Published online Nov 7, 2020. doi: 10.3748/wjg.v26.i41.6378
Peer-review started: June 18, 2020
First decision: August 22, 2020
Revised: September 7, 2020
Accepted: September 29, 2020
Article in press: September 29, 2020
Published online: November 7, 2020
The expression of macrophage inhibitory factor-1 (MIC-1) is increased in peripheral blood of patients with chronic hepatitis and liver cirrhosis. However, whether MIC-1 gene polymorphism is correlated with relevant diseases is not yet reported.
To explore the correlation between gene polymorphism in MIC-1 exon region and chronic hepatitis C virus (HCV) infection.
This case-control study enrolled 178 patients with chronic hepatitis C (CHC) in the case group, and 82 healthy subjects from the same region who had passed the screening examination comprised the control group. The genotypes of rs1059369 and rs1059519 loci in the MIC-1 gene exon were detected by DNA sequencing. Also, the MIC-1 level, liver function metrics, liver fibrosis metrics, and HCV RNA load were determined. Univariate analysis was used to compare the differences and correlations between the two groups with respect to these parameters. Multivariate logistic regression was used to analyze the independent relevant factors of CHC.
The plasma MIC-1 level in the CHC group was higher than that in the control group (P < 0.05), and it was significantly positively correlated with alanine aminotransferase, aspartate aminotransferase (AST), type III procollagen N-terminal peptide (known as PIIINP), type IV collagen, and HCV RNA (P < 0.05), whereas negatively correlated with total protein and albumin (P < 0.05). The genotype and allele frequency distribution at the rs1059519 locus differed between the two groups (P < 0.05). The allele frequency maintained significant difference after Bonferroni correction (Pc < 0.05). Logistic multiple regression showed that AST, PIIINP, MIC-1, and genotype GG at the rs1059519 locus were independent relevant factors of CHC (P < 0.05). Linkage disequilibrium (LD) was found between rs1059369 and rs1059519 loci, and significant difference was detected in the distribution of haplotype A-C between the CHC and control groups (P < 0.05). Meanwhile, we found the MIC-1 level trend to increase among rs1059519 genotypes (P = 0.006) and the level of MIC-1 in GG genotype to be significantly higher than CC genotype (P = 0.009, after Bonferroni correction).
Plasma MIC-1 level was increased in CHC patients and correlated with liver cell damage, liver fibrosis metrics, and viral load. The polymorphism at the MIC-1 gene rs1059519 locus was correlated with HCV infection, and associated with the plasma MIC-1 level. G allele and GG genotype may be an important susceptible factor for CHC.
Core Tip: In this study, the relationship between the polymorphisms in the exon region of macrophage inhibitory factor-1 (MIC-1), plasma MIC-1 level and chronic hepatitis C virus (commonly known as HCV) infection were preliminarily investigated. We found that the genotype at rs1059519 could influence the plasma MIC-1 level and both were correlated with chronic hepatitis C (CHC), and the G allele at rs1059519 was an independent risk factor for CHC. Therefore, the baseline MIC-1 level of plasma and rs1059519 polymorphisms can be used as a predictive marker for HCV infection.
- Citation: Yang XJ, Wang XO, Chen Y, Ye SD. Associations of content and gene polymorphism of macrophage inhibitory factor-1 and chronic hepatitis C virus infection. World J Gastroenterol 2020; 26(41): 6378-6390
- URL: https://www.wjgnet.com/1007-9327/full/v26/i41/6378.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i41.6378
Hepatitis C is a global epidemic. All people are susceptible to the hepatitis C virus (HCV) infection, regardless of sex, age, and race. According to statistics from the World Health Organization, the global infection rate of HCV is about 2.8%, and about 185 million people are infected[1]. China has the largest number of infections, with 14.67 million people infected with HCV[2]. Although the transmission routes of HCV have been identified clearly, the infection of HCV cannot be detected easily in the early stage since it has an imperceptible onset process with a high degree of chronicity. In the patients infected with HCV, only 20%-30% can clear the virus spontaneously in the acute stage, while 70%-80% will suffer a persistent chronic infection. Eventually, about 30% of patients with chronic hepatitis C (CHC) will be subject to a disease progression into cirrhosis and hepatocellular carcinoma, resulting in about 350000 deaths each year[3,4]. The predictors of chronicity of HCV infection include male gender, age > 25 years at the time of infection, no obvious symptoms after infection, human immunodeficiency virus (HIV) infection, immunosuppression, and so on. The chronicity, manifested after HCV infection, is the result of combined effects of host immunity, genetic susceptibility, and virus[5,6]; however, additional studies are essential to elucidate the specific mechanism.
Macrophage inhibitory factor-1 (MIC-1), also known as growth differentiation factor-15, is a member of the transforming growth factor-β superfamily. A previous study showed that the expression level of MIC-1 is low in healthy individuals, which is increased under pathological or stress effects, such as inflammation and trauma[7]. The contribution of MIC-1 to the damage of various organs and progression of various diseases, such as cardiovascular disease[8-10], malignant tumor[11-13], and diabetes[14], could be realized via regulation of an inflammatory reaction and apoptosis pathways. Some recent studies demonstrated that the expression of MIC-1 is increased in patients with chronic viral hepatitis, cirrhosis, or small hepatic cell cancer, and MIC-1 could be used for the diagnosis and prediction of viral hepatitis complications[15-18]. The human MIC-1 gene is localized on chromosome 19p12-13.1 and consists of two exons and one intron. Hitherto, dozens of single nucleotide polymorphism (SNP) loci have been found, among which rs1059519 and 1059369, in the exon region, are under intensive focus. However, current studies on these two loci are mainly focused on cardiovascular disease[19,20] and malignant tumor[21], while correlation with the infection of hepatitis virus has not yet been reported. Thus, the present study aimed to explore the correlation between MIC-1 and the chronic infection of HCV by determining the expression level of MIC-1 and the gene polymorphism in the exon region in CHC patients and healthy subjects who cleared the screening examination. These findings would provide a basis for the diagnosis and treatment of such diseases.
CHC group: A total of 178 CHC patients who visited the Department of Infectious Diseases at the Second Affiliated Hospital of Wenzhou Medical University from September 2016 to August 2019 comprised the CHC group by retrospective recruitment. This cohort fulfilled the diagnostic criteria of the “Guideline for Prevention and Treatment of Hepatitis C (2015)”[22]. Inclusion criteria were age 16-75 years; HCV infection duration > 6 mo, or epidemiological history 6 mo ago, anti-HCV- and HCV RNA-positive, histopathological examination results of liver meeting the diagnostic criteria for chronic hepatitis, and not receiving antiviral treatment or immunomodulation or within past 3 mo. Exclusion criteria included the following: Accompaniment by other viral hepatitis, such as chronic hepatitis B, alcoholic liver disease, drug-induced liver injury, autoimmune hepatitis or other liver diseases; HIV infection; malignant tumor; severe cardiovascular or cerebrovascular disease; hematological disease or thyroid disease; diabetes; or complete case data unavailable.
Control group: A total of 82 healthy subjects enrolled from the Physical Examination Center at the Second Affiliated Hospital of Wenzhou Medical University comprised the control group. Exclusion criteria included the following: accompaniment by other viral hepatitis, such as chronic hepatitis B, alcoholic liver disease, drug-induced liver injury, autoimmune hepatitis, and other liver diseases; HIV infection; malignant tumor; severe cardiovascular and cerebrovascular diseases; hematological disease or thyroid disease; or diabetes. All study subjects were unrelated Han Chinese individuals from Zhejiang Province, China. This study was approved by the Ethics Committee of the Second Affiliated Hospital of Wenzhou Medical University (LCKY2016-128), and all patients signed the informed consent before participation in the study.
A volume of 4-5 mL venous blood sample was collected from all fasting patients with EDTA-K2 in the morning. After the routine blood test (for platelet (PLT) count), the plasma was separated from the blood cells by centrifugation at 3000 rpm for 5 min and stored at -80 °C. The plasma was used to determine the parameters such as alanine aminotransferase (ALT), aspartate aminotransferase (AST) activity, total protein (TP), albumin (ALB), total bilirubin (TBIL), type III procollagen N-terminal peptide (PIIINP), type IV collagen (CIV), MIC-1, and HCV RNA, while the DNA was extracted from the blood cells to analyze the MIC-1 gene polymorphism.
Genomic — DNA extraction and amplification: The extraction was performed using a Blood Genomic DNA Extraction Kit (SK8224; Sangon Biotech Co., Ltd., Shanghai, China), according to the instructions for use of the kit. The amplification was performed using a polymerase chain reaction (PCR) amplification kit (SK2072; Sangon Biotech) in Veriti® 96-well PCR instrument (Applied Biosystems Inc., Foster City, CA, United States). The primers were synthesized by Sangon Biotech. The PCR reaction system consisted of DNA template at 1.0 μL, forward and reverse primers at 0.5 μL each, dNTPs at 0.5 μL, Taq enzyme at 0.2 μL, Taq buffer at 2.5 μL, and ddH2O at 20 μL. The PCR reaction conditions were as follows: pre-denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C, final extension at 72 °C for 10 min, and storage at 4 °C. Primers: rs1059369-rs1059519-F 5’-TACCTTCTGGCGTGAGTATCCG-3’, rs1059369-rs1059519-R 5’-CAGGCGGAGACGGACAAAGT-3’.
Product purification and sequencing: The amplified product was purified using a PCR product purification and recovery kit (SK1141; Sangon Biotech). The DNA was quantified using a NanoDrop 2000C UV spectrophotometer (ThermoScientific, Waltham, MA, United States). The sequencing was carried out on a 3730XL sequencing instrument (Applied Biosystems Inc.), and SeqMan software was used for the comparative analysis of the sequencing map.
Determination of plasma MIC-1 level: The operation was completed using the double-antibody sandwich ELISA method, according to the instructions for use of the kit (Jingmei Biotechnology Co., Ltd., Jiangsu, China). Anthos 2010 Microplate Reader (Shanghai Bioscience Technology Co., Ltd., Shanghai, China) was used to detect the absorbance of samples and reference substances; the standard curve was drawn to calculate the MIC-1 level of the samples. In the event where OD values of the samples were beyond the upper limit of the standard curve, the determination was repeated with the appropriate dilution of the samples.
Quantitative analysis of HCV: RNA was extracted by real-time quantitative fluorescence (FQ)-PCR according to the protocol of the kit (Piji Bioengineering Co., Ltd., Shenzhen, China), and detected using the ABI 7500 FQ-PCR Instrument (Applied Biosystems Inc.). The normal reference ranges were defined as < 5.0 × 102 copies/mL for HCV RNA.
Determination of plasma ALT, AST, TP, ALB, and TBIL levels: The parameters were assessed on an ADVIA2400 Automatic Biochemical Analyzer (Siemens, Malvern, PA, United States) using original reagents and calibrators. The normal reference ranges were defined as 9-50 U/L for ALT, 15-40 U/L for AST, 65-85 g/L for TP, 40-55 g/L for ALB, 6.8-34.2 mol/L for TBIL, and 100-300 × 109/L for PLT.
Determination of plasma PIIINP and CIV levels: These measurements were carried out by chemiluminescence immunoassay on a MAGLUMI 2000 Automatic Chemiluminescence Immunoanalyzer (New Industries Biomedical Engineering Co., Ltd., Shenzhen, China) using original reagents and calibrators. The normal reference range was defined as 0.5-30 ng/mL for PIIINP and 5.0-30 ng/mL for CIV.
An independent clinical case database was established in a mode of double entries by two investigators. SPSS 23.0 for Windows was used for statistical analysis, wherein the enumeration data were described as the composition ratio. Pearson’s chi-square test or Fisher’s exact probability test was used for intergroup comparison. Normal distribution data were expressed as mean ± standard deviation, with t-test or ANOVA used for intergroup comparison, while non-normal distribution data were expressed by median (M), 5th percentile (P5), and 95th percentile (P95), with rank-sum test used for intergroup comparison. The website of CHWE (https://www.genecalculators.net/pq-chwe-polypicker.html) was used for the analysis of heterozygosity and polymorphic information content (PIC) in all studied groups to determine whether the SNP was polymorphic enough for doing statistical analysis in the Chinese population. We used the CHWE website to perform heterozygosity and PIC analysis for all groups, which aims to determine whether SNPs are sufficiently polymorphic for analysis in the Chinese population. The software SHEsis (http://analysis.bio-x.cn/myAnalysis.php) was used for the analysis of difference in genotypes and allele frequency, Hardy-Weinberg equilibrium test, calculation of odds ratio (OR) and 95% confidence interval (CI), genetic LD analysis, and haplotype construction. Multivariate logistic regression was used for the analysis of independent relevant factors of CHC. The two-tailed test was adopted for all the analyses, and P < 0.05 indicated statistically significant differences. Multiple comparisons and genotypes were corrected for multiple comparisons using Bonferroni correction.
The CHC group included 99 males and 79 females, aged 42.46 ± 10.23 (range: 18-74) years, and the control group included 47 males and 35 females, aged 41.09 ± 9.30 (20-69) years. No statistically significant difference was detected in the sex ratio and age distribution between the two groups. After the rank-sum test of two independent samples, the plasma ALT, AST, TBIL, PIIINP, CIV, and MIC-1 levels were significantly higher, and the TP, ALB, and PLT levels were significantly lower in the CHC group than those in the control group (P < 0.05; Table 1).
| Parameter | Control group, n = 82 | CHC group, n = 178 | χ2 or Z | P value |
| Sex, M/F | 47/35 | 99/79 | χ2 = 0.066 | 0.798 |
| Age in yr, ≤ 40/> 40 | 63/61 | 44/38 | χ2 = 0.161 | 0.688 |
| ALT in U/L, M/P5/P95 | 25.5/13.6/43.3 | 44.6/14.3/128.3 | Z = -6.641 | < 0.001 |
| AST in U/L, M/P5/P95 | 21.6/11.3/35.8 | 41.4/19.3/115.2 | Z = -8.925 | < 0.001 |
| TP in g/L, M/P5/P95 | 74.5/66.7/84.7 | 69.2/54.2/85.8 | Z = -2.690 | 0.008 |
| ALB in g/L, M/P5/P95 | 45.6/38.3/55.8 | 42.5/27.5/55.7 | Z = -3.638 | < 0.001 |
| TBIL in mol/L, M/P5/P95 | 17.4/10.0/32.4 | 17.9/11.3/42.5 | Z = -2.030 | 0.035 |
| PIIINP in ng/mL, M/P5/P95 | 16.7/9.4/24.6 | 27.6/9.6/120.4 | Z = -7.348 | < 0.001 |
| CIV in ng/mL, M/P5/P95 | 17.5/11.0/25.5 | 28.9/11.8/142.6 | Z = -8.294 | < 0.001 |
| PLT as 109/L, M/P5/P95 | 204.0/121.5/286.4 | 177.0/56.0/289.0 | Z = -2.780 | 0.003 |
| MIC-1 in pg/mL, M/P5/P95 | 324.6/146.7/725.8 | 407.7/149.2/1535.5 | Z = -3.072 | 0.001 |
Spearman’s rank correlation analysis of plasma MIC-1 level and other biochemical parameters in CHC patients revealed that the MIC-1 level showed a significantly positive correlation with ALT, AST, PIIINP, CIV, and HCV RNA (r = 0.219, 0.169, 0.247, 0.239, and 0.304, respectively; P < 0.05) and a significantly negative correlation with TP and ALB (r = -0.371 and -0.391, respectively, P < 0.05), while no significant correlation was established with TBIL or PLT (P > 0.05).
Three genotypes were identified at rs1059369 locus (AA, AT, and TT) and rs1059519 locus (CC, CG, and GG) of MIC-1 gene by sequencing (Figure 1). The distribution of all genotypes were in accordance with the Hardy-Weinberg equilibrium in controls (all P values were > 0.05). Heterozygosity and PIC calculation showed that both SNPs were polymorphic enough for statistical analysis in the Chinese population (all values were > 0.3). The chi-square test showed that there was no statistically significant difference in the genotype and allele frequency distribution at rs1059369 locus between the CHC and control groups (P > 0.05), while a significant difference was detected at rs1059519 locus between the two groups (P = 0.028, 0.004, ORC/G = 0.570). Only the allele frequency distribution of rs1059519 locus was still significant after Bonferroni correction (Pc = 0.004 × 2 < 0.05). The genetic LD test showed a LD between rs1059369 and rs1059519 loci (D’ = 0.99, r2 = 0.314). Haplotypes (A-C, A-G, and T-G) of two SNP loci of the MIC-1 gene were constructed according to the LD results, where in the distribution of A-C type showed a significant difference between the two groups (P < 0.05; Table 2).
| Genotype | Control group, n = 82 | CHC group, n = 178 | χ2 | P value | P value1 | OR value (95%CI) |
| rs1059369 gene | ||||||
| AA | 32 (0.390) | 58 (0.326) | 4.142 | 0.126 | 0.252 | |
| AT | 42 (0.512) | 85 (0.478) | ||||
| TT | 8 (0.098) | 35 (0.197) | ||||
| A | 106 (0.646) | 201 (0.565) | 3.102 | 0.078 | 0.156 | 0.710 (0.484-1.040) |
| T | 58 (0.354) | 155 (0.435) | ||||
| Het | 0.4574 | 0.4916 | ||||
| PIC | 0.3528 | 0.3707 | ||||
| rs1059519 gene | ||||||
| CC | 16 (0.195) | 17 (0.096) | 7.191 | 0.028 | 0.056 | |
| CG | 33 (0.402) | 63 (0.354) | ||||
| GG | 33 (0.402) | 98 (0.551) | ||||
| C | 65 (0.396) | 97 (0.272) | 8.032 | 0.004 | 0.008 | 0.570 (0.386-0.843) |
| G | 99 (0.604) | 259 (0.728) | ||||
| Het | 0.4784 | 0.396 | ||||
| PIC | 0.364 | 0.3176 | ||||
| Haplotype | ||||||
| AC | 65.00 (0.396) | 97.00 (0.272) | 8.033 | 0.005 | 0.570 (0.386-0.843) | |
| AG | 41.00 (0.250) | 104.00 (0.292) | 0.991 | 0.319 | 1.238 (0.813-1.886) | |
| TG | 58.00 (0.354) | 155.00 (0.435) | 3.102 | 0.078 | 1.409 (0.961-2.066) | |
| TC | 0.00 (0.000) | 0.00 (0.000) | ||||
Parameters with statistically significant difference between CHC patients and healthy individuals were included in a multivariate logistic regression model. Since ALT/AST, TP/ALB, and PIIINP/CIV were correlated to each another, only one of each combination was selected. Logistic regression analysis showed that AST, PIIINP, MIC-1, and GG genotype at rs1059519 locus were independent relevant factors for CHC (P < 0.05; Table 3).
| Parameters | B value | SD | Wald χ2 value | P value | OR value (95%CI) |
| AST in U/L | 0.127 | 0.022 | 34.365 | < 0.001 | 1.135 (1.082%CI) |
| ALB in g/L | -0.030 | 0.029 | 1.068 | 0.301 | 0.971 (0.917%CI) |
| TBIL in mol/L | 0.015 | 0.030 | 0.230 | 0.631 | 1.015 (0.956-1.077) |
| PIIINP in ng/mL | 0.157 | 0.035 | 16.016 | 0.000 | 1.170 (1.093-1.252) |
| PLT as 109/L | -0.001 | 0.003 | 0.198 | 0.657 | 0.999 (0.992-1.005) |
| MIC-1 in pg/mL | 0.002 | 0.001 | 5.369 | 0.020 | 1.002 (1.000-1.004) |
| rs1059519 CC | |||||
| rs1059519 CG | 0.067 | 0.414 | 0.026 | 0.872 | 1.070 (0.474-2.410) |
| rs1059519 GG | 1.402 | 0.593 | 5.598 | 0.026 | 4.065 (1.272-12.987) |
CHC patients (n = 178) were subdivided into two groups based on sex, age, ALT, TBIL, ALB, CIV, PLT, and HCV RNA levels, respectively. The results of the chi-square test did not reveal any statistically significant difference in the genotype and allele frequency distribution at rs1059519 locus between the two subgroups (P > 0.05) (Table 4).
| Group | n | Genotype | χ2 | P value | Allele | χ2 | P value | OR value (95%CI) | |||
| CC | CG | GG | C | G | |||||||
| Sex | |||||||||||
| M | 99 | 10 (0.101) | 40 (0.404) | 49 (0.495) | 2.906 | 0.233 | 60 (0.303) | 138 (0.697) | 2.102 | 0.147 | 0.703 (0.437-1.133) |
| F | 79 | 7 (0.089) | 23 (0.291) | 49 (0.620) | 37 (0.234) | 121 (0.766) | |||||
| Age in yr | |||||||||||
| ≤ 40 | 84 | 8 (0.095) | 31 (0.369) | 45 (0.536) | 0.166 | 0.920 | 47 (0.280) | 121 (0.720) | 0.085 | 0.770 | 0.933 (0.585-1.488) |
| > 40 | 94 | 9 (0.096) | 32 (0.340) | 53 (0.564) | 50 (0.266) | 138 (0.734) | |||||
| ALT in U/L | |||||||||||
| ≤ 50 | 82 | 9 (0.094) | 33 (0.344) | 54 (0.562) | 0.122 | 0.941 | 51 (0.266) | 141 (0.734) | 0.099 | 0.753 | 1.07 (0.6759-1.720) |
| > 50 | 96 | 8 (0.098) | 30 (0.366) | 44 (0.537) | 46 (0.280) | 118 (0.720) | |||||
| TBIL in mol/L | |||||||||||
| ≤ 34.2 | 144 | 12 (0.083) | 49 (0.340) | 83 (0.576) | 2.480 | 0.289 | 73 (0.253) | 215 (0.747) | 2.746 | 0.097 | 1.606 (0.914-2.823) |
| > 34.2 | 34 | 5 (0.147) | 14 (0.412) | 15 (0.441) | 24 (0.353) | 44 (0.647) | |||||
| ALB in g/L | |||||||||||
| ≤ 40 | 82 | 9 (0.110) | 25 (0.305) | 48 (0.585) | 1.692 | 0.429 | 43 (0.262) | 121 (0.738) | 0.162 | 0.687 | 1.101 (0.689-1.760) |
| > 40 | 96 | 8 (0.083) | 38 (0.396) | 50 (0.521) | 54 (0.281) | 138 (0.719) | |||||
| CIV in ng/mL | |||||||||||
| ≤ 30 | 120 | 14 (0.117) | 44 (0.367) | 62 (0.517) | 2.664 | 0.264 | 42 (0.300) | 168 (0.700) | 2.816 | 0.093 | 0.641 (0.380-1.080) |
| > 30 | 58 | 3 (0.052) | 19 (0.328) | 36 (0.621) | 25 (0.216) | 91 (0.784) | |||||
| PLT as 109/L | |||||||||||
| ≤ 100 | 43 | 5 (0.116) | 16 (0.372) | 22 (0.512) | 0.465 | 0.793 | 26 (0.302) | 60 (0.698) | 0.510 | 0.475 | 0.823 (0.483-1.404) |
| > 100 | 135 | 12 (0.089) | 47 (0.348) | 76 (0.563) | 71 (0.263) | 199 (0.737) | |||||
| HCV RNA as copies/mL | |||||||||||
| ≤ 2 × 106 | 69 | 9 (0.130) | 21 (0.304) | 39 (0.565) | 2.266 | 0.322 | 39 (0.283) | 99 (0.717) | 0.117 | 0.733 | 0.920 (0.571-1.483) |
| > 2 × 106 | 109 | 8 (0.073) | 42 (0.385) | 59 (0.541) | 58 (0.266) | 160 (0.734) | |||||
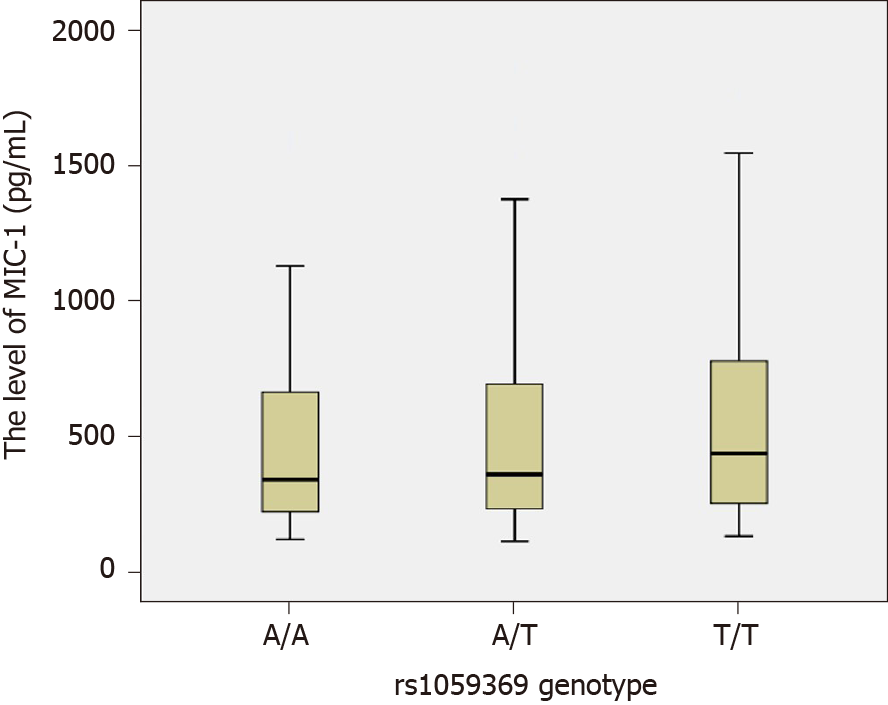
The MIC-1 levels among each rs1059519 genotype in the study subjects (combined CHC group and control group) were as follows: CC genotype (n = 33, M = 265.10, P5 = 117.45, P95 = 1271.05), CG genotype (n = 99, M =344.40, P5 = 146.40, P95 = 1373.30) and GG type (n = 128, M = 401.65, P5 = 175.41, P95 = 1170.86). By rank-sum test of multiple independent samples, there were statistically significant differences in MIC-1 level between rs1059519 genotypes (P = 0.006). Among them, GG genotype was significantly higher than CC genotype (P = 0.009, after Bonferroni correction), as shown in Figure 2.
The comparison between CHC patients and healthy individuals displayed that the plasma ALT, AST, TBIL, PIIINP, CIV, and MIC-1 levels were higher in the CHC group while that the TP, ALB, and PLT levels were lower. Also, a statistically significant difference was detected in the genotype and allele frequency distribution at rs1059519 locus between the two groups (P < 0.05). Therefore, correlation analysis and multivariate regression analysis were performed on these significant variables. The results showed that AST (ALT), PIIINP (CIV), MIC-1, and genotype GG at the rs1059519 locus were not only positively correlated with CHC but were also independent risk factors (P < 0.05).
MIC-1 was first cloned and identified from an activated macrophage cell line by Bootcov et al[23] in 1997. It mainly participated in the growth, differentiation, and development of the organs. Hsiao et al[24] demonstrated that when the liver was damaged, the MIC-1 level was increased significantly. Furthermore, Lee et al[17] reported that the serum MIC-1 level was significantly increased in patients with liver cirrhosis and those with hepatocellular carcinoma. In addition, the expression of MIC-1 in the liver cells was significantly higher than that in the normal liver tissue and that adjacent to the cancer tissue. In the study by Lu et al[15], the expression of MIC-1 was investigated in patients with chronic hepatitis B and cirrhosis, and the results showed that the serum MIC-1 level in chronic hepatitis B and cirrhosis groups was increased significantly, and the expression in the cirrhosis group was significantly higher than that in chronic hepatitis B group. Si et al[25] investigated the correlation between MIC-1 and HCV replication and found that MIC-1 promoted the replication of HCV by changing the signal transduction and growth of host liver cells, and was associated with primary liver cancer caused by HCV. Furthermore, Zhang et al[26] demonstrated that the serum MIC-1 level in CHC patients was positively correlated with the left atrial inner diameter and left ventricular posterior wall thickness and negatively correlated with ejection fraction and that the detection of MIC-1 was valuable for assessing the cardiovascular damage. The current study demonstrated that the plasma MIC-1 level in CHC patients was significantly higher than that in the normal population, which was relatively consistent with the study by Halim et al[18]. The present study also showed that the plasma MIC-1 level was significantly positively correlated with ALT, AST, PIIINP, CIV, and HCV RNA and significantly negatively correlated with TP and ALB. ALT and AST were sensitive indicators that indicated liver cell damage; TP and ALB were related to liver synthesis function and could reflect chronic liver damage and hepatic parenchymal cell reserve function; PIIINP and CIV were closely related to the degree of liver fibrosis and were relatively specific and sensitive indicators that indicated liver fibrosis[27]. High-sensitivity quantitative detection of HCV RNA is not only crucial for determining HCV infection but also a critical parameter for observing interferon efficacy in patients with hepatitis C[28]. The results of the present study indicated that MIC-1 may participate in HCV replication, liver cell damage, and liver fibrosis processes, and that the elevated plasma MIC-1 level may be used as a potential diagnostic marker for HCV infection. However, MIC-1, as a protein that is induced via stress, has significant differences between individuals and at different stages of disease. It requires multiple measurements and dynamic observation to identify the clinical value. Moreover, the specific mechanism of MIC-1 in HCV infection and the determinants MIC level are still unclear. These problems may be explained at the molecular level through the study of MIC-1 polymorphism. In addition, research for genetic susceptibility contributes to screening of vulnerable populations and improvement of prevention measures.
In our study, three genotypes have been identified at the rs1059369 locus, i.e. AA, AT, and TT. A previous study showed a correlation between the polymorphism at the locus and the formation of collateral circulation in myocardial infarction patients with non-ST-segment elevation[19]. Some other studies speculated that this locus did not have any effect on the expression of MIC-1 protein and no significant association with the diseases, such as cardiovascular disease and tumor[29,30]. According to the present study, no statistically significant difference was detected in the genotype and allele frequency distribution at rs1059369 locus between the CHC and control groups (P > 0.05), and no statistically significant difference was detected in the plasma MIC-1 level among different genotypes (P > 0.05; Figure 3). Furthermore, three genotypes were identified at the rs1059519 locus, i.e. CC, CG, and GG. Some studies showed a correlation between the gene polymorphism at this locus and the susceptibility to chronic Keshan disease[31]. Another study demonstrated a difference in plasma MIC-1 level among different genotypes, and that the gene polymorphism at this locus may be correlated with the severity of disease affecting at the plasma level[32]. The present study revealed a statistically significant difference in the allele frequency distribution at rs1059519 locus between the CHC and control groups (P < 0.05). However, no statistically significant difference was detected in the CHC subgroups divided based on sex, age, ALT, TBIL, ALB, CIV, PLT, and HCV RNA levels (P > 0.05), suggesting that the polymorphism at rs1059519 locus was associated to CHC. Intriguingly, no significant correlation was established between the genotype and HCV viral load or liver damage severity. On the contrary, there were significant differences in plasma MIC-1 levels between different genotypes at rs1059519 sites, from genotypes CC, CG to GG, and MIC-1 levels increased gradually, with the GG genotype group having significantly higher than the CC genotype group. Therefore, we speculated that rs1059519 polymorphism might be related to CHC susceptibility by affecting the expression level of MIC-1.
This study has preliminarily explored the correlation between MIC-1 gene polymorphism and chronic HCV infection. Since the subjects in this study were from Zhejiang Province, China, the sample size was small, and we did not include the number of spontaneous viral clearance (SVC) in this study for not meeting the statistical requirements, there may be selection deviation, and the role of MIC-1 in natural HCV clearance cannot be explored[33]. Future studies will need to expand the selection area and sample size, and increase the SVC group. Also, the correlation between the expression level and gene polymorphism of MIC-1 and the prognosis of CHC prognosis would need to be investigated.
In summary, the plasma MIC-1 level is increased in CHC patients, which is correlated with liver cell damage, liver fibrosis, and viral load. Polymorphism of the MIC-1 gene rs1059519 locus affects the plasma content of this protein and is associated with HCV infection. The G allele may be a susceptibility factor, while the C allele and haplotype A-C at rs1059369/rs1059519 locus may be protective factors for CHC.
Macrophage inhibitory factor-1 (MIC-1), regulating the inflammatory response and apoptosis pathway, is involved in multiple organ damage and the progress of various diseases. In recent years, scholars have found that the MIC-1 expression is increased in chronic viral hepatitis, liver cirrhosis, and small cell liver cancer patients. However, the correlation between MIC-1 gene polymorphism and hepatitis C virus (HCV) infection and treatment has not been reported.
To investigate whether plasma MIC-1 level is correlated with liver cell injury, liver fibrosis index and viral load in chronic hepatitis C (CHC) patients. To investigate the relationship between the polymorphism of rs1059369 and rs1059519 in the MIC-1 exon region and the expression level of MIC-1 in plasma and the susceptibility to CHC.
The study aimed to explore the correlation between MIC-1 and the chronic infection of HCV by determining the expression level of MIC-1 and the gene polymorphism in the exon region in CHC patients and healthy subjects who cleared the screening examination. These findings will provide a basis for the diagnosis and treatment of such diseases.
This case-control study enrolled 178 patients with CHC and 82 healthy subjects. The genotypes of rs1059369 and rs1059519 loci in MIC-1 gene exon were detected by DNA sequencing. Also, the MIC-1 level, liver function metrics, liver fibrosis metrics, and HCV RNA load were determined. Univariate analysis was used to compare the differences and correlations between the two groups with respect to these parameters. Multivariate logistic regression was used to analyze the independent relevant factors of CHC.
Compared with healthy subjects, CHC patients had higher plasma levels of MIC-1 and alanine aminotransferase (ALT), aspartate aminotransferase (AST), type III procollagen N-terminal peptide (PIIINP), type IV collagen (CIV), HCV RNA, and lower levels of total protein and albumin. The genotype and allele frequency distribution of rs1059519 were significantly different between CHC group and control group. Logistic multivariate regression revealed that AST (ALT), PIIINP (CIV), the MIC-1 level, rs1059519 GG genotype related to CHC, are independent risk factors. There were statistically significant differences in plasma MIC-1 level between different rs1059519 genotypes (P = 0.006), and GG genotype was significantly higher than CC genotype (P = 0.009).
The plasma MIC-1 level in CHC patients is correlated with liver cell damage, liver fibrosis, and viral load. The polymorphism of MIC-1 gene at rs1059519 locus affects plasma MIC-1 level and is associated with HCV infection.
As the number of spontaneous viral clearance (SVC) was far less than 20% among the CHC cases that we have collected and did not meet the statistical requirements, we had to exclude this group in this study for the time being. However, we will continue to collect more cases in the future, and add the SVC group to further study the relationship between MIC-1 and SVC. In addition, the correlation between MIC-1 and antiviral efficacy and prognosis of CHC is also worth further exploration.
We thank Lin W and other doctors from the Department of Infectious Diseases at The Second Affiliated Hospital of Wenzhou Medical University for their support and assistance in the recruitment, clinical diagnosis, and efficacy judgment of the chronic hepatitis C cases.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: El-Bendary M, Tsukanov V S-Editor: Yan JP L-Editor: Filipodia P-Editor: Liu JH
| 1. | World Health Organization. Guidelines for the screening, care and treatment of persons with hepatitis C infection. 2014. Available from: https://www.who.int/hepatitis/publications/hepatitis-c-guidelines/en/. [Cited in This Article: ] |
| 2. | Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61:S45-S57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1325] [Cited by in F6Publishing: 1311] [Article Influence: 131.1] [Reference Citation Analysis (0)] |
| 3. | Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333-1342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1770] [Cited by in F6Publishing: 1796] [Article Influence: 163.3] [Reference Citation Analysis (0)] |
| 4. | Lemoine M, Nayagam S, Thursz M. Viral hepatitis in resource-limited countries and access to antiviral therapies: current and future challenges. Future Virol. 2013;8:371-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 5. | Heim MH. Innate immunity and HCV. J Hepatol. 2013;58:564-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 6. | Irshad M, Mankotia DS, Irshad K. An insight into the diagnosis and pathogenesis of hepatitis C virus infection. World J Gastroenterol. 2013;19:7896-7909. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 46] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (3)] |
| 7. | Bonaterra GA, Zügel S, Thogersen J, Walter SA, Haberkorn U, Strelau J, Kinscherf R. Growth differentiation factor-15 deficiency inhibits atherosclerosis progression by regulating interleukin-6-dependent inflammatory response to vascular injury. J Am Heart Assoc. 2012;1:e002550. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 8. | Anand IS, Kempf T, Rector TS, Tapken H, Allhoff T, Jantzen F, Kuskowski M, Cohn JN, Drexler H, Wollert KC. Serial measurement of growth-differentiation factor-15 in heart failure: relation to disease severity and prognosis in the Valsartan Heart Failure Trial. Circulation. 2010;122:1387-1395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 229] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 9. | Arkoumani M, Papadopoulou-Marketou N, Nicolaides NC, Kanaka-Gantenbein C, Tentolouris N, Papassotiriou I. The clinical impact of growth differentiation factor-15 in heart disease: A 2019 update. Crit Rev Clin Lab Sci. 2020;57:114-125. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Zhu ZD, Sun T. Association between growth differentiation factor-15 and chronic heart failure in coronary atherosclerosis patients. Genet Mol Res. 2015;14:2225-2233. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Vocka M, Langer D, Fryba V, Petrtyl J, Hanus T, Kalousova M, Zima T, Petruzelka L. Growth/differentiation factor 15 (GDF-15) as new potential serum marker in patients with metastatic colorectal cancer. Cancer Biomark. 2018;21:869-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 12. | Wollmann W, Goodman ML, Bhat-Nakshatri P, Kishimoto H, Goulet RJ Jr, Mehrotra S, Morimiya A, Badve S, Nakshatri H. The macrophage inhibitory cytokine integrates AKT/PKB and MAP kinase signaling pathways in breast cancer cells. Carcinogenesis. 2005;26:900-907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Arfsten H, Cho A, Freitag C, Raderer M, Goliasch G, Bartko PE, Wurm R, Strunk G, Gisslinger H, Marosi C, Kornek G, Zielinski C, Hülsmann M, Pavo N. GDF-15 in solid vs non-solid treatment-naïve malignancies. Eur J Clin Invest. 2019;49:e13168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 14. | Carlsson AC, Nowak C, Lind L, Östgren CJ, Nyström FH, Sundström J, Carrero JJ, Riserus U, Ingelsson E, Fall T, Ärnlöv J. Growth differentiation factor 15 (GDF-15) is a potential biomarker of both diabetic kidney disease and future cardiovascular events in cohorts of individuals with type 2 diabetes: a proteomics approach. Ups J Med Sci. 2020;125:37-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 15. | Lu YY, Tao XD, Zhou LF. Expression of growth differentiation factor-15 in chronic hepatitis B and chronic hepatitis B-associated liver cirrhosis. Zhonghua Linchuang Ganranbing Zazhi. 2014;7:250-252. [DOI] [Cited in This Article: ] |
| 16. | Liu X, Chi X, Gong Q, Gao L, Niu Y, Chi X, Cheng M, Si Y, Wang M, Zhong J, Niu J, Yang W. Association of serum level of growth differentiation factor 15 with liver cirrhosis and hepatocellular carcinoma. PLoS One. 2015;10:e0127518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 17. | Lee ES, Kim SH, Kim HJ, Kim KH, Lee BS, Ku BJ. Growth Differentiation Factor 15 Predicts Chronic Liver Disease Severity. Gut Liver. 2017;11:276-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 18. | Halim MH, Abdulla NA, Kamel A, El Maksoud NA, Ragab HM. Significance of growth differentiation factor 15 in chronic HCV patients. J Genet Eng Biotechnol. 2017;15:403-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 19. | Jing R, Liu Q, Xie Q, Qian Z. Correlation between GDF 15 gene polymorphism and the collateral circulation in acute non-ST segment elevated myocardial infarction. Int J Clin Exp Med. 2015;8:14383-14387. [PubMed] [Cited in This Article: ] |
| 20. | Chen XP, Shang XS, Wang YB, Fu ZH, Gao Y, Feng T. Correlation between GDF-15 gene polymorphism and the formation of collateral circulation in acute ST-elevation myocardial infarction. Rev Assoc Med Bras (1992). 2017;63:1049-1054. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Wang J, Li W, Wang Y, Li C, Ding M, Zhang H, Lai M. The H6D genetic variation of GDF15 is associated with genesis, progress and prognosis in colorectal cancer. Pathol Res Pract. 2015;211:845-850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Chinese Society of Hepatology; Chinese Medical Association; Wei L. [The guideline of prevention and treatment for hepatitis C: a 2015 update]. Zhonghua Gan Zang Bing Za Zhi. 2015;23:906-923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 37] [Reference Citation Analysis (0)] |
| 23. | Bootcov MR, Bauskin AR, Valenzuela SM, Moore AG, Bansal M, He XY, Zhang HP, Donnellan M, Mahler S, Pryor K, Walsh BJ, Nicholson RC, Fairlie WD, Por SB, Robbins JM, Breit SN. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc Natl Acad Sci USA. 1997;94:11514-11519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 810] [Cited by in F6Publishing: 844] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 24. | Hsiao EC, Koniaris LG, Zimmers-Koniaris T, Sebald SM, Huynh TV, Lee SJ. Characterization of growth-differentiation factor 15, a transforming growth factor beta superfamily member induced following liver injury. Mol Cell Biol. 2000;20:3742-3751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 222] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 25. | Si Y, Liu X, Cheng M, Wang M, Gong Q, Yang Y, Wang T, Yang W. Growth differentiation factor 15 is induced by hepatitis C virus infection and regulates hepatocellular carcinoma-related genes. PLoS One. 2011;6:e19967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Zhang J, Guo ZX, Su SH. Expression and significance of growth differentiation factor-15 in chronic hepatitis C patients. Ganzang. 2018;23:629-631. [Cited in This Article: ] |
| 27. | Chen J, Liu C, Chen H, Liu Q, Yang B, Ou Q. Study on noninvasive laboratory tests for fibrosis in chronic HBV infection and their evaluation. J Clin Lab Anal. 2013;27:5-11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Sarrazin C, Shiffman ML, Hadziyannis SJ, Lin A, Colucci G, Ishida H, Zeuzem S. Definition of rapid virologic response with a highly sensitive real-time PCR-based HCV RNA assay in peginterferon alfa-2a plus ribavirin response-guided therapy. J Hepatol. 2010;52:832-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Zhao ML, Guo JT, Zhou J, Zhao ZG. A study of correlation between growth differentiation factor-15 gene polymorphism loci +157A/T and unstable angina pectoris. Linchuang He Shiyan Yixue Zazhi. 2014;13:171-174. [Cited in This Article: ] |
| 30. | Wang X, Yang X, Sun K, Chen J, Song X, Wang H, Liu Z, Wang C, Zhang C, Hui R. The haplotype of the growth-differentiation factor 15 gene is associated with left ventricular hypertrophy in human essential hypertension. Clin Sci (Lond). 2009;118:137-145. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 31. | He Q, Feng HQ, Liu H, Xu BN, Zhang SP, Fu SB. Relationship between polymorphism of GDF-15 gene and susceptibility to chronic Keshan disease. Zhongguo Difangbingxue Zazhi. 2018;37:960-964. [DOI] [Cited in This Article: ] |
| 32. | Chen R. Growth differentiation factor-15 genotype and plasma level: Associations with coronary artery disease. Ph.D. Thesis, Tsinghua University, Peking Union Medical College, and Chinese Academy of Medical Sciences. 2012. Available from: http://cdmd.cnki.com.cn/Article/CDMD-10023-1013312248.htm. [Cited in This Article: ] |
| 33. | El-Bendary M, Neamatallah M, Elalfy H, Besheer T, Elkholi A, El-Diasty M, Elsareef M, Zahran M, El-Aarag B, Gomaa A, Elhammady D, El-Setouhy M, Hegazy A, Esmat G. The association of single nucleotide polymorphisms of Toll-like receptor 3, Toll-like receptor 7 and Toll-like receptor 8 genes with the susceptibility to HCV infection. Br J Biomed Sci. 2018;75:175-181. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |