Published online Jun 21, 2019. doi: 10.3748/wjg.v25.i23.2863
Peer-review started: March 2, 2019
First decision: April 16, 2019
Revised: April 24, 2019
Accepted: May 8, 2019
Article in press: May 8, 2019
Published online: June 21, 2019
Molecular mechanisms associated with inflammation-promoted tumorigenesis have become an important topic in cancer research. Various abnormal epigenetic changes, including DNA methylation, histone modification, chromatin remodeling, and noncoding RNA regulation, occur during the transformation of chronic inflammation into colorectal cancer (CRC). These changes not only accelerate transformation but also lead to cancer progression and metastasis by activating carcinogenic signaling pathways. The NF-κB and STAT3 signaling pathways play a particularly important role in the transformation of inflammation into CRC, and both are critical to cellular signal transduction and constantly activated in cancer by various abnormal changes including epigenetics. The NF-κB and STAT3 signals contribute to the microenvironment for tumorigenesis through secretion of a large number of pro-inflammatory cytokines and their crosstalk in the nucleus makes it even more difficult to treat CRC. Compared with gene mutation that is irreversible, epigenetic inheritance is reversible or can be altered by the intervention. Therefore, understanding the role of epigenetic inheritance in the inflammation-cancer transformation may elucidate the pathogenesis of CRC and promote the development of innovative drugs targeting transformation to prevent and treat this malignancy. This review summarizes the literature on the roles of epigenetic mechanisms in the occurrence and development of inflammation-induced CRC. Exploring the role of epigenetics in the transformation of inflammation into CRC may help stimulate futures studies on the role of molecular therapy in CRC.
Core tip: Chronic inflammation can promote the occurrence and progression of colorectal cancer (CRC), drawing more attention to the role of pro-inflammatory cytokines and immune cells in this cancer. By regulating the expression of various inflammatory signaling pathways and pro-inflammatory cytokines, epigenetic inheritance not only participates in the transformation of inflammation into CRC, but also facilitates invasion, metastasis, and drug resistance of CRC. Compared with gene mutation that is irreversible, epigenetic inheritance is reversible or can be altered by the intervention. Therefore, the tumor microenvironment can be regulated by modulating epigenetic modifications, which may be novel alternatives to prevent and treat CRC.
- Citation: Yang ZH, Dang YQ, Ji G. Role of epigenetics in transformation of inflammation into colorectal cancer. World J Gastroenterol 2019; 25(23): 2863-2877
- URL: https://www.wjgnet.com/1007-9327/full/v25/i23/2863.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i23.2863
In recent years, accumulating evidence indicates that chronic inflammation leads to the occurrence and development of many tumors[1,2]. The relationship between in-flammation and cancer has long been investigated. Two thousand years ago, the Greek physician Galen described the similarities between cancer and inflammation and believed that cancer might evolve from inflammatory lesions[3]. In 1863, Virchow identified inflammatory cell infiltration in tumor tissues and explained it as a reaction to the origination of cancer at sites of chronic inflammation, proposing the hypothesis of the inflammation-cancer transformation[4]. Colorectal cancer (CRC) is the third most common malignant tumor and the fourth leading cause of cancer-related mortality worldwide[5]. The pathogenesis of CRC is complex. Previously, most scholars believed that CRC was a genopathy and a highly heterogeneous tumor resulting from an accumulation of genetic abnormalities, the failure of cancer defense mechanisms, and the activation of carcinogenic pathways[6]. However, increasing evidence suggests that CRC is a typical inflammation-dependent cancer. The risk of developing CRC increases in patients with inflammatory bowel disease (IBD), such as ulcerative colitis or Crohn's disease, which is more likely to be caused by chronic inflammation of the intestinal mucosa than by any definitive genetic predisposition[7-9]. In addition, chronic inflammation plays an important role in the occurrence and development of sporadic CRC, and the expression of interleukin-1 (IL-1), IL-6, IL-17A, and IL-23 is increased in most sporadic CRC cases[10,11].
Various proinflammatory signaling pathways participate in the transformation of inflammation into CRC, including the NF-κB, IL-6/STAT3, cyclooxygenase-2 (COX-2)/PGE2, and IL-23/Th17 pathways, which induce the production of inflammatory mediators, upregulate the expression of antiapoptotic genes, stimulate cell pro-liferation and angiogenesis, and thereby contribute to tumorigenesis[12]. The NF-κB signaling pathway includes both classical and non-canonical pathways. The classical pathway is activated by pro-inflammatory cytokines, pathogen-associated or damage-associated molecular patterns. The non-canonical pathway is activated by a small subset of cytokines including lymphotoxin, receptor activator of NF-κB ligand, CD40 ligand, and B cell activating factor of the tumor necrosis factor (TNF) family[13]. Activation of NF-κB not only affects DNA damage and carcinogenic mutations, but also causes tumorigenesis by promoting the production of reactive oxygen species (ROS) and reactive nitrogen. It can also cause chromosomal instability, aneuploidy, and epigenetic changes, leading to tumorigenesis and development[14,15]. NF-κB and STAT3 are nuclear transcription factors required for the regulation of tumor proliferation, survival, angiogenesis, and invasion; their target genes encode the critical cancer-promoting inflammatory mediators[13-19]. NF-κB and STAT3 signaling contributes to the tumorigenic microenvironment by mediating the secretion of various proinflammatory cytokines, and the crosstalk of these pathways in the nu-cleus makes CRC even more difficult to treat[18,20,21].
To date, abundant evidence has indicated that epigenetic changes play an important role in the transformation of inflammation into CRC as well as in the occurrence, development, invasion, metastasis, and drug resistance of this cancer. These epigenetic changes include DNA methylation, histone modification, and noncoding RNA (ncRNA) alterations. Molecular mechanisms associated with inflammation-promoted tumorigenesis are currently an important branch of cancer research; therefore, understanding the role of epigenetic inheritance in the occurrence and development of CRC may elucidate the pathogenesis of this cancer and promote the development of innovative drugs targeting transformation for CRC treatment.
DNA methylation is an important epigenetic modification related to gene expression and mediated by DNA methyltransferases (DNMTs), and an imbalance in genomic methylation leads to tumors. Approximately half of human gene promoters are rich in C-G sequences, also called CpG loci because of the phosphodiester bond linking the C and G nucleotides. If they are present in DNA sequences, CpG islands are likely located in genetic regulatory elements[22,23] and are usually defined as regions with a length greater than 200 base pairs and a G + C content greater than 50%[24,25]. DNA methylation starts at one end of the islands and continues to gene promoters and initiation sites, altering the three-dimensional configuration of the DNA and inhibiting its interaction with transcription factors, ultimately silencing gene ex-pression. In contrast, hypomethylation promotes gene expression[23,26]. In CRC, the commonly observed types of methylation include hypermethylation of antioncogene DNA and hypomethylation of oncogene DNA. More importantly, DNA methylation can be stably inherited by progeny cells through histone marks at methylation sites, leading to hereditary effects without changes in DNA sequences[27].
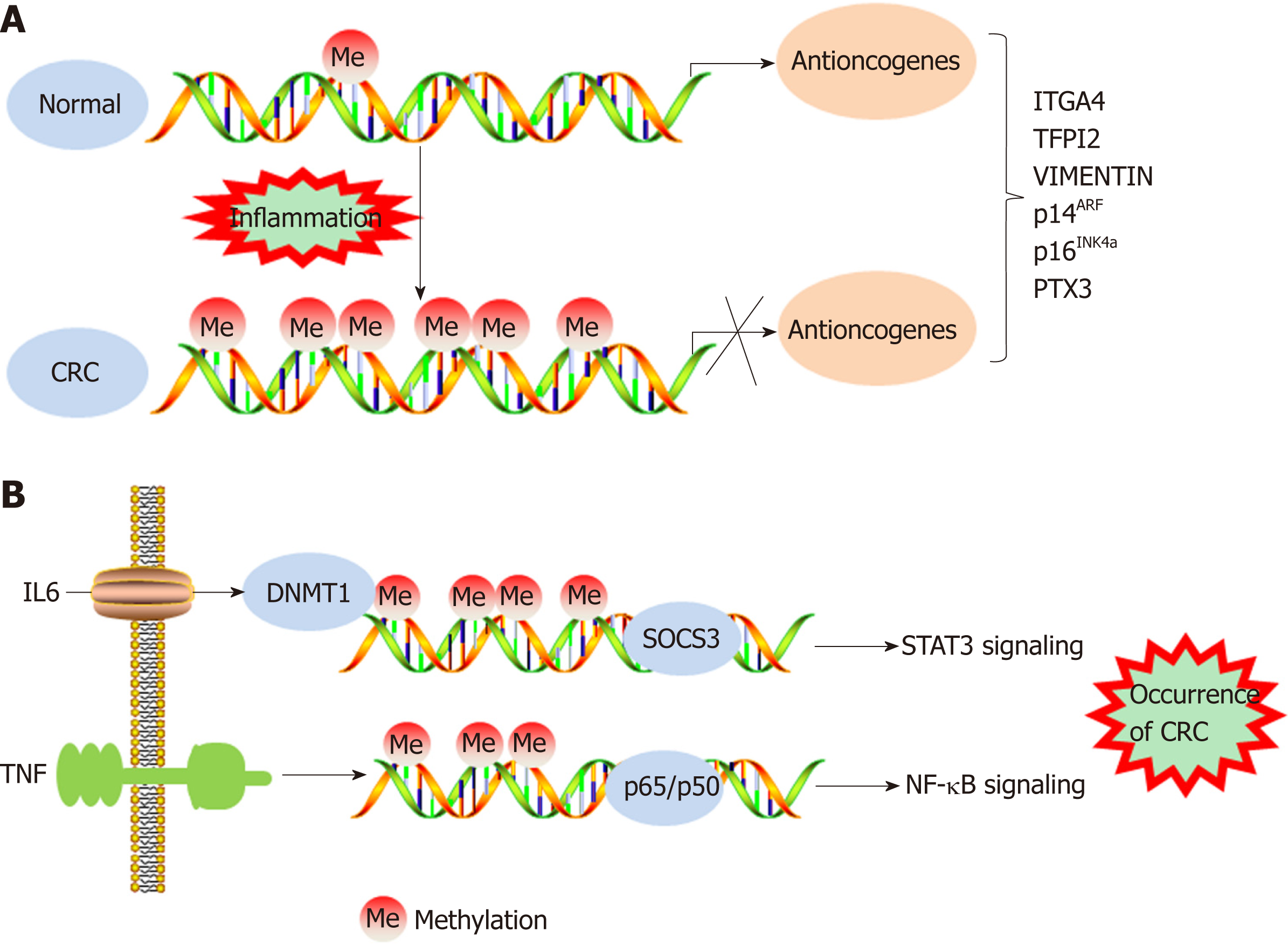
IBD has been demonstrated to increase the risk of CRC incidence. A meta-analysis of population-based cohort studies indicated that the incidence of CRC within 10, 20, and > 20 years was 1%, 2%, and 5%, respectively, in patients with IBD[28,29]. In addition, DNA methylation promotes the tumorigenesis and progression of colitis-associated CRC (CAC). The expression of DNMT1 is appreciably higher in CAC samples than in tumor tissues of patients with sporadic CRC, indicating an increased level of DNA methylation in CAC tissues[30]. However, the frequency of chromosomal instability and microsatellite instability in CAC is generally the same as that in sporadic CRC. In approximately 46% of patients with microsatellite instability-high CAC, the hMLH1 gene is hypermethylated[31]. The cell cycle inhibitor gene p16INK4a, which has been reported to be negatively associated with the occurrence of sporadic CRC, is often methylated in tumor samples from CAC patients[32,33]. In addition, the p14ARF gene can indirectly regulate the expression of the p53 protein, and methylation of p14ARF is a relatively common early event in ulcerative colitis-associated colorectal car-cinogenesis[34]. Furthermore, the methylation levels of the ITGA4, TFPI2, and VIM gene promoters are increased in inflamed colon tissues, which may imply a high risk of CAC development[35] (Figure 1A). Recent genome-wide studies have provided important insights into the characteristics of DNA methylation in tumors[36]. Using monoalkyl methylation profiles in a colitis-induced mouse colon cancer model, Abu-Remaileh et al[37] identified a novel epigenetic modification characterized by hypermethylation of the DNA methylation valley (DMV), which leads to the silencing of DMV-related genes, thus facilitating inflammation-induced cell transformation.
Studies have shown that the methylation of specific genes is associated with inflammatory conditions, dysplasia, and malignant transformation, indicating that epigenetic modifications are involved in inflammation-induced carcinogenesis[38-40]. Many proinflammatory cytokines secreted as a result of NF-κB and STAT3 signaling pathway activation are activated and promote the transformation of inflammation into CRC[20]. For example, IL-6 silences the expression of suppressor of cytokine signaling 3 (SOCS 3) by inducing high expression levels of DNMT1. SOCS3 is an important negative regulator of cytokine-induced STAT3 signaling, and its silencing ultimately contributes to the occurrence of CRC[41]. TNF molecules depend on the NF-κB signaling pathway to silence the gene encoding the proapoptotic protein kinase cδ-binding protein through gene promoter methylation, which facilitates the growth of cancer cells[42]. IL-6 has been demonstrated to increase the methylation of the promoter regions of genes related to tumor inhibition, cell adhesion, and apoptosis resistance, and this increase could be prevented by treatment with the DNMT 1 inhibitor 5-azadeoxycytidine[30] (Figure 1B). IL-6 produced during intestinal inflammation can modulate the expression of CYP2E1 and CYP1B1 (cytochrome P450 enzymes) via transcriptional and epigenetic mechanisms, altering the metabolic capability of epithelial cells. Indeed, one study suggested that IL-6 reduces the expression of miR27b, which targets CYP1B1, through a DNA methylation mechanism, thereby increasing dietary carcinogen activation and DNA injury, which leads to the occurrence of CRC[43]. As an important component of natural humoral immunity, PTX3 activates and regulates the complement cascade by interacting with C1q and factor H and plays a role in the regulation of inflammation. PTX3 has been considered an exogenous antioncogene, and PTX3 deficiency increases sensitivity to epithelial carcinogenesis[44,45]. An analysis of epigenomic data revealed high methylation levels in the PTX3 gene promoter in CRC[46,47]. Prostaglandin, a signaling molecule with important pro- and anti-inflammatory effects, is synthesized from arachidonic acid through the prostaglandin endoperoxide synthase (PTGS; also called cyclooxygenase or COX) pathway. PTGS2 (also called COX-2), one of the key enzymes in the pathway, is overexpressed in CRC, leading to oversecretion of the downstream metabolite prostaglandin E2 (PGE-2)[48]. Deregulation of the COX-2/PGE2 signaling pathway is associated with many tumors, including CRC, and the expression levels of COX-2 and PGE2 are closely related not only to metastasis and poor prognosis in patients with CRC but also to chemotherapeutic resistance in tumors[49-52]. Indeed, a study showed that high methylation rates of select gene promoters stimulate the production of PGE2, block the production of other bioactive prostaglandins, and ultimately promote the development of CRC[53]. Moreover, the results of this study suggest that the antitumor effects of nonsteroidal anti-inflammatory drugs (NSAIDs) may be related to the ability of these drugs to inhibit COX-2. FXR regulates bile acid metabolism and inhibits the production of the secondary bile acid cholic acid; therefore, FXR performs anticancer functions. In CRC, the expression of FXR is negatively associated with the degree of tumor malignancy and with poor clinical outcomes[54,55]. The APC gene is typically mutationally inactivated in the pathogenesis of CRC[56]. Loss of function of APC silences FXR expression through CpG methylation in mouse colonic mucosa and human colon cells, decreasing the expression of downstream bile acid-binding proteins and heterodimers and increasing the expression of related genes (COX-2 and c-MYC) in inflammation and CRC[57]. Recent studies demonstrated that vitamin D (VD) deficiency is associated with the occurrence of CRC. VD, an anti-inflammatory agent, regulates adipocytes and their functions via the VD receptor (VDR), resulting in decreased expression of proinflammatory cytokines[58-61]. Using blood and visceral adipose tissues collected from CRC patients and healthy controls, Castellano-Castillo et al[62] explored the relationship among the levels of serum 25-hydroxyvitamin D [25(OH)D], expression of the VDR gene in adipose tissue, levels of proinflammatory markers, expression of the epigenetic factor DNMT3A, and methylation of the VDR promoter. These results suggest that adipose tissue may be a critical factor in the occurrence of CRC and that low expression levels of 25(OH)D and high expression levels of VDR may partially mediate this relationship by modulating DNA methylation and promoting inflammation[62]. In addition, inflammatory mediators such as ROS and reactive nitrogen species may lead to genomic instability, which contributes to carcinogenesis via the mutation of protooncogenes and tumor suppressor genes[63]. Vitamin C (VC) and vitamin E (VE) are antioxidants that can scavenge free radicals[64,65]. One study suggests that VE antagonizes high glucose-induced oxidative stress, exhibiting beneficial effects on gene promoter methylation and gene expression in the CRC cell line Caco-2[66]. In addition, the results of an in vitro experiment indicated that VC could enhance antitumor drug-induced DNA hydroxymethylation and reactivate epigenetically silenced expression of the tumor suppressor CDKN1A in CRC cells[67]. Therefore, supplementation with related vitamins may be an alternative approach to treat CRC. Moreover, black raspberry (BRB) anthocyanins, which can modulate changes in inflammation and SFRP2 gene methylation, have been reported as agents for CRC prevention[68]. In summary, DNA methylation facilitates the transformation of inflammation into CRC in both the local environment of intestinal inflammation and systemic inflammation.
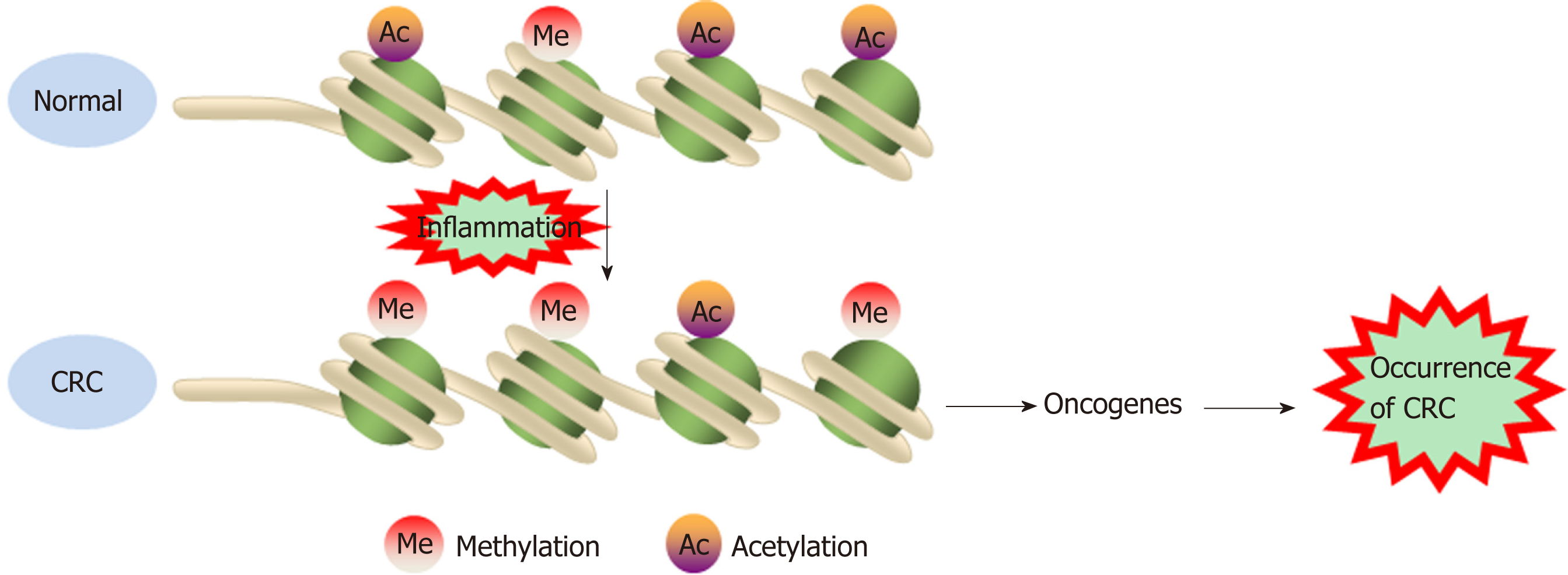
Chromatin is a macromolecular complex composed of DNA, RNA, and proteins. Histones, which regulate DNA strand compaction and gene expression, are the main protein component of chromatin[23]. The core histones are composed of four major families-H2A, H2B, H3, and H4[69]. The nucleosome is a chromatin unit consisting of 150 to 200 base pairs of DNA wrapped closely around a cylindrical histone core. Posttranslational covalent modification of the histone tail constitutes an epigenetic mechanism that regulates chromatin structure and gene expression in human cancers. Histone tail modifications include phosphorylation, methylation, acetylation, ubiquitination, glycosylation, deamination, and ribosylation[70,71]. Various mo-difications alter the three-dimensional structure of nucleosomes and affect the transcriptional control of related genes by inducing either an “inactive” tight heterochromatin or an “active” open euchromatin conformation. Insight into histone modification is not as deep as that into DNA methylation; the only well-studied histone modifications are the acetylation/deacetylation and methylation/ demethylation of lysine and arginine residues in the histone tail[72,73]. These bivalent histone modifications are mediated by polycomb group proteins (transcriptional repressors), which contribute to silencing a specific set of anti-oncogenes in human cancers. Polycomb repressive complexes (PRCs) include PRC1 and PRC2, which silence genes alone or in cooperation[74,75].
Histone modification abnormalities arise during the transformation of inflammation into CRC. Profiles for active enhancers using H3K27ac histone mo-dification identified several cancer markers, such as the LYZ, S100P, and NPSR1 proteins, which were elevated in CAC[76]. In a mouse model, deletion of the Giα2 protein led to spontaneous colitis and right multifocal CRC, which is similar to human CRC with defective mismatch repair. Moreover, MLH1 and PMS2 expression are reduced in the colonic epithelium of Giα2-/- mice after the onset of hypoxic colitis. Notably, MLH1 is epigenetically silenced by reduced histone acetylation. These data connect chronic hypoxic inflammation, histone modulation, and CRC development[77]. The Wnt/β-catenin signaling pathway exerts either pro- or anti-inflammatory functions by activating or inhibiting NF-κB signals, respectively, playing an important role in the occurrence and development of CRC[78]. DACT3, the negative regulator of Wnt/beta-catenin signaling, is transcriptionally suppressed in CRC in a manner related to bivalent histone modification. However, DACT3 expression can be restored after combined administration of drugs targeting histone methylation and deacetylation, resulting in strong inhibition of Wnt/beta-catenin signal transduction and massive CRC cell apoptosis. Therefore, DACT3 may be an important factor in the treatment of CRC through epigenetic mechanisms[79]. EZH2, a catalytic subunit of PRC2, is essential for maintaining the integrity and homeostasis of the epithelial cell barrier in inflammatory states. EZH2 expression is downregulated in IBD patients, and EZH2 inactivation in the intestinal epithelium increases the sensitivity of mice to dextran sodium sulfate (DSS)- and 2,4,6-trinitrobenzenesulfonic acid-induced experimental colitis. One study indicated that EZH2 deficiency could stimulate the expression of TRAF2/5 and enhance the NF-κB signaling induced by TNF-α, which led to an uncontrolled inflammatory reaction and ultimately contributed to tu-morigenesis[80]. Researchers identified a new epigenetic mechanism underlying the preventive effects of aspirin in CAC. Aspirin reduced the activity of histone deacetylases and fully restored H3K27ac. Moreover, aspirin depressed azoxy-methane/DSS-induced H3K27ac accumulation in the promoters of the inducible nitric oxide synthase, TNF-α, and IL-6 genes and suppressed the production of proin-flammatory cytokines, playing a role in the prevention of cancer[81] (Figure 2). Although few studies have investigated the role of histone modification in the transformation of inflammation into CRC, notably, histone modification may interact with DNA methylation to induce epigenetic silencing and promote tumorigenesis.
Most of the human genome is transcribed into RNAs, which are classified as RNAs with coding potential or RNAs without. The latter RNAs are also called ncRNAs. NcRNAs were historically considered “transcriptional waste”, but accumulating evidence indicates that ncRNAs strongly impact many molecular mechanisms[82]. MicroRNAs are single-stranded ncRNAs with a length of 20 nucleotides that function primarily to negatively regulate gene expression by binding to target RNAs and inducing the degradation or inhibiting the translation of those RNAs[83].
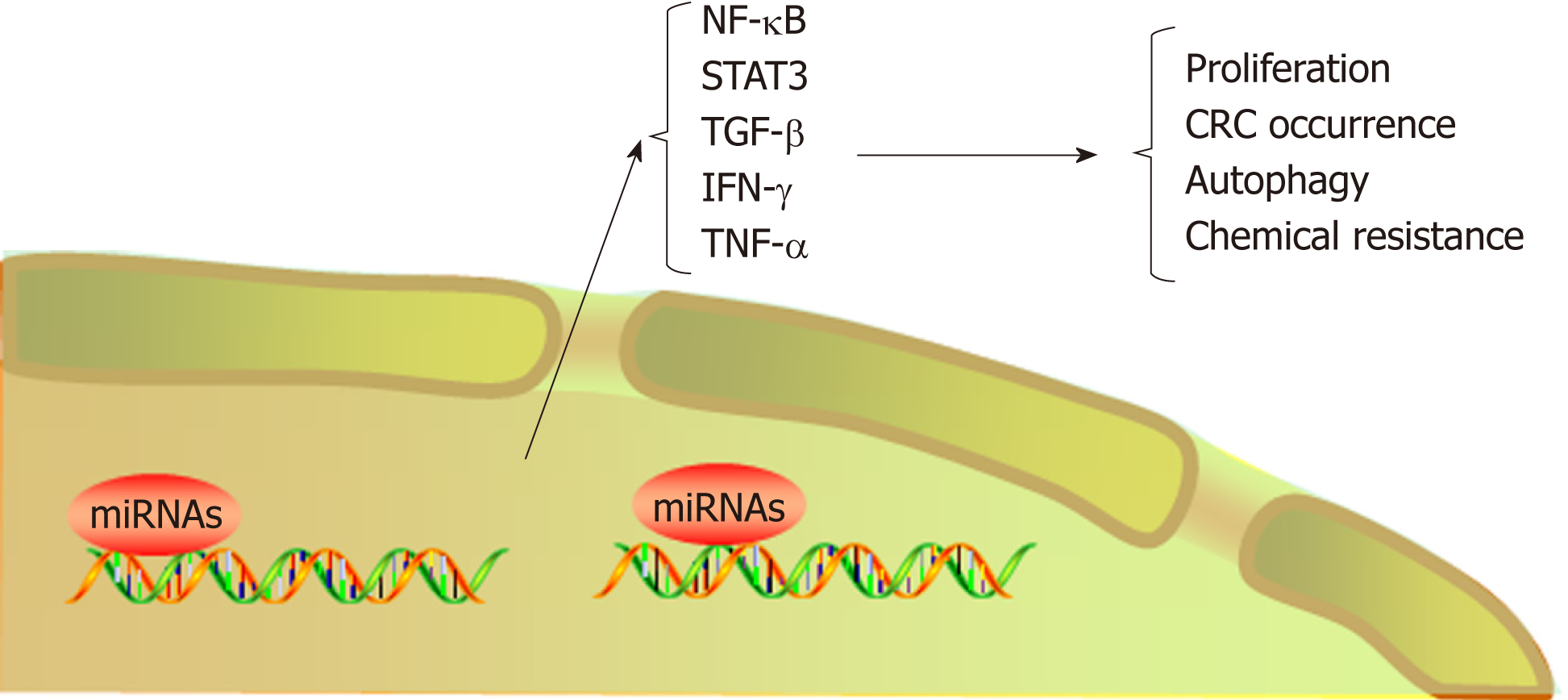
The NF-κB and STAT3 signaling pathways play an important role in the trans-formation of inflammation into cancer[13], and numerous microRNAs promote transformation by participating in these signaling pathways. Slattery et al[84] analyzed the expression profiles of genes and associated microRNAs in the NF-κB signaling pathway between CRC and normal mucosa, providing new insight into therapeutic targets for CRC. TNF-α has been shown to increase the expression of miR-105, which targets RAP2C, activate NF-κB signal transduction by IKK, and ultimately contribute to CRC progression[85]. Another study demonstrated that TNF-α leads to high expression of miR-19a, which can also activate NF-κB signaling to facilitate the occurrence of colitis and CAC[86]. STAT3 not only is a downstream target of IL-6 but also interacts with miR-21, miR-181b-1, PTEN, and CYLD, which implicates the epigenetic switch that links inflammation to cancer[87]. In addition, by activating miR-21, NR2F2 inhibits Smad7 expression and promotes TGF-β-dependent epithelial-mesenchymal transition (EMT) in CRC[88]. In primary CRC samples and cell lines, increased STAT3 expression levels are accompanied by elevated miR-572 and decreased MOAP-1 levels, and miR-572 has been found to reduce the expression of the proapoptotic protein MOAP-1 and to contribute to CRC progression[89]. Öner et al[90] demonstrated that combined inactivation of TP53 and miR-34a promotes CRC metastasis by elevating the levels of IL-6R and PAI1, implying that modifying these processes might be alternative approaches to treat CRC. Immune cells play a dual role in the occurrence and development of CRC, and natural killer (NK) cells promote tumor cell apoptosis by secreting high levels of cytokines, such as IFN-γ and TNF-α[91]. The level of miR-24 was reported to be increased in NK cells from CRC patients, a characteristic that decreased the levels of cytokines, including IFN-γ and TNF-α, by suppressing Paxillin expression and inhibiting the cytotoxic effect of NK cells on CRC cells[92].
MicroRNAs not only promote the transformation of inflammation into CRC but also lead to chemotherapeutic resistance. For example, in CRC patients treated with oxaliplatin, miR-34a expression decreased significantly but Smad4 and TGF-β expression increased. In addition, the expression levels of Smad4 and miR-34a were negatively correlated in CRC patients. Further investigation demonstrated that miR-34a targeted Smad4 through the TGF-β/Smad4 pathway and ultimately inhibited cellular autophagy[93]. Ren et al found overexpression of miR-196b-5p in recurrent CRC tissues, which was associated with a poor prognosis. Further investigation showed that miR-196b-5p activated STAT3 signaling and promoted the chemical resistance of CRC cells to 5-fluorouracil (5-FU) by targeting the negative regulators SOCS1 and SOCS3 in the STAT3 signaling pathway[94]. Clarification of the role of microRNAs in CRC resistance mechanisms can improve the efficacy of chemotherapy by targeting the corresponding microRNAs. Indeed, microRNAs overexpressed during the transformation of inflammation into CRC are often associated with disease progression. In addition, certain microRNAs are silenced or downregulated during the occurrence and development of CRC; these microRNAs generally inhibit the transformation of inflammation into cancer. For instance, miR-148a, miR-6869-5p, and miR-139-5p inhibit transformation by suppressing the NF-κB signaling pathway[95-97]; miR-1299, miR-149, and miR-214 inhibit tumor formation by suppressing the STAT3 signaling pathway[98-100]; and miR-329 inhibits CRC occurrence by targeting TGF-β1[101]. MiR15A and miR16-1, which might act as tumor suppressors, were found to be downregulated in CRC[102]. Animal experiments showed that deficiency of miR15A and miR16-1 led to an accumulation of immunosuppressive IgA+ B cells in intestinal cancer tissues, thereby inhibiting the proliferation and functions of CD8+ T cells with antitumor immunity and ultimately promoting tumor progression[103].
Exosomes are small vesicles released by cells into the extracellular environment. These vesicles transmit information to adjacent or distant cells by transferring RNAs and proteins, thus affecting signaling pathways in various physiological and path-ological conditions[104,105]. Recently, microRNAs enriched in exosomes have been found to interact with immune cells and proinflammatory cytokines and to participate in the progression and metastasis of CRC. For example, exosome-mediated miR-200b was indicated to promote the proliferation of CRC cells upon TGF-β1 exposure[106]. In addition, it was reported that the level of miR-10b was significantly higher in exosomes derived from CRC cells than in those derived from normal colorectal epithelial cells and that CRC-derived exosomes could promote CRC progression[107]. Tumor-associated macrophages (TAMs), a biomarker of solid tumors, are usually associated with a poor prognosis. A recent study has shown that CRC cells carrying mutant p53 genes selectively shed miR-1246-enriched exosomes. These exosomes can be captured by neighboring TAMs, which then secrete numerous proinflammatory cytokines, such as IL-10, TGF-β, and MMPs, to promote the immune suppression, invasion, and metastasis of CRC cells[108] (Figure 3, Table 1).
| MicroRNAs | Target genes | Functions in CRC |
| MiR-105, miR-19a, | NF-κB | Promoted CRC progression |
| MiR-21, miR-181b-1, miR-572 | STAT3 | Promoted CRC progression |
| MiR-21, miR-200b, miR-1246 | TGF-β | Promoted proliferation |
| MiR-34a | IL-6R/PAI1 | Promoted CRC progression |
| MiR-24 | IFN-γ/TNF-α | Inhibited the cytotoxic effect of NK cells |
| MiR-34a | TGF-β/Smad4 | Inhibited autophagy |
| MiR-196b-5p | STAT3 | Promoted chemical resistance |
| MiR-148a, miR-6869-5p, miR-139-5p | NF-κB | Inhibited CRC occurrence |
| MiR-1299, miR-149, miR-214 | STAT3 | Inhibited tumor formation |
| MiR-329 | TGF-β1 | Inhibited CRC occurrence |
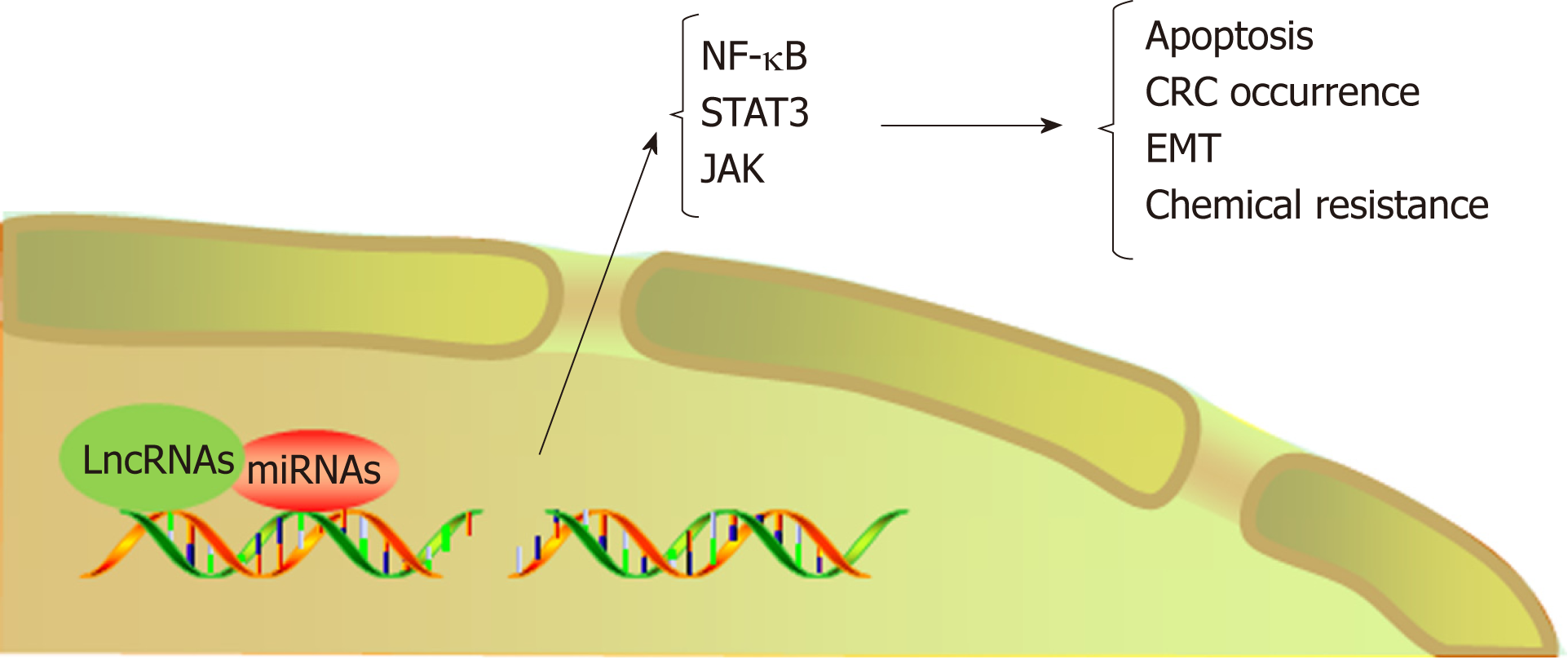
LncRNAs refer to noncoding transcripts of more than 200 nucleotides[109]. LncRNAs are important participants in cancer biology and generally cause the abnormal expression of gene products, leading to the progression of various human tumors[110,111]. In addition, lncRNAs are involved in the transformation of chronic inflammation into CRC. For example, a study indicated that the interaction between lncRNA PRINS and miR-491-5p regulated the proapoptotic factor PMAIP1 and enhanced the antiapoptotic effect of TFF3 against the proapoptotic effects of IFN-γ and TNF-α in CRC cells[112]. LncRNA FEZF1-AS1 expression was higher in CRC tissue than in normal tissue and was associated with a poor prognosis in CRC. FEZF1-AS1 can bind to and increase the stability of the pyruvate kinase 2 (PKM2) protein, which can increase PKM2 levels in the cytoplasm and nucleus and promote pyruvate kinase activity and lactic acid production. Upregulation of nuclear PKM2 induced by FEZF1-AS1 was found to further activate STAT3 signal transduction and accelerate the transformation of inflammation to cancer[113]. In addition, researchers found that lncRNA AB073614 induced EMT in CRC cells by regulating the JAK/STAT3 pathway[114]. Recent studies have shown that lncRNAs not only participate in the transformation of inflammation to tumors but also induce the resistance of CRC to chemotherapy by regulating inflammatory signaling pathways. Abnormal expression of HOTAIR is positively correlated with progression, survival, and poor prognosis in different types of cancers, such as breast cancer, gastric cancer, and CRC[115-117]. Li et al[118] revealed that HOTAIR silenced the expression of the miR-218 gene by recruiting EZH2 for binding to the miR-218 promoter. Silencing of miR-218 resulted in chemotherapeutic resistance of CRC to 5-FU by promoting VOPP1 expression and eventually activating the NF-kB/TS signaling pathway. HOTAIR can thus be used as a novel prognostic indicator and therapeutic target for CRC. Inhibiting HOTAIR may be a future approach to improve the sensitivity of 5-FU chemotherapy (Figure 4, Table 2).
| LncRNA | Target genes | Functions in CRC |
| PRINS | miR-491-5p/PMAIP1/TFF3 | Inhibited apoptosis |
| FEZF1-AS1 | PKM2/STAT3 | Accelerated CRC occurrence |
| AB073614 | JAK/STAT3 | Induced EMT |
| HOTAIR | MiR-218/EZH2/NF-kB | Chemotherapeutic resistance |
Due to the inflammatory basis of CRC, anti-inflammatory agents may be candidates for treating or preventing the disease. NSAIDs are non-selective inhibitors of COX-2[119]. COX-2 is highly expressed in many tumor types, including CRC[120]. NSAIDs play a striking role in the prevention of CRC. A preliminary study on patients with familial adenomatous polyposis indicated that after 1 year of treatment with the NSAID sulindac, patients tended to exhibit a decrease in polyps[121]. A large-scale ob-servational study in 1991 reported that the use of NSAIDs reduced the risk of fatal CRC[122]. Retrospective studies have demonstrated that NSAID treatment is associated with a decreased risk of recurrence of colorectal polyps and tumors. It has been reported that patients who use low-dose aspirin for more than 5 years show a decrease in overall risk of CRC by 40%-50%, and NSAIDs have a positive effect on advanced CRC[123,124]. NSAID therapy can also inhibit the tumor-promoting pathway by inhibiting Wnt signaling[125]. However, a meta-analysis of NSAID treatment to prevent the transformation of IBD into CRC shows that there is a lack of high-quality evidence that anti-inflammatory drugs can be used to prevent CRC in patients with IBD[126]. Most scholars believe that the anti-tumor mechanism of NSAIDs is reflected in two aspects. On the one hand, the cytokines released during inflammation play an important role in reprogramming adult stem cells into malignant cells, and NSAIDs can prevent this process[127]. On the other hand, the mechanism is related to activation of the Wnt pathway by prostaglandins[128]. However, the side effects of NSAIDs must be taken into consideration. NSAID treatment can increase the risk of gastrointestinal bleeding, even at low doses[129]. Caution should be exercised to prevent bleeding when using anti-inflammatory drugs in patients with fragile blood vessels[130]. In addition to NSAIDs, monoclonal antibodies to cytokines such as IL-6 and TNF inhibitors have been investigated in a large number of anti-tumor studies, but most of them are still in the experimental stage[131,132]. The treatment hazard of using anti-cytokines to treat tumors is the increase in the risk of infection[133,134].
In conclusion, chronic inflammation can promote the occurrence and progression of CRC, a finding that is attracting increased attention to the role of proinflammatory cytokines and immune cells in cancer. By regulating the expression of various in-flammatory signaling pathways and proinflammatory cytokines, epigenetic inheritance not only participates in the transformation of inflammation into CRC but also facilitates CRC invasion, metastasis, and drug resistance. However, epigenetic inheritance accelerates the transformation of chronic inflammation into tumors through various modifications influenced by the inflammatory environment. An in-depth understanding of this process will allow us to clarify the pathogenesis of CRC, and some epigenetic modifications can be used as markers for CRC diagnosis. Unlike gene mutations, which are irreversible, epigenetic inheritance is reversible or can be altered by interventions. For example, DNA demethylation promotes tumor sup-pressor gene expression to re-establish tumor prevention and reduce expression of pro-inflammatory cytokines by regulation of histone modifications or ncRNAs, ultimately reducing inflammation infiltration of tumor microenvironment. In the future, this new anti-tumor drug may be used in combination with immunotherapy, chemotherapy, and targeted cancer therapy for the treatment of CRC. Exploring the role of epigenetics in the transformation of inflammation into CRC may help stimulate futures studies on the role of molecular therapy in CRC.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Das U, Sterpetti AV, Vetvicka V, Zheng YW S-Editor: Yan JP L-Editor: Wang TQ E-Editor: Ma YJ
| 1. | Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42985] [Article Influence: 3306.5] [Reference Citation Analysis (4)] |
| 2. | Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10123] [Cited by in F6Publishing: 10570] [Article Influence: 480.5] [Reference Citation Analysis (0)] |
| 3. | Reedy J. Galen on cancer and related diseases. Clio Med. 1975;10:227-238. [PubMed] [Cited in This Article: ] |
| 4. | Balkwill F, Mantovani A. Inflammation and cancer: Back to Virchow? Lancet. 2001;357:539-545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5245] [Cited by in F6Publishing: 5425] [Article Influence: 235.9] [Reference Citation Analysis (0)] |
| 5. | Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-1502. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1965] [Cited by in F6Publishing: 2133] [Article Influence: 213.3] [Reference Citation Analysis (1)] |
| 6. | Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30:363-371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 7. | Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807-1816. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 738] [Cited by in F6Publishing: 779] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 8. | Pedersen N, Duricova D, Elkjaer M, Gamborg M, Munkholm P, Jess T. Risk of extra-intestinal cancer in inflammatory bowel disease: Meta-analysis of population-based cohort studies. Am J Gastroenterol. 2010;105:1480-1487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 9. | Bernstein CN, Blanchard JF, Rawsthorne P, Yu N. The prevalence of extraintestinal diseases in inflammatory bowel disease: A population-based study. Am J Gastroenterol. 2001;96:1116-1122. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 495] [Cited by in F6Publishing: 475] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 10. | Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, Galon J. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71:1263-1271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 774] [Cited by in F6Publishing: 837] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 11. | Schetter AJ, Nguyen GH, Bowman ED, Mathé EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878-5887. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 12. | Luo C, Zhang H. The Role of Proinflammatory Pathways in the Pathogenesis of Colitis-Associated Colorectal Cancer. Mediators Inflamm. 2017;2017:5126048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 13. | Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: Coming of age. Nat Rev Immunol. 2018;18:309-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1132] [Cited by in F6Publishing: 1214] [Article Influence: 202.3] [Reference Citation Analysis (0)] |
| 14. | Karin M, Greten FR. NF-kappaB: Linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749-759. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2227] [Cited by in F6Publishing: 2298] [Article Influence: 120.9] [Reference Citation Analysis (0)] |
| 15. | Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33-40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 799] [Cited by in F6Publishing: 814] [Article Influence: 54.3] [Reference Citation Analysis (0)] |
| 16. | Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740-749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 881] [Cited by in F6Publishing: 863] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 17. | Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97-105. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1723] [Cited by in F6Publishing: 1755] [Article Influence: 87.8] [Reference Citation Analysis (0)] |
| 18. | Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: Role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1322] [Cited by in F6Publishing: 1366] [Article Influence: 80.4] [Reference Citation Analysis (0)] |
| 19. | Bassères DS, Baldwin AS. Nuclear factor-kappaB and inhibitor of kappaB kinase pathways in oncogenic initiation and progression. Oncogene. 2006;25:6817-6830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 517] [Cited by in F6Publishing: 534] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 20. | West NR, McCuaig S, Franchini F, Powrie F. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol. 2015;15:615-629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 260] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 21. | Grivennikov SI, Karin M. Dangerous liaisons: STAT3 and NF-kappaB collaboration and crosstalk in cancer. Cytokine Growth Factor Rev. 2010;21:11-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 748] [Cited by in F6Publishing: 820] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 22. | Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442-1460.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 175] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 23. | Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology. 2015;149:1204-1225.e12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 424] [Cited by in F6Publishing: 484] [Article Influence: 53.8] [Reference Citation Analysis (1)] |
| 24. | Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261-282. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2342] [Cited by in F6Publishing: 2283] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 25. | Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol. 2011;8:686-700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 432] [Cited by in F6Publishing: 475] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 26. | Graff JR, Herman JG, Myöhänen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cells implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322-22329. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 229] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 27. | Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Curr Opin Cell Biol. 2007;19:257-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 96] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Beaugerie L, Itzkowitz SH. Cancers Complicating Inflammatory Bowel Disease. N Engl J Med. 2015;373:195. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 29. | Lutgens MW, van Oijen MG, van der Heijden GJ, Vleggaar FP, Siersema PD, Oldenburg B. Declining risk of colorectal cancer in inflammatory bowel disease: An updated meta-analysis of population-based cohort studies. Inflamm Bowel Dis. 2013;19:789-799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 279] [Cited by in F6Publishing: 308] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 30. | Foran E, Garrity-Park MM, Mureau C, Newell J, Smyrk TC, Limburg PJ, Egan LJ. Upregulation of DNA methyltransferase-mediated gene silencing, anchorage-independent growth, and migration of colon cancer cells by interleukin-6. Mol Cancer Res. 2010;8:471-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 114] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 31. | Fleisher AS, Esteller M, Harpaz N, Leytin A, Rashid A, Xu Y, Liang J, Stine OC, Yin J, Zou TT, Abraham JM, Kong D, Wilson KT, James SP, Herman JG, Meltzer SJ. Microsatellite instability in inflammatory bowel disease-associated neoplastic lesions is associated with hypermethylation and diminished expression of the DNA mismatch repair gene, hMLH1. Cancer Res. 2000;60:4864-4868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 32. | Wang FY, Arisawa T, Tahara T, Takahama K, Watanabe M, Hirata I, Nakano H. Aberrant DNA methylation in ulcerative colitis without neoplasia. Hepatogastroenterology. 2008;55:62-65. [PubMed] [Cited in This Article: ] |
| 33. | Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998;58:3942-3945. [PubMed] [Cited in This Article: ] |
| 34. | Sato F, Harpaz N, Shibata D, Xu Y, Yin J, Mori Y, Zou TT, Wang S, Desai K, Leytin A, Selaru FM, Abraham JM, Meltzer SJ. Hypermethylation of the p14(ARF) gene in ulcerative colitis-associated colorectal carcinogenesis. Cancer Res. 2002;62:1148-1151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 84] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 35. | Gerecke C, Scholtka B, Löwenstein Y, Fait I, Gottschalk U, Rogoll D, Melcher R, Kleuser B. Hypermethylation of ITGA4, TFPI2 and VIMENTIN promoters is increased in inflamed colon tissue: Putative risk markers for colitis-associated cancer. J Cancer Res Clin Oncol. 2015;141:2097-2107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 36. | Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2011;44:40-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 494] [Cited by in F6Publishing: 466] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 37. | Abu-Remaileh M, Bender S, Raddatz G, Ansari I, Cohen D, Gutekunst J, Musch T, Linhart H, Breiling A, Pikarsky E, Bergman Y, Lyko F. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75:2120-2130. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 73] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 38. | Hahn MA, Hahn T, Lee DH, Esworthy RS, Kim BW, Riggs AD, Chu FF, Pfeifer GP. Methylation of polycomb target genes in intestinal cancer is mediated by inflammation. Cancer Res. 2008;68:10280-10289. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 39. | Wehbe H, Henson R, Meng F, Mize-Berge J, Patel T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006;66:10517-10524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 177] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 40. | Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573-3577. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 41. | Li Y, Deuring J, Peppelenbosch MP, Kuipers EJ, de Haar C, van der Woude CJ. IL-6-induced DNMT1 activity mediates SOCS3 promoter hypermethylation in ulcerative colitis-related colorectal cancer. Carcinogenesis. 2012;33:1889-1896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 94] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 42. | Lee JH, Kang MJ, Han HY, Lee MG, Jeong SI, Ryu BK, Ha TK, Her NG, Han J, Park SJ, Lee KY, Kim HJ, Chi SG. Epigenetic alteration of PRKCDBP in colorectal cancers and its implication in tumor cell resistance to TNFα-induced apoptosis. Clin Cancer Res. 2011;17:7551-7562. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 43. | Patel SA, Bhambra U, Charalambous MP, David RM, Edwards RJ, Lightfoot T, Boobis AR, Gooderham NJ. Interleukin-6 mediated upregulation of CYP1B1 and CYP2E1 in colorectal cancer involves DNA methylation, miR27b and STAT3. Br J Cancer. 2014;111:2287-2296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 44. | Bottazzi B, Doni A, Garlanda C, Mantovani A. An integrated view of humoral innate immunity: Pentraxins as a paradigm. Annu Rev Immunol. 2010;28:157-183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 413] [Cited by in F6Publishing: 413] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 45. | Deban L, Jarva H, Lehtinen MJ, Bottazzi B, Bastone A, Doni A, Jokiranta TS, Mantovani A, Meri S. Binding of the long pentraxin PTX3 to factor H: Interacting domains and function in the regulation of complement activation. J Immunol. 2008;181:8433-8440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 46. | Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, Greco C, Feruglio F, Molgora M, Laface I, Tartari S, Doni A, Pasqualini F, Barbati E, Basso G, Galdiero MR, Nebuloni M, Roncalli M, Colombo P, Laghi L, Lambris JD, Jaillon S, Garlanda C, Mantovani A. PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell. 2015;160:700-714. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 218] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 47. | Rubino M, Kunderfranco P, Basso G, Greco CM, Pasqualini F, Serio S, Roncalli M, Laghi L, Mantovani A, Papait R, Garlanda C. Epigenetic regulation of the extrinsic oncosuppressor PTX3 gene in inflammation and cancer. Oncoimmunology. 2017;6:e1333215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 48. | Wang D, Dubois RN. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010;29:781-788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 569] [Cited by in F6Publishing: 606] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 49. | Greenhough A, Smartt HJ, Moore AE, Roberts HR, Williams AC, Paraskeva C, Kaidi A. The COX-2/PGE2 pathway: Key roles in the hallmarks of cancer and adaptation to the tumour microenvironment. Carcinogenesis. 2009;30:377-386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 848] [Cited by in F6Publishing: 891] [Article Influence: 59.4] [Reference Citation Analysis (0)] |
| 50. | Cathcart MC, Lysaght J, Pidgeon GP. Eicosanoid signalling pathways in the development and progression of colorectal cancer: Novel approaches for prevention/intervention. Cancer Metastasis Rev. 2011;30:363-385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 51. | Ma X, Aoki T, Tsuruyama T, Narumiya S. Definition of Prostaglandin E2-EP2 Signals in the Colon Tumor Microenvironment That Amplify Inflammation and Tumor Growth. Cancer Res. 2015;75:2822-2832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 52. | Kunzmann AT, Murray LJ, Cardwell CR, McShane CM, McMenamin UC, Cantwell MM. PTGS2 (Cyclooxygenase-2) expression and survival among colorectal cancer patients: A systematic review. Cancer Epidemiol Biomarkers Prev. 2013;22:1490-1497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 53. | Cebola I, Custodio J, Muñoz M, Díez-Villanueva A, Paré L, Prieto P, Aussó S, Coll-Mulet L, Boscá L, Moreno V, Peinado MA. Epigenetics override pro-inflammatory PTGS transcriptomic signature towards selective hyperactivation of PGE2 in colorectal cancer. Clin Epigenetics. 2015;7:74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 54. | Lax S, Schauer G, Prein K, Kapitan M, Silbert D, Berghold A, Berger A, Trauner M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced in human colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int J Cancer. 2012;130:2232-2239. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 55. | Modica S, Gadaleta RM, Moschetta A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl Recept Signal. 2010;8:e005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 184] [Cited by in F6Publishing: 204] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 56. | Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449-2460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1274] [Cited by in F6Publishing: 1304] [Article Influence: 86.9] [Reference Citation Analysis (2)] |
| 57. | Selmin OI, Fang C, Lyon AM, Doetschman TC, Thompson PA, Martinez JD, Smith JW, Lance PM, Romagnolo DF. Inactivation of Adenomatous Polyposis Coli Reduces Bile Acid/Farnesoid X Receptor Expression through Fxr gene CpG Methylation in Mouse Colon Tumors and Human Colon Cancer Cells. J Nutr. 2016;146:236-242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 58. | Saccone D, Asani F, Bornman L. Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene. 2015;561:171-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 113] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 59. | Song M, Konijeti GG, Yuan C, Ananthakrishnan AN, Ogino S, Fuchs CS, Giovannucci EL, Ng K, Chan AT. Plasma 25-Hydroxyvitamin D, Vitamin D Binding Protein, and Risk of Colorectal Cancer in the Nurses' Health Study. Cancer Prev Res (Phila). 2016;9:664-672. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 60. | Ding C, Gao D, Wilding J, Trayhurn P, Bing C. Vitamin D signalling in adipose tissue. Br J Nutr. 2012;108:1915-1923. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 201] [Cited by in F6Publishing: 214] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 61. | Lira FS, Rosa JC, Cunha CA, Ribeiro EB, do Nascimento CO, Oyama LM, Mota JF. Supplementing alpha-tocopherol (vitamin E) and vitamin D3 in high fat diet decrease IL-6 production in murine epididymal adipose tissue and 3T3-L1 adipocytes following LPS stimulation. Lipids Health Dis. 2011;10:37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 62. | Castellano-Castillo D, Morcillo S, Clemente-Postigo M, Crujeiras AB, Fernandez-García JC, Torres E, Tinahones FJ, Macias-Gonzalez M. Adipose tissue inflammation and VDR expression and methylation in colorectal cancer. Clin Epigenetics. 2018;10:60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 63. | Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: Interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 436] [Cited by in F6Publishing: 468] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 64. | Serbinova E, Kagan V, Han D, Packer L. Free radical recycling and intramembrane mobility in the antioxidant properties of alpha-tocopherol and alpha-tocotrienol. Free Radic Biol Med. 1991;10:263-275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 459] [Cited by in F6Publishing: 371] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 65. | Du J, Cullen JJ, Buettner GR. Ascorbic acid: Chemistry, biology and the treatment of cancer. Biochim Biophys Acta. 2012;1826:443-457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 368] [Cited by in F6Publishing: 475] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 66. | Zappe K, Pointner A, Switzeny OJ, Magnet U, Tomeva E, Heller J, Mare G, Wagner KH, Knasmueller S, Haslberger AG. Counteraction of Oxidative Stress by Vitamin E Affects Epigenetic Regulation by Increasing Global Methylation and Gene Expression of MLH1 and DNMT1 Dose Dependently in Caco-2 Cells. Oxid Med Cell Longev. 2018;2018:3734250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 67. | Gerecke C, Schumacher F, Edlich A, Wetzel A, Yealland G, Neubert LK, Scholtka B, Homann T, Kleuser B. Vitamin C promotes decitabine or azacytidine induced DNA hydroxymethylation and subsequent reactivation of the epigenetically silenced tumour suppressor CDKN1A in colon cancer cells. Oncotarget. 2018;9:32822-32840. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 68. | Chen L, Jiang B, Zhong C, Guo J, Zhang L, Mu T, Zhang Q, Bi X. Chemoprevention of colorectal cancer by black raspberry anthocyanins involved the modulation of gut microbiota and SFRP2 demethylation. Carcinogenesis. 2018;39:471-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 96] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 69. | Andrews AJ, Luger K. Nucleosome structure(s) and stability: Variations on a theme. Annu Rev Biophys. 2011;40:99-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 203] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 70. | Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693-705. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7446] [Cited by in F6Publishing: 7318] [Article Influence: 430.5] [Reference Citation Analysis (0)] |
| 71. | Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074-1080. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7019] [Cited by in F6Publishing: 6672] [Article Influence: 290.1] [Reference Citation Analysis (0)] |
| 72. | Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683-692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3299] [Cited by in F6Publishing: 3283] [Article Influence: 193.1] [Reference Citation Analysis (0)] |
| 73. | Chen HP, Zhao YT, Zhao TC. Histone deacetylases and mechanisms of regulation of gene expression. Crit Rev Oncog. 2015;20:35-47. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 169] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 74. | Hahn MA, Li AX, Wu X, Yang R, Drew DA, Rosenberg DW, Pfeifer GP. Loss of the polycomb mark from bivalent promoters leads to activation of cancer-promoting genes in colorectal tumors. Cancer Res. 2014;74:3617-3629. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 75. | Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846-856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1021] [Cited by in F6Publishing: 991] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 76. | Sarvestani SK, Signs SA, Lefebvre V, Mack S, Ni Y, Morton A, Chan ER, Li X, Fox P, Ting A, Kalady MF, Cruise M, Ashburn J, Stiene J, Lai W, Liska D, Xiang S, Huang EH. Cancer-predicting transcriptomic and epigenetic signatures revealed for ulcerative colitis in patient-derived epithelial organoids. Oncotarget. 2018;9:28717-28730. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 77. | Edwards RA, Witherspoon M, Wang K, Afrasiabi K, Pham T, Birnbaumer L, Lipkin SM. Epigenetic repression of DNA mismatch repair by inflammation and hypoxia in inflammatory bowel disease-associated colorectal cancer. Cancer Res. 2009;69:6423-6429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 78. | Ma B, Hottiger MO. Crosstalk between Wnt/β-Catenin and NF-κB Signaling Pathway during Inflammation. Front Immunol. 2016;7:378. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 272] [Cited by in F6Publishing: 389] [Article Influence: 48.6] [Reference Citation Analysis (0)] |
| 79. | Jiang X, Tan J, Li J, Kivimäe S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, Cheyette BN, Yu Q. DACT3 is an epigenetic regulator of Wnt/beta-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell. 2008;13:529-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 186] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 80. | Liu Y, Peng J, Sun T, Li N, Zhang L, Ren J, Yuan H, Kan S, Pan Q, Li X, Ding Y, Jiang M, Cong X, Tan M, Ma Y, Fu D, Cai S, Xiao Y, Wang X, Qin J. Epithelial EZH2 serves as an epigenetic determinant in experimental colitis by inhibiting TNFα-mediated inflammation and apoptosis. Proc Natl Acad Sci U S A. 2017;114:E3796-E3805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 65] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 81. | Guo Y, Liu Y, Zhang C, Su ZY, Li W, Huang MT, Kong AN. The epigenetic effects of aspirin: The modification of histone H3 lysine 27 acetylation in the prevention of colon carcinogenesis in azoxymethane- and dextran sulfate sodium-treated CF-1 mice. Carcinogenesis. 2016;37:616-624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 82. | ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14285] [Cited by in F6Publishing: 11814] [Article Influence: 984.5] [Reference Citation Analysis (0)] |
| 83. | Castel SE, Martienssen RA. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 713] [Cited by in F6Publishing: 666] [Article Influence: 60.5] [Reference Citation Analysis (0)] |
| 84. | Slattery ML, Mullany LE, Sakoda L, Samowitz WS, Wolff RK, Stevens JR, Herrick JS. The NF-κB signalling pathway in colorectal cancer: Associations between dysregulated gene and miRNA expression. J Cancer Res Clin Oncol. 2018;144:269-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 85. | Shen Z, Zhou R, Liu C, Wang Y, Zhan W, Shao Z, Liu J, Zhang F, Xu L, Zhou X, Qi L, Bo F, Ding Y, Zhao L. MicroRNA-105 is involved in TNF-α-related tumor microenvironment enhanced colorectal cancer progression. Cell Death Dis. 2017;8:3213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 86. | Wang T, Xu X, Xu Q, Ren J, Shen S, Fan C, Hou Y. miR-19a promotes colitis-associated colorectal cancer by regulating tumor necrosis factor alpha-induced protein 3-NF-κB feedback loops. Oncogene. 2017;36:3240-3251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 87. | Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493-506. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 651] [Cited by in F6Publishing: 684] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 88. | Wang H, Nie L, Wu L, Liu Q, Guo X. NR2F2 inhibits Smad7 expression and promotes TGF-β-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem Biophys Res Commun. 2017;485:181-188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 89. | Wang N, He X, Zhou R, Jia G, Qiao Q. STAT3 induces colorectal carcinoma progression through a novel miR-572-MOAP-1 pathway. Onco Targets Ther. 2018;11:3475-3484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 90. | Öner MG, Rokavec M, Kaller M, Bouznad N, Horst D, Kirchner T, Hermeking H. Combined Inactivation of TP53 and MIR34A Promotes Colorectal Cancer Development and Progression in Mice Via Increasing Levels of IL6R and PAI1. Gastroenterology. 2018;155:1868-1882. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 91. | Wang R, Jaw JJ, Stutzman NC, Zou Z, Sun PD. Natural killer cell-produced IFN-γ and TNF-α induce target cell cytolysis through up-regulation of ICAM-1. J Leukoc Biol. 2012;91:299-309. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 158] [Cited by in F6Publishing: 182] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 92. | Zhang LL, Zhang LF, Shi YB. miR-24 inhibited the killing effect of natural killer cells to colorectal cancer cells by downregulating Paxillin. Biomed Pharmacother. 2018;101:257-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 93. | Sun C, Wang FJ, Zhang HG, Xu XZ, Jia RC, Yao L, Qiao PF. miR-34a mediates oxaliplatin resistance of colorectal cancer cells by inhibiting macroautophagy via transforming growth factor-β/Smad4 pathway. World J Gastroenterol. 2017;23:1816-1827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 69] [Cited by in F6Publishing: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 94. | Ren D, Lin B, Zhang X, Peng Y, Ye Z, Ma Y, Liang Y, Cao L, Li X, Li R, Sun L, Liu Q, Wu J, Zhou K, Zeng J. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807-49823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 95. | Zhu Y, Gu L, Li Y, Lin X, Shen H, Cui K, Chen L, Zhou F, Zhao Q, Zhang J, Zhong B, Prochownik E, Li Y. miR-148a inhibits colitis and colitis-associated tumorigenesis in mice. Cell Death Differ. 2017;24:2199-2209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 96. | Yan S, Liu G, Jin C, Wang Z, Duan Q, Xu J, Xu D. MicroRNA-6869-5p acts as a tumor suppressor via targeting TLR4/NF-κB signaling pathway in colorectal cancer. J Cell Physiol. 2018;233:6660-6668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 97. | Zou F, Mao R, Yang L, Lin S, Lei K, Zheng Y, Ding Y, Zhang P, Cai G, Liang X, Liu J. Targeted deletion of miR-139-5p activates MAPK, NF-κB and STAT3 signaling and promotes intestinal inflammation and colorectal cancer. FEBS J. 2016;283:1438-1452. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 98. | Wang AL, Li Y, Zhao Q, Fan LQ. Formononetin inhibits colon carcinoma cell growth and invasion by microRNA‑149‑mediated EphB3 downregulation and inhibition of PI3K/AKT and STAT3 signaling pathways. Mol Med Rep. 2018;17:7721-7729. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 99. | Wang Y, Lu Z, Wang N, Zhang M, Zeng X, Zhao W. MicroRNA-1299 is a negative regulator of STAT3 in colon cancer. Oncol Rep. 2017;37:3227-3234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 100. | Chandrasekaran KS, Sathyanarayanan A, Karunagaran D. miR-214 activates TP53 but suppresses the expression of RELA, CTNNB1, and STAT3 in human cervical and colorectal cancer cells. Cell Biochem Funct. 2017;35:464-471. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 101. | Li B, Huang M, Liu M, Wen S, Sun F. MicroRNA‑329 serves a tumor suppressive role in colorectal cancer by directly targeting transforming growth factor beta‑1. Mol Med Rep. 2017;16:3825-3832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 102. | Huang E, Liu R, Chu Y. miRNA-15a/16: As tumor suppressors and more. Future Oncol. 2015;11:2351-2363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 103. | Liu R, Lu Z, Gu J, Liu J, Huang E, Liu X, Wang L, Yang J, Deng Y, Qian J, Luo F, Wang Z, Zhang H, Jiang X, Zhang D, Qian J, Liu G, Zhu H, Qian Y, Liu Z, Chu Y. MicroRNAs 15A and 16-1 Activate Signaling Pathways That Mediate Chemotaxis of Immune Regulatory B cells to Colorectal Tumors. Gastroenterology. 2018;154:637-651.e7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 104. | Kosaka N, Yoshioka Y, Fujita Y, Ochiya T. Versatile roles of extracellular vesicles in cancer. J Clin Invest. 2016;126:1163-1172. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 206] [Cited by in F6Publishing: 242] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 105. | Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: Overview of the crosstalk between normal and cancer cells. Biomed Res Int. 2014;2014:179486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 167] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 106. | Zhang Z, Xing T, Chen Y, Xiao J. Exosome-mediated miR-200b promotes colorectal cancer proliferation upon TGF-β1 exposure. Biomed Pharmacother. 2018;106:1135-1143. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 47] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 107. | Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F, Zhang J. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer. 2018;105:336-349. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 108. | Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 247] [Cited by in F6Publishing: 320] [Article Influence: 53.3] [Reference Citation Analysis (0)] |
| 109. | Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22:5-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 467] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 110. | Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signalling pathway via suppression of activator protein 2α. Gut. 2016;65:1494-1504. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 254] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 111. | Fu M, Zou C, Pan L, Liang W, Qian H, Xu W, Jiang P, Zhang X. Long noncoding RNAs in digestive system cancers: Functional roles, molecular mechanisms, and clinical implications (Review). Oncol Rep. 2016;36:1207-1218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 112. | Buda A, Jepson MA, Pignatelli M. Regulatory function of trefoil peptides (TFF) on intestinal cell junctional complexes. Cell Commun Adhes. 2012;19:63-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 113. | Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, Yao S, Jin G, Du J, Han W, Yin Y, Huang S, Fei B, Zou J, Huang Z. LncRNA-FEZF1-AS1 Promotes Tumor Proliferation and Metastasis in Colorectal Cancer by Regulating PKM2 Signaling. Clin Cancer Res. 2018;24:4808-4819. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 214] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 114. | Xue J, Liao L, Yin F, Kuang H, Zhou X, Wang Y. LncRNA AB073614 induces epithelial- mesenchymal transition of colorectal cancer cells via regulating the JAK/STAT3 pathway. Cancer Biomark. 2018;21:849-858. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 31] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 115. | Zhang Y, Zhang K, Luo Z, Liu L, Wu L, Liu J. Circulating long non-coding HOX transcript antisense intergenic ribonucleic acid in plasma as a potential biomarker for diagnosis of breast cancer. Thorac Cancer. 2016;7:627-632. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 32] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 116. | Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z, Protivankova M, Vycital O, Liska V, Schwarzova L, Vodickova L, Vodicka P. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35:1510-1515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 194] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 117. | Li CY, Liang GY, Yao WZ, Sui J, Shen X, Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, Zhang WH, Yin LH, Pu YP. Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol. 2016;48:1965-1976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 92] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 118. | Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, Li C, Wang C. lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer. Mol Ther Nucleic Acids. 2017;8:356-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 119. | Huang WW, Hsieh KP, Huang RY, Yang YH. Role of cyclooxygenase-2 inhibitors in the survival outcome of colorectal cancer patients: A population-based cohort study. Kaohsiung J Med Sci. 2017;33:308-314. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 120. | Williams CS, Shattuck-Brandt RL, DuBois RN. The role of COX-2 in intestinal cancer. Expert Opin Investig Drugs. 1999;8:1-12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 121. | Waddell WR, Ganser GF, Cerise EJ, Loughry RW. Sulindac for polyposis of the colon. Am J Surg. 1989;157:175-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 260] [Cited by in F6Publishing: 237] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 122. | Rothwell PM, Wilson M, Elwin CE, Norrving B, Algra A, Warlow CP, Meade TW. Long-term effect of aspirin on colorectal cancer incidence and mortality: 20-year follow-up of five randomised trials. Lancet. 2010;376:1741-1750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 976] [Cited by in F6Publishing: 947] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 123. | Sandler RS, Halabi S, Baron JA, Budinger S, Paskett E, Keresztes R, Petrelli N, Pipas JM, Karp DD, Loprinzi CL, Steinbach G, Schilsky R. A randomized trial of aspirin to prevent colorectal adenomas in patients with previous colorectal cancer. N Engl J Med. 2003;348:883-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 856] [Cited by in F6Publishing: 794] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 124. | Ng K, Meyerhardt JA, Chan AT, Sato K, Chan JA, Niedzwiecki D, Saltz LB, Mayer RJ, Benson AB, Schaefer PL, Whittom R, Hantel A, Goldberg RM, Venook AP, Ogino S, Giovannucci EL, Fuchs CS. Aspirin and COX-2 inhibitor use in patients with stage III colon cancer. J Natl Cancer Inst. 2014;107:345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 102] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 125. | Bos CL, Kodach LL, van den Brink GR, Diks SH, van Santen MM, Richel DJ, Peppelenbosch MP, Hardwick JC. Effect of aspirin on the Wnt/beta-catenin pathway is mediated via protein phosphatase 2A. Oncogene. 2006;25:6447-6456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 117] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 126. | Burr NE, Hull MA, Subramanian V. Does aspirin or non-aspirin non-steroidal anti-inflammatory drug use prevent colorectal cancer in inflammatory bowel disease? World J Gastroenterol. 2016;22:3679-3686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 21] [Cited by in F6Publishing: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 127. | Herfs M, Hubert P, Delvenne P. Epithelial metaplasia: Adult stem cell reprogramming and (pre)neoplastic transformation mediated by inflammation? Trends Mol Med. 2009;15:245-253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 128. | Liu XH, Kirschenbaum A, Weinstein BM, Zaidi M, Yao S, Levine AC. Prostaglandin E2 modulates components of the Wnt signaling system in bone and prostate cancer cells. Biochem Biophys Res Commun. 2010;394:715-720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 129. | Tsoi KK, Chan FC, Hirai HW, Sung JJ. Risk of gastrointestinal bleeding and benefit from colorectal cancer reduction from long-term use of low-dose aspirin: A retrospective study of 612 509 patients. J Gastroenterol Hepatol. 2018;33:1728-1736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 130. | Biffi A, Halpin A, Towfighi A, Gilson A, Busl K, Rost N, Smith EE, Greenberg MS, Rosand J, Viswanathan A. Aspirin and recurrent intracerebral hemorrhage in cerebral amyloid angiopathy. Neurology. 2010;75:693-698. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in F6Publishing: 228] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 131. | Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, Karin M. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1534] [Cited by in F6Publishing: 1664] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 132. | Peyrin-Biroulet L. Anti-TNF therapy in inflammatory bowel diseases: A huge review. Minerva Gastroenterol Dietol. 2010;56:233-243. [PubMed] [Cited in This Article: ] |
| 133. | Ali T, Kaitha S, Mahmood S, Ftesi A, Stone J, Bronze MS. Clinical use of anti-TNF therapy and increased risk of infections. Drug Healthc Patient Saf. 2013;5:79-99. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 157] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 134. | Hoshi D, Nakajima A, Inoue E, Shidara K, Sato E, Kitahama M, Seto Y, Tanaka E, Urano W, Ichikawa N, Koseki Y, Momohara S, Taniguchi A, Nishimoto N, Yamanaka H. Incidence of serious respiratory infections in patients with rheumatoid arthritis treated with tocilizumab. Mod Rheumatol. 2012;22:122-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |