Published online Jun 14, 2021. doi: 10.3748/wjg.v27.i22.3010
Peer-review started: November 14, 2020
First decision: February 28, 2021
Revised: March 8, 2021
Accepted: April 26, 2021
Article in press: April 26, 2021
Published online: June 14, 2021
The intensive crosstalk between the liver and the intestine performs many essential functions. This crosstalk is important for natural immune surveillance, adaptive immune response regulation and nutrient metabolism and elimination of toxic bacterial metabolites. The interaction between the gut microbiome and bile acids is bidirectional. The gut microbiome regulates the synthesis of bile acids and their biological signaling activity and circulation via enzymes. Similarly, bile acids also shape the composition of the gut microbiome by modulating the host’s natural antibacterial defense and the intestinal immune system. The interaction between bile acids and the gut microbiome has been implicated in the pathophysiology of many intestinal and extra intestinal diseases, especially liver diseases. As essential mediators of the gut-liver crosstalk, bile acids regulate specific host metabolic pathways and modulate the inflammatory responses through farnesoid X-activated receptor and G protein-coupled bile acid receptor 1. Several clinical trials have demonstrated the signaling effects of bile acids in the context of liver diseases. We hypothesize the existence of a gut microbiome-bile acids-liver triangle and explore the potential therapeutic strategies for liver diseases targeting the triangle.
Core Tip: This review explores the interaction between the gut microbiome, bile acids and liver disease. Bile acids are involved in liver immune and inflammatory responses via the membrane receptor G protein-coupled bile acid receptor 1 and the intracellular farnesoid-X receptor. The interaction between the gut microbiome and bile acids can control host immunological homeostasis and inhibit liver inflammation.
- Citation: Shao JW, Ge TT, Chen SZ, Wang G, Yang Q, Huang CH, Xu LC, Chen Z. Role of bile acids in liver diseases mediated by the gut microbiome. World J Gastroenterol 2021; 27(22): 3010-3021
- URL: https://www.wjgnet.com/1007-9327/full/v27/i22/3010.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i22.3010
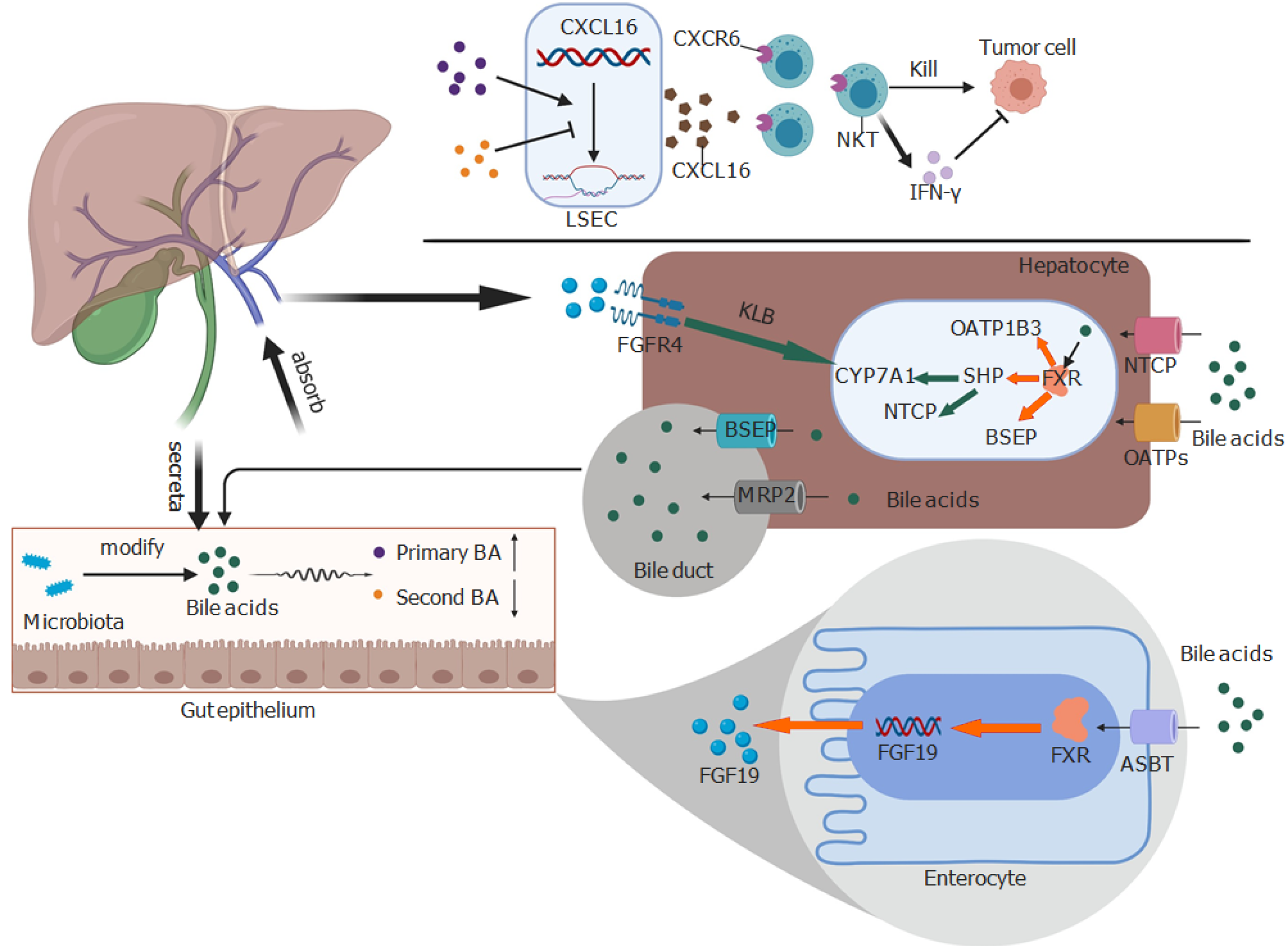
Approximately 70%-75% of the liver blood supply is derived from the intestine, which forms the basis of the gut-liver axis[1]. Liver plays an important role in the immune response of the human body; it serves as the central hub of the crosstalk between the host metabolism and the intestinal microenvironment. Therefore, the liver is exposed to a large number of bacterial components, metabolites and microbiome-derived signals. Recent studies have indicated that bile acids act as pleiotropic signaling molecules that mediate the gut-liver crosstalk[2,3]. The complex relationship between bile acids and the gut microbiome (mutual dependence and inhibition) plays an important role in the maintenance of mammalian homeostasis.
The gut microbiome participates in the conversion of bile acids and regulates the synthesis and reabsorption of bile acids in the liver (Figure 1). Studies have found decreased activity and gene expression levels of cholesterol 7α-hydroxylase (CYP7A1), a rate-limiting enzyme in the classical synthesis pathway of bile acids, in CONV-R mice compared with those in germ-free mice[4,5]. Moreover, the volume of the bile acid pool in CONV-R mice is smaller, which may be related to the decreased expression of ileal sodium-dependent bile acid transporter, which leads to decreased reabsorption of bile acids in distal ileum and increased fecal excretion of bile acids[5]. The 7α-dehydroxylation reaction of the intestinal microbiome is an essential part of biological conversion[6]. The genus Clostridium exhibits 7α-dehydroxylation activity that can transform primary bile acids into secondary bile acids[7]. Most Gram-positive bacteria in the intestine have bile salt hydrolases activity, while only Bacteroides among the Gram-negative bacteria can hydrolyze conjugated bile acids into unconjugated bile acids. The biological functions of bile acids in the body vary depending upon their state[8]. Unconjugated bile acids can eliminate the pH differential across the cell membrane. The consequent disappearance of bio-energy driven by the proton pump can cause direct damage to the cell membrane. Eventually, bile acids inhibit the growth of certain bacteria and participate in the formation of the gut microbiome[9].
Bile acids, as essential signal molecules for bidirectional regulation between the liver and the intestinal tract, are mainly activated by the following two signaling pathways: binding of a signaling molecule to G-protein coupled bile acid receptor 1 (GPBAR1 or TGR5) and activation of the expression of the farnesoid X-activated receptor (FXR). The above two pathways control the balance of energy metabolism, regulate hepatic steatosis and inflammatory response and influence the composition of the gut microbiome by shaping intestinal immunity and some antimicrobial properties of endogenous peptides. Therefore, the use of bile acids as signaling molecules by the gut microbiome may play a role in the pathophysiology of liver diseases[10-13]. Better characterization of the specific sites of action of the gut microbiome and bile acids in different signaling pathways in liver diseases can lay the foundation for novel therapies targeting bile acids.
In a seminal study by Ma et al[10], ABX (primaxin, neomycin and vancomycin) fed mice exhibited fewer and smaller primary or metastatic liver cancer lesions, and this result was not related to the age, strain or sex. These results suggested that regulation of the gut microbiome may usefully alter the growth kinetics of liver tumors.
Several studies have investigated the mechanism linking the gut microbiome and liver tumor immunity and surveillance. In a study, CD8+ T cells and natural killer T (NKT) cells in the liver tissues of ABX-treated EL4-tumor-bearing mice were significantly increased compared with that in the control group. However, in the ABX-treated MYC mice, only NKT cells were increased, and similar results were observed in normal mice without tumors. Moreover, ABX-treated mice showed increased expression of CXCL16 mRNA in the liver sinusoidal endothelial cells (LSEC). Further studies showed that CXCL16 is the only ligand of CXCR6, which can induce the accumulation of CXCR6+ hepatic NKT cells in the liver[14]. NKT cells can directly kill CD1d-expressing tumors (B16, EL4 and A20) and can also suppress liver tumors by secreting interferon-.
To identify the potential link between bile acids and NKT cells, the bile acid profile of ABX-treated mice was determined. The results showed significantly increased primary bile acids compared with that in H2O-treated mice. The findings were further confirmed after treatment of isolated LSECs with various bile acids or a combination of tauro-β-muricholic acid with -muricholic acid or tauro--muricholic acid; the results showed that primary bile acids could indeed upregulate the expression of CXCL16 mRNA. Predictably, the secondary bile acids (lithocholic acid) reversed the ABX-induced suppression of the growth of intrahepatic tumors growth and increased liver surface metastasis.
The above findings suggest that, the level of CXCL16 on LSECs can be upregulated by the gut microbiome through the primary bile acids, which leads to the accumulation of CXCR6+ hepatic NKT cells in the liver, while the secondary bile acids have the opposite effect. Although, ABX kills most of the bacteria in the mice intestine, the regulatory impact of the remaining bacteria on NKT cells cannot be excluded. To address this issue, the above experiment was repeated with germ-free mice. The results showed greater accumulation of NKT cells and increased expression of CXCL16 mRNA in the liver of germ-free mice. Some antibiotics such as cefoperazone and vancomycin were used against Gram-positive bacteria to increase primary bile acids and NKT cells in the liver, while depleting secondary bile acids. The 7-α dehydroxylation reaction is a key step in the transformation of primary bile acids into secondary bile acids[6]; Clostridium XIV among Gram-positive bacteria synthesizes 7-α dehydroxylase[7]. To explore the role of Clostridium in the accumulation of hepatic NKT cells, mice were first fed vancomycin to increase the NKT cells. One week later, the antibiotic treatment was replaced with Clostridium scidens, a type of Clostridium occurring in both humans and mice. On the second day after successful colonization of Clostridium scidens, hepatic NKT levels began to decline with a decrease in primary bile acids. The antibiotic effect was offset when antibiotic-treated mice were gavaged with bile acids metabolizing bacteria or were fed with secondary bile acids. Both the inhibition of intrahepatic tumor growth and accumulation of hepatic NKT cells were reversed in mice with an altered gut microbiome. Clostridium species is a key bacterium for regulating bile acid signal NKT cell accumulation.
LSECs isolated from human samples were treated with chenodeoxycholic acid (CDCA) and taurocholic acid. The results were similar to the mouse study in that primary bile acids were also found to upregulate the expression of CXCL16 mRNA. Normal tissues excised from patients with cholangiocarcinoma and hepatocellular carcinoma showed a positive correlation between CDCA and CXCL16, while secondary bile acids were observed to have opposite results. The above findings indicate that gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells, which is also applicable to the human body.
Nonalcoholic fatty liver disease (NAFLD) is a clinicopathologic syndrome characterized by excessive fat deposition in hepatocytes and other tissues caused by factors other than alcohol and other definitive causes of liver damage. In the absence of effective interventions, NAFLD may progress to fibrosis, cirrhosis and hepatocellular carcinoma. Environmental, genetic and metabolic factors as well as altered gut microbiome have been implicated in the pathogenesis of NAFLD. Bile acids and their metabolites help maintain the homeostasis of glucose, cholesterol and triglycerides in the liver and regulate inflammation; this is considered a potential therapeutic target for NAFLD[15,16].
Recent studies have revealed that the role of bile acids as signaling molecules may affect the development of NAFLD at multiple levels[17]. These regulatory activities are mainly achieved by FXR and TGR5, and different bile acids have different effects on these two signaling pathways[18]. FXR can regulate lipid and glucose metabolism through different pathways, such as by inhibiting the expression of hepatic gluconeogenesis genes and increasing liver glycogen synthesis and insulin sensitivity[19,20]. It can also induce the expression and secretion of liver fibroblast growth factor 21(FGF21), which as a metabolic regulator can stimulate the uptake of glucose in adipose tissue[21]. In addition, FXR activation inhibits lipogenesis, promotes fatty acid oxidation and affects cholesterol transport[22]. Bile acids can also activate the G protein-coupled receptor. TGR5 is expressed in nonparenchymal hepatocytes, monocytes and a variety of macrophages that secrete various inflammatory mediators and play an important role in regulating inflammatory responses[23]. Bile acids can inhibit the lipopolysaccharide-induced secretion of interleukin (IL)-6, IL-1A and IL-1B; in addition, bile acids can inhibit the secretion of tumor necrosis factor by Kupffer cells through a TGR5-cAMP-dependent pathway[24]. TGR5 can also regulate glucose homeostasis by inducing the expression of glucagon-like peptide-1 and inhibiting the activation of Nod-like receptor protein 3 inflammasome. Activation of TGR5 results in increased energy expenditure and weight loss[25,26]. TGR5 or FXR agonists can reduce lipogenesis, improve cholesterolemia, induce energy consumption and reduce liver inflammation in NAFLD patients[27,28].
Clinical studies have implicated disorders of bile acid homeostasis and related signaling pathways in the occurrence of NAFLD[29]. The serum concentration of total bile acids in patients with nonalcoholic steatohepatitis was three times higher than that in healthy individuals. Moreover, the composition of the bile acid pool was also different in these two groups[30]. Bile acids have a direct antibacterial effect; through FXR, they can induce the production of antibacterial peptides, such as angiogenin 1, which participates in the shaping of the gut microbiome[31]. During the development and progression of NAFLD, disruption of bile acid balance is also accompanied by disruption of the gut microbiome. Therefore, the liver-bile acid-gut microbiome triangle is a good entry point for the treatment of NAFLD.
In two human studies, obeticholic acid (OCA), an FXR agonist, was shown to decrease liver fibrosis markers and improve insulin resistance in patients with NAFLD[32]. Patients receiving OCA treatment showed a significant decrease in high-density lipoprotein and an increase in low-density lipoprotein levels. Compared with the placebo-treated group, OCA significantly improved the NAFLD Activity Score while significantly reducing liver fibrosis[33,34]. As a derivative of cholic acid, INT-777 is a selective agonist of TGR5 that increases energy expenditure and induces weight loss in high fat diet-fed mice[35]. McMahan et al[27] found that INT-767, a dual agonist of FXR/TGR5, can reduce the expression of proinflammatory factors, decrease hepatic steatosis and transform monocytes and macrophages to the anti-inflammatory M2 phenotype.
Alcohol-related liver disease (ALD) is caused by chronic consumption of alcohol. It is initially characterized by liver steatosis, which in turn can develop into alcoholic hepatitis, liver fibrosis and cirrhosis. Alcohol can cause damage to multiple target organs, especially the brain, intestines and liver. It worth noting that alcohol intake can alter the structure of the gut microbiome prior to the occurrence of overt liver diseases.
Continued drinking in patients with alcoholic cirrhosis can aggravate gut microbiome disorders, reduce the detection of gut commensal bacteria in feces and worsen the function of the duodenal and colonic mucosa. Chronic alcohol exposure is associated with gut microbiome dysbiosis in preclinical models and the human gut, which is associated with the pathogenesis of ALD. Previous studies have shown altered gut microbiome in patients with alcoholic cirrhosis, which was characterized by an increase in endotoxin-producing bacteria and a decrease in the gut commensal bacteria[36]. Changes in composition of the gut microbiome may alter brain function, and alcohol abuse can also affect the gut-brain axis; this may further aggravate alcohol dependence and induce affective disorders, ultimately, accelerating the development of hepatic encephalopathy. Compared to people who do not have the disease and do not drink alcohol, patients with alcoholic cirrhosis have higher levels of endotoxins and worse gut microbiome dysbiosis even after cessation of alcohol intake[36]. This indicates that alcohol-induced damage to the gut microbiome continues even after cessation of intake and that the damage also extends to the gut-brain axis, leading to cognitive impairment[37].
FXR negatively regulates CYP7A1, which is a rate-limiting enzyme for bile acid synthesis via FGF15. In long-term alcohol-fed mice, ethanol was shown to negate the negative regulation of bile acids by conjugated CDCA through the FXR pathway and increase the expression of CYP7A1 protein in liver cells; this ultimately led to an increased concentration of bile acids in the serum and liver. FXR can induce the expression of antimicrobial molecules in intestinal epithelial cells to prevent alcohol-induced damage to the enteric tight junctions and avoid loss of intestinal barrier integrity[38]. Intragastric administration of the FXR agonist fexaramine caused a decrease in serum alanine aminotransferase levels and hepatic IL-1B and tumor necrosis factor protein. This is because FXR can increase the small heterodimer partner protein to inhibit intestinal inflammation and protect the integrity of the intestinal barrier[11]. The above finding indicates that FXR agonists can negatively regulate the synthesis of bile acids and reduce the concentration of serum bile acids, which can alleviate alcohol-induced steatosis and liver inflammation. Administration of antibiotics to chronic drinkers was found to reduce alcohol-related liver disease. This is because antibiotics can kill gut commensal bacteria, reduce the concentration of bile acids and inhibit the hydrolysis of bile acids; this can only reduce the toxicity of deoxycholic acid to hepatocytes and stabilize the intestinal barrier function.
In summary, alcohol-related changes in the gut microbiome can ultimately alter the bile acid profile. Interventions targeting the bile acid-FXR-FGF15 signaling pathway to regulate the synthesis of CYP7A1 and lipid metabolism can reduce the occurrence of ALD in mice. As a signaling molecule, bile acids can modulate the complex interactions between the gut microbiome and alcohol through the gut-liver-brain axis. Therefore, FGF19 and fexaramine are good candidates for the treatment of ALD.
Chronic cholestasis can lead to liver damage by affecting the expression levels of nuclear receptors and bile acid transporters. Hydrophobic bile acids can induce hepatocyte injury. Bile salt can cause apoptosis at concentrations of 50-200 µmol/L, induce proinflammatory responses at concentration of 200 µmol/L and cause cell necrosis at concentrations of 200-2000 µmol/L[39-42]. The progress of autoimmune cholestatic liver diseases also affects the composition of the gut microbiome, aggravating the development of cholestasis in this interactive cycle[43].
Cholestatic liver diseases are often accompanied by gut microbiome dysbiosis and reduced bacterial diversity[44]. Kummen et al[45] found that the gut microbiome of patients with primary sclerosing cholangitis (PSC) was significantly different from that of patients with ulcerative colitis without liver disease as well as that of healthy individuals. The Veillonella genus was overexpressed only in the intestines of patients with PSC. It is worth noting that Veillonella showed a positive correlation with the pathogenesis of fibrosis, not only in PSC but also in other fibrotic diseases, such as idiopathic pulmonary fibrosis[46]. The changes in gut commensal bacteria are related to the pathogenesis of NAFLD and ALD and to primary biliary cholangitis (PBC) and PSC. This may be related to the abnormal development of immunity caused by an imbalance of the gut microbiome, resulting in imbalanced production of injurious vs cytoprotective metabolites[46].
In previous studies, bile acids have been considered a tissue-damaging factor that promotes inflammation owing to its chemical properties; its detergent effect can destroy the cell and mitochondrial membranes[47]. Overall, there are three important pathways of cytotoxicity induced by bile acids: (1) oxidative stress in the endoplasmic reticulum and mitochondria; (2) direct activation of death receptors Traill2 and Fas; and (3) lysis of the plasma membrane of liver cells. Thus, accumulation of hydrophobic bile acids is the leading cause of cholestatic liver diseases. However, a recent article suggested that two derivatives of lithocholic acid (LCA) (isoalloLCA and 3-oxolCA) can influence the adaptive immune response by regulating the differentiation of T helper (Th)17 and regulatory T (Treg) cells[48]. They are mutually restricted in function, and the change in their ratio has a decisive role in the pathogenesis and clinical prognosis of autoimmune and inflammatory diseases. Many studies have shown an imbalance between Th17 and Treg cells in patients with PBC[49-52]. These patients presented with a defective CD8+ Treg cell subset and preferentially activated Th17 cells[53]. IsoalloLCA can promote the differentiation of Treg cells by increasing mitochondrial reactive oxygen species synthesis and the expression of H3K27ac in the Foxp3 promoter region under the induction of the TGF-β signal[48]. Another LCA derivative, 3-oxoLCA, inhibits Th17 cells differentiation, manifested by significantly reduced IL-17a, thereby inhibiting inflammation[48]. The results demonstrated that LCA derivatives (isoalloLCA and 3-oxoLCA) can regulate the balance between Th17 and Treg cells, which is of great significance for the treatment of cholestatic liver diseases. Moreover, isoalloLCA and 3-oxoLCA are expected to be used for the treatment of autoimmune or inflammatory diseases mediated by Th17/Treg cells imbalance in the future.
Bile acids play an important role in regulating both adaptive and innate immune systems through the gut-bile acids-liver triangle. Among them, ursodeoxycholic acid (UDCA) is currently the only drug that has been approved for the treatment of PBC. It can effectively reduce the retention of toxic bile acids in liver cells and alleviate liver damage[54,55]. However, UDCA has limited efficacy in cholestatic liver disease[56]; in addition, some patients are unable to tolerate the adverse effects of UDCA (such as gastrointestinal symptoms)[57].
Development of effective medical therapy for cholestatic liver disease is a key imperative. 24-norursodeoxycholic acid (norUDCA), a C23 homologue of UDCA with shortened lateral chains, has effective antifibrosis, anticholestatic and anti-inflammatory properties[58,59]. In a phase II clinical study, 12-wk treatment with norUDCA caused a significant dose-dependent reduction in serum alkaline phosphatase levels in PSC patients. A multicenter randomized controlled trial evaluated the efficacy and safety of norUDCA (500 mg/d, 1000 mg/d or 1500 mg/d) compared with placebo in PSC patients. NorUDCA was shown to have an excellent safety profile similar to placebo[60]. Another bile acid, OCA (an FXR agonist), has shown potential benefits for PBC. It has approximately 100 times greater potency in activating FXR than CDCA[61]. OCA protects hepatocytes from the toxic effects of bile acids by activating the FXR receptor, reducing bile acids synthesis and improving choleresis. In addition to the effect of FXR on bile acid homeostasis, OCA monotherapy can improve the secretion of IgM and tumor necrosis factor-α and has direct immunomodulatory, antifibrosis and anti-inflammatory effects[62,63]. In clinical trials, OCA monotherapy caused a significantly greater decrease in alkaline phosphatase and bilirubin levels from baseline as compared to placebo. However, OCA treatment caused a dose-related increase in pruritus[34,57]. In conclusion, OCA may represent a new treatment option for PBC patients who cannot tolerate UDCA. Discovery of new bile acids and understanding the best way to use various bile acids may help develop new treatments for cholestatic liver disease.
Previous studies have shown that gut microbiome changes or dysbiosis in patients with chronic liver disease or cirrhosis are often accompanied by a significant reduction in total bile acids and secondary bile acid/primary bile acid ratio[8]. The dysbiosis is characterized by a decrease in bile acid 7α-dehydroxylating bacteria, a change in Bacteroides/Firmicutes ratio and an increase in pathogenic Gram-negative bacteria[64-66]. During the progression of cirrhosis, there is overgrowth of pathogenic bacteria in the small intestine, due to translocation of lipopolysaccharide, endotoxins and other metabolites and the resultant inflammation. In fact, studies have demonstrated a positive correlation of Enterobacteriaceae with endotoxemia inflammation and CDCA levels in feces. These metabolites derived from oxidative stress and the metabolism of ammonia and aromatic amino acids were positively related to Porphyromonadaceae and Enterobacteriaceae and closely related to the occurrence of hepatic encephalopathy[67].
Kakiyama et al[8] proposed that gut microbiome dysbiosis in cirrhotic patients is partly attributable to the decreased concentration of bile acids in the intestine. For example, the decrease in the number of bile acid 7α-dehydroxylating bacteria is caused by the reduction in the level of primary bile acids in the colon, which serves as an energy source[7,68]. Reduced levels of bile acids entering the small intestine can lead to overgrowth of proinflammatory and pathogenic bacteria and induce the release of inflammatory markers as well as an increase in liver inflammation[69]. Liver inflammation triggers a positive-feedback mechanism, which can further inhibit the synthesis of bile acids[70]. The size and composition of the bile acid pool can significantly regulate the gut microbiome structure and serve as an indicator of the severity of hepatic diseases. In summary, the balance of the liver-bile acid-gut microbiome axis is essential for human health and liver fibrosis.
DCA, the most effective antimicrobial in bile acids, is produced by bile acid 7α-dehydroxylating bacteria[71]. Studies have shown that increasing the DCA/cholic acid ratio in patients with cirrhosis can improve the toxic metabolites from the gut microbiome and increase the incidence of endotoxemia and hepatic encephalopathy, which may be related to the destruction of the intestinal mucosal barrier by DCA[8,72,73]. Compared with DCA, which exacerbates barrier dysfunction, LCA has a much less destructive effect on the gut microbiome due to its insolubility in water and easy excretion with feces[72,73]. TGR5 is a membrane receptor that can be activated by a variety of bile acids, among which LCA is its most potent natural agonist[18]. In the study by Guo et al[74], LCA inhibited the activation of Nod-like receptor protein 3 inflammasome via the TGR5-cAMP-protein kinase A axis, which significantly repressed the maturation of caspase-1 and the secretion of IL-B or IL-18. In addition, LCA was also shown to reduce the release of proinflammatory cytokines induced by lipopolysaccharide and the phagocytic activity of macrophages through TGR5, thus inhibiting liver inflammation[24,75-77]. Recent studies have shown that two different metabolites of LCA can also control host immune responses both in human and mice[78-80]. In clinical studies, LCA content in the stool of patients with advanced cirrhosis was significantly lower than that in patients with early cirrhosis[8]. Previous studies have focused on the cytotoxicity of bile acids. However, recent studies have revealed the anti-inflammatory and immunomodulatory effects of bile acids. Increasing evidence has shown that bile acids are a potential therapeutic target against inflammatory diseases. An appropriate increase in the concentration of bile acids in patients with liver cirrhosis may repress liver inflammation and improve liver fibrosis, which deserves further discussion.
Numerous studies in recent decades have shown that the function of bile acids is beyond that of “digestive surfactants.” At the host level, there is a clear relationship between bile acid signaling and innate immunity in the liver and intestine. In other words, bile acids are the cornerstone of the immune axis between the liver and the gut microbiome. As a mediator in the gut-liver axis, bile acids can regulate the inflammatory response, host metabolism and innate immunity, which are effective therapeutic targets in the context of various hepatic diseases (Table 1). However, most contemporary research on bile acids is based on genetically modified mice models, while the immune system, bile acid metabolism and the gut microbiome of mice are vastly different from that of humans. Recent clinical trials of FXR agonists have yielded promising results. Preclinical data suggest that gut microbiome metabolism of bile acids is also a potential therapeutic target. Changes in the gut microbiome to regulate the composition of bile acids can improve liver health through the use of antibiotics, probiotics, prebiotics and fecal bacteria transplantation[81,82]. The complex interactions between bile acids and host-microbiome in the gut-liver axis are only beginning to be understood. Clinical trials and further in-depth studies can help characterize the different roles of bile acids in healthy individuals and patients with hepatic diseases, allowing their optimal utilization as potential therapeutic targets.
| Title | Ref. | Target | Result |
| Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis | Song et al[83], 2020 | BA nuclear receptors | Restoration of the intestinal BA pool can increase colonic RORγ+ Treg cell counts and ameliorate host susceptibility to inflammatory colitis |
| Bile acid metabolites control Th17 and Treg cell differentiation | Hang et al[48], 2019 | Derivatives of LCA, 3-oxoLCA and isoalloLCA | Administration of 3-oxoLCA and isoalloLCA to mice reduced Th17 cell differentiation and increased Treg cell differentiation, respectively, in the intestinal lamina propria |
| Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome | Guo et al[26], 2016 | Membrane receptor TGR5 | Bile acids inhibited activation of the NLRP3 inflammasome via the TGR5-cAMP-PKA axis. |
| Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells | Ma et al[10], 2018 | Hepatic CXCR6+ NKT cells | Microbiome uses bile acids as a messenger to accumulate NKT cells, which have an activated phenotype and inhibit liver tumor growth. |
| TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading | Pols et al[77], 2011 | Membrane receptor TGR5 | TGR5 activation in macrophages by 6a-ethyl-23(S)-methylcholic acid (6-EMCA, INT-777), a semisynthetic BA, inhibits proinflammatory cytokine production, an effect mediated by TGR5-induced cAMP signaling and subsequent NF-κB inhibition. |
The authors gratefully acknowledge all the people that have made this study.
Manuscript source: Unsolicited manuscript
Specialty type: Food science and technology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ruiz-Margáin A S-Editor: Gong ZM L-Editor: Filipodia P-Editor: Wang LL
| 1. | Abdel-Misih SR, Bloomston M. Liver anatomy. Surg Clin North Am. 2010;90:643-653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 117] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 2. | de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 699] [Cited by in F6Publishing: 771] [Article Influence: 70.1] [Reference Citation Analysis (0)] |
| 3. | Wahlström A, Sayin SI, Marschall HU, Bäckhed F. Intestinal Crosstalk between Bile Acids and Microbiota and Its Impact on Host Metabolism. Cell Metab. 2016;24:41-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1317] [Cited by in F6Publishing: 1477] [Article Influence: 184.6] [Reference Citation Analysis (0)] |
| 4. | Li-Hawkins J, Gåfvels M, Olin M, Lund EG, Andersson U, Schuster G, Björkhem I, Russell DW, Eggertsen G. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 2002;110:1191-1200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 106] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 5. | Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225-235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1367] [Cited by in F6Publishing: 1460] [Article Influence: 132.7] [Reference Citation Analysis (0)] |
| 6. | Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 367] [Cited by in F6Publishing: 325] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 7. | Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1644] [Cited by in F6Publishing: 1789] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 8. | Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, Takei H, Muto A, Nittono H, Ridlon JM, White MB, Noble NA, Monteith P, Fuchs M, Thacker LR, Sikaroodi M, Bajaj JS. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949-955. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 503] [Cited by in F6Publishing: 546] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 9. | Ciocan D, Voican CS, Wrzosek L, Hugot C, Rainteau D, Humbert L, Cassard AM, Perlemuter G. Bile acid homeostasis and intestinal dysbiosis in alcoholic hepatitis. Aliment Pharmacol Ther. 2018;48:961-974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 10. | Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, Agdashian D, Terabe M, Berzofsky JA, Fako V, Ritz T, Longerich T, Theriot CM, McCulloch JA, Roy S, Yuan W, Thovarai V, Sen SK, Ruchirawat M, Korangy F, Wang XW, Trinchieri G, Greten TF. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 549] [Cited by in F6Publishing: 784] [Article Influence: 130.7] [Reference Citation Analysis (0)] |
| 11. | Hartmann P, Hochrath K, Horvath A, Chen P, Seebauer CT, Llorente C, Wang L, Alnouti Y, Fouts DE, Stärkel P, Loomba R, Coulter S, Liddle C, Yu RT, Ling L, Rossi SJ, DePaoli AM, Downes M, Evans RM, Brenner DA, Schnabl B. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67:2150-2166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 147] [Cited by in F6Publishing: 172] [Article Influence: 28.7] [Reference Citation Analysis (0)] |
| 12. | Wehr A, Baeck C, Heymann F, Niemietz PM, Hammerich L, Martin C, Zimmermann HW, Pack O, Gassler N, Hittatiya K, Ludwig A, Luedde T, Trautwein C, Tacke F. Chemokine receptor CXCR6-dependent hepatic NK T Cell accumulation promotes inflammation and liver fibrosis. J Immunol. 2013;190:5226-5236. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 188] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 13. | Jiao N, Baker SS, Chapa-Rodriguez A, Liu W, Nugent CA, Tsompana M, Mastrandrea L, Buck MJ, Baker RD, Genco RJ, Zhu R, Zhu L. Suppressed hepatic bile acid signalling despite elevated production of primary and secondary bile acids in NAFLD. Gut. 2018;67:1881-1891. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 375] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 14. | Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540-550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1747] [Cited by in F6Publishing: 1735] [Article Influence: 86.8] [Reference Citation Analysis (0)] |
| 15. | Arab JP, Karpen SJ, Dawson PA, Arrese M, Trauner M. Bile acids and nonalcoholic fatty liver disease: Molecular insights and therapeutic perspectives. Hepatology. 2017;65:350-362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 383] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 16. | Copple BL, Li T. Pharmacology of bile acid receptors: Evolution of bile acids from simple detergents to complex signaling molecules. Pharmacol Res. 2016;104:9-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 153] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 17. | Chiang JY. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191-1212. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 834] [Article Influence: 75.8] [Reference Citation Analysis (1)] |
| 18. | Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol. 2014;11:55-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 451] [Cited by in F6Publishing: 474] [Article Influence: 47.4] [Reference Citation Analysis (0)] |
| 19. | Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006-1011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 743] [Cited by in F6Publishing: 702] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 20. | De Fabiani E, Mitro N, Gilardi F, Caruso D, Galli G, Crestani M. Coordinated control of cholesterol catabolism to bile acids and of gluconeogenesis via a novel mechanism of transcription regulation linked to the fasted-to-fed cycle. J Biol Chem. 2003;278:39124-39132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 151] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Cyphert HA, Ge X, Kohan AB, Salati LM, Zhang Y, Hillgartner FB. Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21. J Biol Chem. 2012;287:25123-25138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 22. | Watanabe M, Houten SM, Wang L, Moschetta A, Mangelsdorf DJ, Heyman RA, Moore DD, Auwerx J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408-1418. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 865] [Cited by in F6Publishing: 942] [Article Influence: 47.1] [Reference Citation Analysis (0)] |
| 23. | Gruenbacher G, Gander H, Rahm A, Dobler G, Drasche A, Troppmair J, Nussbaumer W, Thurnher M. The Human G Protein-Coupled ATP Receptor P2Y11 Is Associated With IL-10 Driven Macrophage Differentiation. Front Immunol. 2019;10:1870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Keitel V, Donner M, Winandy S, Kubitz R, Häussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78-84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 281] [Cited by in F6Publishing: 293] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 25. | Ding L, Sousa KM, Jin L, Dong B, Kim BW, Ramirez R, Xiao Z, Gu Y, Yang Q, Wang J, Yu D, Pigazzi A, Schones D, Yang L, Moore D, Wang Z, Huang W. Vertical sleeve gastrectomy activates GPBAR-1/TGR5 to sustain weight loss, improve fatty liver, and remit insulin resistance in mice. Hepatology. 2016;64:760-773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 26. | Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L, Zheng M, Zhang X, Xia D, Ke Y, Lu L, Wang D. Bile Acids Control Inflammation and Metabolic Disorder through Inhibition of NLRP3 Inflammasome. Immunity. 2016;45:802-816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 330] [Cited by in F6Publishing: 428] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 27. | McMahan RH, Wang XX, Cheng LL, Krisko T, Smith M, El Kasmi K, Pruzanski M, Adorini L, Golden-Mason L, Levi M, Rosen HR. Bile acid receptor activation modulates hepatic monocyte activity and improves nonalcoholic fatty liver disease. J Biol Chem. 2013;288:11761-11770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 28. | Jadhav K, Xu Y, Li Y, Xu J, Zhu Y, Adorini L, Lee YK, Kasumov T, Yin L, Zhang Y. Reversal of metabolic disorders by pharmacological activation of bile acid receptors TGR5 and FXR. Mol Metab. 2018;9:131-140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 29. | Chu H, Duan Y, Yang L, Schnabl B. Small metabolites, possible big changes: a microbiota-centered view of non-alcoholic fatty liver disease. Gut. 2019;68:359-370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 195] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 30. | Gottlieb A, Bechmann L, Canbay A. The Presence and Severity of Nonalcoholic Steatohepatitis Is Associated with Specific Changes in Circulating Bile Acids. Ann Hepatol. 2018;17:340-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 31. | Parséus A, Sommer N, Sommer F, Caesar R, Molinaro A, Ståhlman M, Greiner TU, Perkins R, Bäckhed F. Microbiota-induced obesity requires farnesoid X receptor. Gut. 2017;66:429-437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 308] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 32. | Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, Adorini L, Sciacca CI, Clopton P, Castelloe E, Dillon P, Pruzanski M, Shapiro D. Efficacy and safety of the farnesoid X receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 2013; 145: 574-82. e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 656] [Cited by in F6Publishing: 680] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 33. | Musso G, Cassader M, Gambino R. Non-alcoholic steatohepatitis: emerging molecular targets and therapeutic strategies. Nat Rev Drug Discov. 2016;15:249-274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 314] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 34. | Hirschfield GM, Mason A, Luketic V, Lindor K, Gordon SC, Mayo M, Kowdley KV, Vincent C, Bodhenheimer HC Jr, Parés A, Trauner M, Marschall HU, Adorini L, Sciacca C, Beecher-Jones T, Castelloe E, Böhm O, Shapiro D. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology 2015; 148: 751-61. e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 391] [Cited by in F6Publishing: 387] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 35. | Pellicciari R, Gioiello A, Macchiarulo A, Thomas C, Rosatelli E, Natalini B, Sardella R, Pruzanski M, Roda A, Pastorini E, Schoonjans K, Auwerx J. Discovery of 6alpha-ethyl-23(S)-methylcholic acid (S-EMCA, INT-777) as a potent and selective agonist for the TGR5 receptor, a novel target for diabesity. J Med Chem. 2009;52:7958-7961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 193] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 36. | Bajaj JS, Heuman DM, Hylemon PB, Sanyal AJ, White MB, Monteith P, Noble NA, Unser AB, Daita K, Fisher AR, Sikaroodi M, Gillevet PM. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60:940-947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 659] [Cited by in F6Publishing: 713] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 37. | Ahluwalia V, Wade JB, Moeller FG, White MB, Unser AB, Gavis EA, Sterling RK, Stravitz RT, Sanyal AJ, Siddiqui MS, Puri P, Luketic V, Heuman DM, Fuchs M, Matherly S, Bajaj JS. The etiology of cirrhosis is a strong determinant of brain reserve: A multimodal magnetic resonance imaging study. Liver Transpl. 2015;21:1123-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 38. | Arroyo V, Moreau R, Kamath PS, Jalan R, Ginès P, Nevens F, Fernández J, To U, García-Tsao G, Schnabl B. Acute-on-chronic liver failure in cirrhosis. Nat Rev Dis Primers. 2016;2:16041. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 263] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 39. | Jansen PL, Ghallab A, Vartak N, Reif R, Schaap FG, Hampe J, Hengstler JG. The ascending pathophysiology of cholestatic liver disease. Hepatology. 2017;65:722-738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 195] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 40. | Chen W, Owsley E, Yang Y, Stroup D, Chiang JY. Nuclear receptor-mediated repression of human cholesterol 7alpha-hydroxylase gene transcription by bile acids. J Lipid Res. 2001;42:1402-1412. [PubMed] [Cited in This Article: ] |
| 41. | Allen K, Jaeschke H, Copple BL. Bile acids induce inflammatory genes in hepatocytes: a novel mechanism of inflammation during obstructive cholestasis. Am J Pathol. 2011;178:175-186. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 314] [Cited by in F6Publishing: 357] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 42. | Schoemaker MH, Conde de la Rosa L, Buist-Homan M, Vrenken TE, Havinga R, Poelstra K, Haisma HJ, Jansen PL, Moshage H. Tauroursodeoxycholic acid protects rat hepatocytes from bile acid-induced apoptosis via activation of survival pathways. Hepatology. 2004;39:1563-1573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 172] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 43. | Li Y, Tang R, Leung PSC, Gershwin ME, Ma X. Bile acids and intestinal microbiota in autoimmune cholestatic liver diseases. Autoimmun Rev. 2017;16:885-896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 134] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 44. | Woodhouse CA, Patel VC, Singanayagam A, Shawcross DL. Review article: the gut microbiome as a therapeutic target in the pathogenesis and treatment of chronic liver disease. Aliment Pharmacol Ther. 2018;47:192-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 104] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 45. | Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, Trøseid M, Marschall HU, Schrumpf E, Moum B, Røsjø H, Aukrust P, Karlsen TH, Hov JR. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut. 2017;66:611-619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 257] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 46. | Santiago P, Scheinberg AR, Levy C. Cholestatic liver diseases: new targets, new therapies. Therap Adv Gastroenterol. 2018;11:1756284818787400. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 51] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 47. | Trauner M, Meier PJ, Boyer JL. Molecular pathogenesis of cholestasis. N Engl J Med. 1998;339:1217-1227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 579] [Cited by in F6Publishing: 497] [Article Influence: 19.1] [Reference Citation Analysis (1)] |
| 48. | Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, Zheng Y, Longman RS, Rastinejad F, Devlin AS, Krout MR, Fischbach MA, Littman DR, Huh JR. Bile acid metabolites control TH17 and Treg cell differentiation. Nature. 2019;576:143-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 456] [Cited by in F6Publishing: 631] [Article Influence: 126.2] [Reference Citation Analysis (0)] |
| 49. | Rong G, Zhou Y, Xiong Y, Zhou L, Geng H, Jiang T, Zhu Y, Lu H, Zhang S, Wang P, Zhang B, Zhong R. Imbalance between T helper type 17 and T regulatory cells in patients with primary biliary cirrhosis: the serum cytokine profile and peripheral cell population. Clin Exp Immunol. 2009;156:217-225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 50. | Acosta-Rodriguez EV, Rivino L, Geginat J, Jarrossay D, Gattorno M, Lanzavecchia A, Sallusto F, Napolitani G. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat Immunol. 2007;8:639-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1373] [Cited by in F6Publishing: 1410] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 51. | Yamazaki T, Yang XO, Chung Y, Fukunaga A, Nurieva R, Pappu B, Martin-Orozco N, Kang HS, Ma L, Panopoulos AD, Craig S, Watowich SS, Jetten AM, Tian Q, Dong C. CCR6 regulates the migration of inflammatory and regulatory T cells. J Immunol. 2008;181:8391-8401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 369] [Cited by in F6Publishing: 391] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 52. | Lim HW, Lee J, Hillsamer P, Kim CH. Human Th17 cells share major trafficking receptors with both polarized effector T cells and FOXP3+ regulatory T cells. J Immunol. 2008;180:122-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 188] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 53. | Fenoglio D, Bernuzzi F, Battaglia F, Parodi A, Kalli F, Negrini S, De Palma R, Invernizzi P, Filaci G. Th17 and regulatory T lymphocytes in primary biliary cirrhosis and systemic sclerosis as models of autoimmune fibrotic diseases. Autoimmun Rev. 2012;12:300-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 54. | Lindor K. Ursodeoxycholic acid for the treatment of primary biliary cirrhosis. N Engl J Med. 2007;357:1524-1529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 55. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of cholestatic liver diseases. J Hepatol. 2009;51:237-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1227] [Cited by in F6Publishing: 1134] [Article Influence: 75.6] [Reference Citation Analysis (1)] |
| 56. | Hirschfield GM, Heathcote EJ, Gershwin ME. Pathogenesis of cholestatic liver disease and therapeutic approaches. Gastroenterology. 2010;139:1481-1496. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 187] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 57. | Kowdley KV, Luketic V, Chapman R, Hirschfield GM, Poupon R, Schramm C, Vincent C, Rust C, Parés A, Mason A, Marschall HU, Shapiro D, Adorini L, Sciacca C, Beecher-Jones T, Böhm O, Pencek R, Jones D; Obeticholic Acid PBC Monotherapy Study Group. A randomized trial of obeticholic acid monotherapy in patients with primary biliary cholangitis. Hepatology. 2018;67:1890-1902. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 182] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 58. | Moustafa T, Fickert P, Magnes C, Guelly C, Thueringer A, Frank S, Kratky D, Sattler W, Reicher H, Sinner F, Gumhold J, Silbert D, Fauler G, Höfler G, Lass A, Zechner R, Trauner M. Alterations in lipid metabolism mediate inflammation, fibrosis, and proliferation in a mouse model of chronic cholestatic liver injury. Gastroenterology 2012; 142: 140-151. e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 59. | Sombetzki M, Fuchs CD, Fickert P, Österreicher CH, Mueller M, Claudel T, Loebermann M, Engelmann R, Langner C, Sahin E, Schwinge D, Guenther ND, Schramm C, Mueller-Hilke B, Reisinger EC, Trauner M. 24-nor-ursodeoxycholic acid ameliorates inflammatory response and liver fibrosis in a murine model of hepatic schistosomiasis. J Hepatol. 2015;62:871-878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 60. | Fickert P, Hirschfield GM, Denk G, Marschall HU, Altorjay I, Färkkilä M, Schramm C, Spengler U, Chapman R, Bergquist A, Schrumpf E, Nevens F, Trivedi P, Reiter FP, Tornai I, Halilbasic E, Greinwald R, Pröls M, Manns MP, Trauner M; European PSC norUDCA Study Group. norUrsodeoxycholic acid improves cholestasis in primary sclerosing cholangitis. J Hepatol. 2017;67:549-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 172] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 61. | Pellicciari R, Fiorucci S, Camaioni E, Clerici C, Costantino G, Maloney PR, Morelli A, Parks DJ, Willson TM. 6alpha-ethyl-chenodeoxycholic acid (6-ECDCA), a potent and selective FXR agonist endowed with anticholestatic activity. J Med Chem. 2002;45:3569-3572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 565] [Cited by in F6Publishing: 566] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 62. | Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E; NASH Clinical Research Network. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956-965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1569] [Cited by in F6Publishing: 1596] [Article Influence: 177.3] [Reference Citation Analysis (0)] |
| 63. | Beuers U, Trauner M, Jansen P, Poupon R. New paradigms in the treatment of hepatic cholestasis: from UDCA to FXR, PXR and beyond. J Hepatol. 2015;62:S25-S37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in F6Publishing: 331] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 64. | Bajaj JS, Ridlon JM, Hylemon PB, Thacker LR, Heuman DM, Smith S, Sikaroodi M, Gillevet PM. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168-G175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 373] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 65. | Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei D, Wang Y, Zhu B, Li L. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 674] [Cited by in F6Publishing: 701] [Article Influence: 53.9] [Reference Citation Analysis (1)] |
| 66. | Quigley EM, Quera R. Small intestinal bacterial overgrowth: roles of antibiotics, prebiotics, and probiotics. Gastroenterology. 2006;130:S78-S90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 205] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 67. | Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, Noble NA, White MB, Fisher A, Sikaroodi M, Rangwala H, Gillevet PM. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 302] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 68. | Ridlon JM, Hylemon PB. Identification and characterization of two bile acid coenzyme A transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J Lipid Res. 2012;53:66-76. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 69. | Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773-1781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 632] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 70. | Ridlon JM, Alves JM, Hylemon PB, Bajaj JS. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4:382-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 227] [Cited by in F6Publishing: 245] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 71. | Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1071] [Cited by in F6Publishing: 1113] [Article Influence: 58.6] [Reference Citation Analysis (0)] |
| 72. | Stenman LK, Holma R, Korpela R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J Gastroenterol. 2012;18:923-929. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 120] [Cited by in F6Publishing: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 73. | Stenman LK, Holma R, Eggert A, Korpela R. A novel mechanism for gut barrier dysfunction by dietary fat: epithelial disruption by hydrophobic bile acids. Am J Physiol Gastrointest Liver Physiol. 2013;304:G227-G234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 149] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 74. | Guo H, Callaway JB, Ting JP. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med. 2015;21:677-687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1797] [Cited by in F6Publishing: 2141] [Article Influence: 237.9] [Reference Citation Analysis (0)] |
| 75. | Haselow K, Bode JG, Wammers M, Ehlting C, Keitel V, Kleinebrecht L, Schupp AK, Häussinger D, Graf D. Bile acids PKA-dependently induce a switch of the IL-10/IL-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol. 2013;94:1253-1264. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 105] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 76. | Perino A, Schoonjans K. TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends Pharmacol Sci. 2015;36:847-857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 98] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 77. | Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, Auwerx J, Schoonjans K. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747-757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 402] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 78. | Kakiyama G, Muto A, Takei H, Nittono H, Murai T, Kurosawa T, Hofmann AF, Pandak WM, Bajaj JS. A simple and accurate HPLC method for fecal bile acid profile in healthy and cirrhotic subjects: validation by GC-MS and LC-MS. J Lipid Res. 2014;55:978-990. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 95] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 79. | Sakai K, Makino T, Kawai Y, Mutai M. Intestinal microflora and bile acids. Effect of bile acids on the distribution of microflora and bile acid in the digestive tract of the rat. Microbiol Immunol. 1980;24:187-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 14] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 80. | Robben J, Caenepeel P, Van Eldere J, Eyssen H. Effects of intestinal microbial bile salt sulfatase activity on bile salt kinetics in gnotobiotic rats. Gastroenterology. 1988;94:494-502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 34] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 81. | David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5625] [Cited by in F6Publishing: 5967] [Article Influence: 542.5] [Reference Citation Analysis (0)] |
| 82. | Dawson PA, Hubbert M, Haywood J, Craddock AL, Zerangue N, Christian WV, Ballatori N. The heteromeric organic solute transporter alpha-beta, Ostalpha-Ostbeta, is an ileal basolateral bile acid transporter. J Biol Chem. 2005;280:6960-6968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 277] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 83. | Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, Kasper DL. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature. 2020;577:410-415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 514] [Article Influence: 102.8] [Reference Citation Analysis (0)] |