Published online Feb 14, 2018. doi: 10.3748/wjg.v24.i6.680
Peer-review started: September 28, 2017
First decision: October 18, 2017
Revised: December 25, 2017
Accepted: January 18, 2018
Article in press: January 18, 2018
Published online: February 14, 2018
To determine the variability/conservation of the domain of hepatitis B virus (HBV) preS1 region that interacts with sodium-taurocholate cotransporting polypeptide (hereafter, NTCP-interacting domain) and the prevalence of the rs2296651 polymorphism (S267F, NTCP variant) in a Spanish population.
Serum samples from 246 individuals were included and divided into 3 groups: patients with chronic HBV infection (CHB) (n = 41, 73% Caucasians), patients with resolved HBV infection (n = 100, 100% Caucasians) and an HBV-uninfected control group (n = 105, 100% Caucasians). Variability/conservation of the amino acid (aa) sequences of the NTCP-interacting domain, (aa 2-48 in viral genotype D) and a highly conserved preS1 domain associated with virion morphogenesis (aa 92-103 in viral genotype D) were analyzed by next-generation sequencing and compared in 18 CHB patients with viremia > 4 log IU/mL. The rs2296651 polymorphism was determined in all individuals in all 3 groups using an in-house real-time PCR melting curve analysis.
The HBV preS1 NTCP-interacting domain showed a high degree of conservation among the examined viral genomes especially between aa 9 and 21 (in the genotype D consensus sequence). As compared with the virion morphogenesis domain, the NTCP-interacting domain had a smaller proportion of HBV genotype-unrelated changes comprising > 1% of the quasispecies (25.5% vs 31.8%), but a larger proportion of genotype-associated viral polymorphisms (34% vs 27.3%), according to consensus sequences from GenBank patterns of HBV genotypes A to H. Variation/conservation in both domains depended on viral genotype, with genotype C being the most highly conserved and genotype E the most variable (limited finding, only 2 genotype E included). Of note, proline residues were highly conserved in both domains, and serine residues showed changes only to threonine or tyrosine in the virion morphogenesis domain. The rs2296651 polymorphism was not detected in any participant.
In our CHB population, the NTCP-interacting domain was highly conserved, particularly the proline residues and essential amino acids related with the NTCP interaction, and the prevalence of rs2296651 was low/null.
Core tip: Simultaneous analysis of both viral and host features important for hepatitis B virus (HBV) entry into hepatocytes provided locally relevant preliminary information in a population previously uncharacterized in this regard. In-house developed next-generation sequencing was successfully used to investigate the variability of the preS1 region of the HBV large envelope protein, and real-time PCR melting curve analysis to detect the rs2296651 polymorphism (NTCP variant, S267F) in the HBV cellular receptor, NTCP. Results in a limited sample indicate that the features analyzed would not decrease the effectiveness of new therapies to block NTCP and avert HBV binding to hepatocytes in our particular CHB population.
- Citation: Casillas R, Tabernero D, Gregori J, Belmonte I, Cortese MF, González C, Riveiro-Barciela M, López RM, Quer J, Esteban R, Buti M, Rodríguez-Frías F. Analysis of hepatitis B virus preS1 variability and prevalence of the rs2296651 polymorphism in a Spanish population. World J Gastroenterol 2018; 24(6): 680-692
- URL: https://www.wjgnet.com/1007-9327/full/v24/i6/680.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i6.680
Hepatitis B virus (HBV) infection remains a major health threat, with around 240 million chronically infected individuals worldwide[1]. Persistently infected people are at high risk for the development of cirrhosis and hepatocellular carcinoma, and about 1 million people die each year due to HBV-associated liver disease[2,3]. Currently, the available anti-HBV treatments include conventional or pegylated interferon-α (IFN-α) and nucleos(t)ide analogs (NAs)[4]. Both types of treatments have drawbacks: IFN-based therapies cause significant side effects and yield long-term clinical benefits in less than 40% of treated patients[5], whereas first-line NAs suppress viral activity in more than 80% of patients, but viral eradication is rare[4,6,7].
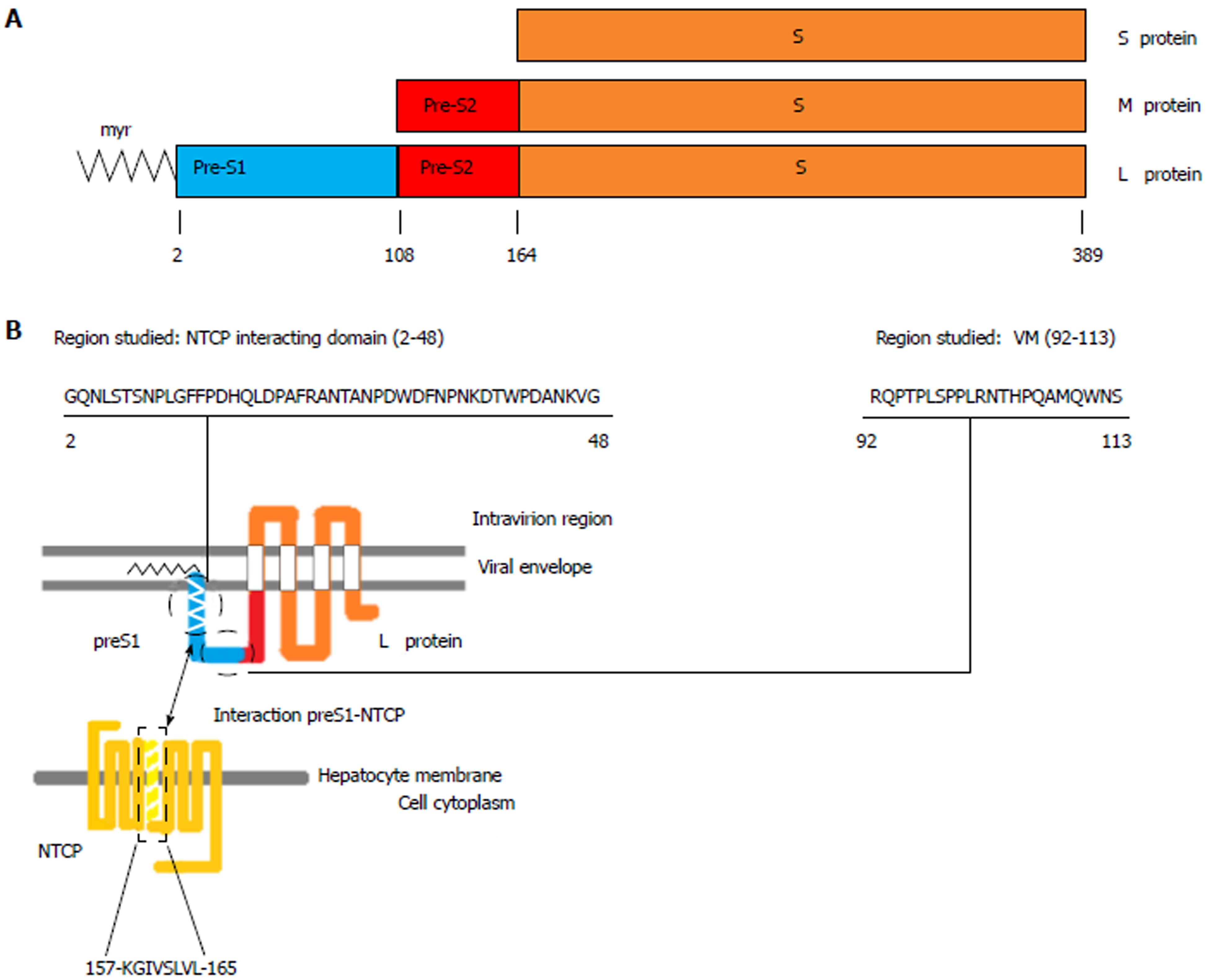
Based on extensive research in the HBV lifecycle and virus-host interactions, several new agents are under development to achieve a functional cure for HBV infection[8,9]. In this sense, identification of sodium-taurocholate cotransporting polypeptide (NTCP), encoded by the SLC10A1 gene and located on chromosome 14[10,11], as a receptor for HBV infection[12] has provided valuable information for the development of inhibitors of HBV entry. NTCP is a multipletransmembrane protein that is predominantly expressed at the basolateral membrane of hepatocytes. The primary role of NTCP is to transport bile salts from the portal blood into hepatocytes[10,11]. Interactions of viral particles with this receptor are mediated by the hepatitis B surface antigen (HBsAg), which is formed by three viral envelope proteins (large, middle, and small) that differ in length at the N-terminal region and share the same C-terminal S region[13,14]. The HBV large envelope proteins (LHBs), which include the preS1, preS2, and S regions of the surface open reading frame of the HBV genome, interact with NTCP through specific binding of a 47 amino acids (aa) domain in the N-terminal end of the preS1 region[15-18] (hereafter referred to as the NTCP-interacting domain), as is shown in Figure 1.
It is reasonable to think that the degree of sequence variability in the NTCP-interacting domain, which is an indication of the extent to which sequence conservation is important to maintain its function[19], may have an impact on the response to inhibitors of HBV entry based on synthetic myristoylated lipopeptides[20] that share the same aa sequence with the NTCP-interacting domain (i.e., therapy to block NTCP). For example, sequence variability in the NTCP-interacting domain might change its affinity to attach to NTCP, changing the dynamics of the competition with synthetic myristoylated lipopeptide analogues, and making them less effective in patients showing such variation.
Furthermore, several single nucleotide polymorphisms (SNPs) that may change the physiological function of NTCP as the key transporter for bile salt homeostasis and affect HBV entry have been identified in SLC10A1[21], and most of them show an ethnicity-dependent profile[22,23]. The rs2296651 SNP in SLC10A1 (g.69778476G>A, GenBank accession number NC_000014.9), which causes the S267F variant, has been identified in Asian populations at a prevalence of 3.1% to 9.2%[22]. Previous studies focusing on rs2296651 and HBV infection have yielded conflicting results[24,25], and the role of this SNP in cirrhosis and hepatocellular carcinoma remains uncertain[26]. However, as synthetic myristoylated lipopeptides share the same aa sequence with the NTCP-interacting domain, it is reasonable to think that the presence of this SNP might influence the effectiveness of these therapies. This variant has not been found in some Caucasian populations (European Americans and Hispanic Americans)[22], but to our knowledge, there are no studies investigating the prevalence of rs2296651 in our country (Spain), where a Caucasian population mainly of European Mediterranean origin is prevalent.
The main aims of this study were to analyze the variability and conservation of the HBV preS1 region NTCP-interacting domain, involved in HBV entry into hepatocytes, and to determine the prevalence of the rs2296651 SNP, causing the S267F NTCP variant, in an HBV patient population from our area (Barcelona, Spain).
The study included 246 individuals recruited from the population attending the outpatient clinic of Vall Hebron University Hospital (Barcelona, Spain). The study was approved by the hospital ethics committee and all individuals gave informed written consent for participation at enrollment. Participants were divided into 3 groups: group A, patients with chronic hepatitis B infection (n = 41, CHB); group B, patients testing negative for HBsAg, but positive for antibodies against the hepatitis B core antigen (anti-HBc) (n = 100, Resolvers); and group C, an HBV-uninfected control group (HBsAg-negative, antiHBc-negative) (n = 105, Controls). Participants from groups B and C were all Caucasian. Ethnic heterogeneity was greater in Group A, with 30/41 (73%) Caucasian, 5/41 Asian (12%), and 6/41 sub-Saharan (15%) participants (Table 1).
| Group A, CHB (n = 41) | Group B, Resolvers (n = 100) | Group C, Controls (n = 105) | P value1 | |
| Gender, n males (%) | 22 (54) | 54 (54) | 54 (51) | NS |
| Age, yr, mean ± SD | 54.8 ± 19.1 | 62.1 ± 13.0 | 60.3 ± 13.5 | NS |
| Origin, n (%) | ||||
| Caucasian | 30 (73) | 100 (100) | 100 (100) | - |
| Asian | 5 (12) | 0 (0) | 0 (0) | - |
| Sub-Saharan | 6 (15) | 0 (0) | 0 (0) | - |
Two main analyses were performed. First, the variability and conservation of the preS1 region of LHBs was analyzed in the 18 patient samples from Group A with a viral load greater than > 4 logIU/mL. This is the sensitivity limit of the PCR to amplify that region. Then we determined the sequence of preS1 by next-generation sequencing (NGS) based on ultra-deep pyrosequencing (UDPS) on the GS-Junior platform (454 Life sciences-Roche, Branford, United States). All 18 patients were treatment-naive and tested negative for hepatitis D virus (HDV), hepatitis C virus (HCV), and human immunodeficiency virus (HIV). The demographic, biochemical, and virological characteristics of these patients are shown in Table 2. Second, the rs2296651 SNP (S267F) was determined in all 246 individuals comprising the 3 groups following a previously described in-house developed protocol to detect SNPs in human serum samples[27]. To avoid the discomfort of additional blood drawing, 1 mL of serum from samples obtained for routine clinical analysis was used for all analyses.
| ID | Gender | Age | Origin | HBV Genotype1 | HBV DNA (logIU/mL) | ALT (IU/L) | HBeAg |
| 1 | Female | 68 | Caucasian | D | 5.0 | 29 | Negative |
| 2 | Male | 55 | Caucasian | A | 5.8 | 46 | Negative |
| 3 | Female | 40 | Caucasian | D | 4.2 | 29 | Negative |
| 4 | Male | 47 | Caucasian | F | 5.8 | 88 | Negative |
| 5 | Female | 47 | Caucasian | D | 6.3 | 170 | Negative |
| 6 | Female | 28 | Caucasian | D | 5.4 | 22 | Negative |
| 7 | Male | 28 | Asian | C | > 8 | 94 | Positive |
| 8 | Female | 41 | Caucasian | A | 4.7 | 18 | Negative |
| 9 | Male | 39 | Caucasian | D | 5.0 | 29 | Negative |
| 10 | Male | 45 | Caucasian | F | > 8 | 51 | Positive |
| 11 | Female | 35 | Asian | C | > 8 | 56 | Positive |
| 12 | Female | 46 | Caucasian | H | 5.3 | 23 | Negative |
| 13 | Male | 37 | Sub-Saharan | E | 4.3 | 37 | Negative |
| 14 | Female | 54 | Caucasian | A | 4.3 | 21 | Negative |
| 15 | Female | 36 | Asian | C | > 8 | 19 | Positive |
| 16 | Female | 20 | Sub-Saharan | E | 5.1 | 22 | Positive |
| 17 | Male | 73 | Caucasian | B | > 8 | 221 | Negative |
| 18 | Male | 49 | Asian | B | 5.6 | 181 | Negative |
Serological markers for HBV (HBsAg, HBeAg, anti-HBe, and anti-HBc) and anti-HCV antibodies were tested using commercial enzyme immunoassays mounted on a COBAS 8000 analyzer (Roche Diagnostics, Rotkreuz, Switzerland). Antibodies against HDV were tested using the HDV Ab kit (Dia.Pro Diagnostic Bioprobes, Sesto San Gioviani, Italy), and anti-HIV antibodies were tested by the Liaison XL murex HIV Ab/Ag kit (DiaSorin, Saluggia, Italy). HBV-DNA was quantified by real-time PCR with a detection limit of 20 IU/mL (COBAS TaqMan HBV V2.0, Roche Diagnostics, Mannheim, Germany). HBV genotypes were determined by Sanger sequencing and phylogenetic analysis with reference sequences from HBV genotypes A to H of the preS1 region (Supplementary Table 1), and supported by NGS analysis of the same region.
In this study, we explored a fragment of the HBV genome including the entire preS1 region and the N-terminal end of preS2. The HBV genotype-specific insertions and deletions occurring along the HBV genome change the nucleotide (nt) numbering of the fragment. Hence, the fragment includes nt positions 2844 to 56 (434 bp) in genotype A (genome size 3221 nt), positions 2838 to 56 in genotypes B, C, D, E and H [434 bp in B, C, and H (genome size 3215 nt in all of them), 401 bp in D (genome size 3182 nt), and 431 bp in E (genome size 3212 nt)] and positions 2837 to 56 (434 bp) in genotype F (genome size 3215 nt).
A detailed description of the molecular amplification, NGS procedures carried out, and subsequent bioinformatics filtering of the sequencing data is provided in the Supplementary Materials and Methods (Supplementary Protocol 1). Briefly, molecular amplification was performed by nested PCR. In the final PCR products (amplicons), the technique incorporated M13 universal adaptor sequences (forward and reverse), a unique identifier that enabled grouping of the sequences derived from each sample [multiplex identifier sequences (MID)], and sequences A and B (adaptors for the elements of the UDPS system). The amplicons were purified and pooled at equimolecular concentrations. The pool was UDPS-analyzed following the manufacturer’s protocol. The sequencing data underwent a bioinformatics filtering procedure based on an in-house-developed pipeline[28], with all computations done in the R environment and language[29]. UDPS reads (sequences from each individual amplicon) were demultiplexed according to their MID sequence, and primers were trimmed. After a quality filter step, reads with the same nt sequence were collapsed into haplotypes, that is, unique sequences covering the full amplicon observed on the clean set of sequences[30]. We then selected haplotypes covering the full amplicon and common to both the forward and reverse strands whose sequences were present in frequencies of > 0.25% of the complete set of sequences.
The variability/conservation of the aa sequence in the NTCP-interacting domain was compared to that of a conserved domain with a pivotal function in virion morphogenesis[31]. This domain, located in the preS1 C-terminal (Figure 1), was selected for comparison purposes as a “sequence conservation control” because of its high degree of conservation in other orthohepadnaviruses, such as woodchuck hepatitis virus[31].
The NTCP-interacting domain includes 47 aa from the N-terminal end of the preS1 region (aa 13-59 in HBV genotypes A, B, C, F and H, aa 2-48 in genotype D, and aa 12-58 in genotype E), whereas the virion morphogenesis domain includes 22 aa lying between the C-terminal end of preS1 and the first 5 aa from the N-terminal end of preS2 (aa 103-124 in HBV genotypes A, B, C, F and H, aa 92-103 in genotype D, and aa 102-123 in genotype E).
The first step of this analysis was to classify the haplotypes from each patient according to their HBV genotype by phylogenetic analysis in order to differentiate sequence variations from genotype-related polymorphisms. First, we selected 86 full-length HBV genome sequences representative of HBV genotypes A to H, obtained from GenBank (accession numbers in Supplementary table 1) and extracted the region from nt 2837, 2838, or 2844 (depending on viral genotype) to 56. We then determined the maximum genetic distances between sequences from the same HBV genotype in this region and the minimum genetic distances between sequences from different genotypes, in order to set a sequence identity threshold. The threshold was then used to cluster the individual haplotypes from each patient; sequences with an identity above the identity threshold were grouped together in the same cluster and their frequencies were added up. The master sequence (the most abundant haplotype) from each cluster and the sequences obtained from the 86 full-length genomes then underwent phylogenetic analysis to determine the HBV genotype. Genetic distances between master sequences and the remaining sequences in their clusters were below the minimum distance between sequences from different genotypes; thus, all the sequences in the cluster were considered to belong to the same HBV genotype as the master sequence. All individual haplotypes were then separated into different fasta files according to the genotype assigned to each of them and translated into aa sequences. The aa haplotypes with the same sequence were recollapsed and their frequencies were updated.
The multiple alignments of aa haplotypes obtained were used to detect point mutations, each of which was considered a separate variant, and to determine their abundance in the two regions studied.
Finally, an overall image of aa conservation/variation in preS1 for each HBV genotype was obtained by calculating the information content (IC) of each position. The IC of an aa position is related to the number of binary decisions (number of questions with a yes/no answer) required to find the correct aa in a given position among a set of 20 possibilities per position in the multiple alignment of haplotypes. For example, if the probabilities of finding any of the 20 aa (pa) were equal in a given position, the number of binary questions required to find the correct aa in this position would be log2 (20) = 4.322. However, since the different aa have different unknown probabilities of occurring in each position of the multiple alignment, an uncertainty measure, -∑a∈aapa log2 (pa)[32], also known as “entropy”, must be added to the IC calculation. Thus the IC of each aa position was calculated using the following equation: aa (Site) = log2 (20)-∑a∈aapa log2 (pa) If only one aa were found in a given position from the multiple alignment (maximum conservation), there would be no uncertainty in that position and the probability of finding that aa would be 20/20 thus -∑a∈aapa log2 (pa) = 0 and aa = log2 (20)-0 = 4.322. On the other hand, if all aa had an equal possibility of occurring at a given position (maximum variability), the degree of uncertainty of that position would be the highest and the probability of finding any aa in that position would be 1/20 thus -∑a∈aapa log2 (pa) = 4.322 and aa = log2 (20)- 4.322 = 0. These calculations in the regions of NTCP interaction (47 aa) and virion morphogenesis (22 aa) were also represented as sequence logos created using the R language package, motifStack[33].
The statistical and bioinformatics methods used in this study were reviewed by Dr. Josep Gregori from the liver disease-viral hepatitis laboratory (Vall d’Hebron Institut Recerca-Hospital Universitari Vall d’Hebron), CIBERehd and Roche Diagnostics SL.
The rs2296651 SNP in the SLC10A1 gene, causing the NTCP S267F variant, was determined using a new in-house developed real-time PCR method based on fluorescence resonance energy transfer (FRET) probes, on the LightCycler 2.0 analyzer (Roche Diagnostics, Rotkreuz, Switzerland). PCR primers were designed to flank a 111-bp region of SLC10A1, including the SNP. The results obtained by this method were validated by direct sequencing (Sanger method) in genomic DNA extracted from blood in 18 samples from the 3 groups: A (12), B (4), and C (2). A detailed explanation of the rs2296651 detection procedure by both real-time PCR and Sanger sequencing is provided in the Supplementary Materials and Methods (Supplementary Protocol 2).
In the regions from nt 2837, 2838, or 2844 (depending on viral genotype) to 56, extracted from the 86 full-length HBV genome sequences representative of genotypes A to H, analysis of the maximum genetic distance in each genotype and the minimum genetic distance between different genotypes (data not shown) resulted in a sequence identity threshold between genotypes of 95%. Therefore, for each patient, haplotypes with a sequence identity > 95% were clustered together, and the master sequence from each cluster was included in the phylogenetic analysis to determine the HBV genotype.
The NTCP-interacting domain (47 aa) and virion morphogenesis domain (22 aa) were analyzed in the 18 patients in group A with viremia levels > 4 logIU/mL. In total, 118779 quality-filtered sequences were obtained from the samples, and a median of 5546 (1903 to 13126, interquartile range, 3585) sequences containing both domains were analyzed per sample.
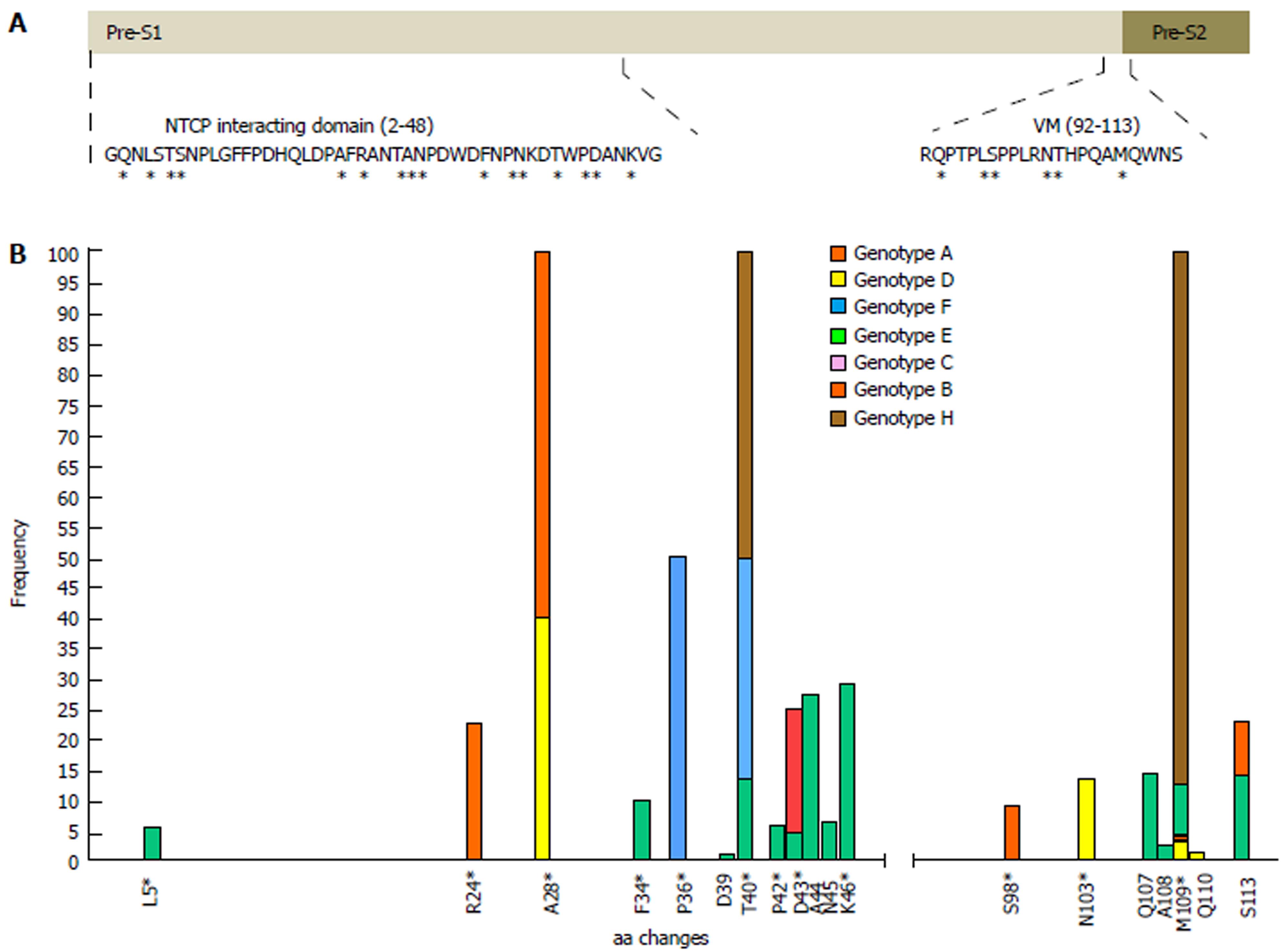
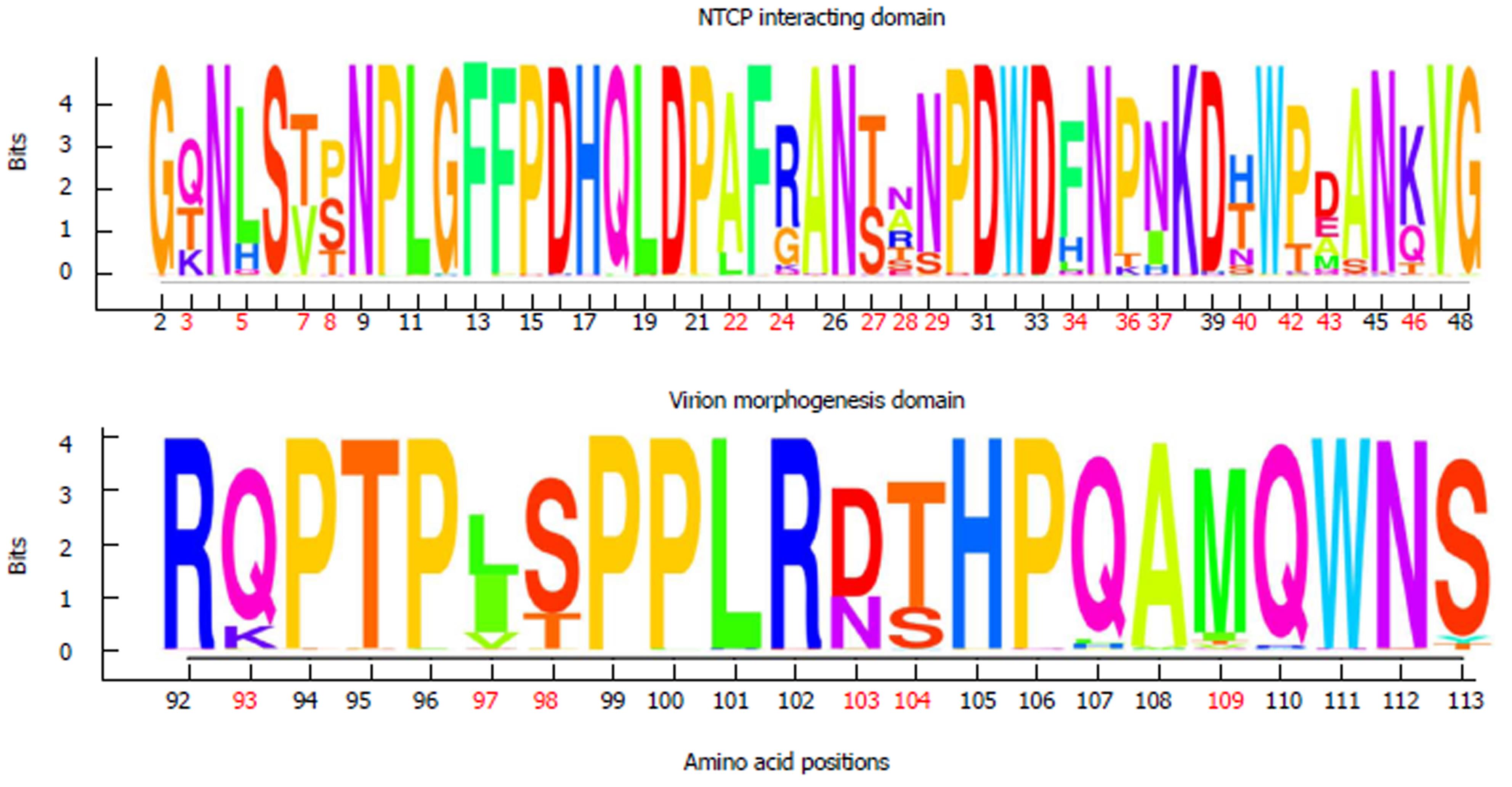
In general, aa residues from the NTCP-interacting domain were conserved within each HBV genotype among the different viral genomes examined. Only 12 of 47 (25.5%) aa positions showed genotype-unrelated changes occurring at > 1% of the HBV quasispecies, a somewhat lower percentage than in the virion morphogenesis domain (7 of 22 aa positions, 31.8% with genotype-unrelated changes) (Figure 2B). Interestingly, according to the consensus sequences of HBV genotypes A to H of both domains, obtained from the 86 full-length HBV genome sequences from GenBank, the virion morphogenesis domain had a lower percentage of aa positions with genotype-associated changes (viral polymorphisms) than the NTCP-interacting domain (6 of 22, 27.3% vs 16 of 47, 34%, respectively) (Figure 2A). On analysis of conservation throughout the NTCP-interacting domain, we observed a high degree of conservation between aa positions 2 to 21 (viral genotype D numeration), in which no aa changes in proportions greater than 1% were found, except in one genotype E patient who showed the H5Q variant in 11.8% of haplotypes (representing 5.9% of all genotype E haplotypes obtained) (Supplementary Table 2). Conservation was especially relevant between aa 9 and 21: very few changes were seen within each HBV genotype (none at > 1%) and wild-type aa were the same in the different genotypes (Figures 2 and 3).
According to the sequence logos layout, which shows the IC analysis for each aa position, HBV genotype C displayed the highest level of conservation: unaltered in the NTCP-interacting domain and only 1 aa change at position 124 in the virion morphogenesis domain (112 according to viral genotype D numeration), in the 4 patients included. HBV genotype E showed the highest variability, although it should be remembered that only 2 patients had this genotype (Supplementary Figure 1). HBV genotypes D and A (the most prevalent in our area)[34] showed moderate variability, with aa 28, 103, and 109 being particularly variable in genotype D, and aa 54 and 120 (43 and 109 according to genotype D numeration, respectively) particularly variable in genotype A (Supplementary Figure 1).
Of note, the sequence logos, including all haplotypes regardless of viral genotype (Figure 3) also indicated that proline (P) residues were highly conserved. Nonetheless, the P residue at position 30 of the NTCP-interacting domain showed a change to lysine (K) in HBV genotype F and to threonine (T) in genotype H, and the P residue in position 36 changed to T in genotype E. Furthermore, there was a notable complete preservation of the five P residues located in the virion morphogenesis domain. Interestingly, serine residues (S) in this domain showed changes to T (positions 109 and 124 in HBV genotype B and positions 98 and 113 in genotype D) or tyrosine (Y) (position 123 in HBV genotype E and position 113 in genotype D) (Figure 3 and Supplementary figure 1); all of these have similar physicochemical characteristics, being the residues most commonly phosphorylated[35]. In general, the S residues of the NTCP-interacting domain were conserved within each viral genotype, but different aa were sometimes seen in this position between the different genotypes (Supplementary figure 1).
Of note, 4 of our patients, harboring haplotypes classified into HBV genotypes A, D, E and H, showed a significant percentage of changes in the preS2 initial methionine residue (position 109 in genotype D) (Supplementary table 2 and Supplementary figure 1). Other relevant aa changes in the NTCP-interacting and virion morphogenesis domains of the patients and their frequencies are shown in Supplementary table 2.
The SLC10A1 gene rs2296651 SNP (S267F) was determined in all 246 individuals recruited (95.53% Caucasian, 2% Asian, and 2.47% Sub-Saharan), which constituted a representative population of patients from the outpatient clinic of our hospital. None of them presented the SNP. Real-time PCR results were also confirmed by direct sequencing in 18 blood samples from the 3 groups: A (12), B (4), and C (2).
Since it was first identified as a specific receptor enabling HBV entry into human hepatocytes, the NTCP bile salt transporter has gained significant attention as a target in antiviral therapy for HBV and HDV (which share the same entry mechanism)[12,36,37]. The interaction between viral particles and this transporter is mediated by the region between aa 2 and 48 (HBV genotype D numeration) of the preS1 N-terminal end of LHBs[15-18], where aa 2 (glycine) must be bound to myristic acid (N-myr) for the viral particles to be infective[38,39]. Identification of this interaction has allowed the development of drugs that can block HBV entry by targeting NTCP[20]. An example of these drugs is Myrcludex-B, a synthetic myristoylated 47-aa lipopeptide derived from aa 2 to 48 of preS1, which has proven to effectively block HBV cellular entry in vitro[18,40], in vivo[41-43], and in patients included in clinical trials[44,45]. In a recent report, Tsukuda et al[46] explored an alternative strategy for blocking HBV entry based on proanthocyanidin and its analogs, which directly act on LHBs. Hence, inhibition of viral cellular entry is becoming consolidated as a viable new therapeutic approach against HBV infection. In this line, analysis of the preS1 N-terminal NTCP-interacting domain and the presence of the rs2296651 SNP (S267F) may be relevant as prognostic markers of the response to this new therapy because of their potential roles in the interaction that enables HBV to infect hepatocytes.
To our knowledge, this is the first study in which the variability/conservation of the essential preS1 NTCP-interacting domain has been investigated by NGS. The results show a high degree of conservation between preS1 aa positions 2 to 21 (HBV genotype D numeration): aa changes in proportions greater than 1% were found in a single position in only 1/18 patients. Of note, among the 20 aa in the N-terminal end of preS1, wild-type aa were found to be conserved between positions 9 and 21 in all genotypes included. These observations suggest an essential function of this segment of preS1, which would agree with the results of a study by Glebe et al[17] In that report, hepatitis B attachment site mapping by infection-inhibiting amino-terminally acylated preS1-derived lipopeptides highlighted the essential role of aa 9 to 18 for viral particle binding to NTCP. In addition, the findings from that study[17] indicated that aa sequences in positions 29 to 48 would play an accessory role, a fact that seems to justify the more significant variability in this region observed in our NGS analysis. Therefore, conservation/variability is not homogenous throughout the entire preS1 NTCP-interacting domain: whereas segments with a previously described essential role in the NTCP interaction show a highly conserved sequence, positions with accessory roles that are nonetheless needed for strongest blocking show greater variability. preS1 structural simulations would be needed to help clarify the contribution of each segment in NTCP interactions and the sequence conservation requirements for their respective functions.
After their translation, approximately 50% of LHBs undergo posttranslational topological reorientation, in which their N-terminal end is translocated to the endoplasmic reticulum lumen[47]. This gives rise to two types of LHBs: those in which the preS1 region has an external position in the viral particles and those in which this region has an internal position. Whereas the N-terminal region of external LHBs is involved in NTCP interactions[15], the C-terminal part of internal LHBs between aa 92 and 113 (HBV genotype D numeration) has a pivotal function in virion morphogenesis by directly contacting the nucleocapsid during viral particle budding[31]. Moreover, this latter domain is reported to be highly conserved among Orthohepadnavirus[31], a concept that is supported by the lower percentage of HBV genotype-associated viral polymorphisms in the virion morphogenesis domain than in the NTCP-interacting domain (27.3% vs 34%, respectively) in consensus sequences of genotypes A to H obtained from the 86 HBV genome sequences downloaded from GenBank. However, in the present study, HBV genotype-unrelated changes above 1% of the quasispecies were found in 25.5% of NTCP vs 31.8% of C-terminal positions, with both domains being most highly conserved in genotype C and most highly variable in genotype E (a limited finding because this high variation was observed in 1 of only 2 genotype E patients included). Thus, while the NTCP-interacting domain seems to be more highly conserved within each genotype, the C-terminal virion morphogenesis domain seems more highly conserved between different HBV genotypes. The high proportion of conserved aa positions in both domains within the same viral genotype and between different genotypes seems to confirm that they conduct essential functions for HBV replication. Again, structural simulations would likely be helpful to understand the reasons for the sequence conservation and variability in these two domains.
Interestingly, P and S residues generally showed a high degree of conservation, particularly in the C-terminal virion morphogenesis domain. It must be kept in mind that P is often found at the end of the α helix or in turns or loops, and it contributes to protein folding by stabilizing these structures[48,49], thus being associated with essential structural protein motifs. Conservation of most P residues in the two preS1 domains suggests their structural preservation, which would facilitate interactions between external LHBs and the NCTP cell receptor or between internal LHBs and nucleocapsids in the same manner as a lock and key. In the virion morphogenesis domain, in positions where S was the wild-type aa, we observed that changes with > 1% prevalence within the same HBV genotype or between different genotypes were either to T or to Y. Bearing in mind that S, T, and Y are the main targets for phosphorylation in eukaryotic cells[35], the tendency to keep these specific aa in specific positions of the virion morphogenesis domain suggests that they could be phosphorylated. This phosphorylation could be important for the functionality of the virion morphogenesis domain. Site-directed mutagenesis experiments, with modification of the P and S residues, could clarify the function of these two conserved aa in both the preS1 essential domains analyzed.
In the present study, we also assessed the prevalence of the SLC10A1 gene SNP, rs2296651 (S267F), in all 246 participants. The effect of this SNP on CHB is controversial. Most studies have reported that it is associated with CHB resistance, as it might interfere with ligand binding, thereby preventing HBV from cellular entry[24,26]. Nonetheless, one study has shown that rs2296651 is associated with enhanced HBV infection[25]. The prevalence of rs2296651 varies greatly between different ethnicities and geographic locations[22,23], being identified prevalently in Asian populations, with the highest rates in southern China and Vietnam[23]. This polymorphism has not been detected in Americans from European, African or Hispanic origins[22]. However, its prevalence in our area (typically classified as having an intermediate HBsAg prevalence)[50] where Caucasian populations mainly of European Mediterranean origin are predominant, has never been assessed. In the present study, rs2296651 was not found in any of the HBV patients or controls studied. Considering the substantial prevalence of this SNP in Asian populations where HBV infection is endemic, the low prevalence found in our area may be associated with the lower incidence of HBV infection[51].
Although the rs2296651 SNP seems to be absent in the chronic HBV-infected population attended in our setting, multiple SNPs in SLC10A1 have been found at relatively high allele frequencies in certain ethnic populations[21-26,52]. As an example, Ho et al[22] reported 6 additional ethnicity-dependent SNPs in the 5 exonic regions of SLC10A1 (3 were non-synonymous and caused aa changes: I223T, I279T, K314E). The functionally relevant NTCP polymorphisms would be expected to modify bile acid homeostasis and HBV cellular entry; hence, it cannot be excluded that Caucasian individuals might have additional, still undescribed NTCP SNPs that could interfere with HBV infectivity. Therefore, further studies should be performed to characterize and determine the prevalence of NTCP SNPs in different ethnic populations and their implications in NTCP function.
This study includes a representative sample of CHB patients attended in our center and includes most HBV genotypes (A to F and H). Necessarily, the number of patients carrying the less prevalent genotypes in our area was low. This limitation may have biased the conservation/variability findings in some genotypes due to individual variability in some patients, as was likely the case of the variability in genotype E. Thus, the findings should be considered preliminary, requiring confirmation in further studies with larger populations. Another potential limitation is that the NGS technology used in this study (ultra-deep pyrosequencing in the GS-Junior platform, 454/Roche) has been discontinued by the supplier. When the study was designed, this technique was selected as the best available one for our purposes, as it enabled inclusion of the preS1 regions of NTCP interactions and virion morphogenesis in the same sequence read, using a single amplicon. A suitable alternative to enable future studies aimed at confirming and expanding the results of the present report, and in general to analyze the viral quasispecies, could be Sequencing By Synthesis technology on MiSeq platforms (Illumina, San Diego, United States), as this technique can generate sequence reads with a length similar to that of the fragment analyzed in this study.
In conclusion, NGS analysis of preS1 domains in strains from 7 of the 8 main HBV genotypes showed a high degree of conservation of essential amino acids related with the NTCP interaction. Proline residues in both domains and potential phosphorylation targets in the virion morphogenesis domain were also highly conserved. Given the low to null prevalence of the rs2296651 SNP of the SLC10A1 gene in our patient population, we would not expect interference from NTCP in its interaction with preS1 or preS1-derived synthetic lipopeptide molecules. Thus, these preliminary results indicate that inhibition of HBV entry by NTCP block therapies would be a suitable treatment in our CHB population.
The findings from this exploratory study are locally relevant as they provide preliminary information on a population previously uncharacterized in this regard. However, they have limited robustness for generalizations. Nonetheless, the study illustrates the value of NGS to investigate the variability of the preS1 region of the LHBs, and real-time PCR melting curve analysis to detect the rs2296651 polymorphism causing S267F variant in NTCP, the HBV cellular receptor. Future studies with larger patient samples are needed to support the preS1 results and investigate additional NTCP polymorphisms. Finally, functional and structural studies will help decipher the implications of the high degree of conservation of the preS1 domains on HBV activity.
The preS1 region of the hepatitis B virus (HBV) large envelope protein interacts with its cellular receptor, sodium-taurocholate cotransporting polypeptide (NTCP), to enable HBV infectivity. Identification of this interaction has led to the development of drugs that can block HBV entry by targeting NTCP, such as synthetic myristoylated lipopeptides derived from the domain of the preS1 N-terminal end which interacts with NTCP (hereafter, NTCP-interacting domain), that can dock to this receptor, blocking the HBV entry mechanism. Several clinical trials are currently testing this type of HBV entry inhibitor; for example, Myrcludex-B. Furthermore, HBV cellular entry may also be impaired by the single nucleotide polymorphism (SNP) rs2296651 in the SLC10A1 gene. This SNP causes the NTCP S267F variant, which can affect interactions between the receptor and viral particles. Study of these viral and host features may be relevant to understand the interaction of NTCP with the preS1 NTCP-interacting domain and, reasonably, to provide an indication of the potential effectiveness of treatments with synthetic myristoylated lipopeptides derived from this domain.
Sequence variability in the NTCP-interacting domain might change its affinity to attach to NTCP, and this would alter the dynamics of competition with synthetic myristoylated lipopeptide inhibitors of HBV entry, potentially weakening the effectiveness of these treatments. The presence of the rs2296651 SNP causing the S267F NTCP variant could also impair the effectiveness of HBV treatment, as synthetic myristoylated lipopeptides have the same amino acid (aa) sequence as the NTCP-interacting domain. These factors could be analyzed in specific patient populations to determine the potential effect they may have on treatment with the new therapies designed to block NTCP and avert HBV binding to hepatocytes. This was done in the chronic hepatitis B (CHB) patient population of our area (Barcelona, Spain), mainly Caucasians of European Mediterranean origin, using robust methods that can be applied in other patient populations.
The main objectives of the study were to determine the variability/conservation of NTCP-interacting domain and the prevalence of the rs2296651 SNP (S267F NTCP variant) in chronically infected HBV patients. Using a high-throughput analytical protocol based on next-generation sequencing (NGS) and an in-house developed PCR using fluorescence resonance energy transfer (FRET) probes, these objectives were realized in an exploratory sample of HBV patients from our setting. Analysis of these viral and host features could be relevant to understand the interactions between HBV and its receptor, and to determine their applicability as prognostic markers of response to treatment strategies based on NTCP blocking.
We performed two main analyses in serum samples from 246 individuals in 3 groups: patients with CHB, patients with resolved HBV infection, and HBV-uninfected individuals. First, the variability/conservation of aa sequences in the NTCP-interacting domain was analyzed and compared to that of a highly conserved preS1 C-terminal domain associated with virion morphogenesis. Comparison between the NTCP-interacting domain and the highly conserved virion morphogenesis domain gave an idea of the magnitude of sequence conservation of the former. To perform this analysis, we developed a high-throughput protocol based on NGS. The raw sequencing data obtained underwent a bioinformatics filtering and analysis procedure based on an in-house-developed pipeline with all computations done in R environment and language. The conservation/variation of these sequences was analyzed by calculating the information content of each position, which was represented graphically using sequence logos, as explained in detail in the Materials and Methods section of the main article.
Second, to estimate the prevalence of the rs2296651 polymorphism in our patient population, this SNP was determined in all samples from the patients included. To accomplish this aim we designed a new in-house real-time PCR method based on FRET probes.
High-throughput NGS analysis yielded viral sequences for most HBV genotypes (A to F and H). In general, the NTCP-interacting domain showed a high degree of conservation, which depended on viral genotype, particularly the sequence between aa 9 to 21. In comparison to the virion morphogenesis domain, the NTCP-interacting domain showed a smaller percentage of HBV genotype-unrelated changes, but greater variability between different HBV genotypes, according to consensus sequences from the GenBank patterns of genotypes A to H. Interestingly, proline residues showed a high degree of conservation in both domains, and serine residues were also particularly conserved in the virion morphogenesis domain, where changes above 1% of the quasispecies were always to potentially phosphorylatable aa. Finally, we demonstrated a low to null prevalence of the rs2296651 SNP in HBV patients from our area. The high degree of conservation of the NTCP-interacting and virion morphogenesis domains should to be confirmed in larger patient series, and the role of proline and potentially phosphorylatable residues and their implications on HBV activity should be clarified. The potential effect of additional NTCP polymorphisms on interactions between HBV and this receptor should also be investigated.
This study describes high-throughput NGS analysis of the preS1 NTCP-interacting domain, which showed overall high conservation that depended on HBV genotype, particularly between aa 9 to 21. These findings concur with previous in vitro results demonstrating that these aa are essential for HBV infectivity. In the comparison with the virion morphogenesis domain, we focused on the proline and serine residues in the two domains: proline showed a high degree of conservation and changes in > 1% of the quasispecies in serine residues were always to potentially phosphorylatable aa in the virion morphogenesis domain. Based on the physical-chemical properties of these aa, we hypothesized that proline residues could stabilize the structure of these two preS1 domains, and the tendency to keep phosphorylatable aa in specific positions of the virion morphogenesis domain suggested that they may be phosphorylated. In addition, we developed an in-house real-time PCR method that allowed us to estimate the prevalence of the rs2296651 SNP in our HBV patients, which turned out to be low to null. Bearing in mind that rs2296651 is reported to be prevalent in Asian populations, where HBV infection is endemic, we hypothesized that the low presence of this SNP in our area may be associated with the lower incidence of HBV infection. Taken together, the findings from this exploratory study suggest that inhibition of HBV entry by NTCP block therapies would be suitable treatment in our CHB patient population.
The present study illustrates the value of NGS to investigate the variability/conservation of the preS1 region of the LHBs. The reasons for the high degree of sequence conservation of the NTCP-interacting and virion morphogenesis domains should be investigated by preS1 structural simulations and site-directed mutagenesis experiments, in particular with modification of the proline and serine phosphorylatable residues. However, it should be borne in mind that the sequence conservation results found in both domains should be confirmed in larger patient samples. In addition, the new in-house real-time PCR method based on FRET probes used here provided fast, reliable detection of the rs2296651 SNP in serum samples of all patients, and revealed a low to null prevalence of this SNP in our patient population. Nonetheless, it cannot be excluded that Caucasian individuals might have additional functionally relevant NTCP SNPs, which would be expected to modify bile acid homeostasis and HBV cellular entry. Therefore, further studies should be performed to determine the prevalence of NTCP SNPs in different populations, and characterize their implications in NTCP function.
The authors thank Celine Cavallo for English language support and helpful editing suggestions.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Spain
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P- Reviewer: Bai G, Diefenbach R, Nagahara H, Spunde K S- Editor: Gong ZM L- Editor: A E- Editor: Ma YJ
| 1. | Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546-1555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1806] [Cited by in F6Publishing: 1853] [Article Influence: 205.9] [Reference Citation Analysis (1)] |
| 2. | Kwon H, Lok AS. Hepatitis B therapy. Nat Rev Gastroenterol Hepatol. 2011;8:275-284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 195] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Block TM, Guo H, Guo JT. Molecular virology of hepatitis B virus for clinicians. Clin Liver Dis. 2007;11:685-706, vii. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | European Association for the Study of the Liver; European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2771] [Cited by in F6Publishing: 3132] [Article Influence: 447.4] [Reference Citation Analysis (0)] |
| 5. | Marcellin P, Bonino F, Yurdaydin C, Hadziyannis S, Moucari R, Kapprell HP, Rothe V, Popescu M, Brunetto MR. Hepatitis B surface antigen levels: association with 5-year response to peginterferon alfa-2a in hepatitis B e-antigen-negative patients. Hepatol Int. 2013;7:88-97. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 112] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 6. | Lauret E, González-Diéguez ML, Rodríguez M, González M, Melón S, Rodrigo L, Rodríguez M. Long-term outcome in Caucasian patients with chronic hepatitis B virus infection after HBsAg seroclearance. Liver Int. 2015;35:140-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 28] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 7. | Liu J, Lee MH, Batrla-Utermann R, Jen CL, Iloeje UH, Lu SN, Wang LY, You SL, Hsiao CK, Yang HI. A predictive scoring system for the seroclearance of HBsAg in HBeAg-seronegative chronic hepatitis B patients with genotype B or C infection. J Hepatol. 2013;58:853-860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Phyo WW, Soh AY, Lim SG, Lee GH. Search for a cure for chronic hepatitis B infection: How close are we? World J Hepatol. 2015;7:1272-1281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Lin CL, Kao JH. Review article: novel therapies for hepatitis B virus cure - advances and perspectives. Aliment Pharmacol Ther. 2016;44:213-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 10. | Hagenbuch B, Meier PJ. Molecular cloning, chromosomal localization, and functional characterization of a human liver Na+/bile acid cotransporter. J Clin Invest. 1994;93:1326-1331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 329] [Cited by in F6Publishing: 325] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 11. | Stieger B. The role of the sodium-taurocholate cotransporting polypeptide (NTCP) and of the bile salt export pump (BSEP) in physiology and pathophysiology of bile formation. Handb Exp Pharmacol. 2011;205-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 12. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1280] [Cited by in F6Publishing: 1428] [Article Influence: 119.0] [Reference Citation Analysis (0)] |
| 13. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1094] [Cited by in F6Publishing: 1159] [Article Influence: 48.3] [Reference Citation Analysis (0)] |
| 14. | Nassal M. Hepatitis B Virus Morphogenesis. Morphogenesis and Maturation of Retroviruses. Berlin, Heidelberg: Springer Berlin Heidelberg; 1996; 297-337. [Cited in This Article: ] |
| 15. | Le Seyec J, Chouteau P, Cannie I, Guguen-Guillouzo C, Gripon P. Infection process of the hepatitis B virus depends on the presence of a defined sequence in the pre-S1 domain. J Virol. 1999;73:2052-2057. [PubMed] [Cited in This Article: ] |
| 16. | Blanchet M, Sureau C. Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J Virol. 2007;81:5841-5849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 17. | Glebe D, Urban S, Knoop EV, Cag N, Krass P, Grün S, Bulavaite A, Sasnauskas K, Gerlich WH. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234-245. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 196] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 18. | Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol. 2005;79:1613-1622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 267] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 19. | Roberts K, Mao R, O’Fallon B, Lyon E. Reporting of clinical genomics test results. Clinical Genomics. A guide to clinical next generation sequencing. USA: Elsevier Academic Press; 2015; 219-231. [DOI] [Cited in This Article: ] |
| 20. | Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 21. | Yan H, Peng B, Liu Y, Xu G, He W, Ren B, Jing Z, Sui J, Li W. Viral entry of hepatitis B and D viruses and bile salts transportation share common molecular determinants on sodium taurocholate cotransporting polypeptide. J Virol. 2014;88:3273-3284. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 22. | Ho RH, Leake BF, Roberts RL, Lee W, Kim RB. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. J Biol Chem. 2004;279:7213-7222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 135] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 23. | Pan W, Song IS, Shin HJ, Kim MH, Choi YL, Lim SJ, Kim WY, Lee SS, Shin JG. Genetic polymorphisms in Na+-taurocholate co-transporting polypeptide (NTCP) and ileal apical sodium-dependent bile acid transporter (ASBT) and ethnic comparisons of functional variants of NTCP among Asian populations. Xenobiotica. 2011;41:501-510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 24. | Peng L, Zhao Q, Li Q, Li M, Li C, Xu T, Jing X, Zhu X, Wang Y, Li F. The p.Ser267Phe variant in SLC10A1 is associated with resistance to chronic hepatitis B. Hepatology. 2015;61:1251-1260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 25. | Li N, Zhang P, Yang C, Zhu Q, Li Z, Li F, Han Q, Wang Y, Lv Y, Wei P. Association of genetic variation of sodium taurocholate cotransporting polypeptide with chronic hepatitis B virus infection. Genet Test Mol Biomarkers. 2014;18:425-429. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 26. | Hu HH, Liu J, Lin YL, Luo WS, Chu YJ, Chang CL, Jen CL, Lee MH, Lu SN, Wang LY. The rs2296651 (S267F) variant on NTCP (SLC10A1) is inversely associated with chronic hepatitis B and progression to cirrhosis and hepatocellular carcinoma in patients with chronic hepatitis B. Gut. 2016;65:1514-1521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 27. | Belmonte I, Montoto L, Miravitlles M, Barrecheguren M, Esquinas C, Rodríguez E, Giralt M, Rodríguez-Frías F. Rapid detection of Mmalton α1-antitrypsin deficiency allele by real-time PCR and melting curves in whole blood, serum and dried blood spot samples. Clin Chem Lab Med. 2016;54:241-248. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Ramírez C, Gregori J, Buti M, Tabernero D, Camós S, Casillas R, Quer J, Esteban R, Homs M, Rodriguez-Frías F. A comparative study of ultra-deep pyrosequencing and cloning to quantitatively analyze the viral quasispecies using hepatitis B virus infection as a model. Antiviral Res. 2013;98:273-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 29. | R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing 2016, Vienna Austria. Available from: http://www.r-project.org/. [Cited in This Article: ] |
| 30. | Gregori J, Perales C, Rodriguez-Frias F, Esteban JI, Quer J, Domingo E. Viral quasispecies complexity measures. Virology. 2016;493:227-237. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 76] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 31. | Bruss V. A short linear sequence in the pre-S domain of the large hepatitis B virus envelope protein required for virion formation. J Virol. 1997;71:9350-9357. [PubMed] [Cited in This Article: ] |
| 32. | Schneider TD. Information content of individual genetic sequences. J Theor Biol. 1997;189:427-441. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 237] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 33. | Ou J, Zhu LJ. motifStack: Plot stacked logos for single or multiple DNA, RNA and amino acid sequence. R package version 1.14.0.. 2016; Available from: https://www.bioconductor.org/packages/release/bioc/html/motifStack.html. [Cited in This Article: ] |
| 34. | Sánchez-Tapias JM, Costa J, Mas A, Bruguera M, Rodés J. Influence of hepatitis B virus genotype on the long-term outcome of chronic hepatitis B in western patients. Gastroenterology. 2002;123:1848-1856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 326] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 35. | Hunter T. Why nature chose phosphate to modify proteins. Philos Trans R Soc Lond B Biol Sci. 2012;367:2513-2516. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 248] [Cited by in F6Publishing: 264] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 36. | Lempp FA, Urban S. Inhibitors of hepatitis B virus attachment and entry. Intervirology. 2014;57:151-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 37. | Li W. The hepatitis B virus receptor. Annu Rev Cell Dev Biol. 2015;31:125-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | Bruss V, Hagelstein J, Gerhardt E, Galle PR. Myristylation of the large surface protein is required for hepatitis B virus in vitro infectivity. Virology. 1996;218:396-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 86] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 39. | Gripon P, Le Seyec J, Rumin S, Guguen-Guillouzo C. Myristylation of the hepatitis B virus large surface protein is essential for viral infectivity. Virology. 1995;213:292-299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 145] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 40. | Schulze A, Schieck A, Ni Y, Mier W, Urban S. Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction. J Virol. 2010;84:1989-2000. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 162] [Cited by in F6Publishing: 173] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 41. | Volz T, Allweiss L, Ben MBarek M, Warlich M, Lohse AW, Pollok JM, Alexandrov A, Urban S, Petersen J, Lütgehetmann M. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol. 2013;58:861-867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 250] [Cited by in F6Publishing: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 42. | Lütgehetmann M, Mancke LV, Volz T, Helbig M, Allweiss L, Bornscheuer T, Pollok JM, Lohse AW, Petersen J, Urban S. Humanized chimeric uPA mouse model for the study of hepatitis B and D virus interactions and preclinical drug evaluation. Hepatology. 2012;55:685-694. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 171] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 43. | Petersen J, Dandri M, Mier W, Lütgehetmann M, Volz T, von Weizsäcker F, Haberkorn U, Fischer L, Pollok JM, Erbes B. Prevention of hepatitis B virus infection in vivo by entry inhibitors derived from the large envelope protein. Nat Biotechnol. 2008;26:335-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 309] [Cited by in F6Publishing: 325] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 44. | Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: First results of a phase Ib/IIa study. J Hepatol. 2016;65:490-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 266] [Article Influence: 33.3] [Reference Citation Analysis (1)] |
| 45. | Blank A, Markert C, Hohmann N, Carls A, Mikus G, Lehr T, Alexandrov A, Haag M, Schwab M, Urban S. First-in-human application of the novel hepatitis B and hepatitis D virus entry inhibitor myrcludex B. J Hepatol. 2016;65:483-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 149] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 46. | Tsukuda S, Watashi K, Hojima T, Isogawa M, Iwamoto M, Omagari K, Suzuki R, Aizaki H, Kojima S, Sugiyama M. A new class of hepatitis B and D virus entry inhibitors, proanthocyanidin and its analogs, that directly act on the viral large surface proteins. Hepatology. 2017;65:1104-1116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 51] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 47. | Bruss V, Lu X, Thomssen R, Gerlich WH. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 1994;13:2273-2279. [PubMed] [Cited in This Article: ] |
| 48. | Krieger F, Möglich A, Kiefhaber T. Effect of proline and glycine residues on dynamics and barriers of loop formation in polypeptide chains. J Am Chem Soc. 2005;127:3346-3352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 160] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 49. | Orzáez M, Salgado J, Giménez-Giner A, Pérez-Payá E, Mingarro I. Influence of proline residues in transmembrane helix packing. J Mol Biol. 2004;335:631-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 50. | Hope VD, Eramova I, Capurro D, Donoghoe MC. Prevalence and estimation of hepatitis B and C infections in the WHO European Region: a review of data focusing on the countries outside the European Union and the European Free Trade Association. Epidemiol Infect. 2014;142:270-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 145] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 51. | Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30:2212-2219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1217] [Cited by in F6Publishing: 1250] [Article Influence: 104.2] [Reference Citation Analysis (0)] |
| 52. | Chen X, Wang Y, Chen X, Cheng K, Li J, Lou J, Ke J, Yang Y, Gong Y, Zhu Y. Genetic variants in the regulatory region of SLC10A1 are not associated with the risk of hepatitis B virus infection and clearance. Infect Genet Evol. 2016;44:495-500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 26] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 53. | Urban S. Liver capsule: Entry and entry inhibition of hepatitis B virus and hepatitis delta virus into hepatocytes. Hepatology. 2016;63:633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |