Published online Nov 28, 2019. doi: 10.3748/wjg.v25.i44.6495
Peer-review started: October 10, 2019
First decision: November 10, 2019
Revised: November 20, 2019
Accepted: November 23, 2019
Article in press: November 23, 2019
Published online: November 28, 2019
The human microRNA 375 (MIR375) is significantly downregulated in human colorectal cancer (CRC) and we have previously shown that MIR375 is a CRC-associated miRNA. The metadherin (MTDH) is a candidate target gene of MIR375.
To investigate the interaction and function between MIR375 and MTDH in human CRC.
A luciferase reporter system was used to confirm the effect of MIR375 on MTDH expression. The expression levels of MIR375 and the target genes were evaluated by quantitative RT-PCR (qRT-PCR), western blotting, or immunohistochemistry.
MTDH expression was found to be upregulated in human CRC tissues compared to that in healthy controls. We show that MIR375 regulates the expression of many genes involved in the MTDH-mediated signal transduction pathways [BRAF-MAPK and phosphatidylinositol-4,5-biphosphate-3-kinase catalytic subunit alpha (PIK3CA)-AKT] in CRC cells. Upregulated MTDH expression levels were found to inhibit NF-κB inhibitor alpha, which further upregulated NFKB1 and RELA expression in CRC cells.
Our findings suggest that suppressing MIR375 expression in CRC regulates cell proliferation and angiogenesis by increasing MTDH expression. Thus, MIR375 may be of therapeutic value in treating human CRC.
Core tip: The microRNA 375 (MIR375) is significantly downregulated in human colorectal cancer (CRC) tissues. In this study, we investigated that metadherin (MTDH) is a direct target gene of MIR375 and that MTDH expression levels were upregulated in CRC tissues. Upregulated MTDH expression levels were found to inhibit NF-κB inhibitor alpha expression, which further upregulated NFKB1 and RELA expression in CRC cells. MIR375 also regulate MTDH-mediated BRAF-MAPK and PIK3CA-AKT signal pathways in CRC cells. Consequently, MIR375 regulates cell proliferation, cell migration, and angiogenesis by suppressing MTDH expression in CRC progression.
- Citation: Han SH, Mo JS, Park WC, Chae SC. Reduced microRNA 375 in colorectal cancer upregulates metadherin-mediated signaling. World J Gastroenterol 2019; 25(44): 6495-6507
- URL: https://www.wjgnet.com/1007-9327/full/v25/i44/6495.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i44.6495
Colorectal cancer (CRC) is a common malignant tumor and is the third leading cause of cancer-related mortality worldwide[1,2]. The cause of CRC is multifactorial, which includes genetic variation as well as epigenetic factors[3]. Overall survival of patients with CRC has not much improved relative to significant advances in the management of CRC[4]. Thus, it is most importance to understand the molecular mechanisms underlying CRC tumorigenesis and recognize the fundamental genes responsible for such fatal cancer.
MicroRNAs (miRNAs) are endogenously expressed, small noncoding RNAs that bind at the 3′ untranslated region (3’-UTR) of their target mRNAs and promote mRNA degradation or inhibit translation[5]. miRNAs act as tumor suppressors or oncogenes by targeting the genes involved in cell proliferation, cell survival, apoptosis, and metastasis[6-8].
In humans, microRNA 375 (MIR375) is located on chromosomal band 2q35. MIR375 has been shown to have dual functions: As a tumor suppressor[9,10] and as an oncogene[11,12]. The dual characteristic of MIR375 depends on the target mRNA. In our previous study, we detected MIR375 in CRC[13] and dextran sulphate sodium (DSS)-induced mice colitis[14] via miRNA expression profiling of CRC tissues versus healthy colorectal tissues and DSS-induced colitis versus healthy colons, respectively. We found that MIR375 was significantly downregulated in both CRC and DSS-induced colitis tissue samples[13,14]. Additionally, we have shown that downregulation of MIR375 modulates epidermal growth factor receptor (EGFR) signaling pathways in human CRC cells and tissues by upregulating connective tissue growth factor (CTGF) expression[15].
Metadherin (MTDH, also known as AEG1, LYRIC, or LYRIC/3D3) is located on chromosome 8q22.1 and encodes for a 64 kDa protein. It was first detected as an upregulated transcript in primary human fetal astrocytes infected with human immunodeficiency virus 1 (HIV-1)[16]. Brown and Ruoslahti have shown that metadherin mediates tumor cell localization at the metastatic sites[17]. Several studies have shown the role of MTDH as an oncogene in different types of human malignant tumors[18] and revealed various functions such as increased tumor growth, invasion and metastasis, angiogenesis, and chemoresistance[19]. Furthermore, our previous research has shown that MTDH is one of the putative target genes of MIR375[15].
In this study, we show that MTDH is a target gene of MIR375 in CRC and analyze its functions in CRC tissues and cell lines. Additionally, we reveal that MIR375 regulates cell proliferation and migration in CRC progression by suppressing MTDH-mediated signaling pathways.
The tissue samples used in this study were provided by Biobank of Wonkwang University Hospital, a member of National Biobank of Korea. On approval from the institutional review board and obtaining informed consent (WKIRB-201710-BR-012) from the patients, we collected 19 CRC tissue samples from 16 patients with colon cancer (10 males and 6 females) and 3 patients with rectal cancer (2 males and 1 female). Mean age of the patients with colon cancer and rectal cancer was 68.4 years and 67.0 years, respectively. Ten colon cancer tissue samples and matching healthy colon tissue samples (7 males and 3 females) were investigated to confirm the endogenous expression of MIR375. Additionally, 12 colon cancer tissue samples with matching healthy colon tissue samples and 3 rectal cancer tissue samples with matching healthy rectal tissue samples were assessed for MTDH expression levels. Four colon cancer tissue samples and matching healthy colon tissue samples (3 males and 1 female) were examined for immunohistochemistry analysis.
Human CRC cell lines; Caco2, HT29, LoVo, HCT116, and SW48 were obtained from Korea Cell Line Bank (KCLB, Seoul, South Korea) or American Type Culture Collection (ATCC, Manassas, VA, United States). SW48, HT29, Lovo, and HCT116 cells were cultured in RPMI 1640 (HyClone, Logan, UT, United States) supplemented with 10% FBS while Caco2 cells were cultured in α-MEM (HyClone, Logan, UT, United States) supplemented with 20% FBS in a humidified atmosphere containing 5% CO2 at 37 °C.
MTDH antibody and all secondary antibodies were purchased from Thermo Fisher Scientific (Waltham, MA, United States). NF-κB inhibitor alpha (NFKBIA/IκBα), nuclear factor κ B subunit 1 (NFκB1/p50), RELA (NFkB3/p65), protein kinase B (AKT), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies were obtained from Santa Cruz Biotechnology, Inc. (Dallas, Texas, United States). RAS, BRAF, p44/42 mitogen-activated protein kinase (MAPK) (Erk1/2), phospho-MAPK, phosphatidylinositol-4,5-biphosphate-3-kinase catalytic subunit alpha (PIK3CA), phospho-AKT, and GAPDH antibodies were purchased from Cell Signaling Technology (Danvers, MA, United States). β catenin (CTNNB1) antibody was purchased from Abcam (Cambridge, United Kingdom). Vascular endothelial growth factor A (VEGFA) antibody was purchased from Novus Biologicals (Centennial, CO, United States). Ez-cytox was obtained from DoGenBio (Seoul, South Korea) and dual luciferase reporter assay system was obtained from Promega (Madison, WI, United States). TRIzol and siPORT NeoFx transfection reagents were purchased from Ambion, Inc. (Waltham, MA, United States). Lipofectamine 2000 reagent was purchased from Invitrogen (Waltham, MA, United States) while Viromer blue transfection agent was purchased from Lipocalyx (Weinbergweg, Halle, Germany). RIPA buffer was obtained from Elpis biotech (Daejeon, South Korea) and DAB substrate kit was purchased from Pierce Biotechnology (Waltham, MA, United States).
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR) were performed according to our previously established protocol[13-15]. Total RNA was isolated using TRIzol reagent. After digesting with DNase and performing a sample clean-up, RNA samples were quantified, aliquoted, and stored at -80 °C. qRT-PCR was performed on total RNA samples that were isolated from tissue samples or cultured cells to synthesize cDNA using StepOne Real-time PCR system (Applied Biosystems, Foster City, CA, United States).
Differential miRNA expression patterns were validated by TaqMan qRT-PCR assay (Applied Biosystems, Foster City, CA, United States). qRT-PCR was performed using SYBR Green dye (ELPIS Biotech, Daejeon, Korea) to assess mRNA expression. RNU48 (for TaqMan qRT-PCR) or 5.8S (for SYBR qRT-PCR), and GAPDH served as endogenous controls for qRT-PCR of miRNA and mRNA, respectively. Each sample was analyzed in triplicates by qRT-PCR. Primers for qRT-PCR and TaqMan analysis are listed in Supplementary Table 1.
Endogenous MIR375 mimic [hsa-miR-375, Pre-miR™ miRNA precursor (AM17100)], MTDH small interfering RNA (siRNA), and each of the negative controls were synthesized commercially (Ambion, Austin, TX, United States) and transfected at 50 nM. Transfection was performed according to our previously published protocols[13-15].
Wild-type (WT) or mutant type (MT) fragments of the 3’-UTR of MTDH containing the predicted binding site for MIR375 were amplified using PCR. The primer set used for the experiment is shown in Supplementary Table 1. Plasmid constructions and analysis of the luciferase assay were executed following our previously published protocols[13-15].
Protein extraction and western blot analysis were performed according to our earlier established methods[13-15]. Briefly, membranes were incubated overnight at 4 °C with primary antibodies to MTDH (1:250), NFKBIA (1:100), NFKB1 (1:50), RELA (1:100), KRAS (1:1000), BRAF (1:500), MAPK (1:1000), p-MAPK (1:500), CTNNB1 (1:2500), PIK3CA (1:1000), AKT (1:100), p-AKT (1:500), and VEGFA (1:500). Subsequently, the membranes were incubated with secondary antibodies (1:1000).
For cell proliferation assay, cells (2 × 104 cells/well) were transfected with MIR375 mimic, negative control siRNA, or MTDH siRNA (siMTDH) in 96-well plates. Cell growth was measured at 72 h after transfection using Ez-Cytox cell viability assay kit following manufacturer’s instructions. After incubating for 2 h, absorbance values were measured at 450 nm using SpectraMax (Molecular Devices, CA, United States). Percentage of viable cells was calculated by comparing to the number of viable cells in the untreated controls. Experiments were performed in triplicates. Cell proliferation assay was performed following our previously published protocols[15,20].
Immunohistochemistry assay was performed according to our previously established protocols[14,15]. The tissue slides were blocked with 3% BSA for 2 h at room temperature followed by overnight incubation at 4 °C with primary antibodies against MTDH (1:50) and RELA (1:50). The following day, the slides were incubated with SignalStain® Boost IHC detection reagent (Cell Signaling Technology; Danvers, MA, United States) for 2 h at room temperature. After washing, chromogenic substrate (Thermo Fisher Scientific; Waltham, MA, United States) was applied to visualize the staining of the target proteins. Following counterstaining with hematoxylin, the sections were dehydrated and mounted using a coverslip.
Sample size was estimated using the G*power software (Version 3.1., Heinrich Heine University, Duesseldorf, Germany). Each experiment was repeated at least three times and consistent results were obtained. Data are expressed as mean ± standard deviation (SD). The differences between the groups were evaluated using GraphPad Prism 5.0 statistical software (GraphPad Software Inc., San Diego, CA, United States) or Student’s t-test. Differences with P value less than 0.05 were considered as statistically significant.
Previously, we have shown MIR375 as a colon cancer-associated miRNA using miRNA microarray analysis of colon tumor tissues and matched healthy colon tissues[13]. Additionally, we have shown that MIR375 expression is downregulated in human CRC tissues. To confirm the result, we compared MIR375 expression in 10 human CRC tissues and matched healthy colon tissues by qRT-PCR. We found that MIR375 expression levels were significantly reduced in CRC tissues (P < 0.01; Supplementary Figure 1A).
To determine the endogenous expression levels of MIR375 in different cell lines, we performed qRT-PCR on the total RNA isolated from various cell lines including Caco2, SW480, HT29, HCT116, LoVo, and SW48 cells. As shown in Figure S1B, MIR375 expression level was highest in HT29 cells while it was lower in HCT116 and Caco2 cells (Supplementary Figure 1B).
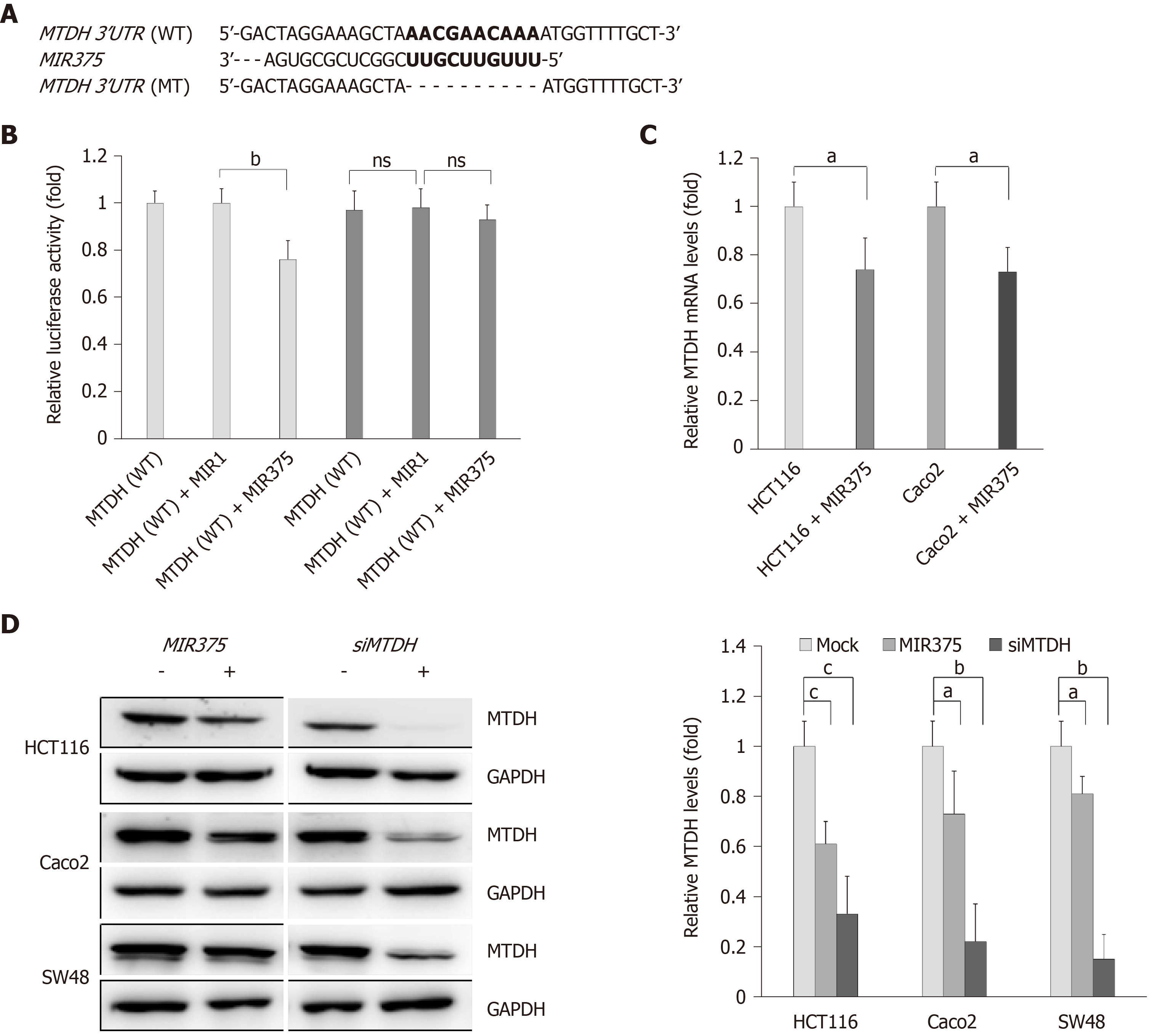
To determine the direct interaction between MTDH 3’-UTR and MIR375, we cloned the WT MTDH 3’-UTR region, the putative target sequence of MIR375, in a luciferase reporter vector (Figure 1A). We observed that luciferase activity was reduced by approximately 24% when cells were co-transfected with pre-MIR375 (P < 0.01, Figure 1B). As a control experiment, we cloned mutated MTDH 3’-UTR sequence which lacked ten of the total complementary bases (Figure 1A). As expected, repression of the luciferase activity was revoked when the interaction between MIR375 and its target 3’-UTR was disrupted (Figure 1B). Additionally, another control experiment was performed where pre-MIR1 (instead of pre-MIR375) was co-transfected with WT and mutated MTDH 3’-UTR constructs. We found that transfection with pre-MIR1 did not affect the luciferase activity of either of the constructs (Figure 1B).
To validate the obtained data, we investigated whether MIR375 regulates MTDH mRNA levels in HCT116 and Caco2 cells. We found that MTDH mRNA expression levels were lower in HCT116 as well as Caco2 cells on transfection with MIR375 mimic compared with that in non-transfected control cells (P < 0.05; Figure 1C). Additionally, we investigated MTDH expression levels in MIR375 mimic or siMTDH-transfected HCT116, Caco2, and SW48 cells and found that cellular MTDH expression was significantly reduced in MIR375-overexpressing HCT116, Caco2, and SW48 cells. Furthermore, MTDH was significantly downregulated by siMTDH transfection (Figure 1D).
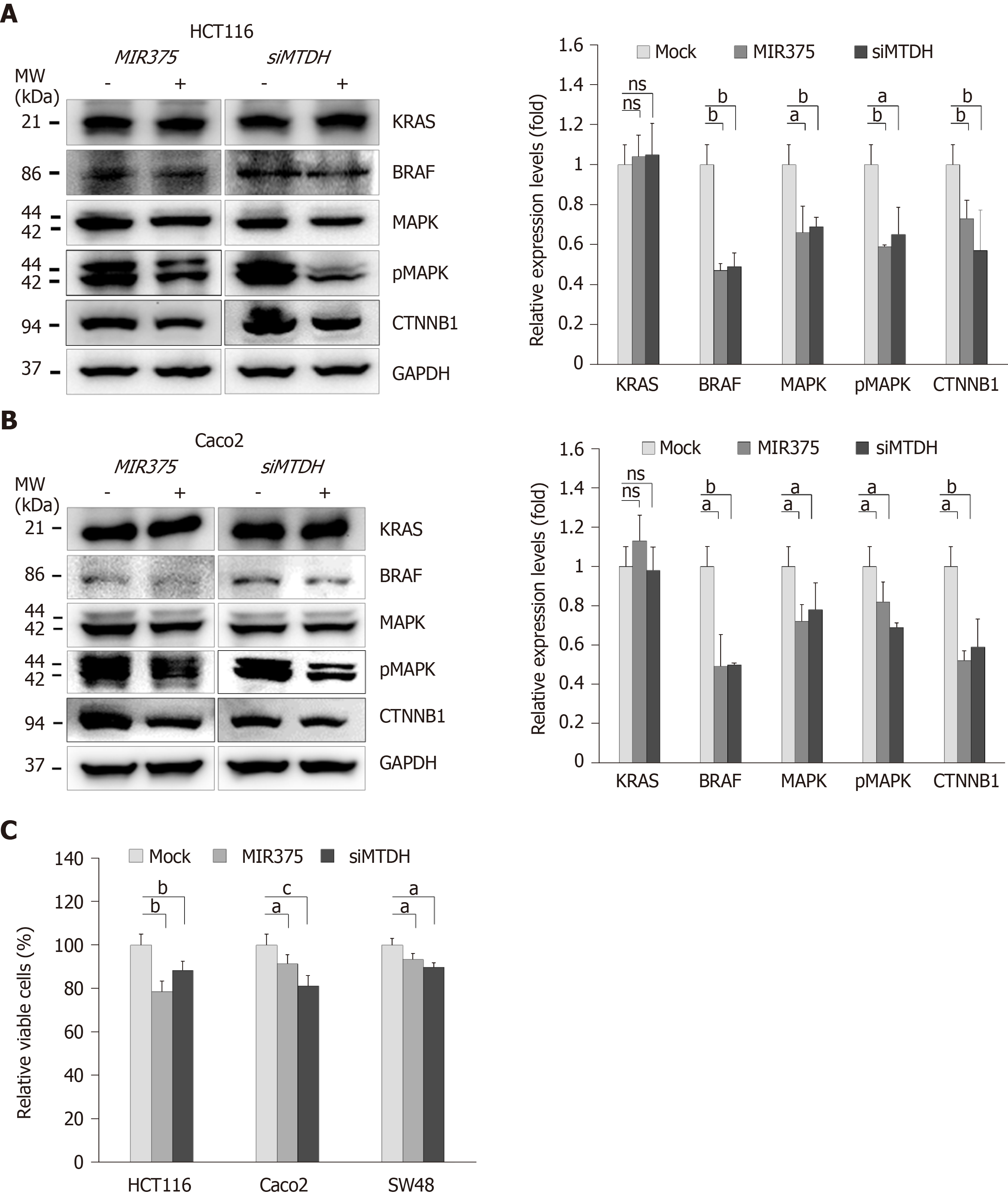
To determine the functional interaction between MIR375 and its target gene MTDH, we analyzed the expression levels of KRAS, BRAF, MAPK, pMAPK, and CTNNB1 in HCT116 and Caco2 cells on MIR375 mimic transfection. Earlier study has shown that HCT116 cells express WT BRAF and mutated KRAS while Caco2 cells express only WT KRAS and WT BRAF[21]. Although KRAS expression level was unaltered on MIR375 transfection, BRAF, MAPK, pMAPK, and CTNNB1 expression levels were significantly downregulated in HCT116 (Figure 2A) and Caco2 (Figure 2B) cells. We observed a similar expression trend in CRC cells on silencing MTDH with siMTDH (Figure 2A and B). These results suggested that MIR375 regulates the MTDH-mediated BRAF-MAPK signal pathway in CRC cells.
We investigated the biological functions of MIR375 in CRC cells. MTT assay showed that cell viability was steadily reduced on MIR375 transfection in the CRC cell lines HCT116 (P < 0.01), Caco2 (P < 0.05), and SW48 (P < 0.05; Figure 2C) cells. Further, we found a similar trend on siMTDH transfection in HCT116 (P < 0.01), Caco2 (P < 0.001), and SW48 cells (P < 0.05; Figure 2C). These results suggested that MIR375 downregulates CRC cell proliferation by inhibiting MTDH expression. The rate of inhibition was lower in SW48 cells compared with that in HCT116 and Caco2 cells. This might be due to relatively high endogenous expression of MIR375 in SW48 cells than that observed in HCT116 and Caco2 cells (Supplementary Figure 1B).
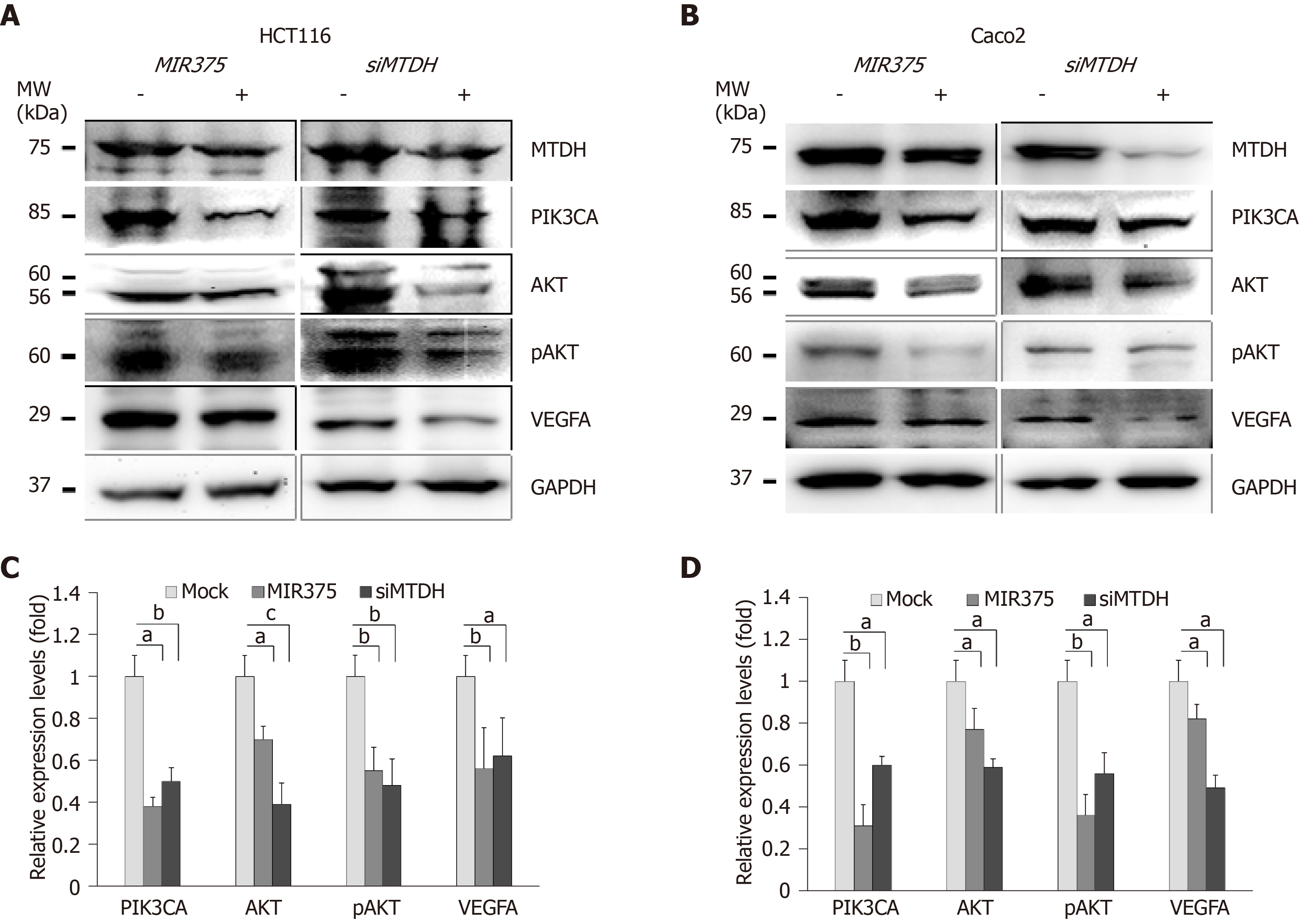
We investigated the functional correlation between MIR375 and MTDH expression in HCT116 and Caco2 cells. HCT116 cells are mutated for PIK3CA whereas Caco2 cells express wild type PIK3CA. We found that PIK3CA, AKT, pAKT, and VEGFA expression levels were downregulated on MIR375 mimic transfection in HCT116 as well as Caco2 cells (Figure 3A and 3B). Similar results were obtained on silencing MTDH in HCT116 and Caco2 cells (Figure 3C and 3D). Thus, the results suggested that MIR375 regulates MTDH-mediated PIK3CA-AKT signaling pathway by inhibiting MTDH expression levels in CRC cells.
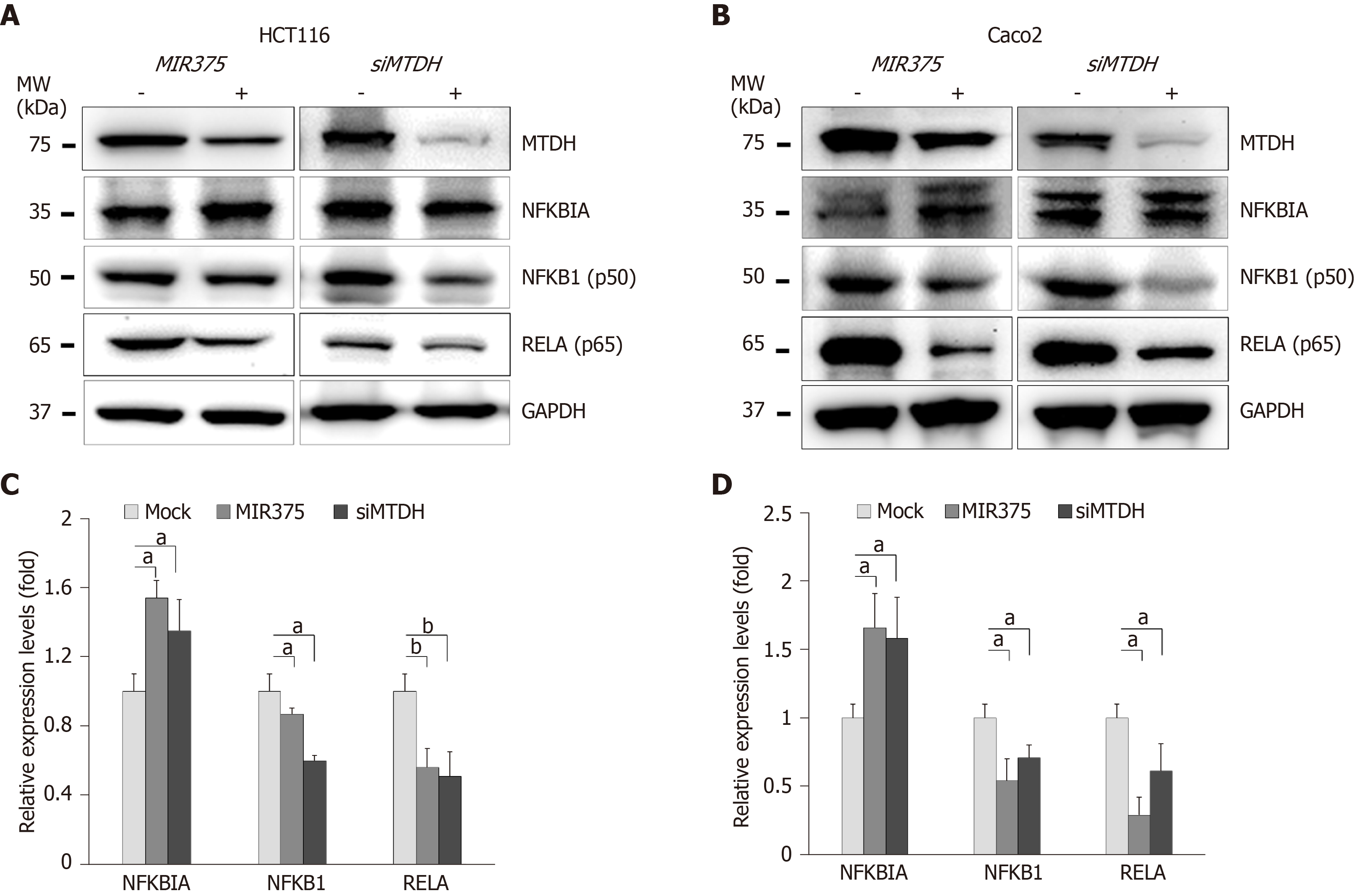
Furthermore, we investigated another MTDH-mediated signaling pathway in CRC cells. To determine the role of MIR375 or MTDH-mediated pathways in NFKB1 signaling, we quantified the expression of relevant proteins in HCT116 and Caco2 cells on MIR375 mimic or siMTDH transfection. NFKB1 and RELA expression levels were found to be significantly downregulated in both the cell lines (HCT116 and Caco2 cells) on MIR375 transfection while NFKBIA expression levels were found to be upregulated (Figure 4A and B). We observed similar results on silencing MTDH using siMTDH in CRC cells (Figure 4C and D). Overall, the results evidently suggested that NFKBIA expression was upregulated on inhibiting MTDH in MIR375-overexpressing CRC cells which further leads to downregulation of NFKB1 and RELA expression in CRC cells. These results showed that MIR375 regulates MTDH-mediated NFKB1 signaling pathway.
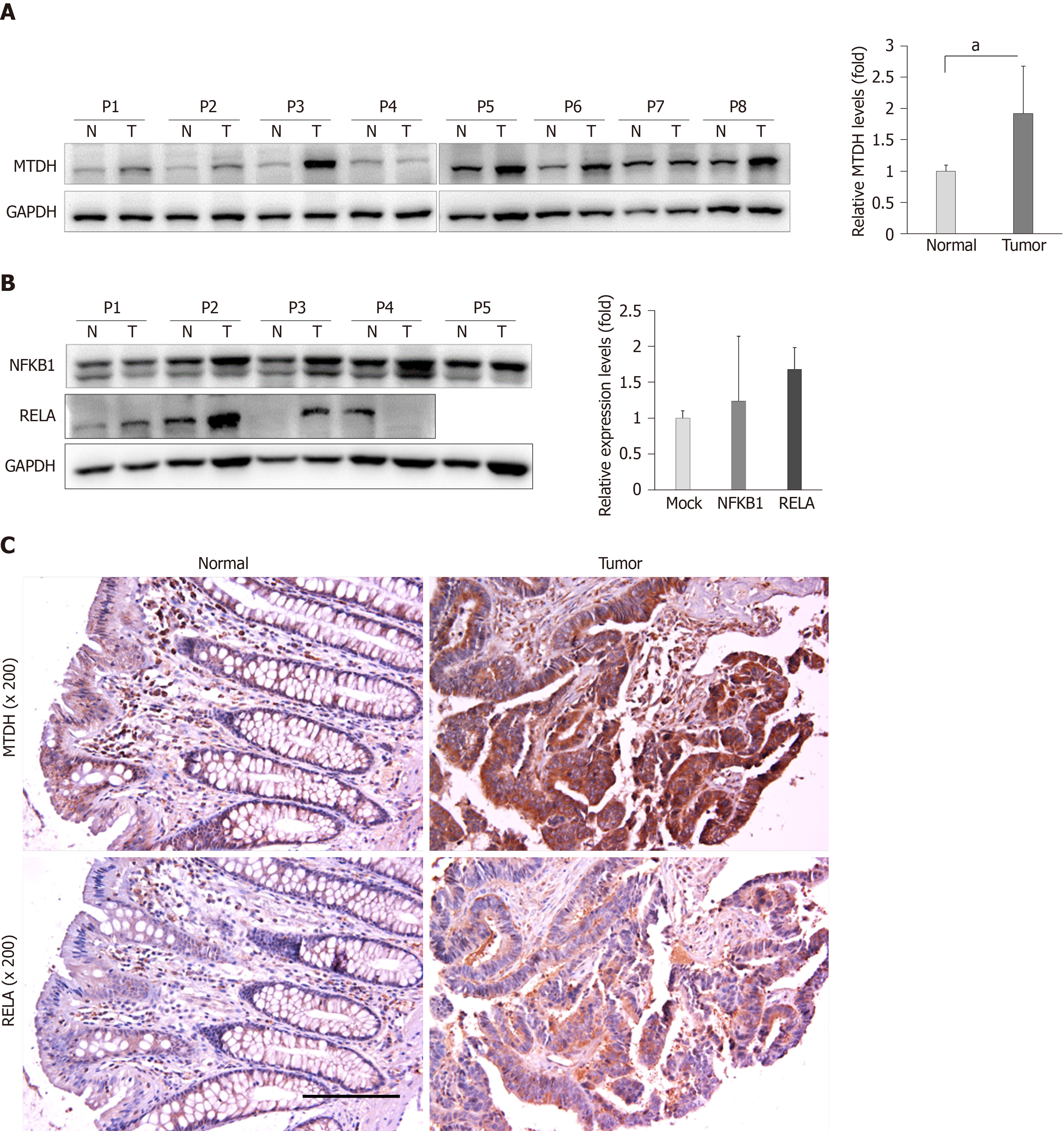
Based on the findings of this study, we evaluated MTDH expression in 15 human CRC tissues and matching healthy colon tissues. Western blot analysis showed that MTDH expression levels were upregulated (12 out of 15 samples) in CRC tissue samples compared with that in healthy colon tissues (P < 0.05, Figure 5A). Further, we investigated NFKB1 and RELA expression levels in five CRC tissues and four CRC tissue samples, respectively. NFKB1 expression level was significantly upregulated in all CRC tissues while RELA expression level was predominantly upregulated in three (75%) CRC tissues (Figure 5B).
Consistent with the results obtained, we investigated MTDH and RELA expression in four human CRC tissues and matching healthy colon tissues using immunohistochemical analysis. MTDH and RELA expression levels were significantly upregulated in CRC tissues (Figure 5C).
Many miRNAs have been detected as associated biomarkers and therapeutic targets in CRC. Several studies have shown that targeting specific miRNAs effectively inhibits cell proliferation and angiogenesis in CRC[22-24]. Thus, a better identification of CRC-associated miRNAs may contribute to the development of efficient miRNA-based therapy for CRC. In our previous study, we used miRNA expression profiling and showed MIR375 to be associated with human CRC tissue[13] as well as DSS-induced mice colitis[14] by comparing the expression pattern in CRC tissues versus matching healthy colorectal tissues and DSS-induced mice colitis versus healthy mice colons, respectively. Hyper-methylation of MIR375 has been demonstrated in CRC cell lines including HCT116 and SW480. Down regulation of MIR375 in HCT116 and SW480 cells compere to HT29 cells is due to hyper-methylation of MIR375 in HCT116 and SW480 cells[25]. In the present study, we confirmed the findings in a larger sample size and showed that MIR375 expression was downregulated in human CRC tissues compared with that in matching healthy colon tissues (Supplementary Figure 1A). The results of the current study are consistent with the earlier research work by Dai et al[26]. In addition to that in our study, MIR375 downregulation has been observed in several other types of cancer such as hepatocellular carcinoma[27], gastric cancer[28], and glioma[29]. However, some reports have revealed that MIR375 is upregulated in tumors of prostate cancer[12] and breast cancer[30]. Primarily, miRNA expression levels were believed to be cell type-specific in many cancer tissues[31].
In our previous study, we found that MIR375 regulates the CTGF-mediated EGFR-PIK3CA-AKT signaling pathway by directly downregulating CTGF expression in CRC cells. However, CTGF is not involved in EGFR-KRAS-BRAF-ERK1/2 signaling[15]. Although KRAS expression was unaffected by MIR375 overexpression, BRAF-ERK1/2 signaling was regulated on MIR375 overexpression in CRC cells[15]. These results guided us to investigate a novel MIR375 target gene that mediates the BRAF-ERK1/2 signaling pathway in CRC cells. In this study, we found that MTDH is a direct target gene of MIR375 in CRC cells (Figure 1). Further, we confirmed that MIR375 regulates the MTDH-mediated BRAF-ERK1/2 (MAPK3/1) signaling pathway (Figure 2A and 2B), which controls proliferation in CRC cells (Figure 2C). It is well known that MTDH contributes to the carcinogenic process of different tissues and organs by regulating multiple signaling pathways such as PI3K/AKT, NF-kB, and MAPK, which subsequently promotes tumorigenesis and metastasis[32,33].
MTDH promotes an invasive phenotype and angiogenesis via the PIK3CA-AKT signaling pathway. In addition, PIK3CA has been proven as a direct target of MIR375 in CRC cells[34]. MTDH expression is upregulated in many types of cancers, and is crucial in oncogenic transformation and angiogenesis[35-37]. The potential role of MTDH in angiogenesis has been correlated with VEGFA expression via the PIK3CA-AKT pathway in head and neck squamous cells[38]. Thus, PIK3CA-AKT-VEGFA signaling is affected on MIR375 overexpression or on VEGFA silencing (siVEGFA). We showed that PIK3CA-AKT-VEGFA signaling is downregulated on MIR375 overexpression and siVEGFA treatment in CRC cells (Figure 3). Our previous study using xenograft mouse model showed that the expression level of the angiogenic marker, CD31 was significantly decreased in xenograft tumors on MIR375 overexpression[15]. Consequently, these results suggest that MIR375 regulates angiogenesis via the MTDH-mediated PIK3CA-AKT-VEGFA signaling pathway in CRC progression.
Furthermore, MTDH has been shown to regulate the anchorage independent growth and invasion of HeLa cells via activation of the NF-κB pathway[35]. MTDH has also been shown to be upregulated during CRC development and liver metastasis through the NF-κB signaling pathway[39]. Similarly, in malignant glioma cells, MTDH has been found to mediate invasion and migration through activation of the NF-κB signaling pathway[40]. In this study, we showed that MIR375 regulates MTDH-mediated NFKB1 and RELA signaling by inhibiting NFKBIA expression in CRC cells (Figure 4).
In this study, MIR375 expression levels were downregulated in CRC tissues (Supplementary Figure 1A). Inversely, MTDH, NFKB1, and RELA expression levels were predominantly upregulated in CRC tumor tissues compared to their expression levels in matched healthy colon tissues (Figure 5A). Additionally, immunohistochemistry staining of the CRC tissues showed that MTDH and RELA expression were upregulated in CRC tumor tissues. Overall, these results suggested that MTDH expression levels were negatively correlated with MIR375 expression in CRC tissues.
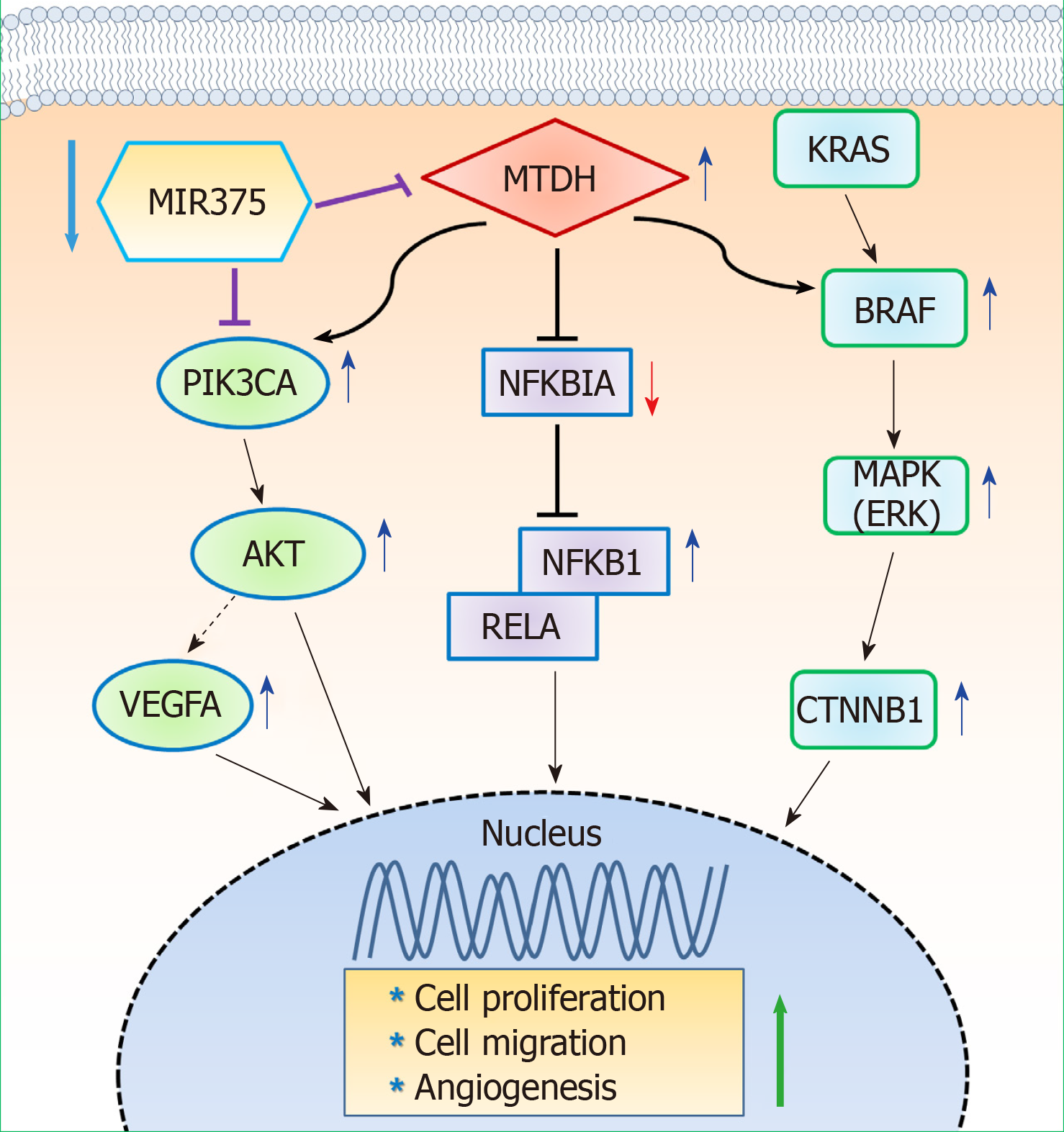
In summary, our study found that MIR375 expression is suppressed in tissues of patients with CRC and that MTDH is a direct target of MIR375. Furthermore, MTDH expression was upregulated in the tumors of CRC tissues on inhibiting MIR375 expression. Overall, our results suggest that MIR375 regulates MTDH-mediated signaling pathways such as MTDH-BRAF-MAPK, MTDH-PIK3CA-AKT, and MTDH-NFKBIA-NFKB1/RELA in CRC progression. Although we did not show MIR375-mediated VEGFA-VEGFR signaling in this study, our previous and present studies suggest that the generated VEGFA by MIR375-mediated PIK3CA-AKT or MTDH-PIK3CA-AKT signaling might effect to endothelial cell’s angiogenesis. Subsequently, MIR375 regulates cell proliferation, cell migration, and angiogenesis in CRC progression (Figure 6). Thus, we propose MIR375 to be a promising therapeutic target in inhibiting CRC tumorigenesis. However, this needs to be further investigated.
Colorectal cancer (CRC) is the third most prevalent type of cancer worldwide. The cause of CRC is multifactorial including genetic variation and epigenetic and environmental factors. However, the precise molecular mechanism underlying the development and progression of CRC remains largely unknown. We previously found that microRNA 375 (MIR375) is significantly downregulated in CRC, and identified metadherin (MTDH) as a candidate target gene of MIR375.
MIR375 and their target MTDH will provide a new therapeutic information for human CRC.
To study the interaction and signaling between MIR375 and MTDH in human CRC pathogenesis.
We constructed luciferase reporter plasmids to confirm the effect of MIR375 on MTDH gene expression. The expression levels of the MIR375 and MTDH were measured by qRT-PCR, Western blot, or immunohistochemistry. The effects of MIR375 on cell growth and angiogenesis were conducted by functional experiments in CRC cells. Assays were performed to explore functional correlation between MTDH and MIR375 in human CRC cells and tissues.
In the present study, we found that the expression levels of MTDH were significantly down-regulated in CRC cells by MIR375 mimic or siMTDH transfection. MTDH expression was up-regulated in human CRC tissues in comparing to match normal colon tissues. Upregulated MTDH expression levels were found to inhibit NF-κB inhibitor alpha (NFKBIA) expression, which further upregulated NFKB1 and RELA expression. We found that MIR375 regulate the expression levels of molecules in MTDH-mediated BRAF-MAPK and PIK3CA-AKT signal pathways in CRC cells.
MIR375 regulates cell proliferation and angiogenesis by regulation of MTDH-mediated signaling pathways such as MTDH-BRAF-MAPK, MTDH-PIK3CA-AKT, and MTDH-NFKBIA-NFKB1/RELA in CRC progression.
This study provides insight into the role of MIR375 in CRC pathogenesis by targeting MTDH. MIR375 might be a new therapeutic target for CRC.
Biospecimens used in this study were provided by Biobank of Wonkwang University Hospital, a member of the National Biobank of Korea which is supported by Ministry of Health and Welfare.
Manuscript source: Invited Manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: South Korea
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Donato R, Tarnawski AS, Yu B S-Editor: Wang J L-Editor: A E-Editor: Qi LL
| 1. | Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2582] [Cited by in F6Publishing: 2409] [Article Influence: 104.7] [Reference Citation Analysis (0)] |
| 2. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13286] [Cited by in F6Publishing: 13414] [Article Influence: 706.0] [Reference Citation Analysis (1)] |
| 3. | Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1163] [Cited by in F6Publishing: 1197] [Article Influence: 92.1] [Reference Citation Analysis (1)] |
| 4. | Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449-2460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1274] [Cited by in F6Publishing: 1304] [Article Influence: 86.9] [Reference Citation Analysis (2)] |
| 5. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25833] [Cited by in F6Publishing: 26768] [Article Influence: 1338.4] [Reference Citation Analysis (0)] |
| 6. | Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1568] [Cited by in F6Publishing: 1543] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 7. | Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-688. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1936] [Cited by in F6Publishing: 1950] [Article Influence: 114.7] [Reference Citation Analysis (0)] |
| 8. | Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 470] [Cited by in F6Publishing: 499] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 9. | Jung HM, Patel RS, Phillips BL, Wang H, Cohen DM, Reinhold WC, Chang LJ, Yang LJ, Chan EK. Tumor suppressor miR-375 regulates MYC expression via repression of CIP2A coding sequence through multiple miRNA-mRNA interactions. Mol Biol Cell. 2013;24:1638-1648, S1-S7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 10. | Wang F, Li Y, Zhou J, Xu J, Peng C, Ye F, Shen Y, Lu W, Wan X, Xie X. miR-375 is down-regulated in squamous cervical cancer and inhibits cell migration and invasion via targeting transcription factor SP1. Am J Pathol. 2011;2580-2588. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 172] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 11. | Giricz O, Reynolds PA, Ramnauth A, Liu C, Wang T, Stead L, Childs G, Rohan T, Shapiro N, Fineberg S, Kenny PA, Loudig O. Hsa-miR-375 is differentially expressed during breast lobular neoplasia and promotes loss of mammary acinar polarity. J Pathol. 2012;226:108-119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Szczyrba J, Nolte E, Wach S, Kremmer E, Stöhr R, Hartmann A, Wieland W, Wullich B, Grässer FA. Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol Cancer Res. 2011;9:791-800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 13. | Mo JS, Alam KJ, Kang IH, Park WC, Seo GS, Choi SC, Kim HS, Moon HB, Yun KJ, Chae SC. MicroRNA 196B regulates FAS-mediated apoptosis in colorectal cancer cells. Oncotarget. 2015;6:2843-2855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 14. | Mo JS, Alam KJ, Kim HS, Lee YM, Yun KJ, Chae SC. MicroRNA 429 Regulates Mucin Gene Expression and Secretion in Murine Model of Colitis. J Crohns Colitis. 2016;10:837-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 15. | Alam KJ, Mo JS, Han SH, Park WC, Kim HS, Yun KJ, Chae SC. MicroRNA 375 regulates proliferation and migration of colon cancer cells by suppressing the CTGF-EGFR signaling pathway. Int J Cancer. 2017;141:1614-1629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 16. | Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao W, Volsky DJ, Fisher PB. Identification and cloning of human astrocyte genes displaying elevated expression after infection with HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid subtraction hybridization, RaSH. Oncogene. 2002;21:3592-3602. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 269] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 17. | Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365-374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 304] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 18. | Ying Z, Li J, Li M. Astrocyte elevated gene 1: biological functions and molecular mechanism in cancer and beyond. Cell Biosci. 2011;1:36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Emdad L, Sarkar D, Su ZZ, Randolph A, Boukerche H, Valerie K, Fisher PB. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 2006;66:1509-1516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 234] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 20. | Mo JS, Han SH, Yun KJ, Chae SC. MicroRNA 429 regulates the expression of CHMP5 in the inflammatory colitis and colorectal cancer cells. Inflamm Res. 2018;67:985-996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 21. | Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis. 2013;2:e71. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 525] [Cited by in F6Publishing: 595] [Article Influence: 54.1] [Reference Citation Analysis (0)] |
| 22. | Li Y, Lauriola M, Kim D, Francesconi M, D'Uva G, Shibata D, Malafa MP, Yeatman TJ, Coppola D, Solmi R, Cheng JQ. Adenomatous polyposis coli (APC) regulates miR17-92 cluster through β-catenin pathway in colorectal cancer. Oncogene. 2016;35:4558-4568. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Ji S, Ye G, Zhang J, Wang L, Wang T, Wang Z, Zhang T, Wang G, Guo Z, Luo Y, Cai J, Yang JY. miR-574-5p negatively regulates Qki6/7 to impact β-catenin/Wnt signalling and the development of colorectal cancer. Gut. 2013;62:716-726. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 99] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Liang L, Gao C, Li Y, Sun M, Xu J, Li H, Jia L, Zhao Y. miR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8:e2968. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 25. | Christensen LL, Holm A, Rantala J, Kallioniemi O, Rasmussen MH, Ostenfeld MS, Dagnaes-Hansen F, Øster B, Schepeler T, Tobiasen H, Thorsen K, Sieber OM, Gibbs P, Lamy P, Hansen TF, Jakobsen A, Riising EM, Helin K, Lubinski J, Hagemann-Madsen R, Laurberg S, Ørntoft TF, Andersen CL. Functional screening identifies miRNAs influencing apoptosis and proliferation in colorectal cancer. PLoS One. 2014;9:e96767. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H. Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep. 2012;5:1299-1304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, Li PY, Song YH, Lin JS. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012;31:3357-3369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 277] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 28. | Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J, Zhou T. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784-793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 275] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 29. | Chang C, Shi H, Wang C, Wang J, Geng N, Jiang X, Wang X. Correlation of microRNA-375 downregulation with unfavorable clinical outcome of patients with glioma. Neurosci Lett. 2012;531:204-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, Malekpour F, Volinia S, Croce CM, Najmabadi H, Diederichs S, Sahin O, Mayer D, Lyko F, Hoheisel JD, Riazalhosseini Y. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175-9184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 216] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 31. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4162] [Cited by in F6Publishing: 4420] [Article Influence: 245.6] [Reference Citation Analysis (0)] |
| 32. | Emdad L, Das SK, Dasgupta S, Hu B, Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: signaling pathways, downstream genes, interacting proteins, and regulation of tumor angiogenesis. Adv Cancer Res. 2013;120:75-111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 33. | Huang Y, Li LP. Progress of cancer research on astrocyte elevated gene-1/Metadherin (Review). Oncol Lett. 2014;8:493-501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 34. | Wang Y, Tang Q, Li M, Jiang S, Wang X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun. 2014;444:199-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 35. | Emdad L, Lee SG, Su ZZ, Jeon HY, Boukerche H, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) functions as an oncogene and regulates angiogenesis. Proc Natl Acad Sci USA. 2009;106:21300-21305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 36. | Li C, Li R, Song H, Wang D, Feng T, Yu X, Zhao Y, Liu J, Yu X, Wang Y, Geng J. Significance of AEG-1 expression in correlation with VEGF, microvessel density and clinicopathological characteristics in triple-negative breast cancer. J Surg Oncol. 2011;103:184-192. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 37. | Long M, Dong K, Gao P, Wang X, Liu L, Yang S, Lin F, Wei J, Zhang H. Overexpression of astrocyte-elevated gene-1 is associated with cervical carcinoma progression and angiogenesis. Oncol Rep. 2013;30:1414-1422. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Zhu GC, Yu CY, She L, Tan HL, Li G, Ren SL, Su ZW, Wei M, Huang DH, Tian YQ, Su RN, Liu Y, Zhang X. Metadherin regulation of vascular endothelial growth factor expression is dependent upon the PI3K/Akt pathway in squamous cell carcinoma of the head and neck. Medicine (Baltimore). 2015;94:e502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | Gnosa S, Shen YM, Wang CJ, Zhang H, Stratmann J, Arbman G, Sun XF. Expression of AEG-1 mRNA and protein in colorectal cancer patients and colon cancer cell lines. J Transl Med. 2012;10:109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 40. | Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478-1484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 193] [Article Influence: 12.1] [Reference Citation Analysis (0)] |