Published online Oct 21, 2020. doi: 10.3748/wjg.v26.i39.5983
Peer-review started: June 10, 2020
First decision: August 22, 2020
Revised: August 30, 2020
Accepted: September 16, 2020
Article in press: September 16, 2020
Published online: October 21, 2020
Ulcerative colitis (UC) is an inflammatory bowel disease that is difficult to diagnose and treat. To date, the degree of inflammation in patients with UC has mainly been determined by measuring the levels of nonspecific indicators, such as C-reactive protein and the erythrocyte sedimentation rate, but these indicators have an unsatisfactory specificity. In this study, we performed bioinformatics analysis using data from the National Center for Biotechnology Information-Gene Expression Omnibus (NCBI-GEO) databases and verified the selected core genes in a mouse model of dextran sulfate sodium (DSS)-induced colitis.
To identify UC-related differentially expressed genes (DEGs) using a bioinformatics analysis and verify them in vivo and to identify novel biomarkers and the underlying mechanisms of UC.
Two microarray datasets from the NCBI-GEO database were used, and DEGs between patients with UC and healthy controls were analyzed using GEO2R and Venn diagrams. We annotated these genes based on their functions and signaling pathways, and then protein-protein interactions (PPIs) were identified using the Search Tool for the Retrieval of Interacting Genes. The data were further analyzed with Cytoscape software and the Molecular Complex Detection (MCODE) app. The core genes were selected and a Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis was performed. Finally, colitis model mice were established by administering DSS, and the top three core genes were verified in colitis mice using real-time polymerase chain reaction (PCR).
One hundred and seventy-seven DEGs, 118 upregulated and 59 downregulated, were initially identified from the GEO2R analysis and predominantly participated in inflammation-related pathways. Seven clusters with close interactions in UC formed: Seventeen core genes were upregulated [C-X-C motif chemokine ligand 13 (CXCL13), C-X-C motif chemokine receptor 2 (CXCR2), CXCL9, CXCL5, C-C motif chemokine ligand 18, interleukin 1 beta, matrix metallopeptidase 9, CXCL3, formyl peptide receptor 1, complement component 3, CXCL8, CXCL1, CXCL10, CXCL2, CXCL6, CXCL11 and hydroxycarboxylic acid receptor 3] and one was downregulated [neuropeptide Y receptor Y1 (NYP1R)] in the top cluster according to the PPI and MCODE analyses. These genes were substantially enriched in the cytokine-cytokine receptor interaction and chemokine signaling pathways. The top three core genes (CXCL13, NYP1R, and CXCR2) were selected and verified in a mouse model of colitis using real-time PCR Increased expression was observed compared with the control mice, but only CXCR2 expression was significantly different.
Core DEGs identified in UC are related to inflammation and immunity inflammation, indicating that these reactions are core features of the pathogenesis of UC. CXCR2 may reflect the degree of inflammation in patients with UC.
Core Tip: Two microarray datasets were used, and differentially expressed genes were analyzed. Seventeen core genes were upregulated, and one was downregulated. These genes were markedly enriched in the cytokine-cytokine receptor interaction and chemokine signaling pathways. The top three core genes [C-X-C motif chemokine ligand 13, neuropeptide Y receptor Y1, C-X-C motif chemokine receptor 2 (CXCR2)] were verified in a dextran sulfate sodium-induced colitis model mice by real-time polymerase chain reaction and showed an increased expression, but only CXCR2 was statistically different. CXCR2 may represent a new biomarker to determine the degree of inflammation or a treatment target in ulcerative colitis.
- Citation: Shi L, Han X, Li JX, Liao YT, Kou FS, Wang ZB, Shi R, Zhao XJ, Sun ZM, Hao Y. Identification of differentially expressed genes in ulcerative colitis and verification in a colitis mouse model by bioinformatics analyses. World J Gastroenterol 2020; 26(39): 5983-5996
- URL: https://www.wjgnet.com/1007-9327/full/v26/i39/5983.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i39.5983
Ulcerative colitis (UC), a nonspecific inflammatory disease that occurs in the colon or rectum, is the most common type of inflammatory bowel disease (IBD) and tends to occur in young and middle-aged people. Symptoms of UC, such as diarrhea, bloody stools with mucus-pus and abdominal pain, substantially affect quality of life[1,2]. Because UC is difficult to cure, readily relapses and has a high risk of cancer, UC has been classified as a refractory disease by the World Health Organization.
Epidemiological studies have confirmed that the annual incidence of UC worldwide is 10.5-14 cases per 100000 people, and the prevalence rate is approximately 246.7 cases per 100000 people[3]. Currently, the number of patients in Europe and the United States accounts for 0.5% of the global population, and the incidence and prevalence rates in urban areas have exceeded those in rural areas. Since the 1990s, the number of children who were initially treated for IBD in Western developed countries has gradually increased. In the past 20 years, the incidence and prevalence of IBD in developed countries have begun to stabilize and plateau, but in other countries an obvious increase in the incidence and prevalence still occurs, particularly in South America and East Asia[4,5]. Recently, a systematic review of 140 studies in 2018 showed that the incidence of IBD in children increased in both developed and developing countries[6].
The pathogenesis of UC is complex, and the interaction of multiple factors may lead to its occurrence. With contributions from the environment, mental factors and intestinal flora, the intestinal barrier of genetically susceptible people is destroyed, and the immune system is dysfunctional, resulting in excessively hyperactive immune reactions and inflammation[7,8]. The combination of C-reactive protein and fecal calprotectin levels may be beneficial for dynamically diagnosing and monitoring the progress of UC at present[9]. However, the limited number of significant and specific biomarkers for UC has become an increasingly prominent problem in its diagnosis and treatment.
Microarray technology reveals numerous genes that are activated in different tissues as well as their physiological and pathological statuses and has been regarded as a novel approach for clarifying the mechanisms underlying different diseases[10]. In recent years with the optimization of gene sequencing platforms, differentially expressed genes (DEGs) have been identified using bioinformatics analyses[11]. To date, several studies have reported the bioinformatics analysis of IBD using arrays or chips[12,13], but the analysis of UC is still lacking. Thus, bioinformatics methods may help us study and more clearly understand the underlying mechanisms of UC[14].
In this study, we applied two databases from Gene Expression Omnibus (GEO), GEO2R and online tools for constructing Venn diagrams to identify the DEGs, including upregulated and downregulated genes. Then, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to analyze the DEGs based on the molecular function (MF), cellular component (CC), and biological process (BP) and different Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways[11]. Third, we constructed a protein-protein interaction (PPI) network using Cytoscape and the Molecular Complex Detection (MCODE) app for further analysis. Using these methods, core genes were chosen, and the KEGG pathway enrichment analysis was repeated. Finally, we verified the top three core genes in the first cluster selected by MCODE in colon tissues from mice with dextran sulfate sodium (DSS)-induced colitis using real-time polymerase chain reaction (PCR).
Microarray data were obtained from the National Center for Biotechnology Information-GEO, a free public database of microarray and gene profiles, and we obtained the gene expression profiles of GSE92415 and GSE87466 in colon mucosal tissues from patients with UC and healthy individuals. Microarray data included in GSE92415 and GSE87466 were obtained from GPL13158 Platforms (HT_HG-U133_Plus_PM] Affymetrix HT HG-U133+ PM Array Plate) and included 162 colonic mucosal tissues from patients with UC (who were treated with the anti-TNF agent golimumab and placebo) and 21 healthy colon mucosal tissues and 87 samples from patients with UC and 21 normal colon mucosal tissues, respectively.
DEGs were identified between specimens from patients with UC and normal colonic mucosa using the GEO2R online tool with a |logFC| > 2 and adjusted P value < 0.05. Then, the raw data in TXT format were imported into the online tool to construct a Venn diagram and identify the common DEGs among the two datasets. The DEGs with a logFC < 0 were considered downregulated genes, while the DEGs with a logFC > 0 were considered upregulated genes.
Gene Ontology (GO) analysis is a commonly used method for identifying the biological properties of high-throughput transcriptome or genome data for genes and their RNA or protein products[15]. KEGG is a collection of databases for genomes, diseases, biological pathways, drugs and chemical materials. DAVID is an online bioinformatics tool designed to identify a large number of gene or protein functions[10]. We used DAVID and performed a GO analysis to visualize the enrichment of DEGs in BP, MF, cellular components and KEGG pathways.
PPI information was evaluated using an online tool, the Search Tool for the Retrieval of Interacting Genes (STRING). A tab-separated file containing the values for the network from STRING was imported into Cytoscape (continuous mapping for columns depended on the frequency of genes; default size of the column = 30.0; upregulated genes were red and downregulated genes were blue), which was applied to detect the potential interactions between these DEGs[16]. In addition, the MCODE app in Cytoscape was used to assess the modules of the PPI network (cutoff = 2, max depth = 100, k-core = 2, node score cutoff = 0.2) and to identify the core genes in the top cluster in UC. Meanwhile, the KEGG pathway enrichment analysis of these core DEGs was repeated.
We complied with the ethics standard for research activity established by the Animal Ethics Committee of Beijing University of Chinese Medicine in accordance with the guidelines issued by the Regulations of Beijing Laboratory Animal Management. Fourteen male specific-pathogen-free (SPF) C57BL/6 mice (weighing 20 ± 2 g) were procured from SPF Biological Technology Co., Ltd., Beijing, China (Certificate No. SCXK [jing] 2019-0010). They were housed in the SPF animal center of Beijing University of Chinese Medicine at a constant temperature of 20-26 °C and humidity of 50% to 60% in a light-controlled environment with a 12-h light/dark cycle. In addition, fodder and sterilized water were supplied. After adaptive feeding for 1 wk, these mice were divided into the control (n = 4) and model (n = 10) groups using the random number table method. Mice in the model group drank 3.5% (weight/volume) DSS (average molecular weight 36000-50000, MP Biomedicals) for 7 consecutive days[17].
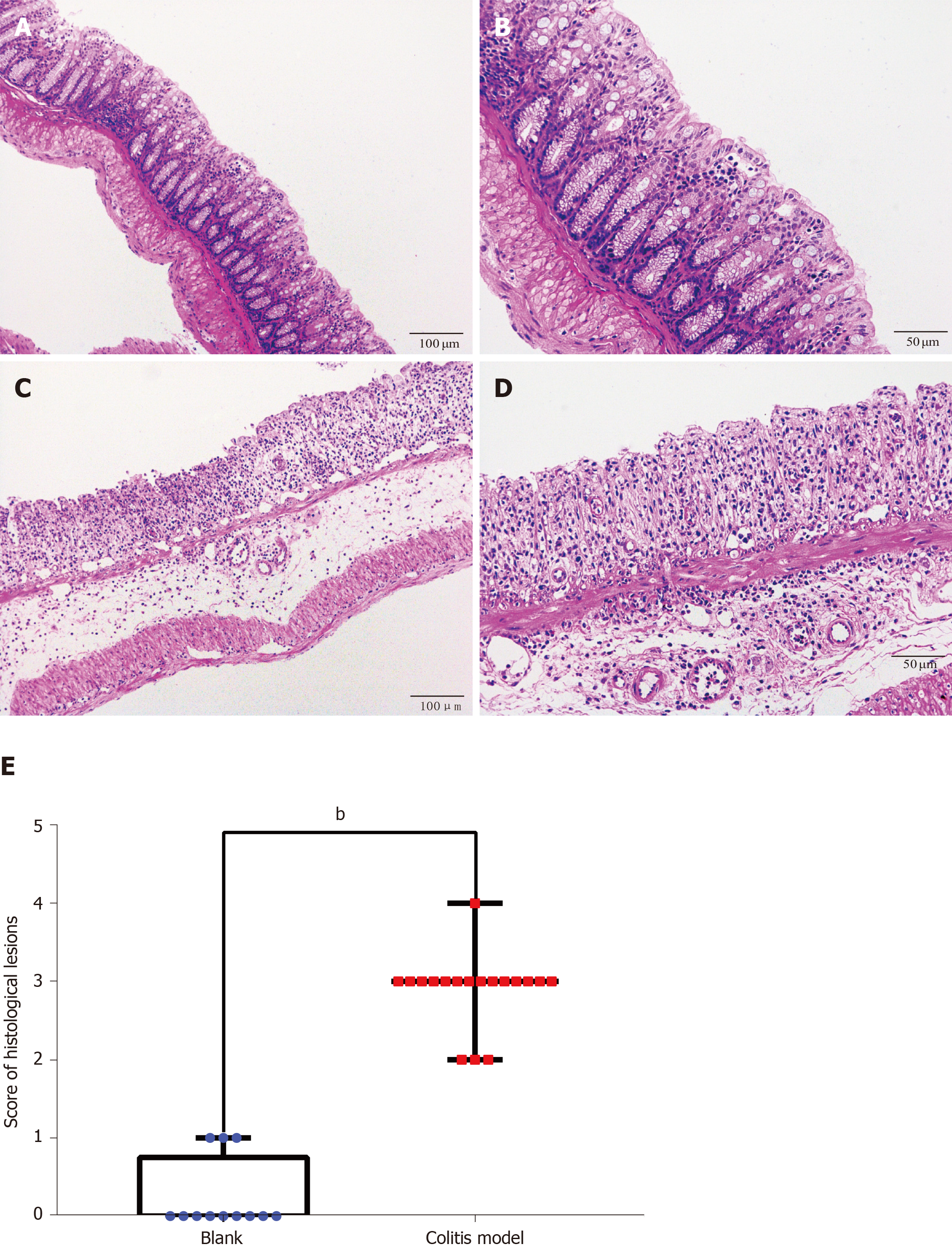
On the 7th d of the experiment, all the mice were fasted for 24 h. On the 8th d, mice were anesthetized by ether inhalation until the superficial reflex disappeared, and the mice were sacrificed by cervical spondylolisthesis. After disinfecting and preparing the skin, the abdomen was incised to separate the colon tissue. Specimens of colon tissues (three 1 cm pieces) from each mouse were fixed with a 10% neutral formaldehyde solution for at least 24 h. The lesion score was judged by the histological criterion after hematoxylin-eosin staining of 4 μm thick sections: 0, no signs of inflammation; 1, a low level of leukocyte infiltration (10-30 leukocytes per high-power field); 2, a moderate level of leukocyte infiltration (31-70 leukocytes per high-power field); 3, a high level of leukocyte infiltration (> 71 leukocytes per high-power field), high vascular density and thickening of the bowel wall; and 4, transmural infiltrations, loss of goblet cells, high vascular density, strong bowel wall thickening, ulcerations and cryptic abscesses[18].
The other tissues were placed in cryotubes, frozen in liquid nitrogen overnight and stored at -80 °C. Total RNA was extracted from the colon tissues using TRIzol reagent (Invitrogen, United States). Specific primers were used to amplify the core genes (Table 1), and the expression of each gene was normalized to β-actin with the standard curve method. Reverse transcription was performed at 42 °C for 60 min, and reverse transcriptase was inactivated at 70 °C for 15 min. The relative quantitative analysis was performed using the 2-ΔΔCT approach.
| Gene | Sequence, 5’ to 3’ |
| β-actin | Forward: GAGATTACTGCTCTGGCTCCTA |
| Reverse: GGACTCATCGTACTCCTGCTTG | |
| CXCL13 | Forward: CCCAACCCACATCCTTGTTCCTT |
| Reverse: CTGAAGTCCATCTCGCAAACCTCT | |
| NPY1R | Forward: CCAGTGAGACCAAGCGAATCAACA |
| Reverse: GCGGTGAGGTGACAGAGCAGAA | |
| CXCR2 | Forward: ATGGTGGAACTTGTGGCTCTAGC |
| Reverse: CCATTCCTGGTCCTGGCATTCC |
The histological lesion score and relative expression levels obtained using real-time PCR were analyzed with SPSS (version 22.0, IBM, Corp., Armonk, NY, United States). Data were distributed normally, and the two groups were compared with an independent-sample t test. A P value < 0.05 indicated a statistically significant difference.
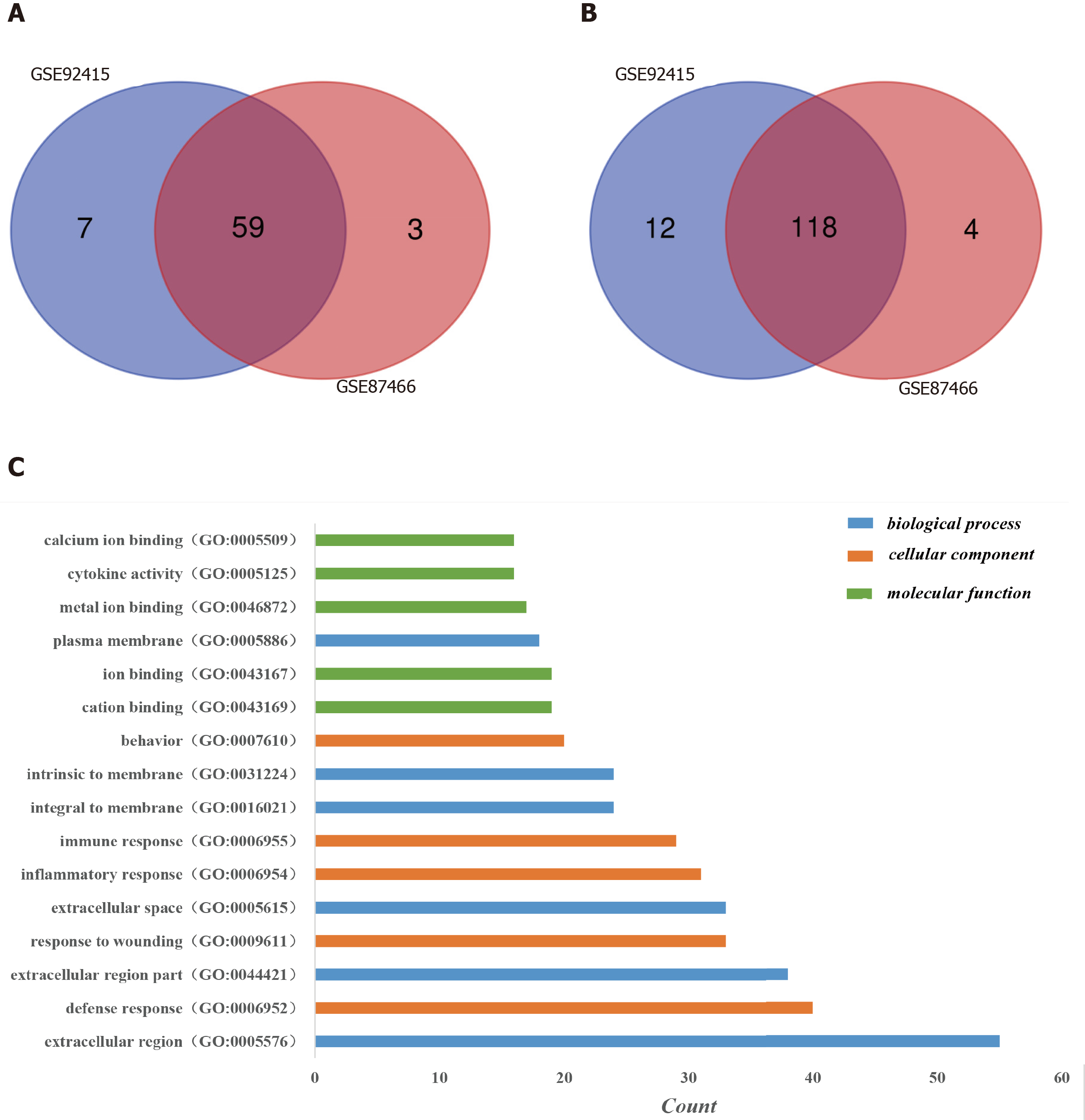
One hundred and ninety-six DEGs were identified in the GSE92415 database, including 130 upregulated and 66 downregulated genes. One hundred and eighty-four DEGs were identified in the GSE87466 database, including 122 upregulated and 62 downregulated genes. Among the two databases, 177 genes (118 upregulated and 59 downregulated) overlapped (Figure 1A and 1B).
Three categories of DEGs, MF, BP and CC, were classified by performing a GO analysis. Among these GO functions, extracellular region (GO: 0005576), defense response (GO: 0006952), extracellular region part (GO: 0044421), response to wounding (GO: 0009611), extracellular space (GO: 0005615) and inflammatory response (GO: 0006954) were the top six terms associated with UC with more than 30 genes identified in each category (Figure 1C). Upregulated genes were mainly enriched in defense response, inflammatory response, response to wounding, extracellular region, extracellular region part, extracellular space, cytokine activity, chemokine activity and chemokine receptor binding and were the top three enriched terms, depending on the P value of the respective categories (Table 2, P < 0.05). Downregulated genes were mainly enriched in transmembrane transport, carboxylic acid transport, organic acid transport, cell fraction, membrane fraction, insoluble fraction, symporter activity, xenobiotic-transporting ATPase activity and multidrug transporter activity and were the top three enriched terms, based on the P value of the respective categories (Table 3, P < 0.05). The KEGG pathway enrichment analysis suggested that DEGs predominantly participated in inflammation-related pathways, including the chemokine signaling pathway, cytokine-cytokine receptor interaction and complement and coagulation cascades (Table 4, P < 0.05).
| Category | Term | Count | Genes |
| BP | Defense response (GO: 0006952) | 40 | CXCL1, KYNU, S100A8, FGR, C3, CXCL3, CXCL2, S100A9, CXCL9, CXCR2, CXCL6, DEFB4A, CXCL11, CXCL10, REG3A, SERPINA3, CSF3R, VNN1, IL1B, LTF, SERPINA1, CFI, SPP1, SELP, CR1, PDPN, CFB, IL1RN, C4BPB, IDO1, CCL18, S100A12, LILRB2, TNFAIP6, PROK2, DEFA6, CXCL13, DEFA5, PLA2G2D, DMBT1. |
| BP | Inflammatory response (GO: 0006954) | 31 | CXCL1, S100A8, C3, CXCL3, CXCL2, S100A9, CXCL9, CXCR2, CXCL6, CXCL11, CXCL10, REG3A, SERPINA3, VNN1, IL1B, SERPINA1, CFI, SPP1, SELP, CR1, PDPN, CFB, IL1RN, C4BPB, IDO1, CCL18, S100A12, TNFAIP6, PROK2, CXCL13, PLA2G2D. |
| BP | Response to wounding (GO: 0009611) | 33 | CXCL1, S100A8, C3, TNC, CXCL3, CXCL2, S100A9, CXCL9, CXCR2, CXCL6, CXCL11, CDH3, CXCL10, REG3A, SERPINA3, VNN1, IL1B, SERPINA1, CFI, SPP1, SELP, CR1, PDPN, CFB, IL1RN, C4BPB, IDO1, CCL18, S100A12, TNFAIP6, PROK2, CXCL13, PLA2G2D. |
| CC | Extracellular region (GO: 0005576) | 55 | REG4, MMP9, MMP7, CXCL11, MMP3, MMP1, CXCL10, REG3A, SERPINA3, CSF3R, IL1B, LTF, SERPINA1, KLK10, CFI, FCGR3B, CYR61, IL13RA2, EGFL6, IL24, TCN1, MMP12, MMP10, PROK2, GZMK, DEFA6, SERPINB5, DEFA5, STC1, PLA2G2D, CXCL1, CXCL5, C3, CXCL3, TNC, CXCL2, CXCL9, CXCL6, DEFB4A, GREM1, TIMP1, THBS2, OLFM1, SPP1, SELP, CFB, IL1RN, CHI3L1, CHI3L2, C4BPB, CCL18, LCN2, CXCL13, PI3, DMBT1. |
| CC | Extracellular region part (GO: 0044421) | 38 | CXCL1, CXCL5, C3, MMP9, TNC, CXCL3, CXCL2, MMP7, CXCL9, CXCL6, CXCL11, MMP3, GREM1, MMP1, CXCL10, TIMP1, REG3A, IL1B, SERPINA1, CFI, OLFM1, SPP1, IL13RA2, SELP, EGFL6, IL1RN, CHI3L1, CHI3L2, IL24, CCL18, MMP12, MMP10, SERPINB5, DEFA6, CXCL13, DEFA5, PI3, STC1. |
| CC | Extracellular space (GO: 0005615) | 33 | CXCL1, CXCL5, C3, MMP9, CXCL3, CXCL2, MMP7, CXCL9, CXCL6, GREM1, CXCL11, MMP3, CXCL10, REG3A, IL1B, SERPINA1, CFI, OLFM1, IL13RA2, SPP1, SELP, EGFL6, IL1RN, CHI3L1, CHI3L2, IL24, CCL18, MMP10, SERPINB5, DEFA6, CXCL13, DEFA5, STC1. |
| MF | Cytokine activity (GO: 0005125) | 16 | CXCL1, NAMPT, CXCL5, CXCL3, IL1RN, CXCL2, CXCL9, CXCL6, IL24, GREM1, CXCL11, CCL18, CXCL10, CXCL13, IL1B, SPP1. |
| MF | Chemokine activity (GO: 0008009) | 10 | CXCL1, CXCL5, CXCL13, CXCL3, CXCL2, CXCL9, CXCL6, CXCL11, CCL18, CXCL10. |
| MF | Chemokine receptor binding (GO: 0042379) | 10 | CXCL1, CXCL5, CXCL13, CXCL3, CXCL2, CXCL9, CXCL6, CXCL11, CCL18, CXCL10. |
| Category | Term | Count | Genes |
| BP | Transmembrane transport (GO: 0055085) | 9 | TRPM6, SLC23A1, SLC16A1, SLC25A34, SLC17A4, ABCB1, AQP7, SLC30A10, SLC26A2. |
| BP | Carboxylic acid transport (GO: 0046942) | 5 | SLC38A4, SLC23A1, SLC16A1, AQP8, SLC3A1. |
| BP | Organic acid transport (GO: 0015849) | 5 | SLC38A4, SLC23A1, SLC16A1, AQP8, SLC3A1. |
| CC | Cell fraction (GO: 0000267) | 12 | HSD3B2, SLC23A1, SLC16A1, AQP8, SLC17A4, CYP2B6, MEP1B, ABCB1, ANPEP, SLC3A1, SLC26A2, PCK1. |
| CC | Membrane fraction (GO: 0005624) | 10 | HSD3B2, SLC23A1, SLC16A1, AQP8, SLC17A4, CYP2B6, MEP1B, ABCB1, SLC3A1, SLC26A2. |
| CC | Insoluble fraction (GO: 0005626) | 10 | HSD3B2, SLC23A1, SLC16A1, AQP8, SLC17A4, CYP2B6, MEP1B, ABCB1, SLC3A1, SLC26A2. |
| MF | Symporter activity (GO: 0015293) | 4 | SLC38A4, SLC23A1, SLC16A1, SLC17A4. |
| MF | Xenobiotic-transporting ATPase activity (GO: 0008559) | 2 | ABCB1, ABCG2. |
| MF | Multidrug transporter activity (GO: 0015239) | 2 | ABCB1, ABCG2. |
| Pathway ID | Term | Genes |
| hsa04062 | Chemokine signaling pathway | CXCL1, FGR, CXCL5, CXCL3, CXCL2, CXCL9, CXCR2, CXCL6, CXCL11, CCL18, CXCL10, CXCL13, JAK3. |
| hsa04060 | Cytokine-cytokine receptor interaction | CXCL1, CXCL5, CXCL3, CXCL2, CXCL9, CXCR2, CXCL6, IL24, CXCL11, CCL18, CXCL10, CXCL13, CSF3R, IL1B. |
| hsa04610 | Complement and coagulation cascades | CR1, C3, CFB, C4BPB, SERPINA1, CFI. |
| hsa04514 | Cell adhesion molecules | SELP, SELL, CD274, CTLA4, CLDN2, CDH3. |
| hsa00830 | Retinol metabolism | CYP2B6, ADH1C, UGT2A3. |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | CYP2B6, ADH1C, UGT2A3. |
| hsa04620 | Toll-like receptor signaling pathway | CXCL9, IL1B, CXCL11, SPP1, CXCL10. |
| hsa00982 | Drug metabolism | CYP2B6, ADH1C, UGT2A3. |
| hsa03320 | PPAR signaling pathway | HMGCS2, AQP7, PCK1. |
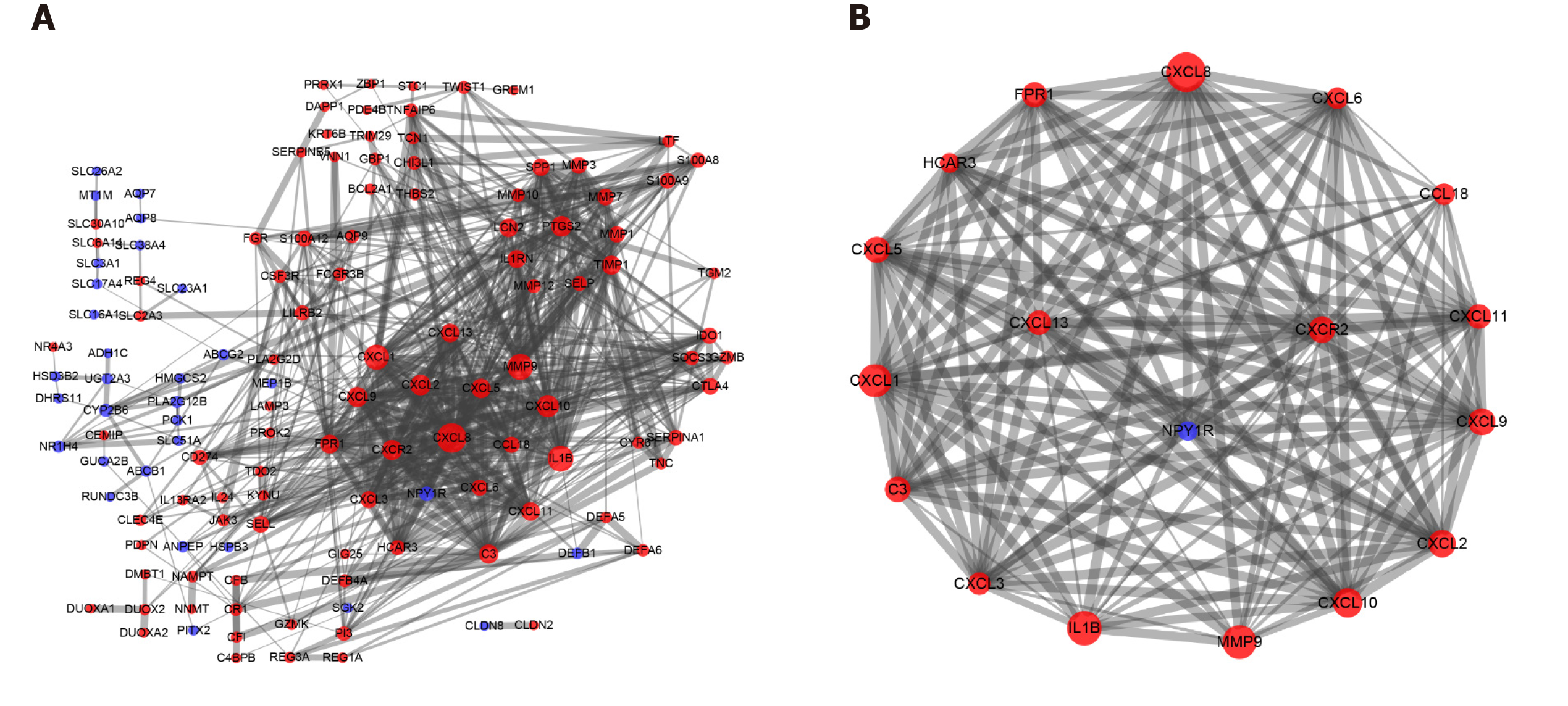
One hundred and seventy-seven DEGs were entered into the PPI network using the Search Tool for the Retrieval of Interacting Genes, including 130 nodes and 639 edges. The thickness of the edges was adjusted based on the level of the combined score. The larger the area of a protein node, the more protein nodes that interacted with it. Upregulated genes were shown in red, and downregulated genes were shown in blue (Figure 2A). Then, we applied the MCODE app for further analysis and identified seven clusters with close interactions in patients with UC. Seventeen core genes were upregulated [C-X-C motif chemokine ligand 13 (CXCL13), C-X-C motif chemokine receptor 2 (CXCR2), CXCL9, CXCL5, C-C motif chemokine ligand 18 (CCL18), interleukin 1 beta (IL1B), matrix metallopeptidase 9 (MMP9), CXCL3, formyl peptide receptor 1 (FPR1), complement component 3 (C3), CXCL8, CXCL1, CXCL10, CXCL2, CXCL6, CXCL11 and hydroxycarboxylic acid receptor 3 (HCAR3)], and one gene was downregulated [neuropeptide Y receptor Y1 (NPY1R)] in the first significant cluster (Figure 2B).
The KEGG pathway enrichment analysis was repeated using the DAVID GO analysis to identify the pathways in which these 18 core DEGs in the top cluster (18 nodes, 142 edges, and a score of 16.706) were involved (P < 0.05). These DEGs were markedly enriched in the cytokine-cytokine receptor interaction and chemokine signaling pathways (Table 5).
| Category | Term | Count, % | Genes |
| KEGG pathway | hsa04060, cytokine-cytokine receptor interaction. | 12, 6.06 | CXCL1, CXCL5, CXCL13, CXCL3, CXCL2, CXCL9, IL1B, CXCR2, CXCL6, CXCL11, CCL18, CXCL10. |
| KEGG pathway | hsa04062, chemokine signaling pathway. | 11, 5.56 | CXCL1, CXCL5, CXCL13, CXCL3, CXCL2, CXCL9, CXCR2, CXCL6, CXCL11, CCL18, CXCL10. |
| KEGG pathway | hsa04620, toll-like receptor signaling pathway. | 4, 2.02 | CXCL9, IL1B, CXCL11, CXCL10. |
| KEGG pathway | hsa04621, NOD-like receptor signaling pathway. | 3, 1.52 | CXCL1, CXCL2, IL1B. |
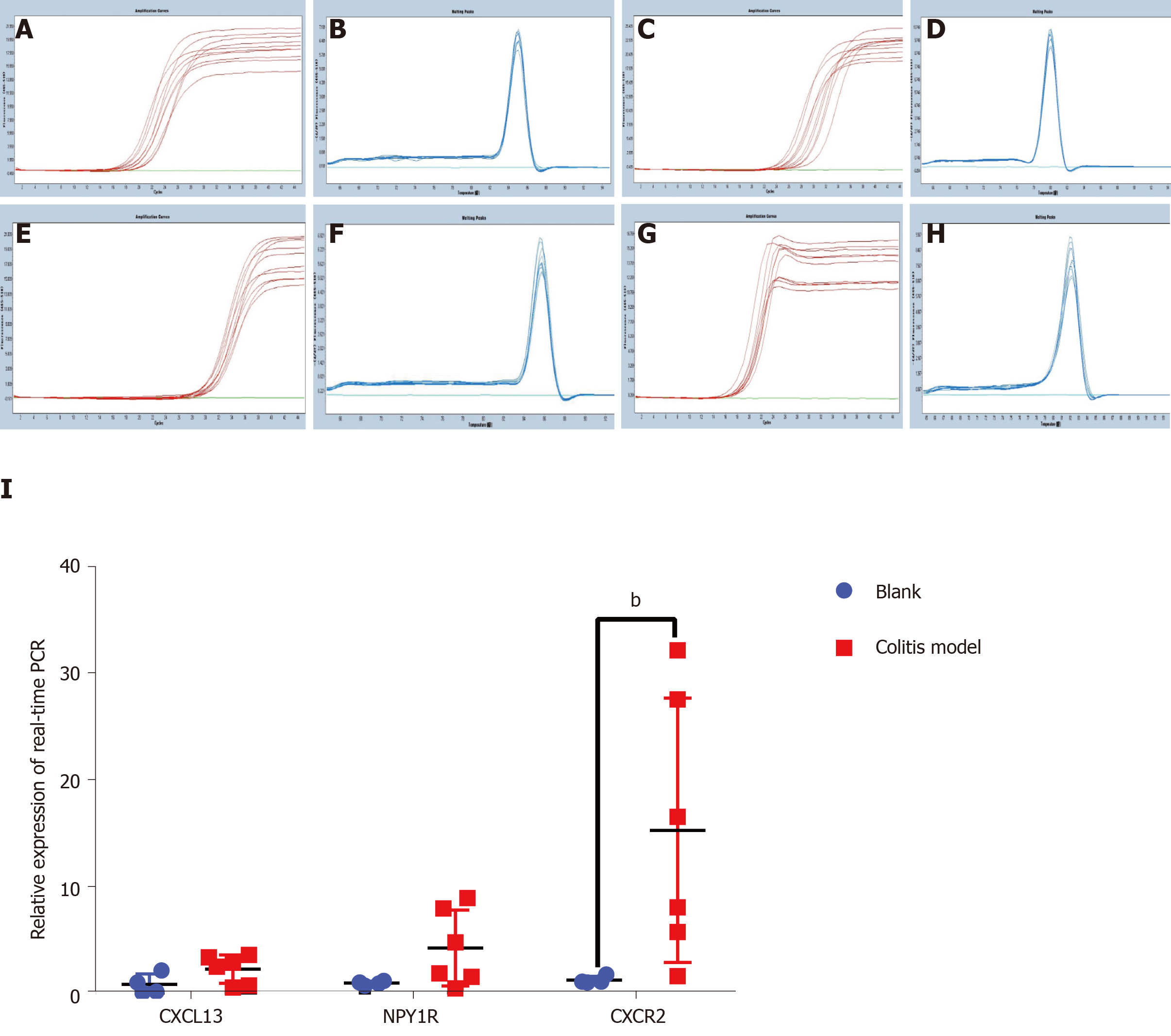
Finally, we verified the top three genes (CXCL13, NPY1R and CXCR2) among the 18 core DEGs in the colitis model mice. Four mice in the model group were sacrificed during the experimental period because of the severity of disease, and the colon tissues of all the remaining mice (control: n = 4; colitis model: n = 6) were observed under a microscope. The normal four-layer structure of intestinal tissues, goblet cells and crypts disappeared, and inflammatory cells had infiltrated the submucosa in the model group compared with the control group (Figure 3A-D). Histological lesions indicated the successful establishment of the colitis model (Figure 3E). The CXCL13 and CXCR2 mRNAs were expressed at higher levels in the colon tissues from the colitis model mice than in the mice from the control group (Figure 4), and the difference in CXCR2 expression was significant (P < 0.01). These manifestations were consistent with the results of the bioinformatics analysis in the present study. Interestingly, higher expression of the NPY1R mRNA was also observed in the colitis model mice (Figure 4), which differed from our bioinformatics results.
The incidence of UC in China and other Asian countries is gradually increasing[19], but due to the complicated pathogenesis, accurate molecular biomarkers for diagnosing UC are still lacking. In the past, biomarkers such as the C-reactive protein level and erythrocyte sedimentation rate were used to judge the degree of inflammation in patients with UC but are generally nonspecific[20], which greatly complicated the determination of the clinical diagnosis, recurrence and prognosis. The lack of biomarkers has increased the medical burden and physical and psychological discomfort of patients and accelerated the waste of public social health resources. Therefore, the identification and excavation of relatively specific molecular biomarkers is a bottleneck problem that must be solved in the diagnosis of UC[21]. Focusing on methods to solve this problem, our study chose the GEO database for an in-depth analysis of biological information.
GEO is an international public repository for high-throughput microarray and next-generation sequencing functional genomic datasets submitted by the research community[22]. We selected two microarray datasets from the GEO database, combined GEO2R and Venn diagram analyses and initially identified 177 DEGs, including 118 upregulated and 59 downregulated genes. Next, we annotated these genes by function and signaling pathways and found that they were mainly related to inflammation, such as chemokines, cytokine receptors and complement proteins. The PPI analysis helped us identify 18 core genes based on their functional annotations, and cytokine-cytokine receptor interactions and chemokine signaling pathways were maximally enriched.
We established colitis model mice and verified the transcript levels of the top three genes in the first cluster in colon tissues using real-time PCR to clarify the expression of these core genes and their main pathophysiological functions. The expression of the CXCL13 and CXCR2 mRNAs, particularly the CXCR2 mRNA, was increased in the colitis model mice, consistent with our bioinformatics analysis.
Chemokines and cytokines play pivotal roles in regulating mucosal inflammation and the immune system by promoting neutrophil migration to sites of inflammation, ultimately leading to tissue damage and destruction[23,24]. Carlsen et al[25] reported the expression of CXCL13 in both healthy individuals and patients with UC, in addition to reports in rodents from published studies; however, differences in the expression of CXCL13 between patients with UC and healthy individuals were unclear. Singh et al[23] reported significantly increased expression of CXCL13 in patients with IBD, including patients with UC and Crohn’s disease, compared to healthy controls.
On the other hand, CXCR2 has been reported to regulate the migration and recruitment of neutrophils to the site of inflammation[26]. A review also suggested that in addition to calprotectin, CXCR2, a neutrophil-related protein, may have potential roles in diagnosis and treatment[27]. As shown in the study by Farooq et al[28], CXCR2-positive mice have more severe symptoms, such as the infiltration of polymorphonuclear neutrophils(PMNs), than CXCR2-deficient mice with DSS-induced colitis. The mechanism of increased CXCR2 expression in colitis has not been conclusively determined, as the infiltration of increased numbers of PMNs in the mucosa or submucosa is a feature of DSS-induced colitis. CXCR2 functions as a PMNchemokine receptor, which would lead to a significantly higher level in individuals with colitis. Our study verified this hypothesis by showing neutrophil infiltration and a high level of the CXCR2 mRNA.
Notably, the expression of the NPY1R mRNA in the colitis model mice differed from our bioinformatics results. NPY is a 36-amino acid peptide with a wide distribution in the central and peripheral nervous system[29] that evokes numerous physiological responses by activating different receptors (Y1, Y2, Y3)[30]. NPY is considered the most potent orexigenic neuropeptide and may be involved in the stress response, anxiety and mood-related disorders as well as the regulation of the immune system and cancer[31].
NPY1R expression was decreased in patients with UC based on the results of the bioinformatics analysis; however, its expression was increased in the colitis model mice as evidenced by real-time PCR, which might be related to the four aspects described below. First, our bioinformatics results were derived from human colon specimens, which were fundamentally heterologous compared to the mice. Second, microarray data were obtained from the human colonic mucosa, while RT-PCR was performed on mouse colon tissues, which contained the mucosa, submucosa, muscle layer and subserosa. Thus, differences in expression were observed between the bioinformatics results and in vivo analysis. Third, because of the current shortage of microarrays containing samples from patients with UC in the same platforms, we chose the GSE92415 microarray dataset, including patients with UC who were treated with a TNF-α inhibitor and placebos, and thus the result potentially did not accurately reflect the detailed scientific features of UC. Additionally, a 3.5% DSS solution was used to establish the colitis model and combined with the histological lesions, the model was much more severe. Further studies are needed to confirm whether dysplasia of the intestinal epithelium occurred, and the NPY/NPY1R system was activated.
In conclusion, the core DEGs identified in patients with UC were CXCL13, NPY1R, CXCR2, CXCL9, CXCL5, CCL18, IL1B, MMP9, CXCL3, formyl peptide receptor 1, C3, CXCL8, CXCL1, CXCL10, CXCL2, CXCL6, CXCL11 and HCAR3. These DEGs are related to inflammation and immune-inflammatory reactions, indicating that inflammation and abnormal activation of the immune system may represent the core features of the pathogenesis of UC. Based on the current data, we propose that CXCR2 may represent a new biomarker for the degree of inflammation or a treatment target, and our study may provide new insights into the diagnosis and treatment of UC.
Ulcerative colitis (UC) tends to occur in young and middle-aged people. It substantially affects the patient’s quality of life because it is difficult to cure, readily relapses and poses a high risk of colon cancer. However, the pathogenesis of UC is complex and multifaceted, and specific biomarkers for UC are currently unavailable.
In recent years, with the optimization of gene sequencing platforms, differentially expressed genes (DEGs) have been identified through bioinformatics analyses by comparing microarrays. To date, several studies have reported the results of bioinformatics analyses of samples from patients with inflammatory bowel disease using arrays or chips, but the analysis of patients with UC is still lacking. The specific molecules or biomarkers of UC are insufficient. Thus, we will apply bioinformatics methods to more clearly elucidate the underlying biomarkers and mechanisms of UC.
To identify UC-related DEGs by performing a bioinformatics analysis and verify them in vivo and to identify novel biomarkers and the underlying mechanisms of UC.
Two microarray datasets from the National Center for Biotechnology Information-Gene Expression Omnibus database were used, and DEGs were analyzed using GEO2R and Venn diagrams. We annotated these genes based on functions and signaling pathways. Then protein-protein interaction (PPI) were constructed using the Search Tool for the Retrieval of Interacting Genes. The data were further analyzed with Cytoscape software and the Molecular Complex Detection (MCODE) app. The core genes were selected, and the Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis was repeated. Finally, colitis model mice were established by administering dextran sulfate sodium, and the top three core genes were verified in colitis mice using real-time polymerase chain reaction.
One hundred and seventy-seven DEGs (118 upregulated genes and 59 downregulated genes) predominantly participated in inflammation-related pathways. Seventeen core genes were upregulated, and one gene was downregulated in the first cluster according to the PPI and MCODE analyses in Cytoscape. These genes were markedly enriched in the cytokine-cytokine receptor interaction and chemokine signaling pathways. The top three core genes showed increased expression compared with the control mice, but only the difference in C-X-C motif chemokine receptor 2 (CXCR2) expression was statistically significant. CXCR2 may reflect the degree of inflammation in patients with UC and serve as an underlying treatment target.
Core DEGs identified in patients with UC are related to inflammation and immune inflammatory reactions, indicating that these reactions are core features of the pathogenesis of UC. CXCR2 may reflect the degree of inflammation in patients with UC.
CXCR2 may represent a new biomarker to determine the degree of inflammation or a treatment target in UC. In the future, the combination of CXCR2 with other biomarkers will potentially improve the ability to diagnose and dynamically monitor UC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ishizawa K, Li C S-Editor: Huang P L-Editor: Filipodia P-Editor: Wang LL
| 1. | da Silva BC, Lyra AC, Rocha R, Santana GO. Epidemiology, demographic characteristics and prognostic predictors of ulcerative colitis. World J Gastroenterol. 2014;20:9458-9467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 167] [Cited by in F6Publishing: 162] [Article Influence: 16.2] [Reference Citation Analysis (1)] |
| 2. | Yu YR, Rodriguez JR. Clinical presentation of Crohn's, ulcerative colitis, and indeterminate colitis: Symptoms, extraintestinal manifestations, and disease phenotypes. Semin Pediatr Surg. 2017;26:349-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 171] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 3. | Shivashankar R, Tremaine WJ, Harmsen WS, Loftus EV Jr. Incidence and Prevalence of Crohn's Disease and Ulcerative Colitis in Olmsted County, Minnesota From 1970 Through 2010. Clin Gastroenterol Hepatol. 2017;15:857-863. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 294] [Article Influence: 42.0] [Reference Citation Analysis (0)] |
| 4. | Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, Panaccione R, Ghosh S, Wu JCY, Chan FKL, Sung JJY, Kaplan GG. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769-2778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2677] [Cited by in F6Publishing: 3169] [Article Influence: 452.7] [Reference Citation Analysis (0)] |
| 5. | Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn's disease. Lancet. 2017;389:1741-1755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1121] [Cited by in F6Publishing: 1355] [Article Influence: 193.6] [Reference Citation Analysis (3)] |
| 6. | Sýkora J, Pomahačová R, Kreslová M, Cvalínová D, Štych P, Schwarz J. Current global trends in the incidence of pediatric-onset inflammatory bowel disease. World J Gastroenterol. 2018;24:2741-2763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 251] [Cited by in F6Publishing: 219] [Article Influence: 36.5] [Reference Citation Analysis (6)] |
| 7. | Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389:1756-1770. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1542] [Cited by in F6Publishing: 1890] [Article Influence: 270.0] [Reference Citation Analysis (1)] |
| 8. | Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, Ashley N, Johnson E, Hublitz P, Bao L, Lukomska J, Andev RS, Björklund E, Kessler BM, Fischer R, Goldin R, Koohy H, Simmons A. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49-55. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 402] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 9. | Brookes MJ, Whitehead S, Gaya DR, Hawthorne AB. Practical guidance on the use of faecal calprotectin. Frontline Gastroenterol. 2018;9:87-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Sobek J, Bartscherer K, Jacob A, Hoheisel JD, Angenendt P. Microarray technology as a universal tool for high-throughput analysis of biological systems. Comb Chem High Throughput Screen. 2006;9:365-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Liang Y, Zhang C, Dai DQ. Identification of differentially expressed genes regulated by methylation in colon cancer based on bioinformatics analysis. World J Gastroenterol. 2019;25:3392-3407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 32] [Cited by in F6Publishing: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 12. | Di Narzo AF, Telesco SE, Brodmerkel C, Argmann C, Peters LA, Li K, Kidd B, Dudley J, Cho J, Schadt EE, Kasarskis A, Dobrin R, Hao K. High-Throughput Characterization of Blood Serum Proteomics of IBD Patients with Respect to Aging and Genetic Factors. PLoS Genet. 2017;13:e1006565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Lee HS, Cleynen I. Molecular Profiling of Inflammatory Bowel Disease: Is It Ready for Use in Clinical Decision-Making? Cells. 2019;8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Feng H, Gu ZY, Li Q, Liu QH, Yang XY, Zhang JJ. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res. 2019;12:35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 15. | Dalmer TRA, Clugston RD. Gene ontology enrichment analysis of congenital diaphragmatic hernia-associated genes. Pediatr Res. 2019;85:13-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Treister A, Pico AR. Identifier Mapping in Cytoscape. F1000Res. 2018;7:725. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Zhang X, Wang Y, Ma Z, Liang Q, Tang X, Hu D, Tan H, Xiao C, Gao Y. Tanshinone IIA ameliorates dextran sulfate sodium-induced inflammatory bowel disease via the pregnane X receptor. Drug Des Devel Ther. 2015;9:6343-6362. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 18. | Kühl AA, Pawlowski NN, Grollich K, Loddenkemper C, Zeitz M, Hoffmann JC. Aggravation of intestinal inflammation by depletion/deficiency of gammadelta T cells in different types of IBD animal models. J Leukoc Biol. 2007;81:168-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Molodecky NA, Soon IS, Rabi DM, Ghali WA, Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema HW, Kaplan GG. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46-54.e42; quiz e30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3134] [Cited by in F6Publishing: 3261] [Article Influence: 271.8] [Reference Citation Analysis (1)] |
| 20. | Walsh AJ, Bryant RV, Travis SP. Current best practice for disease activity assessment in IBD. Nat Rev Gastroenterol Hepatol. 2016;13:567-579. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 139] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 21. | Feigenbaum LZ, Lee D, Ho J. Routine testing of folate levels in geriatric assessment for dementia. J Am Geriatr Soc. 1988;36:755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 24] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 22. | Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991-D995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4527] [Cited by in F6Publishing: 5680] [Article Influence: 473.3] [Reference Citation Analysis (0)] |
| 23. | Singh UP, Singh NP, Murphy EA, Price RL, Fayad R, Nagarkatti M, Nagarkatti PS. Chemokine and cytokine levels in inflammatory bowel disease patients. Cytokine. 2016;77:44-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 195] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 24. | Trivedi PJ, Adams DH. Chemokines and Chemokine Receptors as Therapeutic Targets in Inflammatory Bowel Disease; Pitfalls and Promise. J Crohns Colitis. 2018;12:S641-S652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 72] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Carlsen HS, Baekkevold ES, Johansen FE, Haraldsen G, Brandtzaeg P. B cell attracting chemokine 1 (CXCL13) and its receptor CXCR5 are expressed in normal and aberrant gut associated lymphoid tissue. Gut. 2002;51:364-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 122] [Cited by in F6Publishing: 123] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Steele CW, Karim SA, Leach JDG, Bailey P, Upstill-Goddard R, Rishi L, Foth M, Bryson S, McDaid K, Wilson Z, Eberlein C, Candido JB, Clarke M, Nixon C, Connelly J, Jamieson N, Carter CR, Balkwill F, Chang DK, Evans TRJ, Strathdee D, Biankin AV, Nibbs RJB, Barry ST, Sansom OJ, Morton JP. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2016;29:832-845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 595] [Cited by in F6Publishing: 582] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 27. | Muthas D, Reznichenko A, Balendran CA, Böttcher G, Clausen IG, Kärrman Mårdh C, Ottosson T, Uddin M, MacDonald TT, Danese S, Berner Hansen M. Neutrophils in ulcerative colitis: a review of selected biomarkers and their potential therapeutic implications. Scand J Gastroenterol. 2017;52:125-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 112] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 28. | Farooq SM, Stillie R, Svensson M, Svanborg C, Strieter RM, Stadnyk AW. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulfate-induced colitis. J Pharmacol Exp Ther. 2009;329:123-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 29. | Duarte-Neves J, Pereira de Almeida L, Cavadas C. Neuropeptide Y (NPY) as a therapeutic target for neurodegenerative diseases. Neurobiol Dis. 2016;95:210-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 30. | Wan CP, Lau BH. Neuropeptide Y receptor subtypes. Life Sci. 1995;56:1055-1064. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 41] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 31. | Hofmann S, Bellmann-Sickert K, Beck-Sickinger AG. Chemical modification of neuropeptide Y for human Y1 receptor targeting in health and disease. Biol Chem. 2019;400:299-311. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |