INTRODUCTION
Liver diseases are one of the most common pathologies in the digestive system and have a high incidence. In recent years, the incidence of liver diseases has gradually increased owing to drinking, obesity, viral [hepatitis B virus (HBV) or hepatitis C virus (HCV)] infection, and other reasons[1-3]. Fatty liver and viral hepatitis are common liver diseases that have a prolonged course and often evolve into cirrhosis and even liver cancer. The liver is a complex metabolic and detoxifying organ with significant individual differences[4]. Due to the complex inner microenvironment of the liver, it is difficult to establish a disease model for exploring the occurrence, development, and treatment of liver diseases.
Two-dimensional (2D) cell culture, three-dimensional (3D) cell culture, primary cell culture, and human-animal models have been widely used to study liver physiology, liver disease pathogenesis, and relevant treatments. However, these models often have many shortcomings and limitations in practical application[5]. For example, in 2D and 3D cell cultures, the cells are usually from liver cancer cell lines or liver cell lines transduced with a virus, and their genetic information is relatively constant, so that the individualized liver physiology and pathology cannot be fully displayed[6]. Primary hepatocyte culture can preserve the heterogeneity of genetic information, but its application in the study of liver diseases is limited because of the difficulty of long-term culture in vitro and the lack of a proper hepatocyte microenvironment[7]. The patient-derived xenograft (PDX) model involves transplanting fresh tumor tissue into immunodeficient mice to form tumors in their bodies. The tumor-forming tissue well maintains the biological characteristics and heterogeneity of the tumor[8,9]. The PDX model has been broadly used to study the pathogenesis and treatment of various solid liver tumors including (but not limited to) hepatocellular carcinoma, cholangiocarcinoma, and mixed cell carcinoma[10,11]. However, the PDX model has many deficiencies affecting the study of tumors. For instance, when liver cancer tissue is transplanted into immunodeficient mice, the tumor formation rate is less than 30%, and the process usually involves high expenditures and consumes much time and many resources. Even if the tumor can form in immunodeficient mice, tumor evolution may also ensue, resulting in a greater difference from human tumors[12-14]. Although the above models can be used to study liver diseases, they are not optimal models for research on liver disease occurrence, developmental mechanisms, and treatment.
An organoid culture, a special 3D culture, is made of autologous tissue, pluripotent stem cells (PSCs), adult stem cells and other tissues cultured in vitro by special 3D culture techniques[15]. An organoid culture can stably retain the genetic information of autologous tissue and present the physiological and pathological state of self-tissue. With the development and maturity of culture techniques, organoids have been widely used in the study of stem cell biology and research on the development of various human organs and human diseases, such as the use of human and animal liver organoids for the study of various liver diseases[16,17].
This article mainly describes research progress in the culture of liver organoids (including liver cancer organoids) and their application in liver diseases, points out the existing limitations of liver organoid culture and disease research, and considers what improvements can be made in the future. It is apparent that liver organoids can be better used to study liver diseases.
OVERVIEW AND SOURCES OF ORGANOIDS
Origin and introduction of organoids
Organoids are “microscopic tissues” formed by culturing stem cells in a unique 3D culture system, and their structure and function are largely similar to those in vivo. As early as the 1950s, the literature began to use the term “organoids”[18], and organoids started to gradually appear in various publications in the 1960s. Naturally, a new cell culture method in vitro called “3D organoid” culture emerged, but in the past few decades, there has been no clear and unambiguous definition of organoids. In 2012, organoids were systematically elaborated and defined by Eiraku and Sasai[19]. Organoids, which are “microtissues” formed by special in vitro 3D culture techniques developed in recent years, contain tissues similar to the tissues in vivo. Organoids retain a tissue structure and function similar to the original tissue and are capable of stable long-term culture and passage[19-21]. The stem cell sources of the existing cultured organoids are mainly the following two stem cell lines: (1) PSCs: induced PSCs (iPSCs) and embryonic stem cells (ESCs) are induced to form brain tissue[22], retinal tissue[23], lung tissue[24], stomach tissue[25], small intestinal tissue[26], kidney tissue[27], liver tissue[28], and organoids from other organs by adding substances that contain growth factors [fibroblast growth factor (FGF), WNT, bone morphogenetic protein (BMP), etc.] into the in vitro 3D medium; and (2) adult stem cells (ASCs): stem cells present in mature tissues under specific conditions are induced to differentiate into corresponding tissue organoids by adding substances such as growth factors [epidermal growth factor (EGF), Rspo1, and FGF] from the microenvironment in vivo. For example, by inducing the differentiation of Lgr5+ cells, organoids such as lung tissue[29], small intestine tissue[30], stomach tissue[31], pancreatic tissue[32], and liver tissue[15] are formed. The organoids are cultured in 3D medium through the orderly addition of growth factors and other substances that are in the microenvironment of stem cells in the original tissue. In addition, the directional differentiation of PSCs and ASCs is induced to form a 3D micro-organization similar to the structure and function of the original tissues.
Formation of and latest developments in liver organoids
PSCs differentiate to form liver organoids: Some studies have reported that human PSCs (hPSCs) are successfully induced to form mature hepatocyte-like cells in serum-free medium[33,34]. It was further found that hepatocyte-like cells derived from hPSCs can be cocultured with mesenchymal stem cells and endothelial cells on Matrigel matrix to form 3D hepatic bud organoids[28]. Through immunohistochemistry, the liver bud tissue has been found to contain microvessels. In addition, if the liver bud is transplanted into nude mice, the microvessels will spontaneously communicate with the subcutaneous microvasculature of the nude mice, which provides the blood supply to the 3D liver-bud organoid tissue. The analyzed gene expression was found to have a high similarity with the expression of the original fetal liver genes. In addition, the organoid tissue differentiated more maturely than hepatocytes formed by culturing human iPSCs in a Matrigel-free matrix[28]. Recent studies show that human iPSCs may develop gene mutations or alter gene expression during differentiation, which may even lead to tumorigenesis[35]. Zhang et al[36] added EGF, VEGF, FGF2, Chir99021, and A83-01 separately to induce human iPSCs to differentiate into posterior gut endoderm cells (PGECs), and the progenitor cells of the posterior intestine were further induced to differentiate into liver bud tissue through 3D induction. When implanted into immunodeficient mice, liver bud tissue can differentiate into liver tissue with mature functional liver cells and biliary epithelial cells, with corresponding nutritional angiogenesis[36].
ASCs differentiate into liver organoids: In 2007, Barker et al[37] discovered self-renewing stem cells in the intestinal epithelium, and these stem cells all expressed the Lgr5 protein. An increasing number of researchers believe that Lgr5+ is a marker of stem cells in tissues such as the stomach, small intestine, colon, liver, and pancreas[37]. Lgr5+ cells are adult tissue stem cells of the liver and are rarely observed in the normal physiological state; however, in pathological conditions, the number will increase significantly[15]. For example, when liver damage occurs, Lgr5+ cells are usually bile duct epithelial cells and can differentiate into hepatocytes and biliary epithelial cells in the body, thereby causing spontaneous repair of the liver[15,21,38]. Huch et al[15] extracted Lgr5+ cells, which were induced by CCL4 in injured mouse livers and then were further induced to differentiation in Matrigel matrix to form mature mouse liver organoids. The analysis of the mouse liver organoids revealed that certain liver progenitor cells can differentiate into early hepatocytes and biliary epithelial cells[15]. When the mouse liver organoids were implanted into immunodeficient mice, the liver organoids differentiated into functional liver tissues, and the hepatocytes secreted albumin, cytochrome, bile acids, etc., and partially restored the liver function in the injured mice[15]. In 2015, Huch et al[21] cultured human bile duct-derived bipotent progenitor cells in vitro for a long period and found that they exhibited Lgr5+ cell-like proliferation and differentiation capacity and were able to form organoids in Matrigel matrix. When implanted into immunodeficient mice, these cells can also differentiate into functional liver tissue[21]. Furthermore, progenitor cells can differentiate into organoids in dogs, cats, and other animals by a similar method[39,40]. However, the liver organoids formed by the above methods only differentiate into functional hepatocytes or biliary cells, which are often similar to early fetal liver cells and bile duct epithelial cells during liver development[41], and differ from the physiological function and physiological anatomy of the mature liver. To compensate for these shortcomings, in 2017, Vyas et al[5] cultured human fetal liver progenitor cells by decellularized hepatic extracellular matrix (ECM) scaffolds to form organoids, which exhibit some of the physiological functions (albumin and bilirubin secretion) and the liver-biliary anatomy of mature liver tissue.
Latest culture of liver organoids
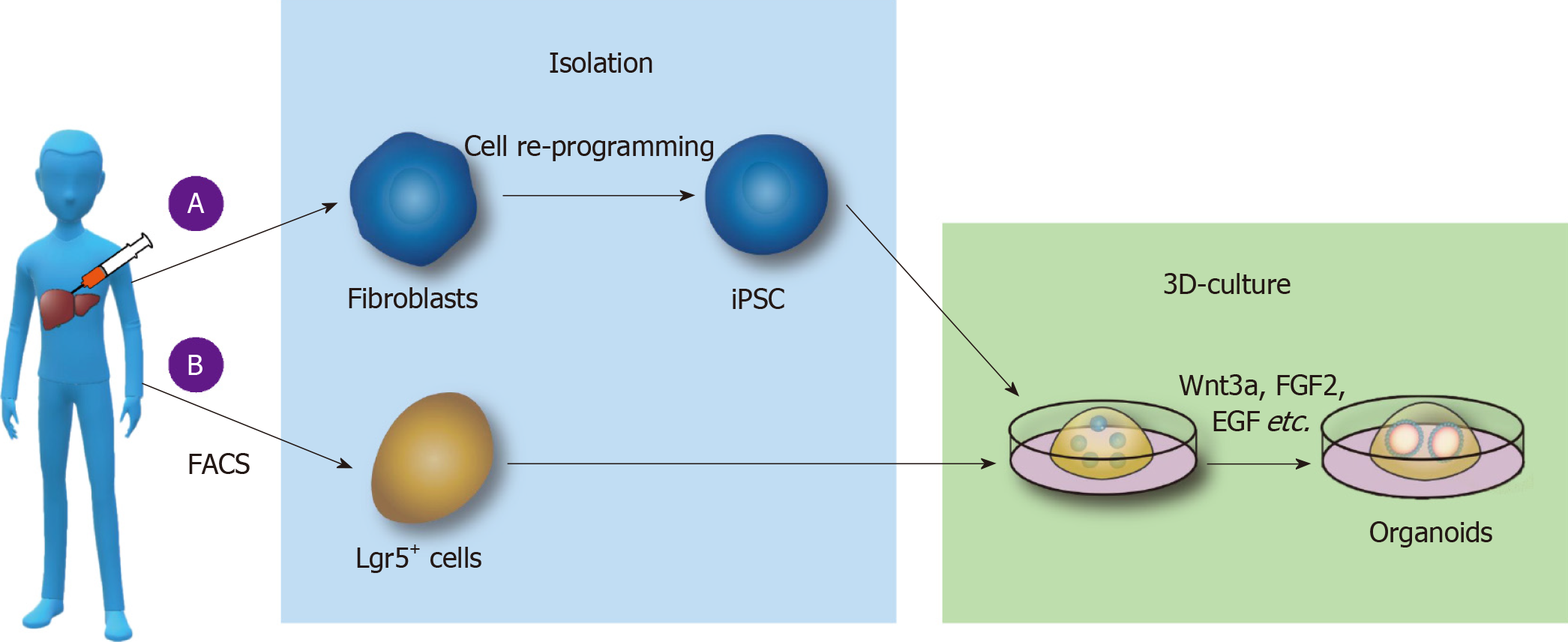
PSC-derived liver organoid culture: At the first stage of culture, laminin-511 E8, activin A, Wnt3a, Rock inhibitor-Y27632, B27, and other substances were added to medium in an orderly manner to culture the PGEC progenitor cells. At the second stage, growth factors such as FGF2, VEGF, EGF, Chir99021, and A83-01 were added to the medium to promote the stable proliferation of PGEC progenitor cells. At the third stage, DM3189, IWP2, PD0325901, RA, A83-01, Bmp4, and other substances were separately added into Matrigel medium to induce differentiation into hepatocytes, and then PGEC progenitor cells were cocultured with interstitial cells and endothelial cells to form livers organoids[36] (Figure 1A).
Figure 1 Culture of liver organoids in vitro.
A: Liver organoids cultured from pluripotent stem cells (PSCs). Somatic cells are reprogrammed into induced PSCs; epidermal growth factor (EGF), fibroblast growth factor 2 (FGF2), and other substances are added to the medium; and the cells are cultured on Matrigel to form liver organoids; B: Liver organoids cultured from adult stem cells. Lgr5+ cells are isolated by fluorescence-activated cell sorting, and Wnt3a, Y27632, EGF, Rspo1, FGF, and other substances are added to form organoids by 3D culture. FACS: Fluorescence-activated cell sorting; iPSCs: Induced pluripotent stem cells; FGF2: Fibroblast growth factor 2; EGF: Epidermal growth factor.
ASC-derived liver organoid culture: First, collagenase was used to dissociate fresh liver tissue, and then, EpCAM+ biliary cells were isolated by fluorescence-activated cell sorting (FACS) and other methods. The biliary epithelium was then cultured and expanded in the serum-free medium including various growth factors (such as EGF, Rspo1, FGF, HGF, N-acetylcysteine, gastrin, and nicotinamide). Additionally, Noggin, Wnt3a, and Y27632 were added in the first 3 d, and the medium did not contain Noggin, Wnt3a, and Y27632 after 3 d. Functional analysis of hepatocytes in the liver organoids revealed that the hepatocytes in the microtissue can take up low-density lipoprotein (LDL), accumulate glycogen, secrete albumin, and synthesize cytochrome P450. Using the above method, the liver tissue organoids were established in the liver tissues of patients with antitrypsin deficiency (A1AT) and Alagille (ALGS) syndrome, and the metabolism of the substances in the liver organoids was similar to that in liver tissues in vivo[42] (Figure 1B).
APPLICATION OF LIVER ORGANOIDS IN BENIGN HEPATIC DISEASES
Liver organoids for studying liver fibrosis (LF)
LF is a chronic liver disease in which the liver undergoes long-term chronic damage (involving viruses, alcohol, poisons, drugs, metabolism, autoimmunity, etc.). Pathology results show that LF is characterized by a significantly increasing accumulation of ECM and the obvious regeneration of hepatic parenchymal fibrous nodules. Most cases of LF will progress to cirrhosis, and even liver cancer, leading to liver failure[43]. At present, LF research models include hepatic stellate cells (HSCs) and hepatocyte 2D coculture, 3D culture, animal LF models, etc.[44]. Using the above disease models to study the pathogenesis and treatment of LF, it was found that an effective antifibrotic drug can be verified in these models but that the antifibrotic effect of such drug is significantly reduced when it is used in clinical practice. Therefore, it is very urgent to establish a disease model that can better reflect the pathological process of LF in humans, to more effectively study the pathogenesis of LF, antifibrotic treatment, and drug resistance mechanisms. It has been reported in the literature that pulmonary organoids and small intestinal organoids are used to establish their respective fibrosis models to study the pathogenesis and mechanism of fibrosis[45-47].
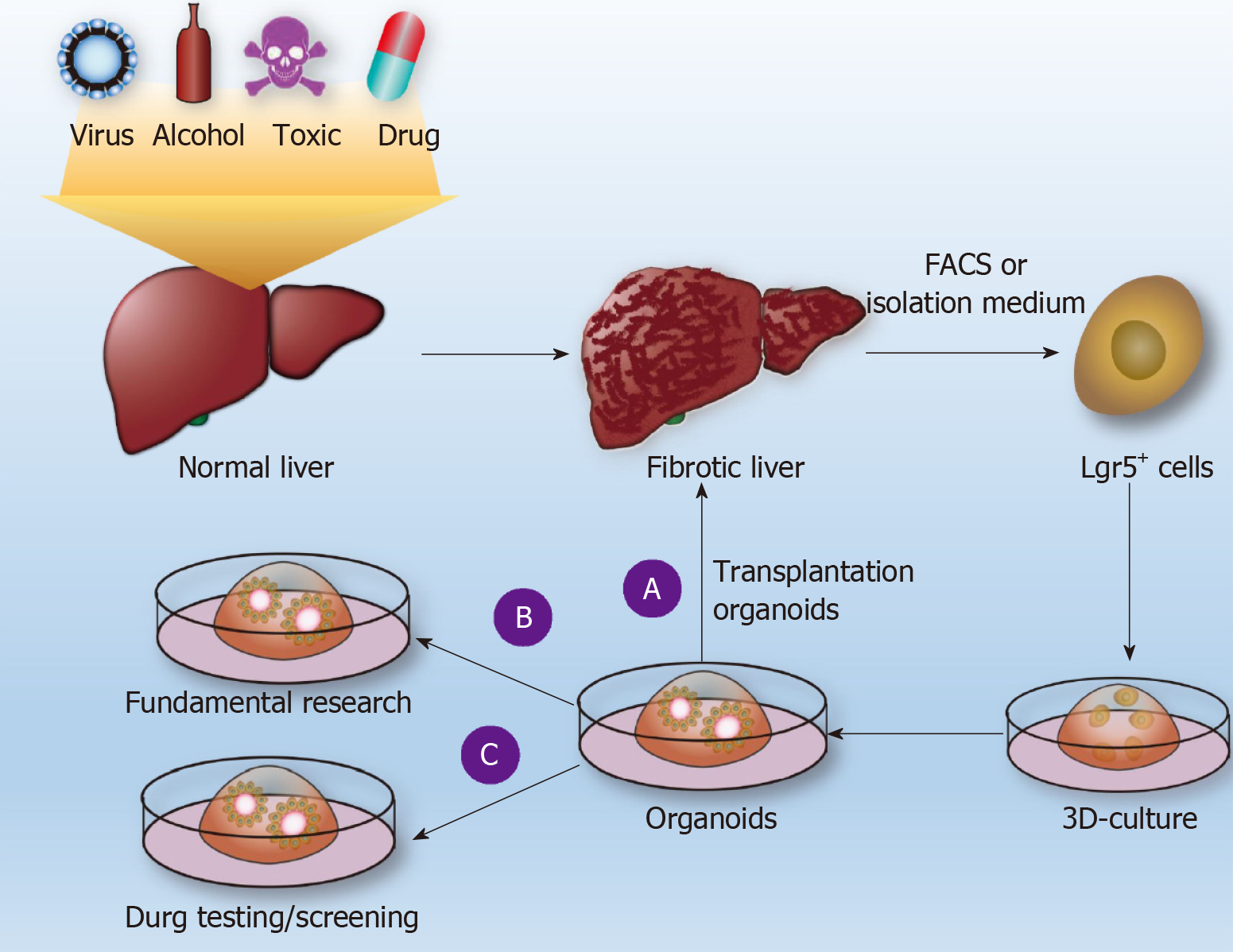
Many drugs (e.g., methotrexate, acetaminophen, and allyl alcohol) can cause LF. The difficulty of studying drug-induced fibrosis is due either to a lack of drug-related fibrosis models or to the fact that the corresponding existing LF presented by the commonly used fibrosis model is significantly different from drug-induced LF in humans. Leite et al[48] reported that HepaRGs cocultured with HSCs developed into liver organoids. LF organoids formed by the coculture of hepatocytes and HSCs can not only maintain the vitality and function of hepatocytes[44] but also exhibit the microenvironment of hepatocytes during the occurrence of LF in the body, such as cell-cell and cell-matrix interactions[44,49]. Through LF organoids, the researchers found that acetaminophen (APAP)-induced fibrosis does not directly activate HSCs but induces hepatocyte damage and then, through hepatocyte injury-dependent HSC activation pathway-induced activation of HSCs, causes an accumulation of ECM[44] (Figure 2B). APAP-induced LF organoid models also display certain features of LF, such as chronic inflammation that can enhance HSC activation, histone deacetylase inhibitors that can inhibit the activation of HSCs, and so on. The researchers established an LF organoid model of APAP and established a model of hepatic fibrosis induced by methotrexate (MTX) and allyl alcohol[44]. In mouse and human livers, Lgr5+ cells are induced always during the process of fibrosis. If Lgr5+ cells were induced to grow out of liver organoids and are implanted into injured livers in mice, the mice can restore some liver function[15,38] (Figure 2A). Although HepaRGs and HSCs have been cocultured into liver organoids, no studies have reported that LF can be induced through the coculturing of Lgr5+ cells with other cells. In the future, Lgr5+ cells in the liver tissue of hepatic fibrosis patients can be isolated by FACS, and Lgr5+-derived LF organoid models can be induced to establish individual-specific LF models. This approach would be helpful for studying the pathogenesis of LF, screening for the best antifibrotic drugs (Figure 2C), repairing liver damage, and improving liver function by orthotopic transplantation of Lgr5+ liver organoids.
Figure 2 Use of liver organoids to study liver fibrosis.
Normal liver tissue is subjected to pathogenic factors (viruses, alcohol, drugs, poisons, etc.) to form liver fibrosis (LF), and Lgr5+ cells are obtained by fluorescence-activated cell sorting or conventional separation to form individualized liver organoids. A: Such organoids are used for transplantation to restore liver function; B: Basic research on LF; C: Drug sensitivity tests. FACS: Fluorescence-activated cell sorting.
Application of liver organoids in metabolic hepatic diseases
The liver is the largest metabolic and detoxifying organ in the human body. However, abnormal metabolism often occurs in the liver, leading to the accumulation of substances and resulting in various liver diseases. Here, hepatic metabolic diseases are classified into gene-deficient metabolic liver diseases, such as A1AT deficiency syndrome caused by antitrypsin deficiency and Wilson’s disease caused by abnormal copper metabolism[15,21,39]; non-gene-deficient metabolic liver diseases; fatty liver diseases caused by abnormal fat metabolism; and other liver diseases. A good model of metabolic diseases requires not only that the tissue structure exhibited by the disease model is similar to the liver tissue structure in the human liver disease but also that the tissue gene expression and tissue metabolism in the disease models are highly similar to the liver tissue in the human liver disease.
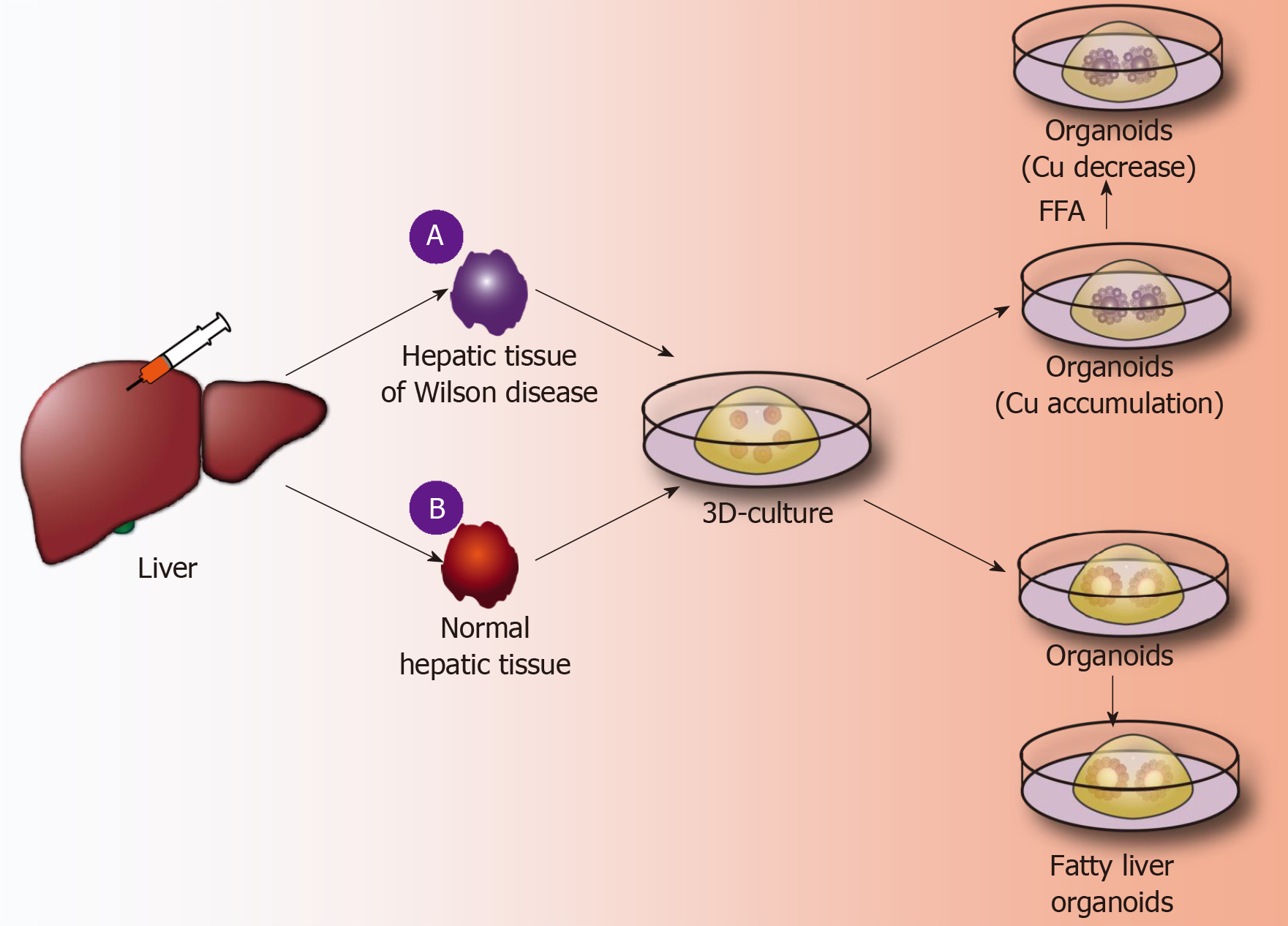
Application of liver organoids in gene-deficient metabolic liver disease: Previous studies of gene-deficient liver diseases have established specific gene-deficient animal models that exhibit certain characteristics of liver disease to some extent. For example, ALGS is a gene dominant genetic disease usually caused by an autosomal jag gene mutation, namely, the Notch signaling pathway jag1 mutation[50]. The main pathological hallmark of ALGS is biliary system damage, which then leads to cholestasis after abnormal bile metabolism, and affected patients finally develop liver function damage after cholestasis. Jag knockout mice exhibit certain symptoms of ALGS. Although the bile duct epithelium was altered, clinical features of ALGS could not be fully demonstrated in mice[51]. Guan et al[52] established liver organoids of patients with ALGS and found that the liver organoids rarely expressed bile duct epithelial markers (KRT19 and KRT17) as analyzed by immunohistochemistry and, moreover, simulated the whole development of ALGS. For example, the bile duct system is normal at birth, and hepatocytes are also fully developed; however, during the growth process, the bile duct system begins to develop chronic damage of the bile duct epithelium[52]. In addition, ATAT, a syndrome caused by a mutation of the SYPANA1 gene, causes antitrypsin deficiency and results in a clinical syndrome characterized by chronic liver disease and chronic obstructive pulmonary disease. Abnormal accumulation of antitrypsin in hepatocytes causes a significant reduction in antitrypsin in the blood[53]. By establishing the liver organoids of patients with ATAT, it was found that the liver organoids can express albumin and take up LDL, etc., like normal liver tissues. However, the hepatocytes in liver organoids have an abnormal accumulation of antitrypsin, and the hepatocellular excretion of antitrypsin is significantly reduced. These characteristics are very similar to the liver metabolism of ATAT syndrome, and therefore, the liver organoid is a good disease model for studying ATAT syndrome[21]. Copper storage disease (Wilson disease) is an autosomal recessive genetic disease caused by the loss of COMMD1. It is characterized by an abnormal accumulation of copper that is caused by abnormal copper metabolism in the liver[54]. Nantasanti et al[39] established liver organoids from copper storage disease and found abnormal copper accumulation in the organoid hepatocytes. In addition, the copper accumulation in the liver-organized organoids was significantly reduced by modifying the COMMD1 gene. Further studies showed that iPSC-derived liver organoids retained the ability to regenerate hepatocytes, intrahepatic bile duct epithelial gene expression, and liver metabolism of patients[15,17,39]. Therefore, the culture of liver organoids may be a more clinically valuable research model for studying genetically deficient metabolic liver diseases in the future (Figure 3A).
Figure 3 Application of liver organoids in benign hepatic diseases.
A: A fine needle is used to puncture Wilson disease tissue and other genetically deficient liver tissues; 3D organoid culture technology is used to obtain the corresponding gene-deficient liver organoids, copper accumulation is identified inside the organoids, and the further using a CRISPR/Cas9 system is used to correct defective genes, thereby significantly reducing copper accumulation in liver organoids; B: A fine needle is used to puncture normal liver tissue, and normal liver organoids are obtained by the 3D organoid culture technique. Free fatty acids are added to the culture medium, and the liver cells in the organoids show lipid accumulation. FFA: Free fatty acids.
Application of liver organoids in non-gene-deficient metabolic liver disease: In the past few decades, the incidence of nonalcoholic fatty liver disease and the number of cases worldwide have risen sharply[55]. It is expected that, by 2025, liver failure induced by nonalcoholic fatty liver disease may be the greatest cause of liver transplantation[56]. Intestinal organoids have reportedly been employed to study intestinal lipid absorption and lipid absorption disorders[57,58]. Liver organoids not only retain the genetic information of the original liver tissues but also reproduce the microenvironment in which the liver cells are naturally located. Nantasnti et al[17] first proposed that liver organoids could be used to study nonalcoholic fatty liver and nonalcoholic hepatitis. In the same year, Kruitwagen et al[40] discovered that the liver organoid model could express marker molecules of the early liver, by culturing the specimens of the livers of cats, dogs, humans, etc. The early biliary epithelial markers (KRT17, KRT19, HNF1A, etc.) were expressed in the expansion medium. When cultured in a differentiation medium, the liver organoids differentiated into mature hepatocytes, which accumulated glycogen, synthesized and secreted albumin and transaminases, and expressed CYP450 molecules[40]. At this time, a sufficient amount of fatty acids was added to the culture medium, and the liver cells ingested a large amount of fatty acids. After a period of culture, it was found that there was lipid accumulation in the liver cells. During the culture process, it was further verified that SREBF1, CPT1A, PPARG, and other genes can promote fat accumulation in hepatocytes, and it was also confirmed that etomoxir or L-carnitine could affect fat metabolism in hepatocytes. Kruitwagen et al[40] found that the liver organoids of dogs and human liver organoids differ in their regulation of lipid metabolic pathways. Studies have shown that canine liver organoids are more prone to lipid accumulation than human liver organoids[40]. It has been verified that the human fatty liver and dog fatty liver markedly differ in pathology and pathogenesis. Currently, normal liver organoids can be cultured, and such organoids can also be cultured to form a fatty liver model. However, the mechanism of fatty liver-related genes, the mechanism of fatty acid uptake by hepatocytes, and the signaling pathway for the regulation of lipid metabolism by hepatocytes have not been studied. In addition, it has not been reported how fatty liver can develop into nonalcoholic cirrhosis or even liver cancer. Therefore, future liver organoids will be a valuable research platform for studying fatty liver (Figure 3B).
Application of liver organoids in chronic viral hepatitis
Chronic viral hepatitis, which has a high incidence in the population, is currently the most prevalent liver disease affecting human health, with approximately 350 million people infected with HBV or HCV worldwide[59]. At present, the most common method for studying hepatitis virus is to culture a transformed cell line or animal model in vitro. The polarity of cells in 2D cell culture is often absent. At the same time, the structure, environmental complexity, and physiological function of tissue composed of multiple cells cannot be presented[60,61]. Animal models are often limited in their application to the pathogenesis and treatment of viral disease due to their long modeling time and high cost. Currently, liver organoids retain the genetic information and expression of liver tissue. Moreover, the molecular phenotypes of the hepatocyte surface in the organoids are similar to those of the original hepatocytes[62]. The development of organoid cultures in vitro provides a new platform for studying the pathogenesis of infectious microbes and diseases[63]. Organoids have been used to explore viral infection and pathogenesis, such as intestinal organoids infected by rotavirus[64] or norovirus[42,65-67] and brain organoids infected by Zika virus. Recent studies suggest that iPSC-derived or ESC-derived hepatocyte-like cells may be infected with HCV[68]. The procedure for generating HCV-infected hepatocytes is a very complex, multistep process involving multiple cytokines. Hepatocytes in humans are usually polarized; however, in today’s 2D cells, hepatocytes usually do not exhibit polarization, and HCV-infected hepatocytes are inefficient. Recently, Baktash et al[69] established 3D liver organoids in which hepatocytes were polarized and able to express viral localization molecules. HCV infection of the hepatocytes showed that the infection efficiency is significantly higher than that of 2D culture[69]. During HCV infection, hepatocytes require colocalization of basement membrane CD81, SR-B1, and EGFR to direct HCV to the junction between the cells, and the virus enters the hepatocytes with the assistance of CLDN1 and OLCN[69]. Baktash et al[69] reproduced the unusual, complex process of HCV infection of hepatocytes by single-molecule imaging in the organoids and unexpectedly found that EGFR is not involved in guiding the localization of HCV but that EGFR- and clathrin-mediated endocytosis is required to achieve particle internalization. However, no studies have reported the mechanism and process of studying HBV-infected hepatocytes by using liver organoids. Liver organoids can reproduce the complex microenvironment of liver tissue (including extracellular factors), hepatocyte gene expression specificity, and immunogenicity on the surface of hepatocytes. Liver organoids will provide a valuable research model for studying the infection mechanism, drug resistance mechanism, and vaccine development for various subtypes of viral hepatitis.
APPLICATION OF LIVER MALIGNANCY ORGANOIDS
Liver malignancy is the fifth most prevalent clinical malignancy in the world but ranks second in global cancer mortality[70]. In recent years, the incidence of malignant liver tumors has increased due to alcohol abuse, viral infection, etc.[71]. Hepatic malignant tumors are divided into two major categories: primary and secondary hepatic malignancies. Among them, primary hepatic malignant tumors are mainly categorized as hepatocellular carcinoma, cholangiocarcinoma, and mixed liver cancer (hereinafter referred to as “liver cancer”), while secondary hepatic malignant tumors are commonly derived from the metastasis of digestive tract malignancies. Genetic analysis shows that liver cancer is usually caused by multiple-gene and multistep mutations in liver tissue, and liver cancer patients rarely have identical oncogenes or the same genes[72,73]. Even in the same patient, there are differences in gene expression between multiple tumor tissues, and even a single tumor tissue is highly heterogeneous[74,75]. The heterogeneity of liver cancer is so high that the importance of individualized precision treatment is self-evident. The main methods of studying liver cancer include culturing a specific liver cancer cell line, primary culture of liver cancer cells, and establishing a PDX model. These approaches are often limited in their application to liver cancer research because of the fixed genotype, difficulty in long-term culture, and high culture cost. The culture technology of tumor organoids has gradually matured. Tumor tissue has been efficiently cultured to produce a variety of tumor organoids, such as organoids derived from colon cancer[76,77], gastric cancer[78], pancreatic cancer[78,79], and liver cancer[80]. Results of genomics and proteomics analyses show that tumor organoids can well represent the histology, superficial molecules, and genetic material of tumors[79,81]. Therefore, tumor organoids are used to study not only the occurrence and progression of tumors and screening of anticancer drugs[82] but also the formation and development of tumor organoids, which provide conditions for the precise treatment of tumors[83]. Liver cancer organoids may provide a good platform for future research on the mechanism of the occurrence of liver cancer as well as its malignant progression and individualized treatment.
Culture of liver malignancy organoids
In 2017, Broutier et al[80] removed Respondin-1, Noggin, and Wnt3a on the basis of a classical culture medium for liver organoids, added dexamethasone and Rho kinase inhibitors, and simultaneously increased the digestion time of liver tumor tissue. In addition, hepatocellular carcinoma organoids, cholangiocarcinoma organoids, and mixed cell carcinoma organoids were successfully cultured in the special medium[80]. By immunohistochemistry and other methods, liver cancer organoids have been shown to stably maintain the tumor tissue structure and protein expression (AFP, EPCAM, KRT19, ALB, Ki-67, etc.) which are similar to those of tumors in vivo in long-term subculture[84]. There are now detailed test methods[21,80,81] to introduce the culture and analysis of mouse and human liver cancer organoids. Fong et al[84] used a 3D macroporous hydrogel instead of the classic Matrigel to culture PDX liver cancer organoids. PDX-derived liver cancer organoids can retain the genetic information and apparent information of the original tumors. Moreover, the liver cancer organoids also have internal heterogeneity[84]. Liver cancer organoids preserve not only the heterogeneity of the liver cancer genome but also that of the liver tumor tissue structure (Figure 4A).
Figure 4 Culture and application of liver cancer organoids.
A: Culture of liver cancer organoids. Primary liver cancer cells are obtained after the enzymatic hydrolysis of collagenase, and similar liver organoids are used to form liver cancer organoids; B: Malignant transformation of normal liver organoids to form liver cancer organoids. Liver organoids are routinely formed, gene editing technology is used to generate multiple-gene, multistep mutations in organoids, and the organoids are orderly cultured to form liver cancer organoids; C: Application of liver cancer organoids. Liver cancer organoids can be used for research of basic mechanism of liver cancer and screening of liver cancer drugs, for example, screening the best chemotherapeutic drugs by comparing the effects of different drugs on the size or number of liver cancer organoids. HCC: Hepatocellular carcinoma; FGF2: Fibroblast growth factor 2; EGF: Epidermal growth factor.
Application of hepatocellular carcinoma organoids
The tumor microenvironment (TME) plays an important role in tumorigenesis and malignant progression[85]. Broutier et al[80] obtained hepatocellular carcinoma organoids by 3D culture of PDX tissue. Hepatocellular carcinoma organoids and primary tumors are not only genetically similar but also reproduce the homogeneous microenvironment in which hepatoma cells are naturally located (Figure 4C)[80]. In addition, invasion and metastasis of hepatocellular carcinoma are important in malignant progression. Wang et al[86] found that liver cancer organoids formed by coculture of hepatoma cells and nonstromal cells can express proteins associated with malignant progression, such as a malignant progression-related protein (MMP-9), an angiogenesis-related protein (VEGFR2), and proteins associated with tumor inflammatory mediators (CXCR4 and CXCL12). For patients who have lost the opportunity for surgery, chemotherapy may be the main treatment for hepatocellular carcinoma. However, hepatocellular carcinoma is very insensitive to conventional systemic chemotherapy. The insensitivity is often caused by the multiple-gene, multistep mutations in liver cancer and by the lack of effective individual preclinical models to screen the systemic chemotherapeutic agents[73] (Figure 4C). Broutier et al[80] have demonstrated the value of liver cancer organoids in drug sensitivity experiments in vitro and in vivo and initially showed that anti-ERK inhibition (SCH772984) may have a significant inhibitory effect on the progression of hepatocellular carcinoma. Further studies found that the establishment of PDX-derived liver cancer organoids verified that CRIPTO expression increased the possibility of resistance to sorafenib in liver cancer patients; furthermore, blocking the CRIPTO/GRP78 signaling pathway could increase the sensitivity of liver cancer patients to sorafenib[87]. In addition, future gene editing techniques will also be applied to the study of liver cancer organoids. The CRISPR/Cas9 system, an emerging gene editing technology, can introduce or knock out related genes to activate or inhibit oncogenes and has been widely used to study some tumors[88]. The study reported that normal intestinal organoids, through CRISPR/Cas9 genome editing, introduced a combination of genes commonly mutated in colorectal cancer (CRC) (APC, KRAS, SMAD4, TP53, etc.), thereby inducing carcinogenesis in normal intestinal organoids[89,90]. Recently, CRISPR/Cas9 and organoids have also been used in combination to study the mechanism of progression of pancreatic cancer[91]. Hepatocellular carcinoma is also a malignant tumor formed through multiple steps involving multiple gene mutations. However, there have been no reports, through CRISPR/Cas9 genome editing, that the appropriate combination of genes was introduced in order to induce canceration in normal liver organoids (Figure 4B). Therefore, hepatocellular carcinoma organoids may provide a valuable preclinical model for future research on the mechanisms of malignant hepatocellular carcinoma progression, gene mutations and signaling pathways of hepatocellular carcinoma, personalized screening of systemic chemotherapy regimens, and gene editing techniques for the treatment of tumors.
Application of cholangiocarcinoma organoids
Cholangiocarcinoma is the second most common tumor among hepatic malignancies. The degree of malignancy of cholangiocarcinoma is extremely high, and early diagnosis is difficult, resulting in the majority of patients being diagnosed in the advanced stage[92,93]. Only approximately one-third of patients can be diagnosed early and receive surgery, but the literature reports that the 5-year survival rate for cholangiocarcinoma is less than 25% after surgery[94]. The mechanisms of pathogenesis and malignant progression of cholangiocarcinoma are still unclear, and cholangiocarcinoma is insensitive to chemotherapeutic drugs. The study of cholangiocarcinoma lacks a preclinical model that can mimic the structure and genetic information of tumors. This preclinical model can be used not only to study the pathogenesis of cholangiocarcinoma but also to screen for chemotherapeutic drugs to which cholangiocarcinoma shows sensitivity. Intrahepatic cholangiocarcinoma organoids can be directly derived from surgical tissue specimens, and can also be generated from PDX tissue. Long-term culture of surgical tissue specimens and PDX tissue can form cholangiocarcinoma organoids, maintaining the primary tumor tissue structure and gene expression specificity. For example, the tumor markers (Ki-67 and CK-19) in intrahepatic cholangiocarcinoma organoids are basically consistent with those in primary tumor tissues[95,96]. It was further found that cholangiocarcinoma organoids form tumors in nude mice and that the tumor tissues are very similar to the primary tumor tissues. To date, cholangiocarcinoma organoids have been gradually used in several applications. In the past, intrahepatic cholangiocarcinoma was mainly caused by carcinogenesis of the intrahepatic bile duct epithelium. However, in recent years, it has been reported that intrahepatic cholangiocarcinoma can also stem from differentiated hepatocytes. Experiments have also shown that cholangiocarcinoma in mice can also be derived from hepatocytes[97]. Through the culture of intrahepatic cholangiocarcinoma organoids, some functions of hepatocytes can be restored, such as the production of mature hepatocytes (exhibiting albumin, bile acids, CYP3A4, HNF4A, etc.). This finding indirectly proves that cholangiocarcinoma can originate from hepatocytes[95]. Extrahepatic cholangiocarcinoma (ECC) accounts for only 10% of cholangiocarcinomas, but its malignancy is always very high, and the prognosis is very poor. At present, there is no corresponding animal model of ECC. By culturing KTC-positive and -negative mouse choledochal cells to form choledochal organoids and planting them under the skin of nude mice, it was found that KTC-positive choledochal organoids had undergone carcinogenesis, which indicates that the KTC gene has an important carcinogenic role in cholangiocarcinoma[98]. Organoids also play an indispensable role in the treatment of cholangiocarcinoma. Lamp is et al[96] through the screening of organoid RNA and 3D drugs for cholangiocarcinoma, found that microRNA21 may be a driver of cholangiocarcinoma resistance to HSP 90 inhibitors. Therefore, the organoid model of cholangiocarcinoma may also be a valuable research model for studying the pathogenesis of cholangiocarcinoma and for screening chemotherapeutic drugs in the future.
Application of secondary liver cancer organoids
Secondary liver cancers are often formed by the metastasis of gastrointestinal malignant tumors, and sometimes the clinical manifestations of liver cancer are mainly found after distant metastasis and a lost opportunity for surgery. The mechanism of liver metastasis in secondary liver cancers is still unclear. Therefore, a metastatic liver cancer model is needed to simulate the liver metastasis process of malignant tumors so that the metastatic mechanism can be well studied. In terms of treatment, chemotherapy for patients with metastatic liver cancer may downgrade the clinical stage of the tumor and may even afford the opportunity to undergo an operation after chemotherapy. However, metastatic liver cancer is usually insensitive to the standard clinical chemotherapeutic regimens, and there is an urgent need for a preclinical model of metastatic liver cancer to screen for the optimal chemotherapy for each patient. Recently, Buzzelli et al[99] obtained metastatic colon cancer organoids from colon cancer liver metastasis tissue; the organoids not only expressed proteins expressed in colon cancer liver metastasis tissues (EpCAM, MUC2, etc.) but also maintained primary colon cancer characteristics. In the susceptibility test, the organoids maintained a similar sensitivity to chemotherapy as that of primary colon cancer[99]. Through gene editing technology, colon cancer organoids with mutated Trp53 were able to cause liver metastasis of colon cancer when they were implanted into nude mice, which further confirmed that Trp53 promotes liver metastasis of colon cancer[100]. When Broutier established secondary liver cancer organoids and implanted them into immunodeficient mice, it was surprisingly found that tumors had also appeared in the liver of the mice[80]. In the future, secondary liver cancer organoids can be used to study tumor metastasis mechanisms and to perform drug susceptibility experiments.