Published online May 14, 2019. doi: 10.3748/wjg.v25.i18.2177
Peer-review started: February 6, 2019
First decision: February 13, 2019
Revised: March 27, 2019
Accepted: March 29, 2019
Article in press: March 30, 2019
Published online: May 14, 2019
The molecular scalpel of clustered regularly interspersed short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) technology may be sharp enough to begin cutting the genes implicated in inflammatory bowel disease (IBD) and consequently decrease the 6.3 billion dollar annual financial healthcare burden in the treatment of IBD. For the past few years CRISPR technology has drastically revolutionized DNA engineering and biomedical research field. We are beginning to see its application in gene manipulation of sickle cell disease, human immunodeficiency virus resistant embryologic twin gene modification and IBD genes such as Gatm (Glycine amidinotransferase, mitochondrial), nucleotide-binding oligomerization domain-containing protein 2, KRT12 and other genes implicated in adaptive immune convergence pathways have been subjected to gene editing, however there are very few publications. Furthermore, since Crohn’s disease and ulcerative colitis have shared disease susceptibility and share genetic gene profile, it is paramount and is more advantageous to use CRISPR technology to maximize impact. Although, currently CRISPR does have its limitations due to limited number of specific Cas enzymes, off-target activity, protospacer adjacent motifs and crossfire between different target sites. However, these limitations have given researchers further insight on how to augment and manipulate enzymes to enable precise gene excision and limit crossfire between target sites.
Core tip: Using clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9 (CRISPR/Cas9) to harness the potential of gene editing technology implicated in inflammatory bowel disease. This revolutionary way of editing genes and its application of gene manipulation is only gaining momentum. Genes such as Gatm (Glycine amidinotransferase, mitochondrial) and nucleotide-binding oligomerization domain-containing protein 2 have been utilized to show CRISPR is able to manipulate these genes precisely, and this is just the beginning.
- Citation: Limanskiy V, Vyas A, Chaturvedi LS, Vyas D. Harnessing the potential of gene editing technology using CRISPR in inflammatory bowel disease. World J Gastroenterol 2019; 25(18): 2177-2187
- URL: https://www.wjgnet.com/1007-9327/full/v25/i18/2177.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i18.2177
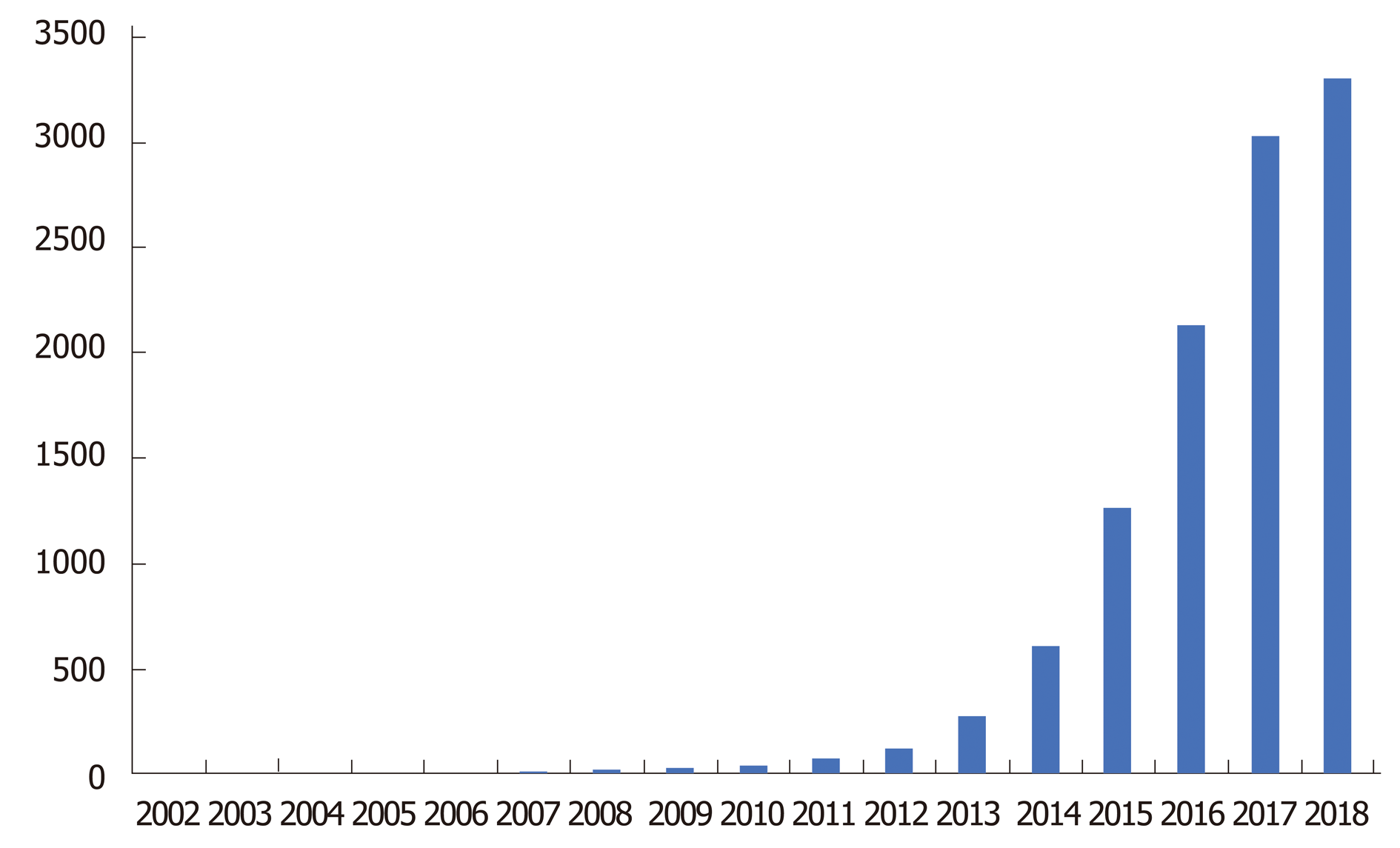
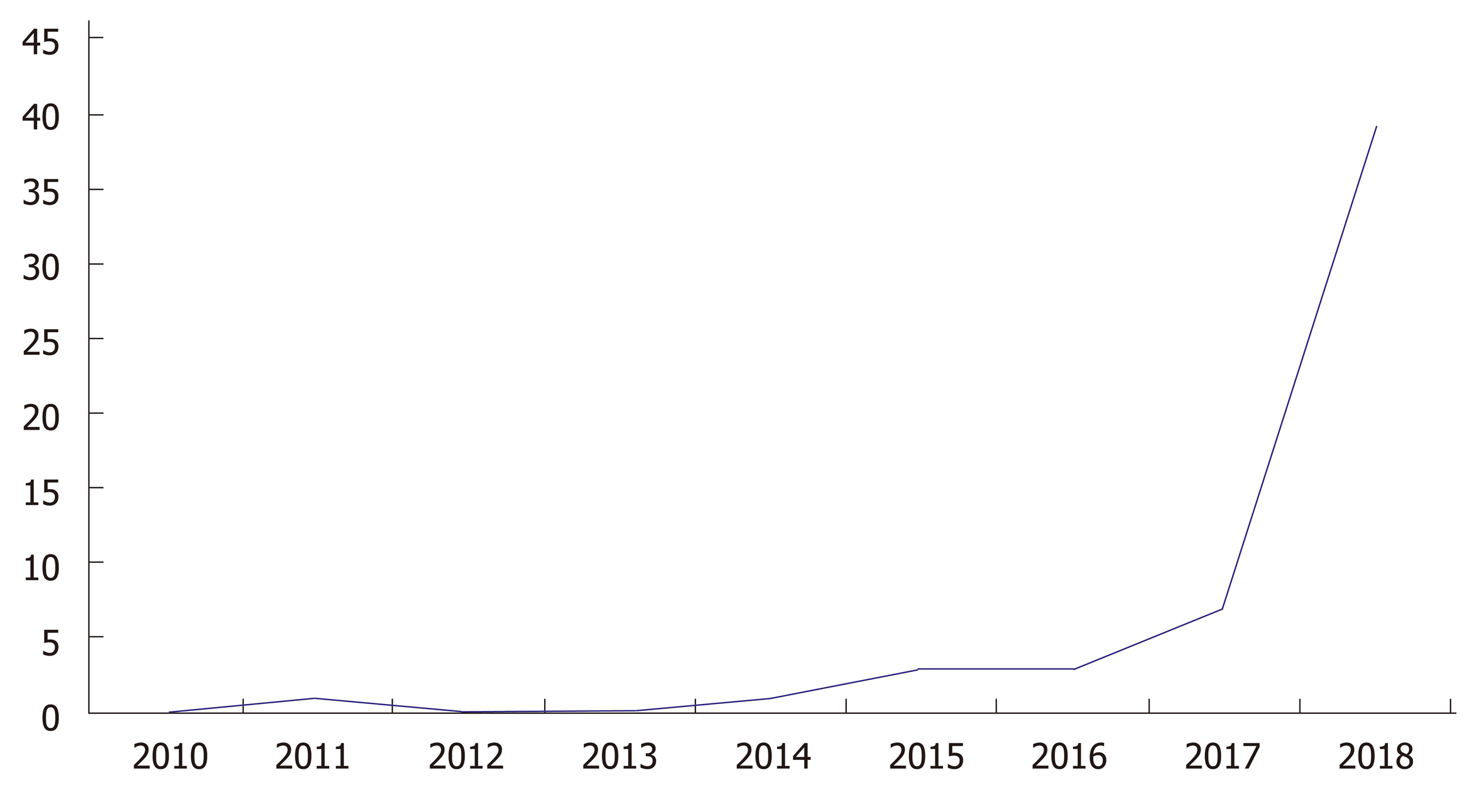
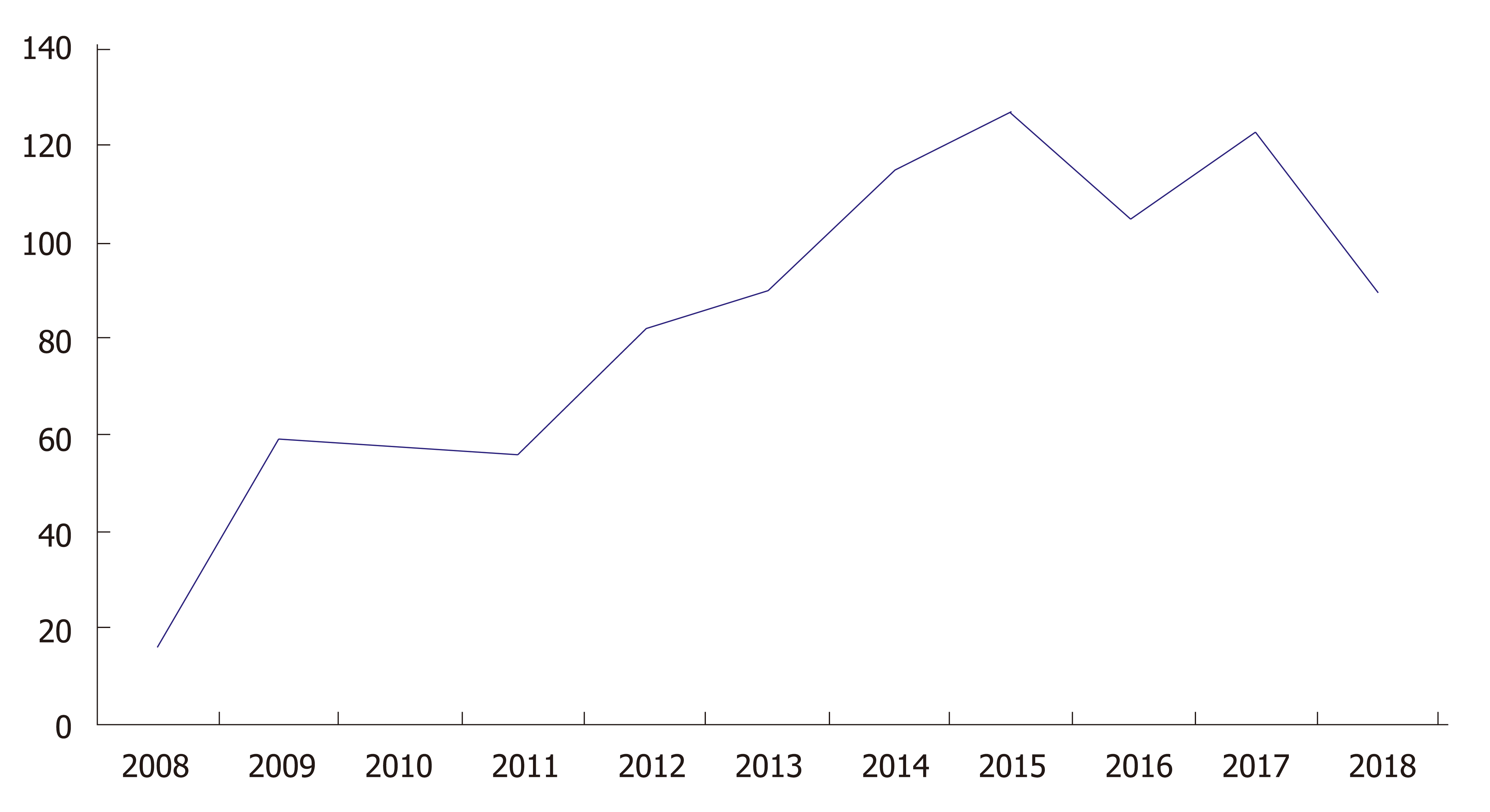
Clustered regularly interspersed short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) technology has drastically revolutionized DNA engineering and biomedical research in the past few years. It has enabled researchers to model disease and advance therapeutic intervention techniques in a pragmatic and clinically efficacious way. The Figure 1 below demonstrates the exponential growth in publications and interest in CRISPR technology over the past few years. Abreast is Figure 2 demonstrating use of CRISPR technology in inflammatory bowel disease (IBD). Strong interest in CRISPR has driven gene therapy publications down in the past few years as seen in Figure 3. Application of CRISPR is gaining momentum and is seen in sickle cell disease gene modification of hematopoietic stem cells, embryonic stem cells, gene-based therapy for HIV-1 infected individuals and other disease gene modification[1-6]. It is a new era in biomedical research for use of CRISPR technology in IBD (Figure 4). These advancements in biomedical research with the use of CRISPR will enable researchers to begin treating IBD and other gene modifying diseases, thereby relieving the healthcare financial burden of IBD.
IBD affects an estimated 1.5 million people in North America[7,8]. These patients have considerable morbidity and the associated financial burden on healthcare system is estimated to be 6.3 billion dollars annually with two thirds of that due to hospitalizations and pharmaceutical therapy alone[9-11]. This does not take into account the financial implications of work and disability of patients and impact on daily life. Even though gene therapy remains expensive in any disease treatment due to pharmaceutical patents, considerable thought should be taken to propel CRISPR use in IBD because the use of this novel technology is less taxing than previously used epigenetic modification[12].
Prior to the discovery of CRISPR/Cas9 enzyme, RNA sequencing[7,8], modular protein recognition DNA such as zinc finger nuclease or TAL effector nuclease were used in epigenetic modification and disease modeling, which are complex and require very specific protein engineering and programming[13]. However, with the revolutionary CRISPR technology, researchers now may edit genomes in a much simpler and precise way as modeled by bacteria[14,15].
CRISPR/Cas9 was found in Streptococcus pyogenes, which uses this mechanism as defense against invading viruses[14]. Upon infection by a virus the bacteria stores the viral DNA sequence in between sections of regularly interspaced palindromic repeated segments that are closely associated with genes that encode CRISPR-associated proteins. In order for the system to confer underlying efficacy and specificity, the CRISPR and the viral DNA is converted to two RNA moieties: TracrRNA and crRNA, the tracrRNA confers enzymatic activity and the crRNA determines substrate specificity[13,16]. Once the moieties bind together, they traverse the cell seeking any genetic material that matches the crRNA. Upon sequence matching, the tracrRNA part of Cas enzyme clips the target DNA in specific nucleotide bases disabling its replication. This incredibly precise system can be utilized in therapeutic advances and programed to target any sequence in the cell. This novel mechanism will be assessed for potential application and targeting of genes implicated in IBD.
CRISPR DNA engineering has advanced quickly and is evolving rapidly. It has already been used in diseases such as sickle cell, beta-thalassemia or HIV resistant human embryos[1,2,6]. Given its exponential application in disease variants, it may suggest that it’s an optimal time to further our understanding of IBD pathogenesis and treatment. Primarily IBD refers to two major categories of chronic relapsing inflammatory intestinal disorders: Ulcerative colitis (UC) and Crohn’s disease (CD). With the advent of genetic research both diseases have seen notable success culminating in the discovery of over 160 susceptible genes, among which, many potentially may be targeted by the CRISPR technology[17,18]. Around one-third of these loci/genes described, confer susceptibility to both CD and UC, which may make targeting and gene editing a viable option[18,19]. The genetic architecture of IBD has shed much light on central themes in IBD and the level of cellular process that the pathogenesis emerges. A putative CD-susceptibility locus has been mapped to chromosome 16 around locus D16S409 and D16S419 which may shed more light etiology of IBD[20]. In CD a common genetic theme is seen in defective processing of intracellular bacteria, autophagy and innate immunity. In UC, genetic evidence demonstrates genes that are responsible for proper barrier function are important in preventing UC. However, when analyzing genetic data in more detail, CD and UC have shared disease susceptibility and shared genetic gene profile that may be targeted[18]. Table 1 demonstrates common genes implicated and their strength of role in IBD[18].
| Gene | Strength of role in inflammatory bowel disease | Pathway | Function |
| NOD2 [42] | +++++ | Cellular Innate Immunity | Intermediate in MDP to NFkB and IL1B production |
| IRGM | ++ | ||
| LRRK2 | + | ||
| ATG5 | + | Autophagy | Phagophore to autophagosome to autophagolysosme creation |
| ATG16L1[42] | ++++ | ||
| IRGM | ++ | ||
| Gatm | +++ | Defective Barrier | Leaky lamina propria allowing antigen entry and T cell response |
| ECM1 | + | ||
| CDH1 | + | ||
| LAMB1 | + | ||
| HNF4A | + | ||
| GNA12 | + | ||
| IL10 | +++++ | Adaptive Immune | Convergence of STAT3, IL10RB, NOD2, ATG16L1 pathway |
| CARD9/15 | ++ | ||
| CCR6 | ++ | ||
| IL2RA | + | ||
| MST1 | + | ||
| TNFSF15 | +++ | ||
| REL | + | ||
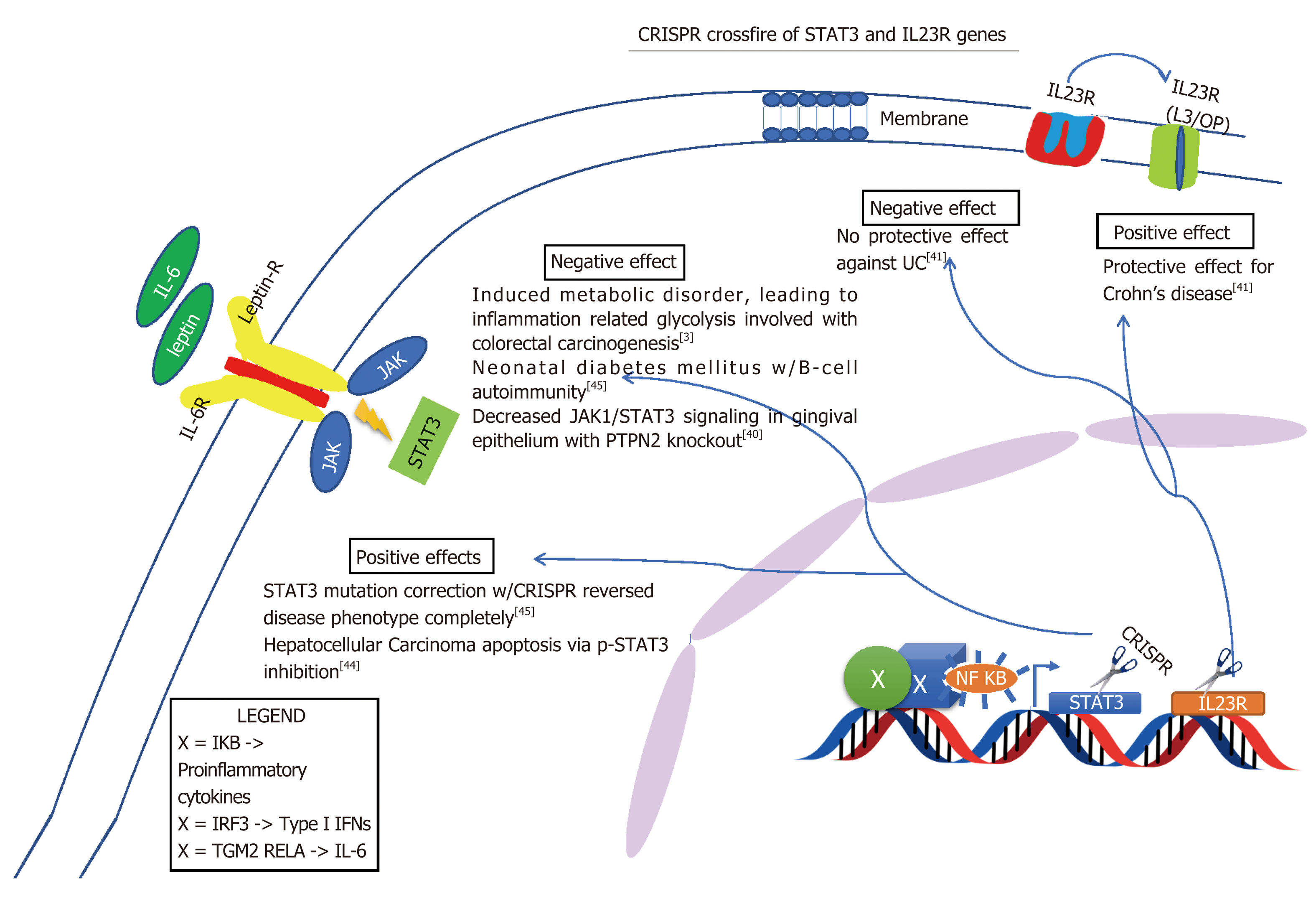
| STAT3 | +++++ | Th17 Mediated | Activation of Th17 adaptive immunity |
| IL23R[42] | +++++ | ||
| IL12B | ++ | ||
| FUT2 | + |
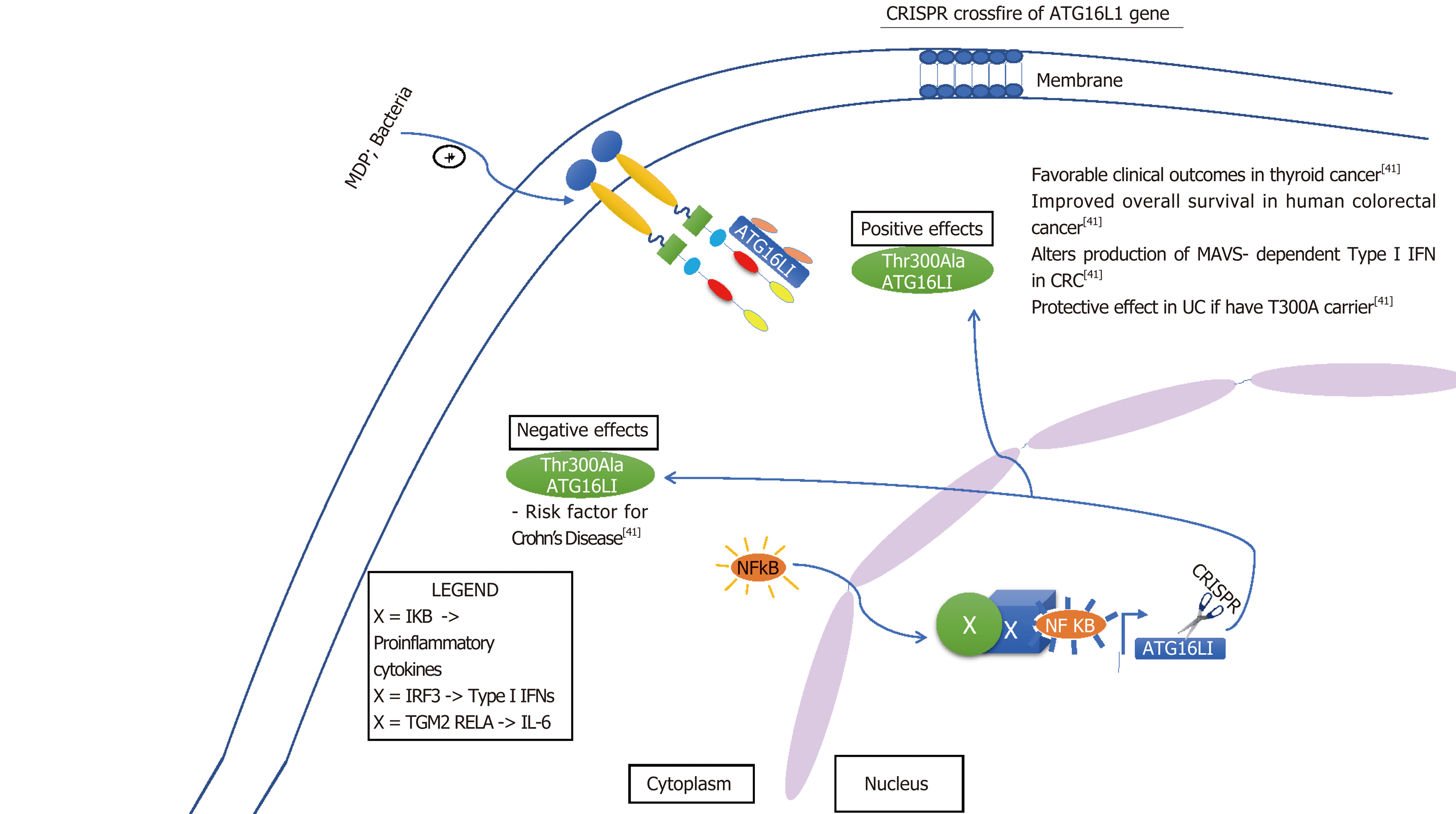
Recent research has identified and linked a gene, Gatm, to IBD using CRISPR/Cas9 gene-editing technology. When the Gatm gene is activated, it induces synthesis of creatine which helps in intestinal mucosal barrier, which helps protect the intestinal wall against inflammation that’s caused by bacteria[21]. When inducing a frame-shift mutation in the Gatm gene via CRISPR/Cas9 there was signs of inflammatory response in the intestinal wall. This demonstrated its important role in mucosal barrier protection and potential manipulation of this gene using CRISPR. Protection from inflammation by an intact barrier is vital to decrease immune response, which is suppressed or increased by enhancers in the immune activation cascade. In the inflammatory response pathway, IL2RA plays a role in signaling T cells to hamper or increase the response. If the enhancers that switch on IL2RA are defective the T cells won’t suppress inflammation and chronic inflammation is associated with 15%-20% of all human malignancies[8,22]. Inflammation also results in autoimmune disorders such as IBD and inflammatory-induced colon cancer mediated by NF-kB pathway[22-24]. Previously it has been demonstrated that single nucleotide polymorphisms -SNPs’ mutation of IL2RA leads to improper activation of T cells and subsequently resulting in autoimmune disorders. These SNP’s may be targeted by CRISPR/Cas9 and repaired with non-homologous end-joining repair. This has been demonstrated in KRT12 mutations-specific targeting of SNP’s as well[25]. Recent advancements in CRISPR/Cas9 specificity and potency of targeted genes demonstrate that SNP’s or genes that have point mutations may be targeted and editing may be attempted.
CD and UC in general have three stages of disease progression, mild-moderate; moderate-severe; and severe. Currently there are no studies indicating potential therapeutic timing when to target affected genes using CRISPR in IBD, however several multicenter trials conducted administering human recombinant IL-10 during active mild/moderate stage of CD or during refractory CD as well as patients undergoing curative ileal or ileocolonic resection[26,27]. However, results did not show significant clinical improvement or higher remission rates secondary to too low IL-10 dose and adverse effects of medication. In addition, IL-10 alone failed to effectively suppress variety of dysregulated pro-inflammatory cytokines[26,27]. In later stages of disease process, significant dysregulation of pro-inflammatory cytokines and redundant pathways occur, such as NF-kB receiving activation from different pathways[28] thus single target impact is futile. Given that CRISPR can simultaneously multiplex several genes, it will aid researchers to devise appropriate intervention timing[29]. We also suggest early intervention is optimal to prevent progression of disease and reduce complications. It is imperative to conduct studies to best identify role of CRISPR in various stages of disease.
Currently, CRISPR is applied in many fields of scientific study. In biotechnology it is used to modify Maize genome in protoplasts. In drug development, it is used to understand modes of drug resistance and drug-target interactions. In epigenetics, it has taken the place of zinc finger nuclease and TALEN in epigenetic modification because the indel frequency is more superior[6]. Since the CRISPR debut, researchers are improving and enhancing the specificity and accuracy of the Cas9. Currently the Cas9 not only cuts the DNA, but can be altered to perform desired functions. The Cas9 protein has a deaminase region that may be altered to increase highly specific alternation of genome sequence, which will allow for broader specific DNA bases manipulation[13]. It can also promote gene transcription using enzyme by deactivating the endonuclease activity and add transcriptional activator to increase transcription. The Cas9 can silence domains that recruit factors so that genes are blocked and they are not transcribed. In general targeting studies, Cas9 can be tagged with fluorescent dye so genes can be followed. Furthermore, Cas9 can be multiplexed with multiple guide RNAs to generate multiple breaks in order to cut out large sequences of DNA in one experiment[29]. This limits time and repetitiveness of experiments conducted and time is of an essence in this race to invent even-more versatile or efficient variations of this powerful enzyme, which greatly simplifies the editing of DNA. Furthermore, very recently successful attempts were made to edit CCR5 gene in human embryos to enable resistance to HIV[30]. Although, ethics and implications of such studies are currently widely debated. Also, recently KRAS oncogenic alleles were modified leading to decreased cancer cell growth without disturbing wild type alleles[31]. Since major oncogenic mutations occur on codon-12 of KRAS exon-2, direct targeting of oncogenic KRAS single-nucleotide missense substitution c.35 G>T mutation using CRISPR/Cas9 system inhibited cancer cell growth and is dependent on efficient target cell transduction[31].
The excitement generated by the new CRISPR technology in the science community is reflected by exponential publications and application in various fields of study, including agriculture, drug development and epigenetic control but its revolutionary genome editing method is far from perfect. The CRISPR is an RNA based DNA recognition system. It is dependent on a guide molecule composed of RNA to recognize a sequence in the DNA that has specific molecular features. The problem is that the original Cas9 can only land on genome segments that have a trio of ‘NGG’ (N being any nucleotide) nucleotide base pairs and cut only limited fraction of the genome. The human genome contains 3.2 billion-bases and only one-sixteenth of the genome where Cas9 can land. This limits the specific target genes of interest and leads to “off-target” mutations however to limit these effects a highly specific guide RNA sequence is selected[15,32]. Also, recently researchers modified the enzyme and developed an xCas9s that has a broader range of three-base landing pads, referred to as protospacer adjacent motifs or PAMs. It works best with NGN sequence, which occurs in one-fourth of the genome. It allows for researches to perform gene knockout studies, which help them, determine what gene is implicated in disease. However, since majority of diseases are associated with “point mutations”, it is difficult to target and repair a mutated gene. Not only that, once the enzyme clips the desired DNA gene, the repair mechanism of the cell is wobbly and during repairs it tends to insert or delete DNA bases. Furthermore, Cas9 enzyme only cuts DNA and only 2% of the genome codes directly from DNA to protein and 98% of genome is regulatory gene sequences. This poses a challenge for precisely modifying RNA. Although, very recently a new enzyme was characterized called Cpf1, which was found in Staphylococcus aureus, and is capable of cleaving both DNA and RNA[33]. This will allow targeting of RNA gain-of-function mutations such as NOD2 or other mutations that can be edited. Furthermore, Cpf1 is also smaller which makes transfection much easier[33].
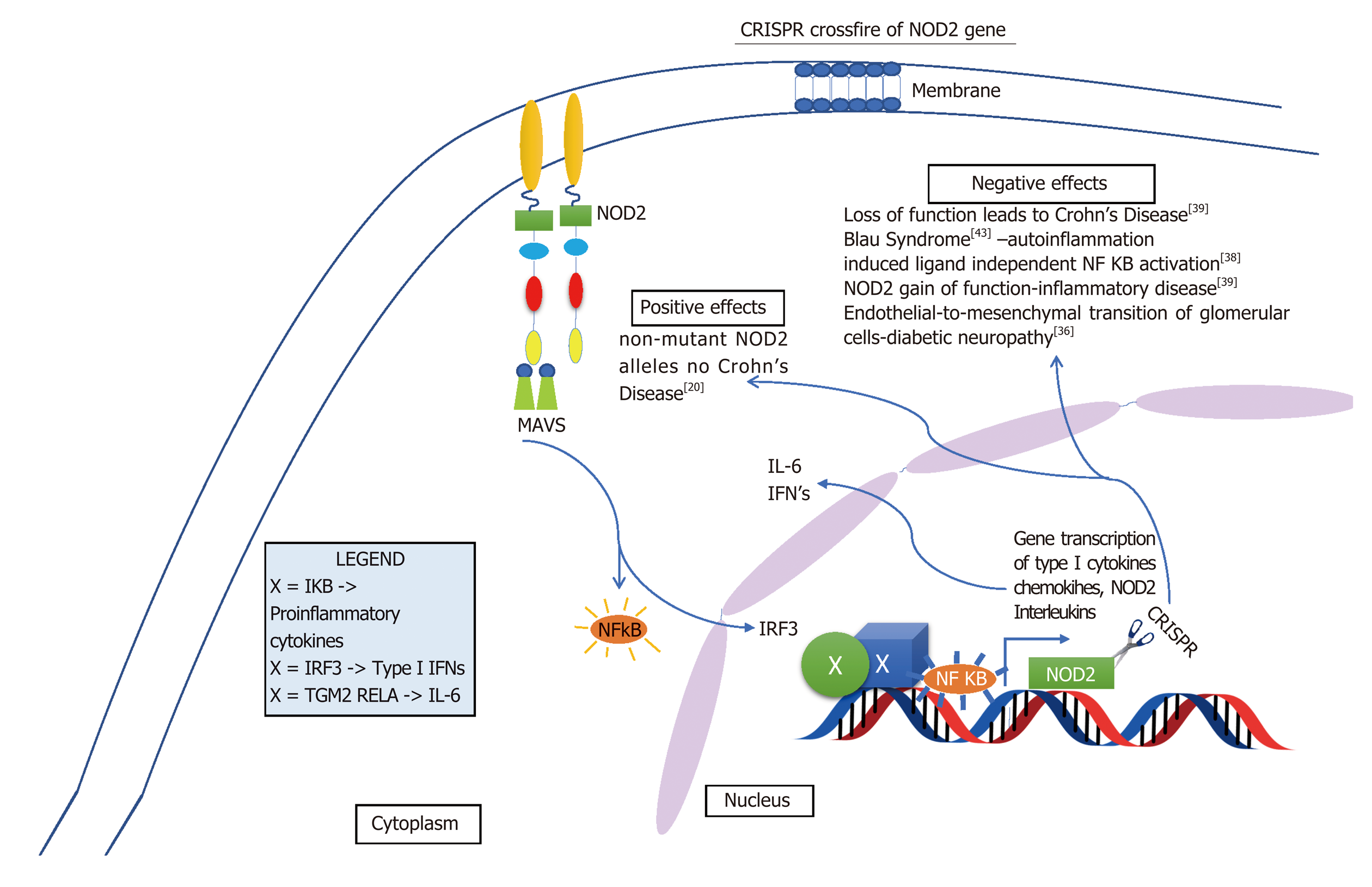
Although some of these limitations of the Cas9 are quickly becoming insightful opportunities and researchers are drastically improving it by altering its capabilities. The new enzymes are more precise than the original Cas9 and now there is xCas9, dCas9 and dCas13 which are capable of editing specific base pairs. For example, dCas13 can convert base A to I in RNA and I is a universal base[34]. Such manipulation of bases is a very appealing target for therapies, particularly inflammation. Furthermore, because RNA is around in the cell for a short period of time before it is degraded repeated administration of RNA base editors would need to be given[34]. Also, sequencing RNA is problematic and laborious[35]. On the other hand this may seem disadvantageous but working with RNA may limit some off target genome mutations. In addition to off target genome mutations, crossfire between different cells may occur. Intended target may be either gain of function mutation or loss of function mutation, either way, altering alleles may have their own detrimental effect. For example, as indicated in Figure 5, NOD2 loss of function leads to Crohn’s and gain of function causes endothelial to mesenchymal transition of glomerular endothelial cells causing diabetic nephropathy[36]. However, some of these cross reactions may not be as detrimental. In Figure 6, altering ATG16L1 allele to T300A ATG16L1 only incases risk for CD type, but not disease onset[37]. Figures 5-7 below demonstrate positive and negative effects of altering main genes implicated in IBD; NOD2, STAT3 ATG16L1, IL23R genes using CRISPR technology[37-41].
Despite CRISPR/Cas technology limitations, as new innovative techniques such as anti-CRISPR proteins and new Cas proteins are developed to advance precise DNA editing, application of this revolutionary mechanism is at its prime time to hone in on genes implicated in IBD. Implementation of CRISPR in IBD research will lead to better outcomes and may decrease financial burden on the health care system.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Cerwenka H, Chiu KW, Hashimoto N S-Editor: Yan JP L-Editor: A E-Editor: Zhang YL
| 1. | Wen J, Tao W, Hao S, Zu Y. Cellular function reinstitution of offspring red blood cells cloned from the sickle cell disease patient blood post CRISPR genome editing. J Hematol Oncol. 2017;10:119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Ye L, Wang J, Tan Y, Beyer AI, Xie F, Muench MO, Kan YW. Genome editing using CRISPR-Cas9 to create the HPFH genotype in HSPCs: An approach for treating sickle cell disease and β-thalassemia. Proc Natl Acad Sci U S A. 2016;113:10661-10665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 137] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 3. | DeWitt MA, Magis W, Bray NL, Wang T, Berman JR, Urbinati F, Heo SJ, Mitros T, Muñoz DP, Boffelli D, Kohn DB, Walters MC, Carroll D, Martin DI, Corn JE. Selection-free genome editing of the sickle mutation in human adult hematopoietic stem/progenitor cells. Sci Transl Med. 2016;8:360ra134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 369] [Cited by in F6Publishing: 317] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 4. | Saayman S, Ali SA, Morris KV, Weinberg MS. The therapeutic application of CRISPR/Cas9 technologies for HIV. Expert Opin Biol Ther. 2015;15:819-830. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 5. | Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, Lv J, Xie X, Chen Y, Li Y, Sun Y, Bai Y, Songyang Z, Ma W, Zhou C, Huang J. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6:363-372. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 715] [Cited by in F6Publishing: 615] [Article Influence: 68.3] [Reference Citation Analysis (0)] |
| 6. | Antony JS, Latifi N, Haque AKMA, Lamsfus-Calle A, Daniel-Moreno A, Graeter S, Baskaran P, Weinmann P, Mezger M, Handgretinger R, Kormann MSD. Gene correction of HBB mutations in CD34+ hematopoietic stem cells using Cas9 mRNA and ssODN donors. Mol Cell Pediatr. 2018;5:9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 7. | Gao M, Zhong A, Patel N, Alur C, Vyas D. High throughput RNA sequencing utility for diagnosis and prognosis in colon diseases. World J Gastroenterol. 2017;23:2819-2825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 14] [Cited by in F6Publishing: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Hollis M, Nair K, Vyas A, Chaturvedi LS, Gambhir S, Vyas D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J Gastroenterol. 2015;21:8284-8292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 87] [Cited by in F6Publishing: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 9. | Murthy SK, James PD, Antonova L, Chalifoux M, Tanuseputro P. High end of life health care costs and hospitalization burden in inflammatory bowel disease patients: A population-based study. PLoS One. 2017;12:e0177211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Niewiadomski O, Studd C, Hair C, Wilson J, McNeill J, Knight R, Prewett E, Dabkowski P, Dowling D, Alexander S, Allen B, Tacey M, Connell W, Desmond P, Bell S. Health Care Cost Analysis in a Population-based Inception Cohort of Inflammatory Bowel Disease Patients in the First Year of Diagnosis. J Crohns Colitis. 2015;9:988-996. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Kappelman MD, Rifas-Shiman SL, Porter CQ, Ollendorf DA, Sandler RS, Galanko JA, Finkelstein JA. Direct health care costs of Crohn's disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907-1913. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 469] [Cited by in F6Publishing: 510] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 12. | Sherkow JS. CRISPR, Patents, and the Public Health. Yale J Biol Med. 2017;90:667-672. [PubMed] [Cited in This Article: ] |
| 13. | Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2175] [Cited by in F6Publishing: 2085] [Article Influence: 208.5] [Reference Citation Analysis (0)] |
| 14. | Deshpande K, Vyas A, Balakrishnan A, Vyas D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Genetic Engineering: Robotic Genetic Surgery. Am J Robot Surg. 2015;2:49-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Fujii M, Clevers H, Sato T. Modeling Human Digestive Diseases With CRISPR-Cas9-Modified Organoids. Gastroenterology. 2019;156:562-576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 89] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 16. | Heussler GE, O'Toole GA. Friendly Fire: Biological Functions and Consequences of Chromosomal Targeting by CRISPR-Cas Systems. J Bacteriol. 2016;198:1481-1486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 17. | Cater D, Vyas A, Vyas D. Robotics in Colonoscopy. Am J Robot Surg. 2014;1:48-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: Common pathways with other diseases. Gut. 2011;60:1739-1753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 406] [Cited by in F6Publishing: 420] [Article Influence: 32.3] [Reference Citation Analysis (1)] |
| 19. | Ek WE, D'Amato M, Halfvarson J. The history of genetics in inflammatory bowel disease. Ann Gastroenterol. 2014;27:294-303. [PubMed] [Cited in This Article: ] |
| 20. | Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard-Jones JE, Cortot A, Colombel JF, Thomas G. Mapping of a susceptibility locus for Crohn's disease on chromosome 16. Nature. 1996;379:821-823. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 649] [Cited by in F6Publishing: 665] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 21. | Turer E, McAlpine W, Wang KW, Lu T, Li X, Tang M, Zhan X, Wang T, Zhan X, Bu CH, Murray AR, Beutler B. Creatine maintains intestinal homeostasis and protects against colitis. Proc Natl Acad Sci U S A. 2017;114:E1273-E1281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 22. | Gambhir S, Vyas D, Hollis M, Aekka A, Vyas A. Nuclear factor kappa B role in inflammation associated gastrointestinal malignancies. World J Gastroenterol. 2015;21:3174-3183. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 76] [Cited by in F6Publishing: 77] [Article Influence: 8.6] [Reference Citation Analysis (2)] |
| 23. | Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, Lituiev DS, Nguyen ML, Gate RE, Subramaniam M, Li Z, Woo JM, Mitros T, Ray GJ, Curie GL, Naddaf N, Chu JS, Ma H, Boyer E, Van Gool F, Huang H, Liu R, Tobin VR, Schumann K, Daly MJ, Farh KK, Ansel KM, Ye CJ, Greenleaf WJ, Anderson MS, Bluestone JA, Chang HY, Corn JE, Marson A. Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature. 2017;549:111-115. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 189] [Cited by in F6Publishing: 191] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 24. | Negroni A, Stronati L, Pierdomenico M, Tirindelli D, Di Nardo G, Mancini V, Maiella G, Cucchiara S. Activation of NOD2-mediated intestinal pathway in a pediatric population with Crohn's disease. Inflamm Bowel Dis. 2009;15:1145-1154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Courtney DG, Moore JE, Atkinson SD, Maurizi E, Allen EH, Pedrioli DM, McLean WH, Nesbit MA, Moore CB. CRISPR/Cas9 DNA cleavage at SNP-derived PAM enables both in vitro and in vivo KRT12 mutation-specific targeting. Gene Ther. 2016;23:108-112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 95] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 26. | Herfarth H, Schölmerich J. IL-10 therapy in Crohn's disease: At the crossroads. Treatment of Crohn's disease with the anti-inflammatory cytokine interleukin 10. Gut. 2002;50:146-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Tilg H, van Montfrans C, van den Ende A, Kaser A, van Deventer SJ, Schreiber S, Gregor M, Ludwiczek O, Rutgeerts P, Gasche C, Koningsberger JC, Abreu L, Kuhn I, Cohard M, LeBeaut A, Grint P, Weiss G. Treatment of Crohn's disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut. 2002;50:191-195. [PubMed] [Cited in This Article: ] |
| 28. | Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812-4818. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1034] [Cited by in F6Publishing: 1011] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 29. | Pankowicz FP, Barzi M, Kim KH, Legras X, Martins CS, Wooton-Kee CR, Lagor WR, Marini JC, Elsea SH, Bissig-Choisat B, Moore DD, Bissig KD. Rapid Disruption of Genes Specifically in Livers of Mice Using Multiplex CRISPR/Cas9 Editing. Gastroenterology. 2018;155:1967-1970.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Xu L, Yang H, Gao Y, Chen Z, Xie L, Liu Y, Liu Y, Wang X, Li H, Lai W, He Y, Yao A, Ma L, Shao Y, Zhang B, Wang C, Chen H, Deng H. CRISPR/Cas9-Mediated CCR5 Ablation in Human Hematopoietic Stem/Progenitor Cells Confers HIV-1 Resistance In Vivo. Mol Ther. 2017;25:1782-1789. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 158] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 31. | Lee W, Lee JH, Jun S, Lee JH, Bang D. Selective targeting of KRAS oncogenic alleles by CRISPR/Cas9 inhibits proliferation of cancer cells. Sci Rep. 2018;8:11879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 32. | Zhang JH, Adikaram P, Pandey M, Genis A, Simonds WF. Optimization of genome editing through CRISPR-Cas9 engineering. Bioengineered. 2016;7:166-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 33. | Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759-771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3250] [Cited by in F6Publishing: 2700] [Article Influence: 300.0] [Reference Citation Analysis (0)] |
| 34. | Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019-1027. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 923] [Cited by in F6Publishing: 1018] [Article Influence: 145.4] [Reference Citation Analysis (0)] |
| 35. | Bianco AM, Girardelli M, Tommasini A. Genetics of inflammatory bowel disease from multifactorial to monogenic forms. World J Gastroenterol. 2015;21:12296-12310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 68] [Cited by in F6Publishing: 62] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 36. | Shang J, Zhang Y, Jiang Y, Li Z, Duan Y, Wang L, Xiao J, Zhao Z. NOD2 promotes endothelial-to-mesenchymal transition of glomerular endothelial cells via MEK/ERK signaling pathway in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:435-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 37. | Duan W, Mehta AK, Magalhaes JG, Ziegler SF, Dong C, Philpott DJ, Croft M. Innate signals from Nod2 block respiratory tolerance and program T(H)2-driven allergic inflammation. J Allergy Clin Immunol. 2010;126:1284-93.e10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 38. | Takada S, Kambe N, Kawasaki Y, Niwa A, Honda-Ozaki F, Kobayashi K, Osawa M, Nagahashi A, Semi K, Hotta A, Asaka I, Yamada Y, Nishikomori R, Heike T, Matsue H, Nakahata T, Saito MK. Pluripotent stem cell models of Blau syndrome reveal an IFN-γ-dependent inflammatory response in macrophages. J Allergy Clin Immunol. 2018;141:339-349.e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 39. | Chirieleison SM, Marsh RA, Kumar P, Rathkey JK, Dubyak GR, Abbott DW. Nucleotide-binding oligomerization domain (NOD) signaling defects and cell death susceptibility cannot be uncoupled in X-linked inhibitor of apoptosis (XIAP)-driven inflammatory disease. J Biol Chem. 2017;292:9666-9679. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Zhang P, Zhang W, Zhang D, Wang M, Aprecio R, Ji N, Mohamed O, Li Y, Ding Y, Wang Q. 25-Hydroxyvitamin D3 -enhanced PTPN2 positively regulates periodontal inflammation through the JAK/STAT pathway in human oral keratinocytes and a mouse model of type 2 diabetes mellitus. J Periodontal Res. 2018;53:467-477. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 41. | Serbati N, Senhaji N, Diakite B, Badre W, Nadifi S. IL23R and ATG16L1 variants in Moroccan patients with inflammatory bowel disease. BMC Res Notes. 2014;7:570. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 42. | Gajendran M, Loganathan P, Catinella AP, Hashash JG. A comprehensive review and update on Crohn's disease. Dis Mon. 2018;64:20-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 243] [Article Influence: 34.7] [Reference Citation Analysis (1)] |
| 43. | Kanazawa N, Okafuji I, Kambe N, Nishikomori R, Nakata-Hizume M, Nagai S, Fuji A, Yuasa T, Manki A, Sakurai Y, Nakajima M, Kobayashi H, Fujiwara I, Tsutsumi H, Utani A, Nishigori C, Heike T, Nakahata T, Miyachi Y. Early-onset sarcoidosis and CARD15 mutations with constitutive nuclear factor-kappaB activation: Common genetic etiology with Blau syndrome. Blood. 2005;105:1195-1197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 404] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 44. | Zhang X, Zhang S, Sun Q, Jiao W, Yan Y, Zhang X. Compound K Induces Endoplasmic Reticulum Stress and Apoptosis in Human Liver Cancer Cells by Regulating STAT3. Molecules. 2018;23:pii: E1482. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 45. | Saarimäki-Vire J, Balboa D, Russell MA, Saarikettu J, Kinnunen M, Keskitalo S, Malhi A, Valensisi C, Andrus C, Eurola S, Grym H, Ustinov J, Wartiovaara K, Hawkins RD, Silvennoinen O, Varjosalo M, Morgan NG, Otonkoski T. An Activating STAT3 Mutation Causes Neonatal Diabetes through Premature Induction of Pancreatic Differentiation. Cell Rep. 2017;19:281-294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 10.6] [Reference Citation Analysis (0)] |