig HG, Zoetendal EG, de Vos WM, Ponsioen CY. The mucosa-associated microbiota of PSC patients is characterized by low diversity and low abundance of uncultured Clostridiales II. J Crohns Colitis. 2015;9:342-348. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 87] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 68. | Tabibian JH, Talwalkar JA, Lindor KD. Role of the microbiota and antibiotics in primary sclerosing cholangitis. Biomed Res Int. 2013;2013:389537. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 69. | Mattner J. Impact of Microbes on the Pathogenesis of Primary Biliary Cirrhosis (PBC) and Primary Sclerosing Cholangitis (PSC). Int J Mol Sci. 2016;17:1864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 70. | Liu Q, Li F, Zhuang Y, Xu J, Wang J, Mao X, Zhang Y, Liu X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019;11:1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 109] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 71. | Deng YD, Peng XB, Zhao RR, Ma CQ, Li JN, Yao LQ. The intestinal microbial community dissimilarity in hepatitis B virus-related liver cirrhosis patients with and without at alcohol consumption. Gut Pathog. 2019;11:58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 72. | Yun Y, Chang Y, Kim HN, Ryu S, Kwon MJ, Cho YK, Kim HL, Cheong HS, Joo EJ. Alterations of the Gut Microbiome in Chronic Hepatitis B Virus Infection Associated with Alanine Aminotransferase Level. J Clin Med. 2019;8:173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 73. | Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1657] [Cited by in F6Publishing: 1670] [Article Influence: 104.4] [Reference Citation Analysis (0)] |
| 74. | Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H, Xie H, Chen X, Shao L, Zhang R, Xu S, Zhang H, Cui G, Sun R, Wen H, Lerut JP, Kan Q, Li L, Zheng S. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68:1014-1023. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 387] [Article Influence: 77.4] [Reference Citation Analysis (0)] |
| 75. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 674] [Cited by in F6Publishing: 701] [Article Influence: 53.9] [Reference Citation Analysis (1)] |
| 76. | Wei X, Yan X, Zou D, Yang Z, Wang X, Liu W, Wang S, Li X, Han J, Huang L, Yuan J. Abnormal fecal microbiota community and functions in patients with hepatitis B liver cirrhosis as revealed by a metagenomic approach. BMC Gastroenterol. 2013;13:175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 77. | Tsai KN, Kuo CF, Ou JJ. Mechanisms of Hepatitis B Virus Persistence. Trends Microbiol. 2018;26:33-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 78. | Betrapally NS, Gillevet PM, Bajaj JS. Gut microbiome and liver disease. Transl Res. 2017;179:49-59. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 79. | Zhao Y, Mao YF, Tang YS, Ni MZ, Liu QH, Wang Y, Feng Q, Peng JH, Hu YY. Altered oral microbiota in chronic hepatitis B patients with different tongue coatings. World J Gastroenterol. 2018;24:3448-3461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 26] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 80. | Kanmani P, Suganya K, Kim H. The Gut Microbiota: How Does It Influence the Development and Progression of Liver Diseases. Biomedicines. 2020;8:501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 81. | Voukelatou P, Vrettos I, Kalliakmanis A. Neurologic symptoms as the only manifestation of B12 deficiency in a young patient with normal hematocrit, MCV, peripheral blood smear and homocysteine levels. Oxf Med Case Reports. 2016;2016:omw091. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 82. | Wang WW, Zhang Y, Huang XB, You N, Zheng L, Li J. Fecal microbiota transplantation prevents hepatic encephalopathy in rats with carbon tetrachloride-induced acute hepatic dysfunction. World J Gastroenterol. 2017;23:6983-6994. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 69] [Cited by in F6Publishing: 63] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 83. | Fasano A. Intestinal permeability and its regulation by zonulin: diagnostic and therapeutic implications. Clin Gastroenterol Hepatol. 2012;10:1096-1100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 214] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 84. | Heidrich B, Steinmann E, Plumeier I, Kirschner J, Sollik L, Ziegert S, Engelmann M, Lehmann P, Manns MP, Pieper DH, Wedemeyer H. Frequent detection of HCV RNA and HCVcoreAg in stool of patients with chronic hepatitis C. J Clin Virol. 2016;80:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 85. | Kandasamy S, Vlasova AN, Fischer D, Kumar A, Chattha KS, Rauf A, Shao L, Langel SN, Rajashekara G, Saif LJ. Differential Effects of Escherichia coli Nissle and Lactobacillus rhamnosus Strain GG on Human Rotavirus Binding, Infection, and B Cell Immunity. J Immunol. 2016;196:1780-1789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 86. | Heidrich B, Vital M, Plumeier I, Döscher N, Kahl S, Kirschner J, Ziegert S, Solbach P, Lenzen H, Potthoff A, Manns MP, Wedemeyer H, Pieper DH. Intestinal microbiota in patients with chronic hepatitis C with and without cirrhosis compared with healthy controls. Liver Int. 2018;38:50-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 87. | Inoue T, Nakayama J, Moriya K, Kawaratani H, Momoda R, Ito K, Iio E, Nojiri S, Fujiwara K, Yoneda M, Yoshiji H, Tanaka Y. Gut Dysbiosis Associated With Hepatitis C Virus Infection. Clin Infect Dis. 2018;67:869-877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 103] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 88. | Preveden T, Scarpellini E, Milić N, Luzza F, Abenavoli L. Gut microbiota changes and chronic hepatitis C virus infection. Expert Rev Gastroenterol Hepatol. 2017;11:813-819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 89. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, Morgan XC, Kostic AD, Luo C, González A, McDonald D, Haberman Y, Walters T, Baker S, Rosh J, Stephens M, Heyman M, Markowitz J, Baldassano R, Griffiths A, Sylvester F, Mack D, Kim S, Crandall W, Hyams J, Huttenhower C, Knight R, Xavier RJ. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382-392. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1945] [Cited by in F6Publishing: 2054] [Article Influence: 205.4] [Reference Citation Analysis (0)] |
| 90. | Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J; MetaHIT consortium, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1208] [Cited by in F6Publishing: 1332] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 91. | Ponziani FR, Putignani L, Paroni Sterbini F, Petito V, Picca A, Del Chierico F, Reddel S, Calvani R, Marzetti E, Sanguinetti M, Gasbarrini A, Pompili M. Influence of hepatitis C virus eradication with direct-acting antivirals on the gut microbiota in patients with cirrhosis. Aliment Pharmacol Ther. 2018;48:1301-1311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 92. | Pérez-Matute P, Íñiguez M, Villanueva-Millán MJ, Recio-Fernández E, Vázquez AM, Sánchez SC, Morano LE, Oteo JA. Short-term effects of direct-acting antiviral agents on inflammation and gut microbiota in hepatitis C-infected patients. Eur J Intern Med. 2019;67:47-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 93. | Cani PD, de Vos WM. Next-Generation Beneficial Microbes: The Case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 507] [Cited by in F6Publishing: 589] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 94. | Bajaj JS, Kassam Z, Fagan A, Gavis EA, Liu E, Cox IJ, Kheradman R, Heuman D, Wang J, Gurry T, Williams R, Sikaroodi M, Fuchs M, Alm E, John B, Thacker LR, Riva A, Smith M, Taylor-Robinson SD, Gillevet PM. Fecal microbiota transplant from a rational stool donor improves hepatic encephalopathy: A randomized clinical trial. Hepatology. 2017;66:1727-1738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 376] [Article Influence: 53.7] [Reference Citation Analysis (0)] |
| 95. | Aly AM, Adel A, El-Gendy AO, Essam TM, Aziz RK. Gut microbiome alterations in patients with stage 4 hepatitis C. Gut Pathog. 2016;8:42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 96. | Kwak DS, Jun DW, Seo JG, Chung WS, Park SE, Lee KN, Khalid-Saeed W, Lee HL, Lee OY, Yoon BC, Choi HS. Short-term probiotic therapy alleviates small intestinal bacterial overgrowth, but does not improve intestinal permeability in chronic liver disease. Eur J Gastroenterol Hepatol. 2014;26:1353-1359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 97. | Bajaj JS, Hylemon PB, Ridlon JM, Heuman DM, Daita K, White MB, Monteith P, Noble NA, Sikaroodi M, Gillevet PM. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol. 2012;303:G675-G685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 375] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 98. | Santiago A, Pozuelo M, Poca M, Gely C, Nieto JC, Torras X, Román E, Campos D, Sarrabayrouse G, Vidal S, Alvarado-Tapias E, Guarner F, Soriano G, Manichanh C, Guarner C. Alteration of the serum microbiome composition in cirrhotic patients with ascites. Sci Rep. 2016;6:25001. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 99. | Bajaj JS, Betrapally NS, Hylemon PB, Heuman DM, Daita K, White MB, Unser A, Thacker LR, Sanyal AJ, Kang DJ, Sikaroodi M, Gillevet PM. Salivary microbiota reflects changes in gut microbiota in cirrhosis with hepatic encephalopathy. Hepatology. 2015;62:1260-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 216] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 100. | Bajaj JS, Vargas HE, Reddy KR, Lai JC, O'Leary JG, Tandon P, Wong F, Mitrani R, White MB, Kelly M, Fagan A, Patil R, Sait S, Sikaroodi M, Thacker LR, Gillevet PM. Association Between Intestinal Microbiota Collected at Hospital Admission and Outcomes of Patients With Cirrhosis. Clin Gastroenterol Hepatol. 2019;17:756-765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 101. | Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, Singh S, Madamba EV, Bettencourt R, Richards L, Yu RT, Atkins AR, Huan T, Brenner DA, Sirlin CB, Downes M, Evans RM, Loomba R. A Universal Gut-Microbiome-Derived Signature Predicts Cirrhosis. Cell Metab. 2020;32:901. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 102. | Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I, Caviglia JM, Khiabanian H, Adeyemi A, Bataller R, Lefkowitch JH, Bower M, Friedman R, Sartor RB, Rabadan R, Schwabe RF. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21:504-516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 854] [Cited by in F6Publishing: 901] [Article Influence: 75.1] [Reference Citation Analysis (0)] |
| 103. | Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397-411. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 571] [Cited by in F6Publishing: 762] [Article Influence: 127.0] [Reference Citation Analysis (0)] |
| 104. | Sheng L, Jena PK, Hu Y, Liu HX, Nagar N, Kalanetra KM, French SW, Mills DA, Wan YY. Hepatic inflammation caused by dysregulated bile acid synthesis is reversible by butyrate supplementation. J Pathol. 2017;243:431-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 105. | Krüttgen A, Horz HP, Weber-Heynemann J, Vucur M, Trautwein C, Haase G, Luedde T, Roderburg C. Study on the association of Helicobacter species with viral hepatitis-induced hepatocellular carcinoma. Gut Microbes. 2012;3:228-233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 106. | Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, Fidelle M, Flament C, Poirier-Colame V, Opolon P, Klein C, Iribarren K, Mondragón L, Jacquelot N, Qu B, Ferrere G, Clémenson C, Mezquita L, Masip JR, Naltet C, Brosseau S, Kaderbhai C, Richard C, Rizvi H, Levenez F, Galleron N, Quinquis B, Pons N, Ryffel B, Minard-Colin V, Gonin P, Soria JC, Deutsch E, Loriot Y, Ghiringhelli F, Zalcman G, Goldwasser F, Escudier B, Hellmann MD, Eggermont A, Raoult D, Albiges L, Kroemer G, Zitvogel L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2493] [Cited by in F6Publishing: 3120] [Article Influence: 445.7] [Reference Citation Analysis (0)] |
| 107. | Russo E, Nannini G, Dinu M, Pagliai G, Sofi F, Amedei A. Exploring the food-gut axis in immunotherapy response of cancer patients. World J Gastroenterol. 2020;26:4919-4932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 14] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (1)] |
| 108. | Yang XA, Lv F, Wang R, Chang Y, Zhao Y, Cui X, Li H, Yang S, Li S, Zhao X, Mo Z, Yang F. Potential role of intestinal microflora in disease progression among patients with different stages of Hepatitis B. Gut Pathog. 2020;12:50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 109. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5625] [Cited by in F6Publishing: 5971] [Article Influence: 542.8] [Reference Citation Analysis (0)] |
| 110. | Ren YD, Ye ZS, Yang LZ, Jin LX, Wei WJ, Deng YY, Chen XX, Xiao CX, Yu XF, Xu HZ, Xu LZ, Tang YN, Zhou F, Wang XL, Chen MY, Chen LG, Hong MZ, Ren JL, Pan JS. Fecal microbiota transplantation induces hepatitis B virus e-antigen (HBeAg) clearance in patients with positive HBeAg after long-term antiviral therapy. Hepatology. 2017;65:1765-1768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 111. | Dhiman RK, Rana B, Agrawal S, Garg A, Chopra M, Thumburu KK, Khattri A, Malhotra S, Duseja A, Chawla YK. Probiotic VSL#3 reduces liver disease severity and hospitalization in patients with cirrhosis: a randomized, controlled trial. Gastroenterology. 2014;147:1327-1337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 233] [Cited by in F6Publishing: 220] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 112. | Dore GJ, Ward J, Thursz M. Hepatitis C disease burden and strategies to manage the burden (Guest Editors Mark Thursz, Gregory Dore and John Ward). J Viral Hepat. 2014;21 Suppl 1:1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 113. | Doskali M, Tanaka Y, Ohira M, Ishiyama K, Tashiro H, Chayama K, Ohdan H. Possibility of adoptive immunotherapy with peripheral blood-derived CD3⁻CD56+ and CD3+CD56+ cells for inducing antihepatocellular carcinoma and antihepatitis C virus activity. J Immunother. 2011;34:129-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |