Published online Jul 14, 2019. doi: 10.3748/wjg.v25.i26.3408
Peer-review started: March 19, 2019
First decision: May 9, 2019
Revised: June 5, 2019
Accepted: June 7, 2019
Article in press: June 8, 2019
Published online: July 14, 2019
Different histological growth patterns (HGPs) of colorectal carcinoma (CRC) liver metastasis are associated with patients’ prognosis and response to antiangiogenic therapy. However, the relationship between HGPs of liver metastasis and clinicopathological and genomic characteristics of primary cancer has not been well established.
To assess whether certain clinicopathological and genomic features of primary CRC could predict the HGPs of liver metastasis.
A total of 29 patients with paired resections of both primary CRC and liver metastasis were divided into two groups: A (15 cases with desmoplastic liver metastasis) and B (14 cases with replacement liver metastasis). Clinical information was obtained from patients’ charts. Mismatch repair proteins, BRAFV600E, and PD-L1 were evaluated by immunohistochemistry. Five cases were selected randomly from each group for whole exome sequencing (WES) analysis.
In the primary tumor, expanding growth pattern, low tumor budding score (TBS), and Crohn’s disease-like response (CDR) were associated with desmoplastic liver metastasis and better overall survival, whereas infiltrating growth pattern alone of primary carcinoma could predict the replacement liver metastasis and worse overall survival (P < 0.05). On WES analysis, primary carcinoma with desmoplastic liver metastasis showed mutations in APC (4/5); TP53 (3/5); KRAS, PIK3CA, and FAT4 (2/5); BRCA-1, BRCA2, BRAF, and DNAH5 (1/5), whereas primary carcinoma with replacement liver metastasis showed mutations in APC and TP53 (3/5); KRAS, FAT4, DNH5, SMAD, ERBB2, ERBB3, LRP1, and SDK1 (1/5).
The HGPs, TBS, and CDR of primary CRC as well as the presence of specific genetic mutations such as those in PIK3CA could be used to predict the HGPs of liver metastasis, response to therapy, and patients’ prognosis.
Core tip: Different histological growth patterns (HGPs) of colorectal carcinoma (CRC) liver metastasis are associated with patients’ prognosis and response to antiangiogenic therapy. The aim of our study was to assess whether certain clinicopathological and genomic features of primary CRC could predict the HGPs of liver metastasis. We found that the HGPs, tumor budding score, and Crohn’s disease-like response of primary CRC as well as the presence of specific genetic mutations such as those in PIK3CA could be used to predict the HGPs of liver metastasis, response to therapy, and patients’ prognosis.
- Citation: Wu JB, Sarmiento AL, Fiset PO, Lazaris A, Metrakos P, Petrillo S, Gao ZH. Histologic features and genomic alterations of primary colorectal adenocarcinoma predict growth patterns of liver metastasis. World J Gastroenterol 2019; 25(26): 3408-3425
- URL: https://www.wjgnet.com/1007-9327/full/v25/i26/3408.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i26.3408
Colorectal carcinoma (CRC) is the third and second most commonly diagnosed cancer in men and women, respectively, with an estimated global incidence of 1.4 million new cases, and 693900 deaths in the world every year[1]. The prognosis after curative resection depends entirely on the development of metastasis, especially liver metastasis. At the time of diagnosis, 25% of patients with CRC had liver metastases (CRCLMs) and another 50% of patients without initial liver metastases had liver metastasis during follow-up[2]. Metastatic CRC is one of the leading causes of cancer-related deaths worldwide[3].
According to the international consensus guidelines for scoring the histological growth patterns (HGPs) of liver metastasis[4], CRCLMs present with distinct HGPs, including desmoplastic, pushing, replacement, and two rarer HGPs (sinusoidal and portal HGPs). The HGPs are defined based on the distinct interfaces between cancer cells and adjacent normal liver parenchyma. Importantly, the HGPs of liver metastases were shown to have prognostic and predictive value by some retrospective studies. Several studies showed that patients with desmoplastic liver metastasis had a longer recurrence free survival than patients with non-desmoplastic liver me-tastasis[5,6]. They attributed the poorer survival in patients with non-desmoplastic growth pattern to the higher recurrence rate. Frentzas et al[7] confirmed that CRC with replacement liver metastasis did not respond well to bevacizumab treatment, due to the fact that these tumors utilize vessel co-option instead of angiogenesis. On the other hand, desmoplastic liver metastasis, which relies on sprouting angiogenesis for its blood supply, showed a better response to bevacizumab. They proposed that the HGPs of CRCLMs could be used as a predictive biomarker for anti-angiogenic therapy.
It is natural to expect that the HGP and other clinicopathological characteristics of primary cancer may have biological, predictive, and important prognostic information. However, the relationship between HGPs of liver metastasis and clinicopathological characteristics of primary cancer have not been well established. In the study of Rajaganeshan et al[8], 69.2% of primary expanding CRCs developed capsulated liver metastases, whereas only 17.2% of infiltrating primary CRCs developed a capsulated phenotype (P < 0.001). Although almost all liver metastases from breast cancer adopt a replacement growth pattern, CRCLMs may present different HGPs[7,9]. Furthermore, the underlying genetic abnormalities and molecular pathways that drive the distinct HGPs remain unknown. In this study, we first analyzed the clinicopathological features of CRC patients with two distinct pattern of liver metastases. We then studied the genomic differences of primary CRCs between these two groups using whole exome sequencing analysis. Our study will provide new insights into the primary tumor in terms of their value in predicting the growth pattern of liver metastasis, response to antiangiogenic therapy, and patients’ prognosis.
With the approval of McGill University Health Center Research Ethics Board (No. 11-196-HGP), formalin-fixed, paraffin-embedded tissue blocks of matched primary CRC and liver metastases from 29 patients were obtained from the archives (2012-2017) of the Department of Pathology, McGill University Health Center. The primary cases were divided into two groups according to HGPs of matched CRCLMs: A (15 cases with matched CRCLMs showing desmoplastic growth pattern [DGP]) and B (14 cases with matched CRCLMs showing replacement growth pattern [RGP]). Tumor tissues of primary CRCs from five randomly selected cases in each group and their matched normal large intestinal mucosa were collected for whole exome sequencing. Clinical variables including age, gender, serum carcinoembryonic antigen (CEA) level, and survival data were obtained from patients’ health records. Pathological characteristics of the primary colorectal tumor including localization, size, gross configuration, histologic grade, tumor depth, lymph node metastasis, tumor budding score (TBS), tumor deposit, lymphovascular invasion, and perineural invasion were retrieved from patients’ pathology reports.
For each case, two pathologists (Gao ZH and Wu JB ) reviewed at least three slides stained with hematoxylin and eosin (H&E) under a light microscope (BX45, Olympus, Tokyo, Japan). Discrepancies of HGPs were resolved by reevaluation and discussion in a multi-head microscope setting.
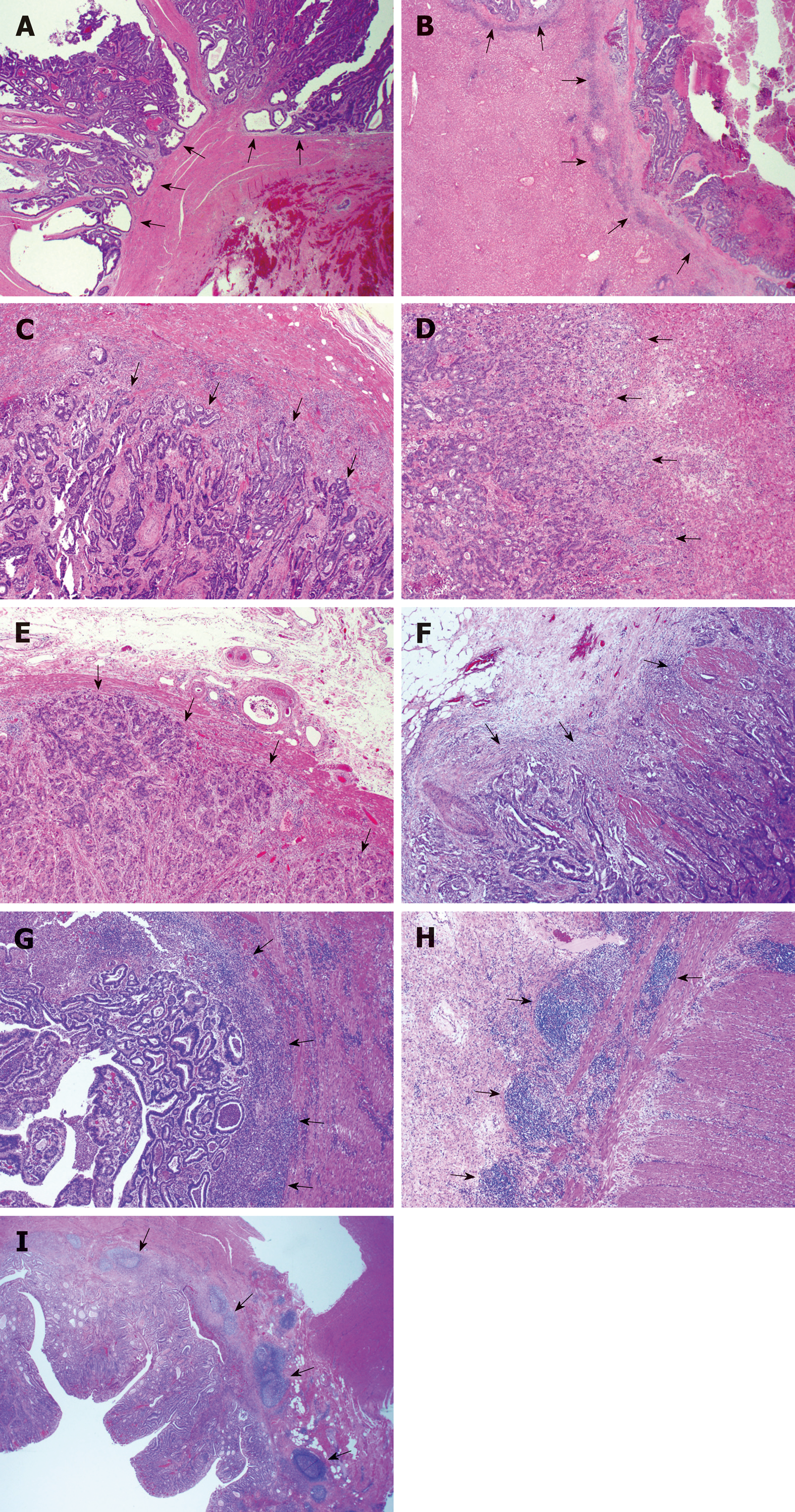
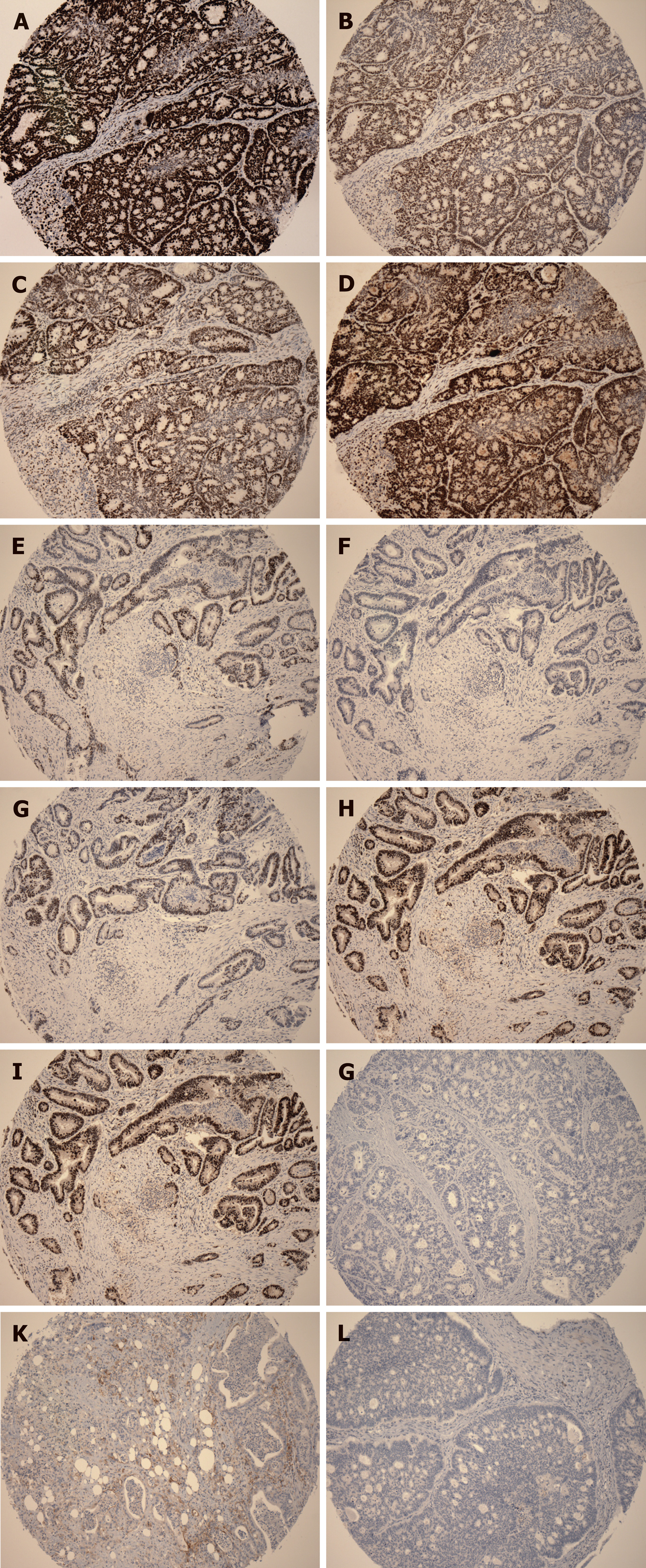
The HGPs of liver metastasis were evaluated according to the international consensus on the HGPs of liver metastasis[4]. In the DGP, the metastatic cancer cells are separated from the liver tissue by a band of desmoplastic tissue. In the RGP, cancer cells form cell plates that are continuous with the hepatocyte plates. The invasive front of the primary cancers were classified as either expanding growth pattern (EGP) or infiltrating growth pattern (IGP) based on the predominant morphology, as defined by Jass et al[10], where the expanding type had been described as the pushing growth type of adenocarcinoma, whereas the infiltrating type had been described as the wide spread streaming form of adenocarcinoma (Figure 1A-D).
The tumor-normal tissue interface was assessed for the degree of ICB and the presence or absence of CDR in H&E stained serial sections (Figure 1E-I). If ICB is present in less than 10% area of the tumor-normal tissue interface, the case was categorized as negative ICB; if between 10% and 50%, the case was categorized with low grade ICB; if ICB was observed in 50% or more of the tumor-normal tissue interface, the case was categorized with high grade ICB. CDR was defined as dense lymphoid aggregates, with or without germinal centers present at the mucosal-submucosal junction, or in the deeper aspects of the bowl wall, including the subserosal adipose tissue (“Crohn’s rosary”)[11].
All 29 formalin-fixed, paraffin-embedded colorectal tumor tissues and one normal intestinal tissue were used for tissue microarray construction, as previously described by Zhang et al[12]. Three 1.5 mm representative punches were included from each tumor tissue.
Automated immunohistochemistry (IHC) was performed on 3-μm-thick TMA slides using a Ventana benchmark machine according to the manufacturer’s instructions (Ventana Medical Systems, Inc., Tucson, AZ, United States). The following commercially available antibodies were used in the IHC: MLH1 [clone ES05, 1:100, Dako (Glostrup, Denmark)], MSH2 (clone FE11, 1:100, Dako), MSH6 (clone EP49, 1:100, Dako), PMS2 (clone EP51, 1:100, Dako), programmed death receptor ligand-1 (PDL1) [clone ES05, SP263, VENTANA (Roche, United States)], and BRAFV600E [clone VE1, 1:100, Abcam (Cambridge, United Kingdom)]. On-slide positive and negative controls were used and the expected reaction was confirmed.
The IHC slides were evaluated independently by two gastrointestinal pathologists blindly using images of full slides obtained with a digital slide scanner (Aperio ScanScope AT Turbo, Leica microsystem, Concord, ON, Canada) and analyzed using the Aperio Image Analyzer. Deletion of MLH1, MSH2, MSH6, or PMS2 in cancerous tissues was defined as the absence of detectable nuclear staining of tumor cells. BRAF V600E staining was considered positive if the cytoplasmic staining was similar to the positive control in each batch. It was determined that any isolated nuclear staining was negative. In line with Zoroquiain et al[13], we defined PDL1 positivity by a threshold of 5% of tumor cells with strong cytoplasmic expression and membrane-accentuation or single membrane pattern. The expression of negative or equivocal protein expression detected in TMA was validated by IHC staining on the corresponding large tumor sections of the resection specimen using the same protocol.
Formalin-fixed, paraffin-embedded tumor tissue blocks and matched normal blocks were microdissected to remove residual normal tissue and enhance neoplastic cellularity. The DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Netherlands) according to the manufacturer’s protocol. The DNA concentration was determined using a Qubit 2.0 Fluorimeter (Invitrogen, Carlsbad, CA, United States). After DNA extraction, we obtained 282 to 6480 ng of DNA per lesion from 10 CRCs and matched normal tissues.
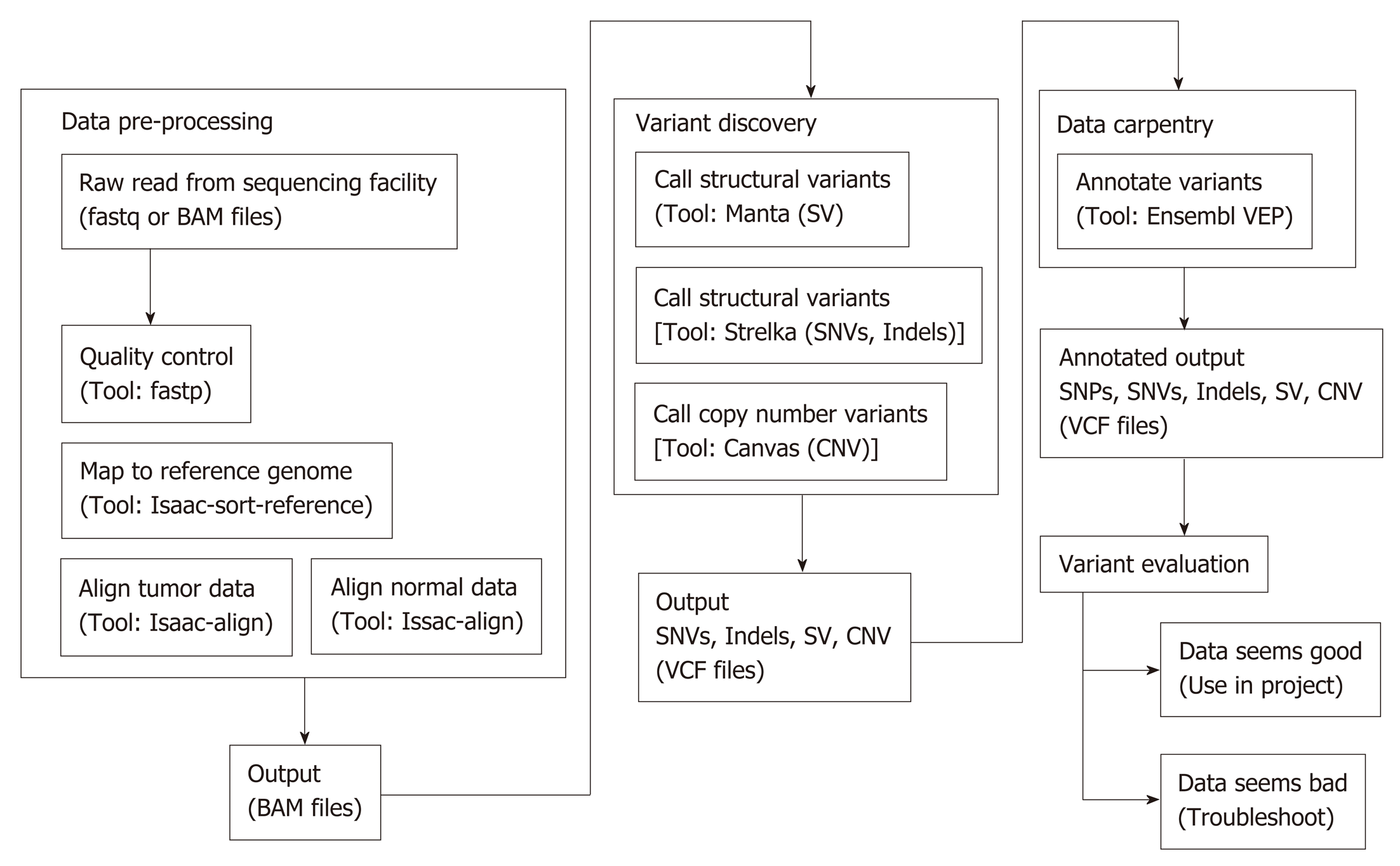
Whole exome sequencing was performed at McGill University and Genome Quebec Innovation Center. The gDNA library was prepared using a TruSeq DNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions, followed by sequencing on an Illumina HiSeq, as previously described[14]. As shown in Figure 2, the raw DNA sequences were aligned, trimmed, and duplicates flagged to the NCBI human genome build 38 version 93, using Isaac aligner (Version: Isaac-04.18.01.19)[15]. Structural variant (SV) analysis calls were generated using Manta (version manta-1.4.0)[16]. Small variants [single nucleotide variant (SNV) and small indels] in germline and somatic variations in tumor/normal sample pairs were achieved using Strelka (Version: strelka-2.9.0)[17]. Copy number variant (CNV) calls were generated using Canvas (Version: Canvas/1.11.0/) with the tumor-normal-enrichment workflow[18]. The callers share output, and therefore there is no double counting of calls. Manta and Strelka were run with Python (Version: 2.7.14). Annotation of the resulting calls was done with the Ensembl Variant Effect Predictor (VEP) using version 93.3 in a Perl (Version 5.22.4) environment[19]. Fastp (version 0.19.4) was used to collect QC metrics of the raw reads[20].
Sequencing result analyses and figures were prepared in the R statistical software suite (version 3.5.1) using standard R functions in custom code[21]. Circlize (version 0.4.4) was used to generate the Circos plots[22]. GGplot2 (version 3.0.0) was used to generate the heat maps. Custom R code was developed and run in RStudio (Version: 1.1.453).
Statistical analyses were performed using Prism 7.00 statistical program (GraphPad, 2015, San Diego, CA, United States); P < 0.05 was considered statistically significant. The statistical review of the study was performed by a biomedical statistician. Comparison of clinicopathological features and immunohistochemical staining results between groups was performed using the t-test, Wilcoxon Signed rank test, and Fisher’s exact test. Survival analysis was performed using the Kaplan-Meier method and the log-rank test. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated to assess the prediction capacity of histologic features and genomic alternations of primary CRC for the growth patterns of liver metastasis.
The clinical characteristics of the study population are shown in Table 1. The statistical comparison of the clinicopathological characteristics between Group A and Group B is shown in Table 2. Cases 1 to 15 belong to Group A, and cases 16 to 29 belong to Group B. With respect to age and gender, no significant differences were identified between Group A and Group B. The mean serum CEA level in Group B (46.9 μg/L) was noticeably higher than that in Group A (15.5 μg/L), although not statistically significant.
| Case No. | Liver metastasis growth pattern | Primary colorectal growth pattern | Tumor budding score | Crohn's disease-like appearance | Liver metastasis (mo) | Follow-up (mo) | Prognosis | Major mutations |
| 1 | DGP | EGP | L | + | 16 | 57 | Alive without tumor | APC, KRAS, BRCA2 |
| 2 | DGP | IGP | M | - | 6 | 25 | Alive with bone metastasis | NA |
| 3 | DGP | IGP | H | - | 46 | 52 | Alive with liver metastasis | NA |
| 4 | DGP | EGP | M | + | 25 | 33 | Alive with liver metastasis | APC, KRAS, PIK3CA, BRCA1 |
| 5 | DGP | EGP | L | + | 39 | 46 | Alive without tumor | NA |
| 6 | DGP | IGP | L | - | 8 | 33 | Alive with liver metastasis | NA |
| 7 | DGP | IGP | L | + | 25 | 32 | Alive without tumor | NA |
| 8 | DGP | EGP | L | - | 6 | 44 | Alive with tubular adenoma | APC, TP53, PIK3CA |
| 9 | DGP | EGP | L | - | 12 | 28 | Alive without tumor | NA |
| 10 | DGP | EGP | L | + | 7 | 21 | Alive without tumor | NA |
| 11 | DGP | EGP | L | - | 7 | 16 | Alive without tumor | APC, TP53, FAT4, BRAF |
| 12 | DGP | IGP | M | + | 5 | 25 | Alive without tumor | NA |
| 13 | DGP | IGP | L | + | 6 | 42 | Alive without tumor | NA |
| 14 | DGP | EGP | M | - | 38 | 54 | Alive with lung metastasis | NA |
| 15 | DGP | EGP | L | - | 17 | 20 | Alive without tumor | TP53, DNAH5, FAT4 |
| 16 | RGP | IGP | H | - | 2 | 17 | Alive without tumor | DNAH5, KRAS, FAT4 |
| 17 | RGP | IGP | H | - | 5 | 72 | Alive without tumor | NA |
| 18 | RGP | IGP | H | - | 16 | 63 | Died of liver metastasis | NA |
| 19 | RGP | IGP | L | - | 9 | 35 | Alive without tumor | NA |
| 20 | RGP | IGP | H | - | 25 | 58 | Alive without tumor | NA |
| 21 | RGP | EGP | L | - | 6 | 23 | Alive without tumor | NA |
| 22 | RGP | IGP | M | - | 28 | 48 | Alive without tumor | APC, TP53, SMAD4 |
| 23 | RGP | IGP | H | - | 40 | 60 | Alive without tumor | APC, CTNNB1 |
| 24 | RGP | IGP | M | - | 6 | 17 | Died of liver metastasis | NA |
| 25 | RGP | IGP | M | - | 6 | 33 | Died of liver metastasis | APC, TP53, KRAS, ERBB2, ERBB3 |
| 26 | RGP | EGP | L | + | 7 | 41 | Died of liver metastasis | NA |
| 27 | RGP | IGP | H | - | 5 | 35 | Died of brain metastasis | NA |
| 28 | RGP | IGP | L | - | 27 | 32 | Died of liver metastasis | NA |
| 29 | RGP | IGP | H | - | 3 | 12 | Alive without tumor | TP53, LRP2, SDK1 |
| Characteristic | Group A | Group B | P-value | |
| Sex | Male | 9 | 10 | 0.6999 |
| Female | 6 | 4 | ||
| Age | Mean ± SD | 67.5 ± 9.6 | 62.3 ± 13.6 | 0.2774 |
| Median | 67 | 60.5 | ||
| Range | 50-82 | 43-82 | ||
| Serum CEA level | Mean ± SD | 15.5 ± 25.2 | 46.9 ± 148.7 | 0.3823 |
| (μg/L) | Median | 3.7 | 5.65 | |
| Range | 0.9-88.0 | 1.1-563.2 | ||
| Localization | Cecum | 3 | 3 | 0.6621 |
| Ascending | 2 | 0 | ||
| Transverse | 1 | 1 | ||
| Descending | 1 | 0 | ||
| Sigmoid | 2 | 3 | ||
| Rectum | 6 | 7 | ||
| Size (cm) | Mean ± SD | 4.2 ± 1.3 | 4.3 ± 2.4 | 0.5814 |
| Median | 4.6 | 4 | ||
| Range | 2.0-6.0 | 2.0-11.0 | ||
| Configuration | Exophytic | 7 | 7 | 0.2643 |
| Endophytic | 8 | 5 | ||
| N/A | 0 | 2 | ||
| Histologic grade | G1 | 5 | 7 | 0.3060 |
| G2 | 8 | 7 | ||
| G3 | 2 | 0 | ||
| Tumor depth | T1 | 0 | 0 | 0.6999 |
| T2 | 0 | 0 | ||
| T3 | 9 | 10 | ||
| T4 | 6 | 4 | ||
| Lymph node metastasis | Positive | 9 | 10 | 0.6999 |
| Negative | 6 | 4 | ||
| Tumor budding score (TBS) | High | 1 | 7 | 0.0275 |
| Intermediate | 4 | 3 | ||
| Low | 10 | 4 | ||
| Negative | 0 | 0 | ||
| Tumor deposits | Positive | 3 | 4 | 0.6817 |
| Negative | 12 | 10 | ||
| Lymphovascular invasion | Positive | 11 | 12 | 0.6513 |
| Negative | 4 | 2 | ||
| Perineural invasion | Positive | 10 | 12 | 0.3898 |
| Negative | 5 | 2 | ||
| Peri-tumoral inflammatory cell band | Negative | 5 | 4 | 0.5974 |
| Low grade | 8 | 6 | ||
| High grade | 2 | 4 | ||
| Crohn’s disease-like response | Positive | 7 | 1 | 0.0352 |
| Negative | 8 | 13 | ||
| Histologic growth pattern | EGP | 9 | 2 | 0.0209 |
| IGP | 6 | 12 |
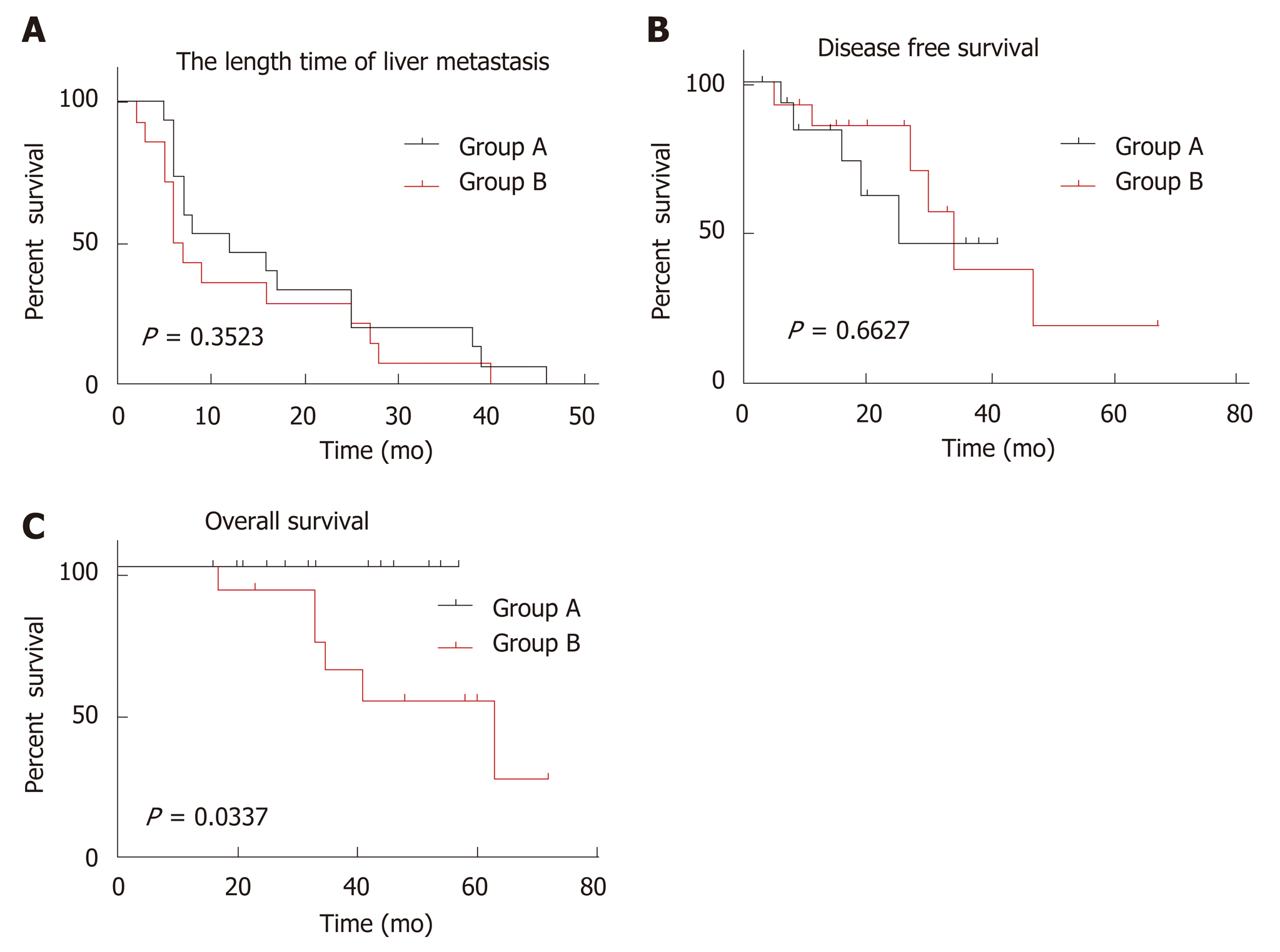
The mean length of follow-up for all patients was 36.8 mo (95%CI: 30.81-42.85 mo). Seventy-nine percent (23/29) patients were alive on completion of the study. There was no death in group A. Five cases were alive with liver, bone, or lung metastasis, the remaining 10 cases were alive without evidence of tumor recurrence or metastasis. In group B, however, 6 cases died of liver, lung, or brain metastasis and the remaining 8 cases had no evidence for tumor recurrence or metastasis. The length time for liver metastasis was calculated from the date of diagnosis of the primary CRC to the date of diagnosis of liver metastasis. The disease-free survival (DFS) was defined as the time from CRCLM resection to recurrence, or to the date of censoring in October, 2018, when the patient had no signs or symptoms of tumor recurrence or metastasis. Overall survival (OS) was calculated from the date of diagnosis of the primary carcinoma to the date of death, or to the date of censoring of live patients in October, 2018. With respect to the length time of liver metastasis and the DFS, there were no significant differences between Group A and Group B (Figure 3A and B, P > 0.05). However, OS was significantly longer in Group A than Group B (Figure 3C, P = 0.0337).
Among 29 cases, 37.9% (11/29) of the primary cancers were EGP and 62.1% (18/19) were IGP. Primary CRC with an EGP were more likely to have desmoplastic phenotype liver metastasis (81.8% vs 33.3%, P < 0.05). On the other hand, primary cancers with an IGP were significantly more likely to form replacement pattern liver metastasis (66.7% vs 18.2 %, P < 0.05)
TBS and tumor deposit were evaluated according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual (Eight Edition). Most of TBS was low (66.7%, 10/15) in Group A, and intermediate or high in group B (71.4%, 10/14). CDR was identified in 46.7% (7/15) of cases in Group A, and only in 7.1% (1/14) in Group B. There was no significant difference between Group A and Group B regarding tumor location, size, gross configuration, histologic grade, tumor depth, lymph node metastasis, tumor deposit, lymphovascular invasion, perineural invasion, or peri-tumoral ICB. With respect to TBS and CDR, there was significant higher TBS in Group B than in Group A (P < 0.05) and significantly more CDR in Group A than in Group B (P < 0.05).
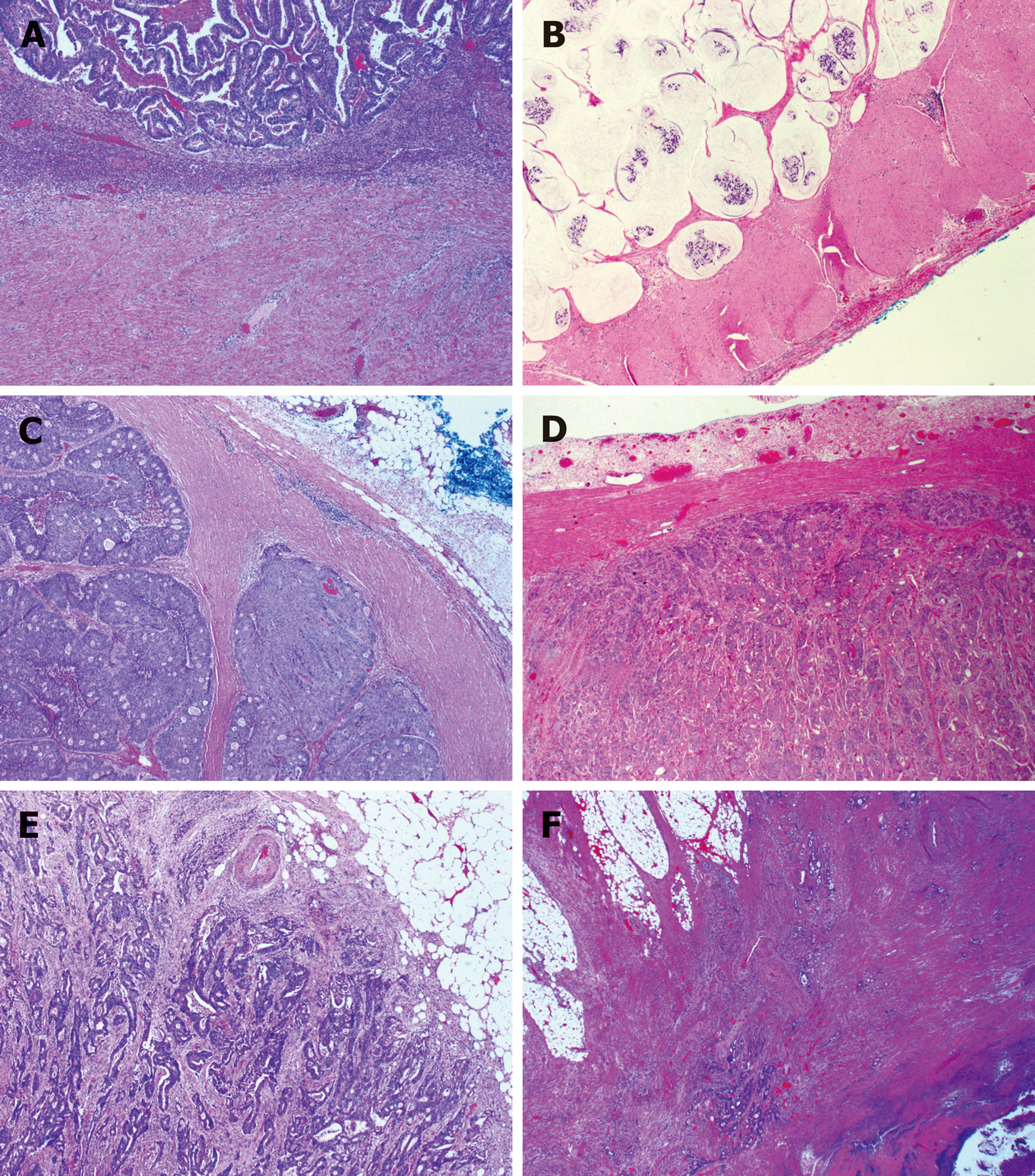
Interestingly, as shown in Figure 4, we observed that the EGP of primary cancer could be divided into four subtypes: (1) Ordinary expanding, where the tumor gland pushes the stroma and forms a sharp dividing line at the interface; (2) Mucinous expanding, where pools of tumor epithelium containing mucin pushes the stroma and forms a sharp dividing line at the interface; (3) Cribriform expanding, where the tumor gland at the invasive front forms a round cribriform structure, and there is a clear dividing line between the tumor and the surrounding stroma; and (4) Micropapillary expanding, where the tumor cells at the invasive front form micropapillary architecture, and there is a clear dividing line between the tumor and the surrounding stroma. On the other hand, infiltrating growth pattern could be divided into two subtypes: (1) Ordinary infiltrating, where tumor cells infiltrate to the stroma and connect to the original tumor mass; and (2) Skip infiltrating, where there is a gap (>0.5 cm) of non-neoplastic tissue between the main tumor bulk and the invasive front.
As shown in Figure 5, there was no significant differences between the two groups in the tumor cell expression of MLH1, MSH2, MSH6, PMS2, BRAFV600E, or PDL1. Interestingly, the three MSI deficiency cases were all MSH2 protein deficient.
EGP, low TBS score, and positive CDR in the primary tumor were shown to have predictive value for the DGPs of CRCLMs. As shown in Table 3, HGP + TBS + CDR is the most sensitive and has the highest negative predictive value, whereas CDR alone is the most specific and has the highest positive predictive value.
| Pathological characteristic | Value | Group A | Group B |
| HGP | SE% (CI) | 60.0 (35.8-80.2) | 85.7 (60.1-97.5) |
| SF% (CI) | 85.7 (60.1-97.5) | 60.0 (35.8-80.2) | |
| PPV% (CI) | 81.8 (52.3-96.8) | 66.7 (43.8-83.7) | |
| NPV% (CI) | 66.7 (43.8-83.7) | 81.8 (52.3-96.8) | |
| TBS | SE% (CI) | 66.7 (41.7-84.8) | 71.4 (45,4-88.3) |
| SF% (CI) | 71.4 (45,4-88.3) | 66.7 (41.7-84.8) | |
| PPV% (CI) | 71.4 (45.4-88.3) | 66.7 (41.7-84.8) | |
| NPV% (CI) | 66.7 (41.7-84.8) | 71.4 (45.4-88.3) | |
| CDR | SE% (CI) | 46.7 (24.8-69.9) | 92.9 (68.5-99.6) |
| SF% (CI) | 92.9 (68.5-99.6) | 46.7 (24.8-69.9) | |
| PPV% (CI) | 87.5 (52.9-99.4) | 61.9 (40.9-79.3) | |
| NPV% (CI) | 61.9 (40.9-79.3) | 87.5 (52.9-99.4) | |
| HGP + TBS | SE% (CI) | 80.0 (54.8-93.0) | 85.7 (60.1-97.5) |
| SF% (CI) | 71.4 (45.4-88.3) | 46.7 (24.8-69.9) | |
| PPV% (CI) | 75.0 (50.5-89.8) | 60.0 (38.7-78.1) | |
| NPV% (CI) | 76.9 (49.7-91.8) | 77.8 (45.3-96.1) | |
| HGP + CDR | SE% (CI) | 80.0 (54.8-93.0) | 92.9 (68.5-99.6) |
| SF% (CI) | 85.7 (60.1-97.5) | 26.7 (10.9-52.0) | |
| PPV% (CI) | 85.7 (60.1-97.6) | 54.2 (35.1-72.1) | |
| NPV% (CI) | 80.0 (54.8-93.0) | 80.0 (37.6-99.0) | |
| TBS + CDR | SE% (CI) | 80.0 (54.8-93.0) | 92.9 (68.5-99.6) |
| SF% (CI) | 71.4 (45.4-88.3) | 33.3 (15.2-58.3) | |
| PPV% (CI) | 75.0 (50.5-89.8) | 56.5 (36.8-74.4) | |
| NPV% (CI) | 76.9 (49.7-91.8) | 83.3 (43.7-99.2) | |
| HGP + TBS + CDR | SE% (CI) | 86.7 (62.1-97.6) | 92.9 (68.5-99.6) |
| SF% (CI) | 71.4 (45.4-88.3) | 20.0 (7.0-45.2) | |
| PPV% (CI) | 76.5 (52.7-90.4) | 52.0 (33.5-70.0) | |
| NPV% (CI) | 83.3 (55.2-97.0) | 75.0 (30.1-98.7) |
IGP, high or intermediate TBS score, and negative CDR were shown to have predictive value for the RGP of CRCLMs. As shown in Table 3, CDR alone or combined with HGP and TBS are the most sensitive; TBS alone is the most specific; HGP or TBS has the highest PPV, whereas CDR has the highest NPV. When combining HGP, TBS, and CDR, although the sensitivity is 92.9%, the specificity is only 20.0%.
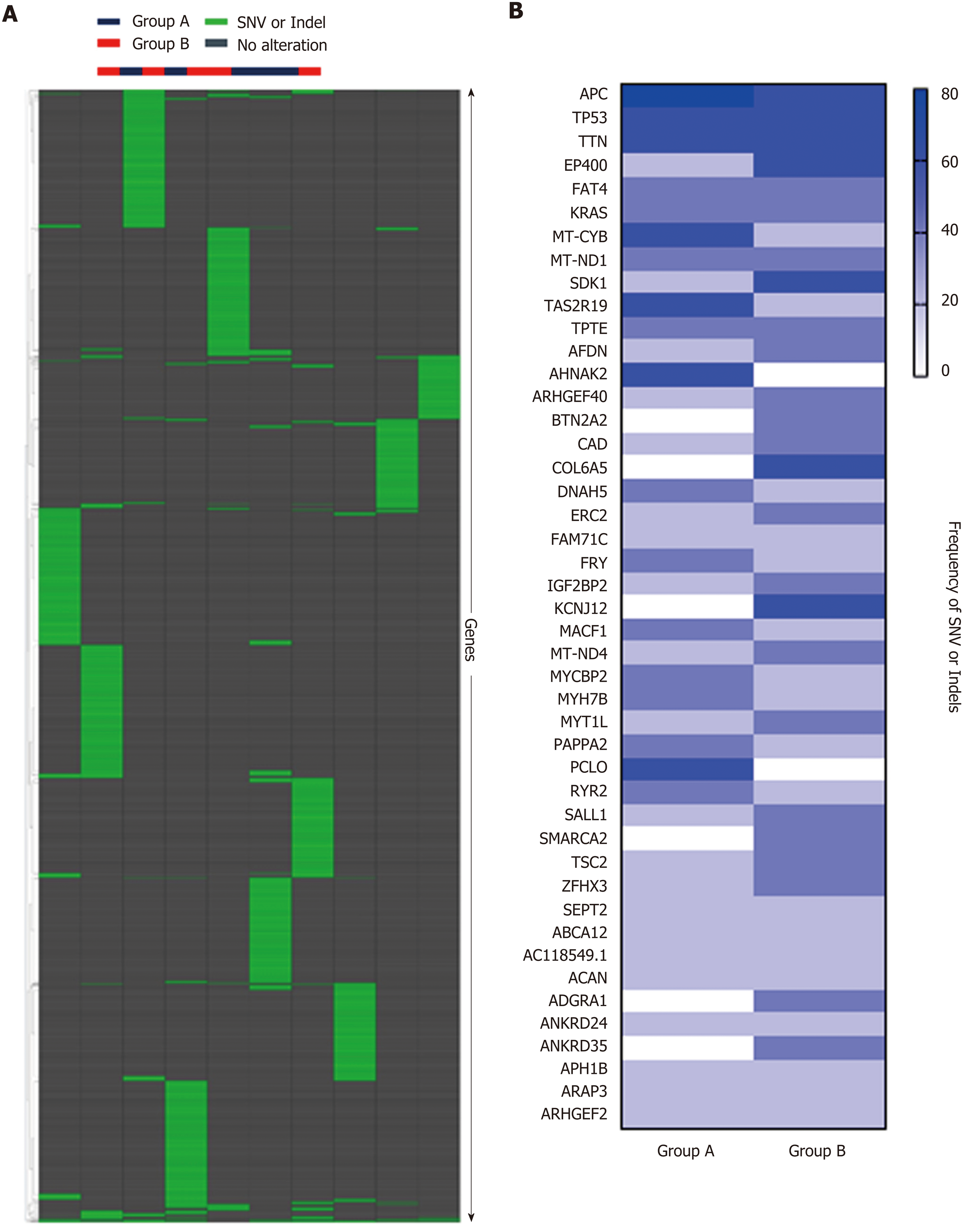
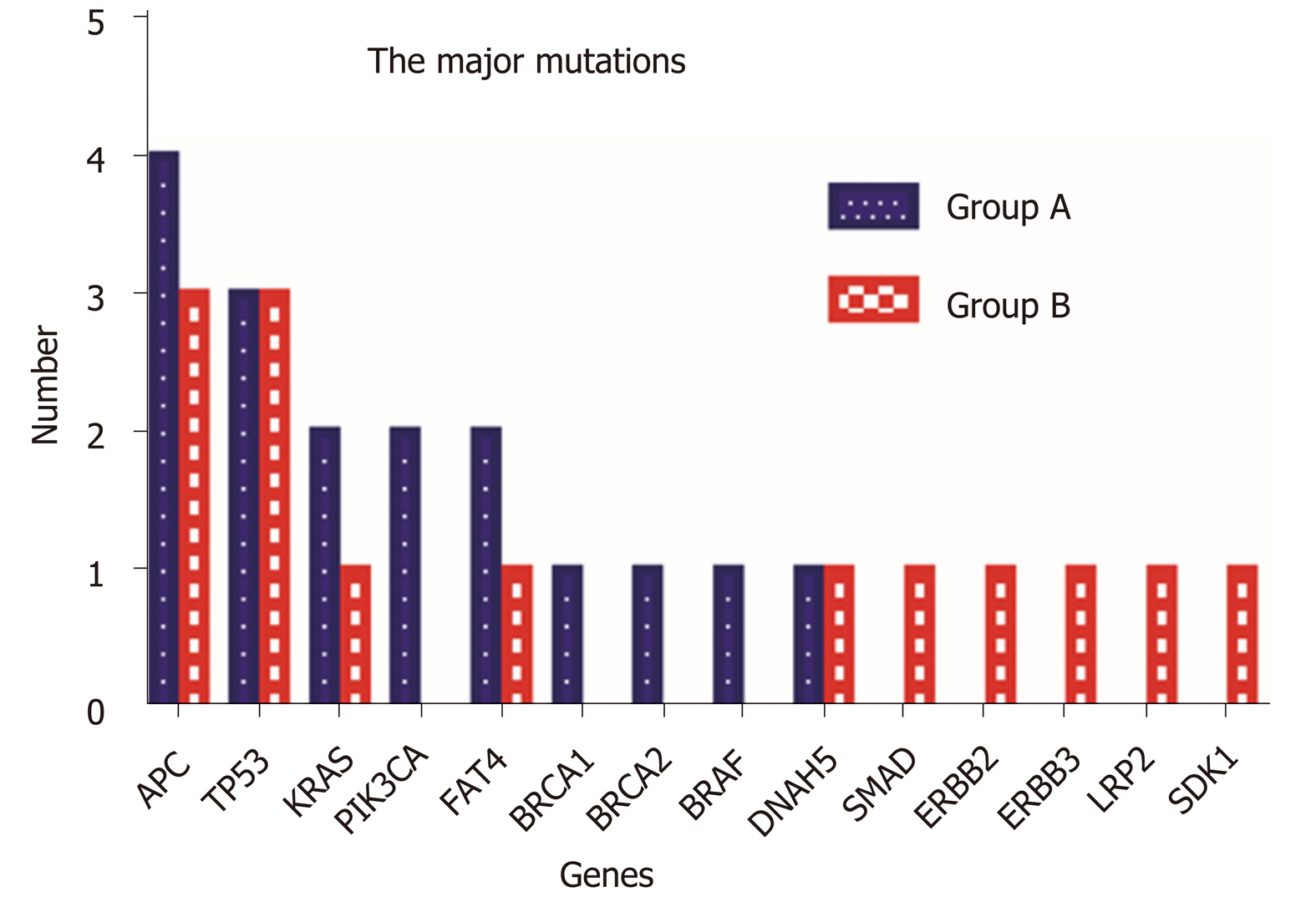
Hierarchical clustering of Groups A and B based on SNV and indels is shown in Figure 6A, and frequency of SNV or indels in Group A and Group B is shown in Figure 6B. There was no statistical difference between the two groups in SNV or indels. However, our whole-exome sequencing revealed 14 major gene mutations, as shown in Table 1 and Figure 7. The most prevalent major mutation occurred in the APC gene, which occurred in 80% (4/5) of Group A cases and 60% (3/5) cases of Group B, followed by TP53 and KRAS mutations. Both Group A and Group B had the same frequency of TP53 and DNAH5 mutations. Interestingly, PIK3CA mutations were showed in 40% (2/5) cases of Group A but no one case in Group B, similarly, the mutations of BRCA1, BRCA2, and BRAF were only observed in Group A, and the mutations of SMAD, ERBB2, ERBB3, LRP2, and SDK1 were identified only in Group B, each in one patient.
Colorectal adenocarcinoma liver metastasis has been categorized into three growth patterns: desmoplastic, pushing, and replacement, each of which has characteristic morphological features[4]. Using this classification, Van den Eynden et al[6] reported that the pushing pattern was an independent predictor of poor survival. In contrast, Nielsen et al showed that patients with an RGP had a death risk 2 to 2.5 times higher than patients with a pushing growth pattern or mixed growth pattern, and almost 4 times more than patients with a DGP[5]. According to Eefsen et al[3], similar findings in chemonaive patients and patients receiving neoadjuvant therapy were identified. Histologically differentiating DGP and invasive growth pattern were more clearly cut with almost no interobserver discrepancies and the clinical relevance was more obvious[4]. For these reasons, only those CRCLMs with well-defined desmoplastic or replacement growth patterns were enrolled in this study.
Our results demonstrated that expanding CRCs tend to develop desmoplastic liver metastasis, whereas infiltrating CRCs tended to develop replacement liver metastasis, which is consistent with previous findings of Rajaganeshan et al[8]. Our study also confirmed that patients with expanding primary cancers had a significant longer OS than those with infiltrating primary tumors, which is in line with the previously reported findings of Cianchi et al[23], Nystrom et al[24], and Pinheiro et al[25]. Therefore, the growth pattern of primary CRC could, to a certain extent, predict the growth pattern of CRCLMs and patients’ prognosis.
During our study, we identified four subtypes of primary expanding CRC and two subtypes of primary infiltrating cancers. Awareness of these morphological variations is critical in order to appropriately subclassify the primary CRCs. Zoning on the epithelial-stromal interface, we found that the infiltrating CRC had higher TBS, whereas the expanding cancer was more prone to form CDR. Our observation is in keeping with the previous report on the prognostic value of TBS and CDR of primary CRC[26-29]. To the best of our knowledge, this study is the first comprehensive morphological sub-classification of the growth patterns of primary CRCs in association with clinical follow-up data and corresponding HGPs of CRCLMs.
Using a standard formula, we analyzed the sensitivity, specificity, PPV, and NPV of selected pathological characteristics (HGP, TBS, and CDR) in predicting the HGPs of CRCLMs. Our results indicated that combined EGP, low TBS, and positive CDR of primary cancer could be used to predict the DGP of CRCLMs, whereas IGP alone of primary cancer could be the best to predict the RGP of CRCLMs. Once validated by larger set of cases, these parameters have the potential for clinical application.
The prognostic value of immunohistochemical markers such as microsatellite instability (MSI), BRAF V600E mutation, and PD-L1 expression has been reported[30-33]. However, there were no studies on the association between the expression of these IHC markers and the HGPs of CRC. Our study did not reveal any significant differences in the tumor cell expression of these IHC markers between the two groups.
It is reasonable to expect that the genomic makeup of the primary CRC plays an important role in determining the HGP of CRCLMs. However, to the best of our knowledge, there has been no report on the specific genomic drivers of the primary tumor that determine the specific growth pattern of liver metastasis. If the growth patterns of liver metastasis could be predicted based on the molecular biomarkers present in the primary CRC and each of the growth patterns could be associated with a different underlying biology, this could have important implications in the stratification of patients for the oncological treatment[34]. We compared the mutation rate of genes related to metastasis (WNA5A, TIMP1, MMP-1, MMP-2, COX-2, and HIF-1α), angiogenesis (VEGF, TGF, EGF, and TNF), epithelial-mesenchymal transition (E-cadherin, FGF, P63, and FOXC2), oncogenes (C-myc, K-ras, and Bcl-2), tumor suppressor genes (p53, APC, and NGX6), and other genes, such as Survivin and CIAPIN1, between the two groups[35-39]. The only gene that stands out is pho-sphoinositide-3-kinase, catalytic, alpha polypeptide (PIK3CA), which was present in 40% of tumors with desmoplastic group pattern in the CRCLMs. PIK3CA is an essential element of the signaling pathway of phosphatidylinositol-3 kinase (PIK3) downstream of EGFR. The PIK3CA mutation activates the PIK3 signaling pathway, improves cell proliferation, and ultimately leads to carcinogenesis[40]. It is associated with angiogenesis as it is essential in endothelial cell migration during vascular development through vascular endothelial growth factor-A (VEGFA) signaling, possibly by regulating RhoA activity[41]. The significantly higher expression of PIK13CA in primary CRC with desmoplastic liver metastasis indicates that metastasis can become vascularized through sprouting angiogenesis, in a process stimulated by VEGFA. In addition to PIK3CA, we also found that BRCA-1, BRCA-2, and BRAF genes were present only in primary CRC with desmoplastic liver metastasis, and SMAD, ERBB-2, ERBB-3, LRP2, and SDK1 genes were present only in CRCs with invasive liver metastasis. Once these findings are validated with a larger set of cases, these growth pattern-related genomic abnormalities could become new targets for precision therapy.
Although the existence of two invasive phenotypes (expanding or infiltrating) in CRC is now well recognized and their prognostic implications proven, the biological mechanisms for their existence remain unexplained. It is unclear why some tumors cause a desmoplastic and angiogenic response while others adopt the replacement growth pattern and acquire nutrition through vessel co-option. In addition to the biology of primary tumor cells, the surrounding liver microenvironment may also play a role in determining the growth pattern or dependence of angiogenesis. One possible explanation for the desmoplastic or replacement liver metastasis is that these HGPs summarize the different responses of the liver to injury[4]. Fibrosis in the desmoplastic liver metastasis may be mediated by the same biological mechanisms which drive liver fibrosis in response to an injury[42]. In addition, replacement HGP is similar to liver regeneration because cancer cells replace liver cells in the same way that new liver cells replace old liver cells during liver regeneration[43]. Another explanation is that the different tumor growth patterns are related to differential gene expression, which may be a driving factor for HGP. Sakariassen et al[44] investigated that vessel co-opting glioblastomas (GBMs) upregulated gene expression associated with fetal development and cell motility, whereas angiogenic GBMs had higher angiogenic regulatory factors, such as VEGF and angiopoetin-2 expression. In the present study, mutation of the PIK3CA gene was present only in the primary CRC with desmoplastic CRCLMs, which further validated its role as a marker for sprouting angiogenesis and a potential target for anti-angiogenic gene therapy.
In summary, primary CRCs with an EGP have a better OS than those with an IGP. Expanding CRCs tend to develop desmoplastic liver metastasis, whereas infiltrating cancers tend to develop replacement liver metastasis. Combined HGP, TBS, and CDR of primary CRC could be used to predict the HGPs of liver metastasis. Up to 40% of primary CRCs with an EGP showed PIK3CA gene mutations in contrast to 0% of primary CRCs with an IGP. These genomic differences, if validated in a larger cohort of cases, have the potential to become not only clinically applicable diagnostic and prognostic biomarkers but also therapeutic targets of genomic engineering.
Different histological growth patterns (HGPs) of colorectal carcinoma (CRC) liver metastasis are associated with patients’ prognosis and response to antiangiogenic therapy.
Through studying the relationship between the different HGPs of liver metastasis and clinicopathological and genomic characteristics of primary cancer, we aimed to evaluate whether certain clinicopathological and genomic features of primary CRC could predict the HGPs of liver metastasis
To understand the biology of the primary CRCs in association with different HGPs of liver metastasis, and to identify histological and biology markers in the primary tumor that could predict the HGPs of liver metastasis.
A total of 29 patients with paired resections of both primary CRC and liver metastasis were divided into two groups: A (15 cases with desmoplastic liver metastasis) and B (14 cases with replacement liver metastasis). Clinical information was obtained from patients’ charts. Mismatch repair proteins, BRAFV600E, and PD-L1 were evaluated by immunohistochemistry. Five cases from each group were randomly selected for WES analysis.
In the primary tumor, expanding growth pattern, low tumor budding score (TBS), and Crohn’s disease-like response (CDR) were associated with desmoplastic liver metastasis and better overall survival, whereas infiltrating growth pattern alone of primary carcinoma could predict the replacement liver metastasis and worse overall survival (P < 0.05). On WES analysis, primary carcinoma with desmoplastic liver metastasis showed mutations in APC (4/5); TP53 (3/5); KRAS, PIK3CA, and FAT4 (2/5); BRCA-1, BRCA2, BRAF, and DNAH5 (1/5), whereas primary carcinoma with replacement liver metastasis showed mutations in APC and TP53 (3/5); KRAS, FAT4, DNH5, SMAD, ERBB2, ERBB3, LRP1, and SDK1 (1/5).
The primary CRCs with an expanding growth pattern have a better overall survival that those with an infiltrating growth pattern. Expanding CRCs tend to develop desmoplastic liver metastasis, whereas infiltrating cancers tend to develop replacement liver metastasis. Combined HGP, TBS, and CDR of primary CRC could be used to predict the HGPs of liver metastasis. Up to 40% of primary CRCs with an expanding growth pattern show PIK3CA gene mutations in contrast to 0% of primary CRCs with an invasive growth pattern.
Multicenter collaborative studies with a larger number of patients and prospective studies to assess the predictive value of the clinicopathological features of primary CRC on the HGPs of its liver metastasis could help to further validate our results. These genomic differences between the two groups of primary CRC, if validated in a larger cohort of case, have the potential to become not only clinically applicable diagnostic and prognostic biomarkers but also therapeutic targets of genomic engineering.
We thank Dr. Logan Walsh at Department of Human Genetics, Rosalind and Morris Goodman Cancer Research Center, McGill University for his assistance in analyzing the whole exome sequencing data, and associate professor Jianfeng Luo at Department of Biostatistics, School of Public Health, Fudan University for the help of biostatistical analysis.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Canada
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Meteoglu I, Xu J S-Editor: Ma RY L-Editor: Wang TQ E-Editor: Zhang YL
| 1. | Fonseca GM, Herman P, Faraj SF, Kruger JAP, Coelho FF, Jeismann VB, Cecconello I, Alves VAF, Pawlik TM, de Mello ES. Pathological factors and prognosis of resected liver metastases of colorectal carcinoma: implications and proposal for a pathological reporting protocol. Histopathology. 2018;72:377-390. [PubMed] [DOI] [Cited in This Article: ] |
| 2. | Brachet D, Lermite E, Rouquette A, Lorimier G, Hamy A, Arnaud JP. Prognostic factors of survival in repeat liver resection for recurrent colorectal metastases: review of sixty-two cases treated at a single institution. Dis Colon Rectum. 2009;52:475-483. [PubMed] [DOI] [Cited in This Article: ] |
| 3. | Eefsen RL, Vermeulen PB, Christensen IJ, Laerum OD, Mogensen MB, Rolff HC, Van den Eynden GG, Høyer-Hansen G, Osterlind K, Vainer B, Illemann M. Growth pattern of colorectal liver metastasis as a marker of recurrence risk. Clin Exp Metastasis. 2015;32:369-381. [PubMed] [DOI] [Cited in This Article: ] |
| 4. | van Dam PJ, van der Stok EP, Teuwen LA, Van den Eynden GG, Illemann M, Frentzas S, Majeed AW, Eefsen RL, Coebergh van den Braak RRJ, Lazaris A, Fernandez MC, Galjart B, Laerum OD, Rayes R, Grünhagen DJ, Van de Paer M, Sucaet Y, Mudhar HS, Schvimer M, Nyström H, Kockx M, Bird NC, Vidal-Vanaclocha F, Metrakos P, Simoneau E, Verhoef C, Dirix LY, Van Laere S, Gao ZH, Brodt P, Reynolds AR, Vermeulen PB. International consensus guidelines for scoring the histopathological growth patterns of liver metastasis. Br J Cancer. 2017;117:1427-1441. [PubMed] [DOI] [Cited in This Article: ] |
| 5. | Nielsen K, Rolff HC, Eefsen RL, Vainer B. The morphological growth patterns of colorectal liver metastases are prognostic for overall survival. Mod Pathol. 2014;27:1641-1648. [PubMed] [DOI] [Cited in This Article: ] |
| 6. | Van den Eynden GG, Bird NC, Majeed AW, Van Laere S, Dirix LY, Vermeulen PB. The histological growth pattern of colorectal cancer liver metastases has prognostic value. Clin Exp Metastasis. 2012;29:541-549. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao ZH, Shi Y, Van den Eynden G, Daley F, Peckitt C, Tan X, Salman A, Lazaris A, Gazinska P, Berg TJ, Eltahir Z, Ritsma L, Van Rheenen J, Khashper A, Brown G, Nystrom H, Sund M, Van Laere S, Loyer E, Dirix L, Cunningham D, Metrakos P, Reynolds AR. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med. 2016;22:1294-1302. [PubMed] [DOI] [Cited in This Article: ] |
| 8. | Rajaganeshan R, Prasad R, Guillou PJ, Chalmers CR, Scott N, Sarkar R, Poston G, Jayne DG. The influence of invasive growth pattern and microvessel density on prognosis in colorectal cancer and colorectal liver metastases. Br J Cancer. 2007;96:1112-1117. [PubMed] [DOI] [Cited in This Article: ] |
| 9. | Stessels F, Van den Eynden G, Van der Auwera I, Salgado R, Van den Heuvel E, Harris AL, Jackson DG, Colpaert CG, van Marck EA, Dirix LY, Vermeulen PB. Breast adenocarcinoma liver metastases, in contrast to colorectal cancer liver metastases, display a non-angiogenic growth pattern that preserves the stroma and lacks hypoxia. Br J Cancer. 2004;90:1429-1436. [PubMed] [DOI] [Cited in This Article: ] |
| 10. | Jass JR, Love SB, Northover JM. A new prognostic classification of rectal cancer. Lancet. 1987;1:1303-1306. [PubMed] [Cited in This Article: ] |
| 11. | Nylund K, Leh S, Immervoll H, Matre K, Skarstein A, Hausken T, Gilja OH, Birger Nesje L, Ødegaard S. Crohn's disease: Comparison of in vitro ultrasonographic images and histology. Scand J Gastroenterol. 2008;43:719-726. [PubMed] [DOI] [Cited in This Article: ] |
| 12. | Zhang D, Salto-Tellez M, Putti TC, Do E, Koay ES. Reliability of tissue microarrays in detecting protein expression and gene amplification in breast cancer. Mod Pathol. 2003;16:79-84. [PubMed] [DOI] [Cited in This Article: ] |
| 13. | Zoroquiain P, Esposito E, Logan P, Aldrees S, Dias AB, Mansure JJ, Santapau D, Garcia C, Saornil MA, Belfort Neto R, Burnier MN. Programmed cell death ligand-1 expression in tumor and immune cells is associated with better patient outcome and decreased tumor-infiltrating lymphocytes in uveal melanoma. Mod Pathol. 2018;31:1201-1210. [PubMed] [DOI] [Cited in This Article: ] |
| 14. | Hosoda W, Chianchiano P, Griffin JF, Pittman ME, Brosens LA, Noë M, Yu J, Shindo K, Suenaga M, Rezaee N, Yonescu R, Ning Y, Albores-Saavedra J, Yoshizawa N, Harada K, Yoshizawa A, Hanada K, Yonehara S, Shimizu M, Uehara T, Samra JS, Gill AJ, Wolfgang CL, Goggins MG, Hruban RH, Wood LD. Genetic analyses of isolated high-grade pancreatic intraepithelial neoplasia (HG-PanIN) reveal paucity of alterations in TP53 and SMAD4. J Pathol. 2017;242:16-23. [PubMed] [DOI] [Cited in This Article: ] |
| 15. | Raczy C, Petrovski R, Saunders CT, Chorny I, Kruglyak S, Margulies EH, Chuang HY, Källberg M, Kumar SA, Liao A, Little KM, Strömberg MP, Tanner SW. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics. 2013;29:2041-2043. [PubMed] [DOI] [Cited in This Article: ] |
| 16. | Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, Cox AJ, Kruglyak S, Saunders CT. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220-1222. [PubMed] [DOI] [Cited in This Article: ] |
| 17. | Kim S, Scheffler K, Halpern AL, Bekritsky MA, Noh E, Källberg M, Chen X, Kim Y, Beyter D, Krusche P, Saunders CT. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018;15:591-594. [PubMed] [DOI] [Cited in This Article: ] |
| 18. | Roller E, Ivakhno S, Lee S, Royce T, Tanner S. Canvas: versatile and scalable detection of copy number variants. Bioinformatics. 2016;32:2375-2377. [PubMed] [DOI] [Cited in This Article: ] |
| 19. | McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. [PubMed] [DOI] [Cited in This Article: ] |
| 20. | Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884-i890. [PubMed] [DOI] [Cited in This Article: ] |
| 21. | Dean CB, Nielsen JD. Generalized linear mixed models: a review and some extensions. Lifetime Data Anal. 2007;13:497-512. [PubMed] [DOI] [Cited in This Article: ] |
| 22. | Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811-2812. [PubMed] [DOI] [Cited in This Article: ] |
| 23. | Cianchi F, Messerini L, Palomba A, Boddi V, Perigli G, Pucciani F, Bechi P, Cortesini C. Character of the invasive margin in colorectal cancer: does it improve prognostic information of Dukes staging? Dis Colon Rectum. 1997;40:1170-5; discussion 1175-6. [PubMed] [Cited in This Article: ] |
| 24. | Nyström H, Naredi P, Berglund A, Palmqvist R, Tavelin B, Sund M. Liver-metastatic potential of colorectal cancer is related to the stromal composition of the tumour. Anticancer Res. 2012;32:5183-5191. [PubMed] [Cited in This Article: ] |
| 25. | Pinheiro RS, Herman P, Lupinacci RM, Lai Q, Mello ES, Coelho FF, Perini MV, Pugliese V, Andraus W, Cecconello I, D'Albuquerque LC. Tumor growth pattern as predictor of colorectal liver metastasis recurrence. Am J Surg. 2014;207:493-498. [PubMed] [DOI] [Cited in This Article: ] |
| 26. | Lugli A, Kirsch R, Ajioka Y, Bosman F, Cathomas G, Dawson H, El Zimaity H, Fléjou JF, Hansen TP, Hartmann A, Kakar S, Langner C, Nagtegaal I, Puppa G, Riddell R, Ristimäki A, Sheahan K, Smyrk T, Sugihara K, Terris B, Ueno H, Vieth M, Zlobec I, Quirke P. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30:1299-1311. [PubMed] [DOI] [Cited in This Article: ] |
| 27. | Marks KM, West NP, Morris E, Quirke P. Clinicopathological, genomic and immunological factors in colorectal cancer prognosis. Br J Surg. 2018;105:e99-e109. [PubMed] [DOI] [Cited in This Article: ] |
| 28. | Graham DM, Appelman HD. Crohn's-like lymphoid reaction and colorectal carcinoma: a potential histologic prognosticator. Mod Pathol. 1990;3:332-335. [PubMed] [Cited in This Article: ] |
| 29. | Väyrynen JP, Sajanti SA, Klintrup K, Mäkelä J, Herzig KH, Karttunen TJ, Tuomisto A, Mäkinen MJ. Characteristics and significance of colorectal cancer associated lymphoid reaction. Int J Cancer. 2014;134:2126-2135. [PubMed] [DOI] [Cited in This Article: ] |
| 30. | Cheng HH, Lin JK, Chen WS, Jiang JK, Yang SH, Chang SC. Clinical significance of the BRAFV600E mutation in Asian patients with colorectal cancer. Int J Colorectal Dis. 2018;33:1173-1181. [PubMed] [DOI] [Cited in This Article: ] |
| 31. | Eachkoti R, Farooq S, Syeed SI, Wani HA, Majid S, Pampori MR. Prevalence and prognostic relevance of BrafV600E mutation in colorectal carcinomas from Kashmir (North India) valley. Mutagenesis. 2018;33:225-230. [PubMed] [DOI] [Cited in This Article: ] |
| 32. | Lee LH, Cavalcanti MS, Segal NH, Hechtman JF, Weiser MR, Smith JJ, Garcia-Aguilar J, Sadot E, Ntiamoah P, Markowitz AJ, Shike M, Stadler ZK, Vakiani E, Klimstra DS, Shia J. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol. 2016;29:1433-1442. [PubMed] [DOI] [Cited in This Article: ] |
| 33. | Droeser RA, Hirt C, Viehl CT, Frey DM, Nebiker C, Huber X, Zlobec I, Eppenberger-Castori S, Tzankov A, Rosso R, Zuber M, Muraro MG, Amicarella F, Cremonesi E, Heberer M, Iezzi G, Lugli A, Terracciano L, Sconocchia G, Oertli D, Spagnoli GC, Tornillo L. Clinical impact of programmed cell death ligand 1 expression in colorectal cancer. Eur J Cancer. 2013;49:2233-2242. [PubMed] [DOI] [Cited in This Article: ] |
| 34. | Eefsen RL, Van den Eynden GG, Høyer-Hansen G, Brodt P, Laerum OD, Vermeulen PB, Christensen IJ, Wettergren A, Federspiel B, Willemoe GL, Vainer B, Osterlind K, Illemann M. Histopathological growth pattern, proteolysis and angiogenesis in chemonaive patients resected for multiple colorectal liver metastases. J Oncol. 2012;2012:907971. [PubMed] [DOI] [Cited in This Article: ] |
| 35. | Budinska E, Popovici V, Tejpar S, D'Ario G, Lapique N, Sikora KO, Di Narzo AF, Yan P, Hodgson JG, Weinrich S, Bosman F, Roth A, Delorenzi M. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63-76. [PubMed] [DOI] [Cited in This Article: ] |
| 36. | Li SR, Dorudi S, Bustin SA. Identification of differentially expressed genes associated with colorectal cancer liver metastasis. Eur Surg Res. 2003;35:327-336. [PubMed] [DOI] [Cited in This Article: ] |
| 37. | Ki DH, Jeung HC, Park CH, Kang SH, Lee GY, Lee WS, Kim NK, Chung HC, Rha SY. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121:2005-2012. [PubMed] [DOI] [Cited in This Article: ] |
| 38. | Ochiai H, Nakanishi Y, Fukasawa Y, Sato Y, Yoshimura K, Moriya Y, Kanai Y, Watanabe M, Hasegawa H, Kitagawa Y, Kitajima M, Hirohashi S. A new formula for predicting liver metastasis in patients with colorectal cancer: immunohistochemical analysis of a large series of 439 surgically resected cases. Oncology. 2008;75:32-41. [PubMed] [DOI] [Cited in This Article: ] |
| 39. | Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A, Donoghue MTA, You D, Viale A, Kemeny N, Segal NH, Stadler ZK, Varghese AM, Kundra R, Gao J, Syed A, Hyman DM, Vakiani E, Rosen N, Taylor BS, Ladanyi M, Berger MF, Solit DB, Shia J, Saltz L, Schultz N. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33:125-136.e3. [PubMed] [DOI] [Cited in This Article: ] |
| 40. | Ariyama J, Suyama M, Ogawa K, Ikari T, Nagaiwa J, Fujii D, Tsuchida A. The detection and prognosis of small pancreatic carcinoma. Int J Pancreatol. 1990;7:37-47. [PubMed] [DOI] [Cited in This Article: ] |
| 41. | Lamalice L, Le Boeuf F, Huot J. Endothelial cell migration during angiogenesis. Circ Res. 2007;100:782-794. [PubMed] [DOI] [Cited in This Article: ] |
| 42. | Bradač GB, Daniele D, Bergui M, Cerrato P, Ferrio MF, Stura G, Coriasco M. Lacunes and other holes: diagnosis, pathogenesis, therapy. Neuroradiol J. 2008;21:35-52. [PubMed] [DOI] [Cited in This Article: ] |
| 43. | Dezső K, Papp V, Bugyik E, Hegyesi H, Sáfrány G, Bödör C, Nagy P, Paku S. Structural analysis of oval-cell-mediated liver regeneration in rats. Hepatology. 2012;56:1457-1467. [PubMed] [DOI] [Cited in This Article: ] |