Published online Jan 21, 2016. doi: 10.3748/wjg.v22.i3.949
Peer-review started: September 2, 2015
First decision: October 14, 2015
Revised: November 4, 2015
Accepted: November 24, 2015
Article in press: November 24, 2015
Published online: January 21, 2016
Processing time: 135 Days and 11.7 Hours
Genetic factors play an important role in the etiology of inflammatory bowel disease (IBD). The launch of genome-wide association study (GWAS) represents a landmark in the genetic study of human complex disease. Concurrently, computational methods have undergone rapid development during the past a few years, which led to the identification of numerous disease susceptibility loci. IBD is one of the successful examples of GWAS and related analyses. A total of 163 genetic loci and multiple signaling pathways have been identified to be associated with IBD. Pleiotropic effects were found for many of these loci; and risk prediction models were built based on a broad spectrum of genetic variants. Important gene-gene, gene-environment interactions and key contributions of gut microbiome are being discovered. Here we will review the different types of analyses that have been applied to IBD genetic study, discuss the computational methods for each type of analysis, and summarize the discoveries made in IBD research with the application of these methods.
Core tip: Computational methods have rapidly progressed during the last a few years, which rendered us the ability to analyze genotype data on a genome-wide level. The application of these methods in inflammatory bowel disease (IBD) genetic study yielded productive results. We discuss the major types of analyses in genome-wide study, and the different computational methods used in each type of analysis. We also show how these computation methods were used in the IBD genetic study and the major findings achieved.
- Citation: Li J, Wei Z, Hakonarson H. Application of computational methods in genetic study of inflammatory bowel disease. World J Gastroenterol 2016; 22(3): 949-960
- URL: https://www.wjgnet.com/1007-9327/full/v22/i3/949.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i3.949
Inflammatory bowel disease (IBD) consists of two major subtypes: Crohn’s disease (CD) and ulcerative colitis (UC) according to the diseases’ clinical features. It is a complex disease, the determinants of which involve both genetic factors and environmental influences. The genetic contribution of IBD was first demonstrated by family studies and twin studies. The occurrence of the disease was observed as being aggregated in families, with a relative risk of 13-36 for CD patients’ siblings and 7-17 for UC[1]. Several studies found a significantly higher proband concordance rate in monozygotic twins (38.5%-63.6%) than that in dizygotic twins (0%-6.7%)[2]. The difference is less pronounced among UC patients, 6.6%-27.9% in monozygotic twins vs 0%-8.0% in dizygotic twins[2]. Similar to other complex diseases, IBD is not a monogenetic disease and does not strictly follow the Mendelian inheritance pattern. Instead, it clearly demonstrates the features of polygenic disease. Linkage based genome-wide scanning studies among IBD families led to the discovery of IBD susceptibility loci IBD1-9[1,3-5], which constitute the early stage of IBD molecular genetics study.
The launch of genome-wide association studies (GWAS) focusing on single nucleotide polymorphism (SNP) then resulted in the discovery of numerous susceptibility loci for complex diseases including IBD. In most IBD GWAS designs, the effect allele frequency of each SNP was compared between cases and controls. In this type of case/control study design, the underlying basic statistical test at each SNP is χ2 test or Fisher’s exact test, and logistic regression which allows for the adjustment of various covariates. IBD is one of the successful examples in utilizing GWAS for revealing of its genetic architecture. A total of 163 genetic loci have been found to be genome-wide significantly associated with IBD[6]. Certainly the rapid development of genotyping technology has made it possible to simultaneously examine millions of SNPs. The advance of computation methods also has made essential contributions to the discovery of such a tremendously large number of genetic determinants. Multiple software tools have been developed for the GWAS studies, among which, PLINK is the most popular one[7]. PLINK provides a compact, comprehensive tool-box for GWAS from basic quality control filtering, SNP association testing to advanced features including gene based analysis, annotation and epistasis test. PLINK has been used as the primary GWAS analytical tool in the vast majority of the IBD genetic studies, from the early finding of loci IL10, ARPC2[8], HNF4A, CDH1, CDH3, LAMB1[9] to the more recent identification of 163 IBD loci[6]. In GWAS, sample structure, which includes both population stratification and hidden relatedness[10], is a common issue resulting in inflation of statistics and false positive results. Sample structure is usually estimated using identical-by-descent analysis implemented in PLINK and principal component analysis implemented in EIGENSTRAT[11,12]. During the recent years, linear mixed model (LMM), which captures both fixed and random effects, has been applied to GWAS. Software based on LMM includes EMMAX[10], FaST[13], and GEMMA[14]. They simultaneously correct for population stratification and hidden relatedness. GEMMA can also be used to analyze multiple correlated phenotypes.
Advance in methodology development of imputation and meta-analysis renders GWAS more power for the identification of genome-wide significant genetic loci. Imputation is to infer genotypes of un-genotyped SNPs based on the SNPs that have been assayed for a group of samples and the haplotype information from a reference population set. The aims of using imputation are to fine-map the associated regions, to boost study power by examining more SNPs, and to meta-analyze cohorts which are genotyped on different platforms[15]. The common practice of applying imputation in GWAS mainly involves three steps: haplotype estimation (pre-phasing), genotype imputation and association testing. The first step pre-phasing is to estimate underlying haplotypes from SNP genotype data. Among several existing methods, software packages fastPHASE[16], IMPUTE2[17], MACH[18], BEAGLE[19] and SHAPEIT[20] all employ coalescent-based methods and hidden Markov models[21]. MACH and IMPUTE2 utilize the hidden states as “template haplotypes”[17,18,21]; fastPHASE employs parsimonious haplotype-clustering for a certain number of clusters[16], and BEAGLE adopts a localized haplotype clustering[19]. SHAPEIT further improves by collapsing haplotypes into a graph structure[20]. Several of these software packages also contain the application of imputation, such as IMPUTE V2, MACH, fastPHASE and BEAGLE[21]. They are the most commonly used software for imputation. For example, IMPUTE V2 carries out haploid imputation for the SNPs that are not genotyped but in the reference panel by conditioning on the haplotypes estimated from the genotyped SNPs[17]. BEAGLE iteratively fits the localized haplotype clustering model to the estimated haplotypes and resamples haplotypes conditional on the genotype and diploid HMM model to derive the probability of the un-genotyped SNP[19]. The review by Marchini J. and Howie B. provides a good summary of the pros and cons of each method[15]. For step 3, association testing, one approach is to convert the genotype probability to the bi-allelic format by specifying a probability threshold. Another more commonly used approach is to take imputation uncertainty into account by frequentist tests such as the score test in SNPTEST[22]. A novel approach of Bayesian method to derive Bayes factor has also been incorporated into SNPTEST and BIMBAM[23]. Software has been developed to work seamlessly for the above three steps, for instance, using SHAPEIT for pre-phasing, and then IMPUTE2 for imputation, which was followed by SNPTEST for association testing. Each of the imputation method discussed above has its own advantage. In the two large cohort studies of IBD, BEAGLE was used as the imputation tool, but other methods have also been widely used in the GWAS study of IBD[6,24].
As another popular approach in GWAS to increase statistical power, meta-analysis combines results from individual studies. Identifications of many IBD genome-wide significant loci have benefited from meta-analysis[24-26], with around 30 susceptibility loci being discovered from each of these large scale meta-analyses. In all these three studies, Z-score based inverse variance weighted meta-analysis was carried out, with the assumption of fixed effects. Other popular approaches include traditional P value based method with sample size as weight[27], random effect method and Bayesian approach[28]. Popular software packages for GWAS meta-analysis include METAL[27], META[22,29], MetABEL[30], GWAMA[31]; PLINK also has its own meta-analysis option. All these software packages have a fixed effects model implemented, but some also contain random-effect models[32]. When conducting meta-analysis, practitioners should consider the direction of effects and heterogeneity between studies.
Though a large number of susceptibility loci have been identified by SNP based GWAS, the disease variance explained by these loci was still limited: 13.6% for CD and 7.5% for ulcerative colitis[6]. At least part of the missing heritability may reside in those genetic loci of moderate effect, which did not reach genome-wide significance due to the limited power in most genetic studies. If genes in these loci function collaboratively, they could make significant contributions to the disease etiology. Pathway studies, which jointly assess the statistical significance of a group of genes, can be applied to identify such loci. From a biological point of view, gene products work cooperatively to carry out certain molecular and cellular functions; and disease etiology is more likely the result of pathway dysregulation. From the statistical point of view, examining the joint effect of a group of genes will allow the identification of those in which each gene member make only a modest contribution. By collapsing SNP statistics to gene level and further pathway level, the number of multiple testing will be reduced. Though challenges and arguments persist in the field of pathway analysis, advance in methodology development has been rapid. The major steps involved in pathway/network analysis are mapping SNPs to genes, aggregating SNPs statistics to gene level, and then assessing the enrichment of pathways or projection of genes into protein-protein interaction (PPI) network. Different methods have been evolved handling the tasks at each step differently. Over 50 pathway analysis software packages have been developed[33]. CD has been used as a common example for the development and application of GWAS-based pathway software. For example, Wang et al[34] applied their software GenGen to the study of the Wellcome Trust Case Control Consortium CD dataset and uncovered the significant associations of pathway IL12/IL23 with CD status in several cohorts genotyped on different SNP chips and of different ethnicities[35]. This is one of the earliest GWAS-based pathway analyses of IBD. Another study by Torkamani et al[36], revealed another interleukin association with CD - IL3 activation and signaling pathway. The software that they used, MetaCore, is a commercial one which is developed by GeneGo. Significant enrichment of “JAK-STAT signaling pathway”, “Cytokine-cytokine receptor interaction” was found by Peng et al[37] using statistical method Simes/FDR. Similar findings of several cytokine signaling pathways were made by Carbonetto and Stephens using a model based approach[38]. In addition to IL/cytokine signaling, the involvement of MHC genes was found by Holmans et al[39] using software ALIGATOR. Similarly, when searching for enrichment of canonical pathways or Gene Ontology (GO) terms, Jostins et al[6] found the highly significant enrichment of GO terms regulation of cytokine production, lymphocyte activation, and pathway JAK-STAT signaling pathway in the largest IBD GWAS so far. Though being common in that these pathways were all interleukin/immune related pathways, the non-overlapping of top significant pathways between different methods could be attributed to different statistical methods and/or the different pathway databases used in their studies. Several non-immune pathways were also found to be significantly associated, such as “calcium signaling”, “ChREBP regulation pathway” identified by Torkamani et al[36]; “ABC transporters” and “Extracellular matrix-receptor interaction” found by Peng et al[37] by Fisher’s exact test. It would be interesting to further experimentally test the biological contributions of genes in these pathways to IBD. A lot of convenient online software tools have been developed for pathway analysis based on GWAS data, such as i-GSEA4GWAS[40] and GSEA-SNP[41]. In addition to the methods discussed above, other pathway analysis tools that were originally designed for gene expression data have also been used after concatenating SNP statistics to gene statistics, such as DAVID[42], GeneTrail[43], and WebGestalt[44].
As an alternative to pathway analysis, another way to assess genes’ joint effect is PPI network, in which the nodes represent the protein products of the genes, and the edges connecting the nodes represent the biochemical interactions between a pair of proteins. After mapping significant genes to PPI networks in various databases, the resulting nodes, edges and the size of the largest connected component was then computed. Cytoscape[45] is the most commonly used comprehensive toolkits for network analysis and visualization. Other online tools include DAPPLE[46], STRINGS[47], and FUNCOUP[48]. Pathway and network analyses can also be combined, which yield significantly enriched pathways within the network. Such pathway analyses are implemented in software like STRINGS and FUNCOUP; and can be integrated into Cytoscape. Network analysis tools, such as DAPPLE, have been used for prioritization of candidate genes in IBD analysis[6].
In addition to pathway analysis in general, gene-gene interaction is also an important topic in IBD research. Gene-gene interaction is often referred to as “epistasis”, for which there are its mathematical definition and its biological interpretation. Simplistically, it implicates a departure from independent effect of each gene per se, but we need to be aware that epistasis in mathematical meaning and biological context are not often interchangeable. Mathematically, epistasis testing for complex human diseases including IBDs, was carried out in two ways - hypothesis driven and hypothesis free. In the former, particular groups of genetic variants were selected and tested; and in the later especially in the context of GWASs, pairs of interactions between all SNPs studied were exhaustively searched[49]. Most traditional studies in the IBD research field, which were limited by sample size and resources, focused on epistasis testing between specific candidate genes, using χ2 test or logistic regression. Because of the huge number of combinations n(n-1)/2 needed to be tested for n SNPs, one can image the tremendous computational burden created to carry out the hypothesis free search. In recent years, the methodology developments have made it possible in human genetics study, which has been summarized in detail in review article by Wei et al[49]. They classified the common methods into several classes, such as regression-based methods, LD- and haplotype-based methods, Bayesian methods, machine-learning and data-mining methods, and data filtering methods[49]. Among these methods, the most commonly used one to detect epistasis interactions in either hypothesis free or hypothesis driven setting is the regression-based methods by comparing the statistical model with interaction terms (saturated model) and the one without (reduced model)[49]. Logistic regression has been commonly used in epistasis testing among candidate genes, such as interactions between CARD8 and NALP3[50]. This is relatively easy for testing interactions between a small number of SNPs of interest. For genome-wide epistasis testing, with the rapid development of modern computational technology, this is not an impossible task anymore. Epistasis test implemented in PLINK is a typical example of using regression based method on SNP alleles[7]. Proper application of approximation methods further speeds up the computing time for initial screening purpose. For example, software package “BOOST” employed the Kirkwood superposition approximation[51] at the initial genome-wide screening stage for epistasis interactions involved in disease traits[52], followed by the classical likelihood ratio test only at the testing stage after eliminating the many non-significant interactions first. In addition to the two-step design and utilization of approximation approach, BOOST also transformed genotype data to Boolean representation which allows for fast logic computing[52]. Now together with joint effect tests[53], BOOST algorithm has been incorporated into PLINK 1.9 (http://pngu.mgh.harvard.edu/~purcell/plink2/epistasis.html). Other algorithms have also been introduced into the field of epistasis testing, such as decision tree learning which is a predictive modeling method of machine-learning and data mining[54]; as well as multifactor dimensionality reduction (MDR)[55,56]. These methods also allow for the identification of higher order interactions. Such methods have been used in epistasis analysis for IBD. Most recent publications in this regard include the one by Wang et al[57] using regression tree to search for models that contain logical SNP interactions that better explain UC disease compared to single SNP combinations. However, their study was limited to the known genome-wide significant 133 UC loci. MDR is a non-parametric method that select and combine high-dimensional genotypic data into a one-dimensional model with a variable of high and low risk classes[55]. By MDR analysis, Okazaki et al[58] found suggestive evidence for high-order interactions between genes IL23R, IBD5 and ATG16L1 variants in a CD case-control cohort.
Here the epistasis testing that we have discussed refers to its mathematic meaning. The epistasis interactions captured in statistical model often are hard to investigate for its biological mechanism in experimental settings.
Gene-environment interaction is another hot topic in IBD research. A broad range of environmental factors have been studied in IBD genetics, including environment factors in traditional definition such as smoking, diet, physical activity, medication, infection and other life style factors[59,60]; and the newly defined “in-vironment” factors, like the microbiome in the gut. In fact, the gut “in-vironment” is affected by environment factors like diet[61]. It has been well recognized that the development of IBD is the result of interplaying between individual’s genetic composition and his/her environmental exposure. To study the gene-environment interactions, the χ2 test and logistic regression methods can be similarly used, just by introducing environmental factors into the regression. Important findings include the interactions between smoking and polymorphisms of important IBD risk genes, NOD2, CYP2A6 and GSTP1[62,63]. New methods have been adopted in this regard too, such as the logic regression[64,65], which fits the regression model with Boolean expression of the predictor variables, similarly as we discussed above for gene-gene interactions. In the work by Wang et al[57], they not only found gene-gene interactions but also identified interactions between genes and smoking in UC cohorts by logic regression. The same group has also applied this method to a similar study of genetic and environmental factors for CD[66]. By far, smoking is the most studied environmental factors for IBD.
It has long been suggested that the interaction between the intestinal immune system and its microbe environment plays key roles in the pathogenesis of IBD. The highly significant enrichment of pathway “response to molecules of bacterial origin” among the significant IBD GWAS loci reinforced such concept in the largest IBD GWAS study to date[6]. Dysbiosis, which refers to the reduction of gut microbiota diversity or abnormal change of composition, has been known to play a major pathological role in IBD etiology[67]. The Dysbiosis of microbiota in IBD patients was investigated using different experimental methods. Two approaches are commonly used in the microbiome investigation. A traditional approach is the targeted amplicon approach in which the taxonomy and the phylogeny of the microbiota were examined by using several marker genes[68]. One most commonly used marker is ribosomal small subunit --16S ribosomal DNA (rDNA) because of its universal existence across species and its composition of both slowly evolving sequences and fast evolving sequences[68]. A more recent approach is the metagenomics approach which is high-throughput sequencing based microbiota genomic DNA profiling[68]. Many computational tools have been developed for analyzing sequencing data from either approach. The major analytical tools for 16S rDNA data include QIIME (quantitative insights into microbial ecology)[69] established using PyCogent toolkit[70], mothur, which incorporates algorithms in several other tools and new features[71], and VAMPS[72]. For metagenomics data analysis, common software packages, including PhymmBL[73] and MEGAN[74], compare high-throughput sequencing reads to known microbiome genome sequences; while others, such as PhyIOTU[75] and MLtreemap[76], derive phylogenetic origin based on marker genes from the large amounts of sequencing data. Both approaches have been used in microbiome study of IBD pathogenesis. Gut microbiota are shaped by various factors including both host genetic factors, and environment factors, like diet, medication and others. Information from research in this field is emerging. A reduction in diversity of fecal microbiome has been reported for CD patients when compared to healthy controls, which is the result of studying 16S rRNA genes[77], and similar results were found for UC patients[78]. Another microarray study using 16S rDNA from CD patients and healthy controls specifically observed a reduction in bacterial populations within the phylum Firmicutes[79]. Reduced diversity has also been reported when comparing fully inflamed tissue to non-flamed tissue for the same CD or UC patient[80]. On the other hand, increased fungal diversity has been found in CD patients compared with controls[81]. A recent large pediatric treatment-naïve CD cohort examining microbiome indicates increase and reduction of abundance in different bacteria species; and also suggests the influence of antibiotic on such changes[82]. Several studies implicate that host genetics, like NOD2, ATG16L1 variants have an impact on abundance of mucosal microbiota abundance[83,84]. A study focusing on bacterial and metaproteomic analysis of the mucosal-luminal interface has emphasized the host-microbe interactions as one of the important underlying factors for IBD[85].
IBD is a type of auto-immune disease (AID) and genetic studies have found it shares susceptibility loci, signaling pathways with other immune diseases and non-immune diseases, despite their distinct clinical features. Lees et al[86] provided a good summary of susceptibility loci common to IBD and other diseases. Pleiotropy is a common phenomenon among auto-immune diseases. Signaling pathways like cytokine signaling and innate immunity have been linked to several AIDs[87-89], including IBD[90,91]. Results from more recent studies of cohorts genotyped on the immunochip provide further evidence for the genetic sharing among different types of immune-related diseases. Traditionally, when a cohort comprising patients of multiple related diseases were studied, pooled analysis or meta-analysis is employed, including both fixed effects meta-analysis and random effects meta-analysis. However, inherent heterogeneity in these cohorts makes it loose power to find subtypes specific signals or signals that are of opposite directions of effect in related diseases. To compensate for these disadvantages, several methods have been developed. Cotsapas et al[92] developed the cross-phenotype meta-analysis method which tests for the deviation from the null hypothesis that there is no additional association beyond the known ones; and the alternative hypothesis involves associations with two or more phenotypes. They applied this method for the test of pleiotropic effects of 107 SNPs which are known for immune-disease association among 7 diseases including CD; and found evidence of association with more than one disease for 47 SNPs. Bhattacharjee et al[93] presented a subset-based meta-analysis, which exhaustively investigates combinations of diseases subsets against the shared control set or the rest of all the samples; logistic regression was iteratively applied for each combination to find the best subset combination that would yield strongest association for each SNP; and the discrete local maxima method[94] was used to get the estimation of P value upper boundary. Other methods made extensions to the weighted sum of the univariate test statistics[95], reflecting effect heterogeneity among phenotypes[96,97]. Newly proposed methods like GPA (Genetic analysis incorporating Pleiotropy and Annotation)[98] employed a two-groups model[99] and an Expectation-Maximization (EM) algorithm[100] for estimation of parameters in the GPA model. They also incorporated annotation enrichment into the analysis to prioritize identification of functional signals. Schifano et al[101] developed a novel approach SMAT for evaluating shared effect of genetic variants on several secondary continuous phenotypes in a case-control cohort by ways of a scaled marginal model.
Genetics study is aimed to identify high-risk populations for early prevention and future targeted treatment. Comparing to numerous GWAS on IBD, the number of risk prediction studies using genetic markers is limited. Recently, Wei et al[102] used an advanced machine-learning technique and the large IBD Genetic Consortium’s cohort of more than 60000 samples. They developed a step-wise risk assessment model. The model achieved the highest areas under the curve (AUCs) so far for CD and UC, which are 0.86 and 0.83 respectively. Compared to previous studies, this is a large improvement. Li et al[103] proposed a risk prediction method integrating pleiotropic effects between diseases by using a bivariate ridge regression method. They demonstrated improved prediction accuracy when CD and UC datasets were jointly analyzed.
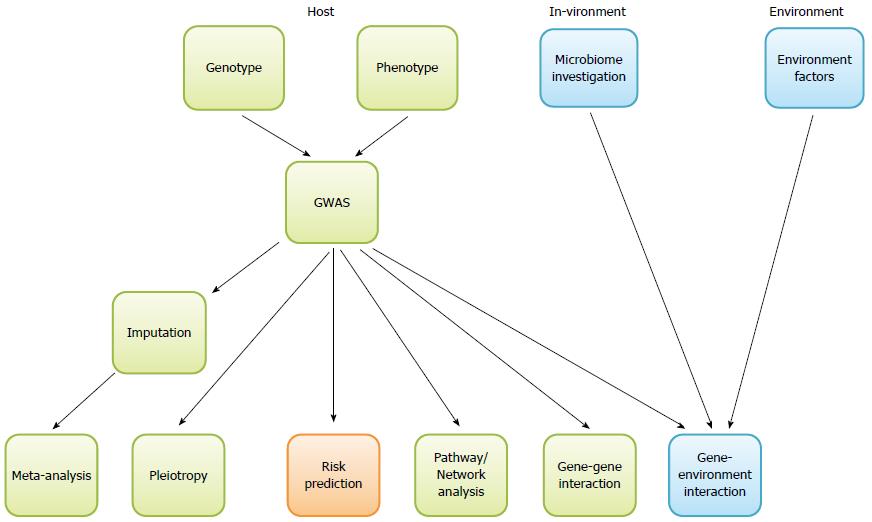
We listed the commonly used software package for each type of analysis in Table 1 and Figure 1 and summarized the major points of our discussion above into the Table 2 as take-home messages.
| Take home messages |
| GWAS is an unbiased method to identify common genetic variants that are significantly associated with complex human diseases. Sample structure needs to be carefully handled to avoid false positive results |
| Imputation is often employed to infer un-genotyped SNPs based on those genotyped ones, followed by meta-analysis to combine results from multiple studies, in order to increase the power in GWAS |
| Pathway analyses help to identify genetic variants that have modest individual effect but jointly make significant contribution to disease susceptibility |
| Both gene-gene interactions and gene-environment interactions are important underlying factors for IBD |
| Pleiotropy studies aim to identify genetic loci shared by IBD and other immune diseases |
| Risk prediction is one of the ways to translate GWAS discoveries to clinics - to identify patients at high risk |
Genome-wide genotyping results provide a huge reservoir of genetic data to explore. From single variant to gene and to pathway, analyses can be carried out at multiple levels. How host factors interact with environment is another important aspect to consider, especially for IBD. The ultimate goal of genetic studies is to translate discoveries to clinical applications. Thus it is important to develop good methods for risk prediction so as to identify patients at high risk for early prevention. Other applications should also be considered to maximize the value of discoveries from genetic studies.
Through the rapid development of computation methods and other technologies, tremendous advance has been made in the field IBD genetic studies; yet much remains to be explored and investigated.
As a successful example of GWAS, 163 susceptibility loci have been discovered for IBD[6], however, much remains unknown how these loci contribute to IBD etiology, especially the majority of these loci were not in the coding region of any gene. Many questions in this regard need to be addressed, for example, whether these SNPs directly affect gene expression level or gene function, whether they tag for any other SNP. More resources are becoming available for functional annotation of genetic variants, even those in the gene desert regions, such as the ENCODE[104], Epigenome Roadmap[105] for chromatin markers, and GTEx[106] for eQTL. It is expected that integrated analysis of genetic, epigenetic and eQTL data will provide more insight into the understanding of these genetic variants, and will help to prioritize the genetic variants for experimental studies.
In addition to common variants, rare variants can also contribute to IBD susceptibility. The current methods for rare variants identification include exome array, whole exome sequencing and whole genome sequencing. Compared to the well-established pipeline for GWAS and commonly accepted threshold standards to determine GWAS significant loci, much remains to be explored in the field of rare variants study. Rare variants were thought to confer larger effects than common variants, based on experience from Mendelian disease and early findings of complex disease such as variants I1307K and E1317Q in gene APC associated with colorectal tumors[107,108], and rare variants in gene PCSK9 associated with coronary heart disease[109]. More studies revealed that the genetic risks attributed to many rare variants are also modest which makes large sample size a necessary factor for such studies, and the effects of rare variants may need to be aggregated for analysis at the gene level to conquer the low power in rare variant analysis[110]. Another feature of rare variants is population specificity, which makes population stratification an outstanding issue in rare variant studies[110]. Because of these features, assumptions in analytical methods for common variants may not hold true for rare variants. Cautions need to be taken when utilizing existing methods that were used for common variants; and new statistical and computational methods development need to meet such demands from the studies of rare variants.
With the combined study for both common variants and rare variants, and integrated functional annotation, more susceptibility and causality loci will be identified for IBD, which will pave the way for the development of suitable treatments for each patient.
P- Reviewer: Cheon JH, Lakatos PL S- Editor: Ma YJ L- Editor: Filipodia E- Editor: Liu XM
| 1. | Ahmad T, Satsangi J, McGovern D, Bunce M, Jewell DP. Review article: the genetics of inflammatory bowel disease. Aliment Pharmacol Ther. 2001;15:731-748. [PubMed] |
| 2. | Ek WE, D'Amato M, Halfvarson J. The history of genetics in inflammatory bowel disease. Ann Gastroenterol. 2014;27:294-303. [PubMed] |
| 3. | Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996;379:821-823. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 598] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 4. | Satsangi J, Parkes M, Louis E, Hashimoto L, Kato N, Welsh K, Terwilliger JD, Lathrop GM, Bell JI, Jewell DP. Two stage genome-wide search in inflammatory bowel disease provides evidence for susceptibility loci on chromosomes 3, 7 and 12. Nat Genet. 1996;14:199-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 476] [Cited by in RCA: 458] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 5. | Cooney R, Jewell D. The genetic basis of inflammatory bowel disease. Dig Dis. 2009;27:428-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 6. | Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119-124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3979] [Cited by in RCA: 3603] [Article Influence: 277.2] [Reference Citation Analysis (0)] |
| 7. | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24221] [Cited by in RCA: 24246] [Article Influence: 1347.0] [Reference Citation Analysis (0)] |
| 8. | Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 471] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 9. | Barrett JC, Lee JC, Lees CW, Prescott NJ, Anderson CA, Phillips A, Wesley E, Parnell K, Zhang H, Drummond H. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330-1334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 417] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 10. | Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348-354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2365] [Cited by in RCA: 1944] [Article Influence: 129.6] [Reference Citation Analysis (0)] |
| 11. | Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7107] [Cited by in RCA: 7391] [Article Influence: 389.0] [Reference Citation Analysis (0)] |
| 12. | Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3782] [Cited by in RCA: 3607] [Article Influence: 189.8] [Reference Citation Analysis (0)] |
| 13. | Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 807] [Cited by in RCA: 781] [Article Influence: 55.8] [Reference Citation Analysis (0)] |
| 14. | Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nat Methods. 2014;11:407-409. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 637] [Cited by in RCA: 542] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 15. | Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1366] [Cited by in RCA: 1140] [Article Influence: 76.0] [Reference Citation Analysis (0)] |
| 16. | Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1480] [Cited by in RCA: 1416] [Article Influence: 74.5] [Reference Citation Analysis (0)] |
| 17. | Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2978] [Cited by in RCA: 3172] [Article Influence: 198.3] [Reference Citation Analysis (0)] |
| 18. | Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816-834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1637] [Cited by in RCA: 1558] [Article Influence: 103.9] [Reference Citation Analysis (0)] |
| 19. | Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2236] [Cited by in RCA: 2300] [Article Influence: 127.8] [Reference Citation Analysis (0)] |
| 20. | Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1332] [Cited by in RCA: 1394] [Article Influence: 99.6] [Reference Citation Analysis (0)] |
| 21. | Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nat Rev Genet. 2011;12:703-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 491] [Cited by in RCA: 413] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 22. | Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906-913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2029] [Cited by in RCA: 2083] [Article Influence: 115.7] [Reference Citation Analysis (0)] |
| 23. | Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 24. | Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118-1125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2110] [Cited by in RCA: 2007] [Article Influence: 133.8] [Reference Citation Analysis (0)] |
| 25. | Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246-252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1095] [Cited by in RCA: 1048] [Article Influence: 74.9] [Reference Citation Analysis (1)] |
| 26. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955-962. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2114] [Cited by in RCA: 2047] [Article Influence: 120.4] [Reference Citation Analysis (0)] |
| 27. | Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190-2191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3066] [Cited by in RCA: 3808] [Article Influence: 253.9] [Reference Citation Analysis (0)] |
| 28. | Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661-678. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7741] [Cited by in RCA: 7160] [Article Influence: 397.8] [Reference Citation Analysis (0)] |
| 29. | de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122-R128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 451] [Cited by in RCA: 418] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 30. | Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1390] [Cited by in RCA: 1376] [Article Influence: 76.4] [Reference Citation Analysis (0)] |
| 31. | Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 379] [Cited by in RCA: 505] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 32. | Evangelou E, Ioannidis JP. Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet. 2013;14:379-389. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 422] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 33. | Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends Genet. 2014;30:390-400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 34. | Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 657] [Cited by in RCA: 684] [Article Influence: 38.0] [Reference Citation Analysis (0)] |
| 35. | Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 227] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 36. | Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 37. | Peng G, Luo L, Siu H, Zhu Y, Hu P, Hong S, Zhao J, Zhou X, Reveille JD, Jin L. Gene and pathway-based second-wave analysis of genome-wide association studies. Eur J Hum Genet. 2010;18:111-117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 187] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 38. | Carbonetto P, Stephens M. Integrated enrichment analysis of variants and pathways in genome-wide association studies indicates central role for IL-2 signaling genes in type 1 diabetes, and cytokine signaling genes in Crohn’s disease. PLoS Genet. 2013;9:e1003770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 39. | Holmans P, Green EK, Pahwa JS, Ferreira MA, Purcell SM, Sklar P, Owen MJ, O’Donovan MC, Craddock N. Gene ontology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Am J Hum Genet. 2009;85:13-24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 330] [Cited by in RCA: 308] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 40. | Zhang K, Cui S, Chang S, Zhang L, Wang J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 2010;38:W90-W95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 41. | Holden M, Deng S, Wojnowski L, Kulle B. GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics. 2008;24:2784-2785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 42. | Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24735] [Cited by in RCA: 27977] [Article Influence: 1748.6] [Reference Citation Analysis (0)] |
| 43. | Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Müller R, Meese E, Lenhof HP. GeneTrail--advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186-W192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 282] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 44. | Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741-W748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1342] [Cited by in RCA: 1463] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 45. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33583] [Article Influence: 1599.2] [Reference Citation Analysis (0)] |
| 46. | Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 412] [Cited by in RCA: 378] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 47. | Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808-D815. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3099] [Cited by in RCA: 3361] [Article Influence: 280.1] [Reference Citation Analysis (0)] |
| 48. | Schmitt T, Ogris C, Sonnhammer EL. FunCoup 3.0: database of genome-wide functional coupling networks. Nucleic Acids Res. 2014;42:D380-D388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 49. | Wei WH, Hemani G, Haley CS. Detecting epistasis in human complex traits. Nat Rev Genet. 2014;15:722-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 278] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 50. | Roberts RL, Topless RK, Phipps-Green AJ, Gearry RB, Barclay ML, Merriman TR. Evidence of interaction of CARD8 rs2043211 with NALP3 rs35829419 in Crohn’s disease. Genes Immun. 2010;11:351-356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 86] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 51. | Matsuda H. Physical nature of higher-order mutual information: intrinsic correlations and frustration. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics. 2000;62:3096-3102. [PubMed] |
| 52. | Wan X, Yang C, Yang Q, Xue H, Fan X, Tang NL, Yu W. BOOST: A fast approach to detecting gene-gene interactions in genome-wide case-control studies. Am J Hum Genet. 2010;87:325-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 53. | Ueki M, Cordell HJ. Improved statistics for genome-wide interaction analysis. PLoS Genet. 2012;8:e1002625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 54. | Rokach L, Maimon O. Data mining with decision trees: theory and applications. World Scientific Pub Co Inc.. 2008;. |
| 55. | Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1426] [Cited by in RCA: 1332] [Article Influence: 55.5] [Reference Citation Analysis (0)] |
| 56. | Winham SJ, Motsinger-Reif AA. An R package implementation of multifactor dimensionality reduction. BioData Min. 2011;4:24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 57. | Wang MH, Fiocchi C, Zhu X, Ripke S, Kamboh MI, Rebert N, Duerr RH, Achkar JP. Gene-gene and gene-environment interactions in ulcerative colitis. Hum Genet. 2014;133:547-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 58. | Okazaki T, Wang MH, Rawsthorne P, Sargent M, Datta LW, Shugart YY, Bernstein CN, Brant SR. Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn’s disease risk in a population-based case-control study: evidence of gene-gene interactions. Inflamm Bowel Dis. 2008;14:1528-1541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 59. | Rogler G, Vavricka S. Exposome in IBD: recent insights in environmental factors that influence the onset and course of IBD. Inflamm Bowel Dis. 2015;21:400-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 60. | Wang MH, Achkar JP. Gene-environment interactions in inflammatory bowel disease pathogenesis. Curr Opin Gastroenterol. 2015;31:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 61. | Fiocchi C. Genes and ‘in-vironment’: how will our concepts on the pathophysiology of inflammatory bowel disease develop in the future? Dig Dis. 2012;30 Suppl 3:2-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 62. | Helbig KL, Nothnagel M, Hampe J, Balschun T, Nikolaus S, Schreiber S, Franke A, Nöthlings U. A case-only study of gene-environment interaction between genetic susceptibility variants in NOD2 and cigarette smoking in Crohn’s disease aetiology. BMC Med Genet. 2012;13:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 63. | Ananthakrishnan AN, Nguyen DD, Sauk J, Yajnik V, Xavier RJ. Genetic polymorphisms in metabolizing enzymes modifying the association between smoking and inflammatory bowel diseases. Inflamm Bowel Dis. 2014;20:783-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 64. | Ruczinski I, Kooperberg C, LeBlanc ML. Logic regression. J Comput Graph Statis. 2003;12:475-511. |
| 65. | Ruczinski I, Kooperberg C, LeBlanc ML. Exploring interactions in high-dimensional genomic data: an overview of logic regression, with applications. J Multivar Anal. 2004;90:178-195. |
| 66. | Wang MH, Fiocchi C, Ripke S. A model integrating genetic and environmental factors interactions can predict Crohn’s disease risk. Am J Gastroenterol. 2014;S503. |
| 67. | Satokari R. Contentious host-microbiota relationship in inflammatory bowel disease--can foes become friends again? Scand J Gastroenterol. 2015;50:34-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 68. | Noval Rivas M, Burton OT, Wise P, Zhang YQ, Hobson SA, Garcia Lloret M, Chehoud C, Kuczynski J, DeSantis T, Warrington J. A microbiota signature associated with experimental food allergy promotes allergic sensitization and anaphylaxis. J Allergy Clin Immunol. 2013;131:201-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 334] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 69. | Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335-336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29318] [Cited by in RCA: 23743] [Article Influence: 1582.9] [Reference Citation Analysis (0)] |
| 70. | Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, Eaton M, Hamady M, Lindsay H, Liu Z. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 71. | Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537-7541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14372] [Cited by in RCA: 13803] [Article Influence: 862.7] [Reference Citation Analysis (0)] |
| 72. | Huse SM, Dethlefsen L, Huber JA, Mark Welch D, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 680] [Cited by in RCA: 632] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 73. | Brady A, Salzberg S. PhymmBL expanded: confidence scores, custom databases, parallelization and more. Nat Methods. 2011;8:367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 74. | Mitra S, Rupek P, Richter DC, Urich T, Gilbert JA, Meyer F, Wilke A, Huson DH. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinformatics. 2011;12 Suppl 1:S21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 75. | Sharpton TJ, Riesenfeld SJ, Kembel SW, Ladau J, O’Dwyer JP, Green JL, Eisen JA, Pollard KS. PhylOTU: a high-throughput procedure quantifies microbial community diversity and resolves novel taxa from metagenomic data. PLoS Comput Biol. 2011;7:e1001061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 76. | von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 248] [Cited by in RCA: 222] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 77. | Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1677] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 78. | Martinez C, Antolin M, Santos J, Torrejon A, Casellas F, Borruel N, Guarner F, Malagelada JR. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 146] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 79. | Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis. 2010;16:2034-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 278] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 80. | Sepehri S, Kotlowski R, Bernstein CN, Krause DO. Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:675-683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 180] [Article Influence: 10.0] [Reference Citation Analysis (2)] |
| 81. | Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol. 2008;43:831-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 322] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 82. | Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1945] [Cited by in RCA: 2356] [Article Influence: 214.2] [Reference Citation Analysis (0)] |
| 83. | Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44-54. [PubMed] |
| 84. | Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, Zhu W, Sartor RB, Boedeker EC, Harpaz N. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179-184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 499] [Cited by in RCA: 465] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 85. | Presley LL, Ye J, Li X, Leblanc J, Zhang Z, Ruegger PM, Allard J, McGovern D, Ippoliti A, Roth B. Host-microbe relationships in inflammatory bowel disease detected by bacterial and metaproteomic analysis of the mucosal-luminal interface. Inflamm Bowel Dis. 2012;18:409-417. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 86. | Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. 2011;60:1739-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 454] [Article Influence: 32.4] [Reference Citation Analysis (3)] |
| 87. | Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363-391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 224] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 88. | Rai E, Wakeland EK. Genetic predisposition to autoimmunity--what have we learned? Semin Immunol. 2011;23:67-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 89. | Richard-Miceli C, Criswell LA. Emerging patterns of genetic overlap across autoimmune disorders. Genome Med. 2012;4:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 114] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 90. | Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1545] [Cited by in RCA: 1978] [Article Influence: 179.8] [Reference Citation Analysis (1)] |
| 91. | Gersemann M, Wehkamp J, Stange EF. Innate immune dysfunction in inflammatory bowel disease. J Intern Med. 2012;271:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 120] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 92. | Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, Wallace C, Abecasis GR, Barrett JC, Behrens T, Cho J. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 408] [Cited by in RCA: 411] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 93. | Bhattacharjee S, Rajaraman P, Jacobs KB, Wheeler WA, Melin BS, Hartge P, Yeager M, Chung CC, Chanock SJ, Chatterjee N. A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am J Hum Genet. 2012;90:821-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 194] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 94. | Taylor JE, Worsley KJ, Gosselin F. Maxima of discretely sampled random fields, with an application to ‘bubbles’. Biometrika. 2007;1-18. |
| 95. | O’Brien PC. Procedures for comparing samples with multiple endpoints. Biometrics. 1984;40:1079-1087. [PubMed] |
| 96. | Xu X, Tian L, Wei LJ. Combining dependent tests for linkage or association across multiple phenotypic traits. Biostatistics. 2003;4:223-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 97. | Yang Q, Wu H, Guo CY, Fox CS. Analyze multivariate phenotypes in genetic association studies by combining univariate association tests. Genet Epidemiol. 2010;34:444-454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 98. | Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 153] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 99. | Efron B. Microarrays, empirical Bayes and the two-groups model. Statistical Science. 2008;1-22. |
| 100. | Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol. 1977;1-38. |
| 101. | Schifano ED, Li L, Christiani DC, Lin X. Genome-wide association analysis for multiple continuous secondary phenotypes. Am J Hum Genet. 2013;92:744-759. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 58] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 102. | Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, Kim C, Mentch F, Van Steen K, Visscher PM. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. 2013;92:1008-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 135] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 103. | Li C, Yang C, Gelernter J, Zhao H. Improving genetic risk prediction by leveraging pleiotropy. Hum Genet. 2014;133:639-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 59] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 104. | ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14554] [Cited by in RCA: 12955] [Article Influence: 996.5] [Reference Citation Analysis (0)] |
| 105. | Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5261] [Cited by in RCA: 4461] [Article Influence: 446.1] [Reference Citation Analysis (0)] |
| 106. | GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4162] [Cited by in RCA: 3822] [Article Influence: 382.2] [Reference Citation Analysis (0)] |
| 107. | Frayling IM, Beck NE, Ilyas M, Dove-Edwin I, Goodman P, Pack K, Bell JA, Williams CB, Hodgson SV, Thomas HJ. The APC variants I1307K and E1317Q are associated with colorectal tumors, but not always with a family history. Proc Natl Acad Sci USA. 1998;95:10722-10727. [PubMed] |
| 108. | Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci USA. 2004;101:15992-15997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 145] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 109. | Cohen JC, Boerwinkle E, Mosley TH, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354:1264-1272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2574] [Cited by in RCA: 2374] [Article Influence: 124.9] [Reference Citation Analysis (0)] |
| 110. | Lettre G. Rare and low-frequency variants in human common diseases and other complex traits. J Med Genet. 2014;51:705-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |