Published online Aug 14, 2015. doi: 10.3748/wjg.v21.i30.9021
Peer-review started: April 28, 2015
First decision: May 18, 2015
Revised: June 2, 2015
Accepted: July 8, 2015
Article in press: July 8, 2015
Published online: August 14, 2015
Although thousands of DNA damaging events occur in each cell every day, efficient DNA repair pathways have evolved to counteract them. The DNA repair machinery plays a key role in maintaining genomic stability by avoiding the maintenance of mutations. The DNA repair enzymes continuously monitor the chromosomes to correct any damage that is caused by exogenous and endogenous mutagens. If DNA damage in proliferating cells is not repaired because of an inadequate expression of DNA repair genes, it might increase the risk of cancer. In addition to mutations, which can be either inherited or somatically acquired, epigenetic silencing of DNA repair genes has been associated with carcinogenesis. Gastric cancer represents the second highest cause of cancer mortality worldwide. The disease develops from the accumulation of several genetic and epigenetic changes during the lifetime. Among the risk factors, Helicobacter pylori (H. pylori) infection is considered the main driving factor to gastric cancer development. Thus, in this review, we summarize the current knowledge of the role of H. pylori infection on the epigenetic regulation of DNA repair machinery in gastric carcinogenesis.
Core tip: Considering the relevance of DNA repair mechanisms in the maintenance of genome integrity and the role of epigenetics in its regulation on gastric carcinogenesis, in this review, we highlight the effects of Helicobacter pylori infection on the modulation of epigenetics mechanisms regulating DNA repair pathways associated with gastric carcinogenesis.
-
Citation: Santos JC, Ribeiro ML. Epigenetic regulation of DNA repair machinery in
Helicobacter pylori -induced gastric carcinogenesis. World J Gastroenterol 2015; 21(30): 9021-9037 - URL: https://www.wjgnet.com/1007-9327/full/v21/i30/9021.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i30.9021
It is now believed that gastric cancer is a disease that has a primarily epigenetic origin. It has been shown that Helicobacter pylori (H. pylori) infection plays an important role in the development of this disease and elicits a pathological progression in the gastric mucosa that starts with chronic gastritis and progresses to atrophic gastritis, intestinal metaplasia, dysplasia and eventually gastric cancer. The H. pylori infection initiates an activation of immunologic and inflammatory cascades through the host’s immune responses. The inflammatory response generated by the bacteria causes both oxidative DNA damage and changes in cell turnover. In addition, it has been shown that the proliferation of gastric cells is associated with the appearance of somatic mutations due to errors in replication and/or inappropriate DNA repair capacity.
In response to the DNA damage , there are four repair systems: (1) base excision repair (BER); (2) nucleotide excision repair (NER); (3) mismatch repair (MMR); and (4) double-strand break repair (DSBR). It is thus believed that repair system failure is an important risk factor in carcinogenesis. Epigenetic changes are important mechanisms that lead to the failure of these repair genes. The most studied epigenetic modification is DNA methylation. In this review, we address the role of H. pylori infection in DNA repair mechanisms, the modulating effect of H. pylori on epigenetic mechanisms (such as DNA methylation and histone modifications) and DNA repair machinery in H. pylori-induced gastric carcinogenesis.
Gastric cancer represents the second highest cause of cancer mortality worldwide[1]. Gastric cancer develops from the accumulation of several genetic and epigenetic changes over the lifetime of a patient, which lead to the activation of oncogenes and/or the inactivation of tumor suppressor genes. An unbalance among cell cycle regulators, alterations in growth factors and cytokines, or failures in DNA repair machinery might lead to genetic instability, cell proliferation and apoptosis reduction. All of these factors may be affected by epigenetic mechanisms that could lead to an altered gene expression pattern favoring tumorigenesis[2].
The vast majority of gastric cancers are sporadic and represent approximately 80% of all cases of gastric cancers. According to Lauren’s histological classification, these neoplasms can be defined as intestinal (well differentiated) or diffuse (poorly differentiated) types[3]. On the one hand, the more frequent intestinal type is characterized by structures of united malignant cells that resemble functional gastrointestinal glands. Intestinal tumors develop through sequential steps, which begin with gastritis and progress to atrophic gastritis and are followed by intestinal metaplasia and gastric cancer. The tumor grows expansively with high vascularization and frequently occurs in old people; these tumors are strongly influenced by the environment[4]. On the other hand, the less common diffuse type comprises non-adherent cells that have diffusely infiltrated into the gastric stroma with little glandular formation. This type of neoplasia is most frequent in young people, is typically hereditary and has no preceding steps; it comprises more intra- and transmural spread and is thus associated with poor prognosis compared with the intestinal type[4].
Hereditary factors play a strong role in gastric cancer development. The most famous familial case is that of Napoleon Bonaparte. There is documented evidence showing that the Emperor had a malignant ulcer and hemorrhage in his stomach. His father, Charles Bonaparte, died from scirrhous carcinoma of the pylorus, and his grandfather, Joseph Bonaparte, also died of gastric cancer. Both of them died at approximately age 40. Moreover, one brother and one sister died of the same malignancy, which supports the hypothesis that genetic factors can increase risk of developing malignancy in the stomach[5].
Several proto-oncogenes are activated in gastric cancer, depending on the histological type. The c-met gene is amplified in 19% of intestinal type and 39% of diffuse type cancers[6], whereas the k-sam gene is preferentially expressed in advanced diffuse tumors[7]. The overexpression of another proto-oncogene, c-erbB2, is correlated with poorer prognosis and liver metastases[8]. Conversely, it has been demonstrated that tumor suppressor genes are frequently lost in gastric cancer by LOH (loss of heterozygosity), missense mutations, frame shift deletions, promoter methylation and post-translational mechanisms[9-11]. Additionally, it has been shown that genetic alterations in the TP53 gene, such as a high frequency of TP53 mutations, LOH, and overexpression of the p53 protein, may lead to a consequent loss of p53 function, which could be an early event in gastric carcinogenesis[12,13]. Furthermore, the abnormal expression of cell cycle regulators may permit the development of gastric cancer. This can be observed in the frequent overexpression of cyclin E and E2F genes and the down-regulation of p27 that are associated with aggressiveness, metastasis and invasiveness of the tumor[14].
Gastric cancer is also associated with high levels of MSI (Microsatellite instability), which is strongly related to the carcinogenic process primarily because of its association with defective MMR. In gastric cancer, MSI occurs in approximately 15% to 30% of all cases[15]. MSI-positive tumors exhibit many differences in clinical, pathological, and molecular characteristics compared to MSI-negative ones, regardless of their hereditary or sporadic origins. MSI leads to a mutator phenotype because frameshift mutations accumulate in repeated sequences that are located in coding regions of target tumor suppressor genes. The cancer with a high level of MSI often shows aberrant epigenetic alterations, such as promoter hypermethylation of MMR genes, which leads to gene inactivation[16].
Although the etiology of gastric cancer is given mainly by gene-environment interaction, consuming diets that are high in salt and nitrates favors gastric malignancy, whereas consuming diets that contain the natural antioxidants found in fruits and vegetables may prevent tumor emergence[17]. Alcohol and smoking are also risk factors for disease, although the main driving factor behind gastric cancer development is H. pylori infection, which leads to both chronic inflammation and molecular alterations that affect epithelial cell regulation, ROS levels, DNA damage, mutations caused by high MSI landscape and epigenetic deregulation[18]. The strong epidemiological association between H. pylori infection and the development of gastric cancer made the World Health Organization to classify the bacterium as a carcinogen class I, a definite carcinogen, in 1994[19].
The bacterium known as H. pylori has been the subject of intense research since its first culture from a gastric biopsy in 1982. From the beginning, this microorganism has provoked the interest of many health professionals, including researchers in the area of oncology. The possibility that a bacterium could cause gastritis, peptic ulcer and possibly cancer was a difficult concept to accept. However, researchers had known of evidence for the involvement of a microorganism in the development of ulcers in experimental animals since the previous century. Bizzozero, in 1893, detected the presence of coiled organisms, known as spirochetes, in gastric glands and in the parietal cells from canine stomachs. Subsequently, in the 1940s, spirochetes were identified in human gastric samples, and most of them were present in patients with gastric ulcer. However, the researchers’ interest in identifying a bacterium as the causative agent of ulcers was decreased by the reports in the literature that highlighted a probable association between viruses and peptic ulcer. Steer and collaborators renewed interest in the association between the development of ulcers and the presence of bacteria in 1975 but failed to isolate and identify the microorganisms observed[20].
Finally, in 1982, Warren and Marshall reported the presence of a spiral bacterium in patients with peptic ulcer and chronic gastritis. There were similarities in both the morphological and biochemical growing conditions between this organism and the genus Campylobacter; thus, the researchers named it the Campylobacter like organism. Later, the microorganism was named Campylobacter pyloridis and was then grammatically corrected to Campylobacter pylori[21,22]. Further studies, however, showed biochemical and molecular differences between this bacterium and the genus Campylobacter, and in 1989, it was recognized as belonging to a new genus, Helicobacter[21].
The infection caused by H. pylori is considered the most frequent chronic infection in humans. It is believed that approximately half the world’s population is infected with H. pylori, which makes infection by this bacterium one of the major pathogens in humans and a serious public health problem[23]. Although there is a high rate of colonization, the presence of bacteria is not always associated with the development of pathologies because 70% of the infected population remains asymptomatic. However, the colonized individuals may develop gastritis, peptic ulcer, gastric cancer and MALT lymphoma[23,24].
The prevalence of infection is higher in developing countries (greater than 80%) and lower in developed countries (less than 50%), with a tendency to decline worldwide due to improvements in sanitation[25,26]. However, because the socioeconomic levels vary among subpopulations within a country, the prevalence in these subgroups may be different[27]. It has been shown that increased prevalence is directly related to lower socioeconomic status, inadequate sanitary conditions, contaminated or untreated water, insufficient hygiene practices, families with large numbers of individuals inhabiting the same household[25,28]. These conditions are usually found in developing countries, where there is indeed a high prevalence of H. pylori infection[29]. In a complementary manner, it was observed that improving the general hygiene conditions decreases the prevalence of infection. This finding suggests the existence of a set of environmental conditions to which children are exposed, particularly in developing areas[30].
It is known that humans ingest many microorganisms daily, but most of these microorganisms cannot colonize the stomach because of its acidic pH, which is one of the most important antibacterial properties that this organ has. Under fasting conditions, the gastric lumen has a pH < 2, which prevents bacterial growth. In the mucus layer, which covers the gastric epithelial cells, is a pH gradient ranging from 2 the luminal surface to 5-6 on the surface of epithelial cells[31]. After entering the stomach, H. pylori enters the gastric mucous layer, which is a less acidic environment[32]. Within the mucosal layer, H. pylori can adhere to the apical surface of gastric epithelial cells and occasionally be internalized[33].
The ability of H. pylori to colonize the human stomach can be attributed to the production of specific bacterial products, which are collectively called colonization factors[34,35]. The bacteria have multiple membrane proteins, such as AlpA, BabA, SabA, and HopZ, that mediate the adherence of the bacteria to the gastric epithelial cells, which results in the inactivation of numerous signaling pathways, and allow toxins and other effector molecules to enter the host cells[36]. Furthermore, the presence of virulence factors enables the bacteria to colonize and remain in the gastric mucosa of the host, thereby provoking an inflammatory response that would lead to a progression of events that can lead to gastric cancer. H. pylori virulence factors play a role in determining the patterns of disease with genetic differences affecting the clinical outcome of infection[37]. The most characterized virulence factor is cag pathogenicity island (cag-PAI), a 40-kb length of chromosomal DNA, which contains approximately 31 genes that encode a type IV secretion system[38]. This system allows CagA (cytotoxin-associated gene A) protein to be injected into the epithelial cell cytosol and then to interact with several intracellular signaling molecules, ultimately causing morphological alterations and inducing higher inflammatory levels[18]. Another virulence factor that is also associated with gastric cancer is the VacA gene, which induces vacuole formation in the host cell, stimulates apoptosis by release of cytochrome c from mitochondria and induces inflammation[39,40]. In addition, lipopolysaccharide (LPS), is recognized as a potent endotoxin capable of increase the proinflammatory cytokine production[41].
H. pylori infection plays a critical role in carcinogenesis, in which a long-term interaction between bacterial inflammatory factors and epithelium progenitor and stem cells of the host culminates in the accumulation of mutations and epigenetic modifications that may lead to neoplasia. H. pylori infection is usually acquired during childhood, persists for several decades, and is followed by progressive mucosal damage because of continued interaction of H. pylori with the mucosa and the consequent chronic inflammatory milieu, which leads to mucosal atrophy, intestinal metaplasia, overall resulting in an environment with enhanced risk of developing dysplasia and carcinoma[42,43]. Additionally, this landscape may lead to hypochlorhydria and permit the overgrowth of other bacteria that may increase carcinogenic potential in the stomach[44].
Once infected by H. pylori, the epithelial cells of gastric mucosa undergo activation of immunologic and inflammatory cascades initiated by immune responses[45]. The cellular signaling changes are characterized by release of cytokines into the mucosa lamina propria to activate macrophages, dendritic cells and other inflammatory cells to release inflammatory mediators such as interleukin-1 (IL-1), IL-6, IL-8, tumor necrosis factor (TNF)-α[46]. IL1-β and TNF-α induce the activation of nuclear factor-κB (NF-κB), a key regulator of inflammation and other cellular cascades that underlie carcinogenesis in epithelial cells, which lead to cell proliferation and the suppression of apoptosis[47]. The activation of NF-κB can also regulate the expression of the pro-inflammatory cyclooxygenase (COX-2) enzyme, which induces TNF-α, interferon-γ and IL-1 and contributes to maintenance of cell proliferation, inhibition of apoptosis, and stimulation of angiogenesis in favor of carcinogenesis[48]. It has been demonstrated in vitro that strains harboring cagA induce the activation of extracellular-signal-regulated kinases (ERKs), such as p38, and MAP kinases, such as c-Jun N-terminal kinase (JNK). Additionally, it has been shown that the exposure of gastric epithelial cells to H. pylori induces the activation of the transcription factor activator protein 1 (AP1) and both the c-fos and c-jun protooncogenes[49,50]. These effects might occur through the activation of ERK/MAP kinase pathways, which results in the phosphorylation of Elk-1 and an increase in the transcription of c-fos[49]. Because MAP kinases regulate cell proliferation, differentiation, apoptosis, oxidative stress and inflammatory response, it has been suggested that the activation of this pathway by cagA-positive H. pylori strains seems to be essential to induce gastric inflammation and cancer[51].
This inflammatory-related stress also results in the increased production of ROS and RNS by neutrophils, which causes cell damage by the formation of oxidative DNA lesion products, including 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxodG), and an increase in gene mutation. In this sense, it is important to note that one of the factors that contribute to the development of gastric cancer is the generation of oxidative stress. Oxidative damage to DNA due to H. pylori infection has been well documented in gastric biopsies, and it seems to be related to the presence of cagA and vacA virulence factors[52-57]. In vitro experiments have indicated that the exposure of gastric epithelial cells to different H. pylori strains induces the generation of pro-inflammatory cytokines and ROS[58]. Moreover, it has been observed that patients with H. pylori-associated gastritis, have increased levels of inducible nitric oxide synthase (iNOS) and COX-2. Bearing in mind that both products are potentially mutagenic, it is believed they could be related to the mutations detected in patients with chronic gastritis with an increased risk of developing gastric carcinoma[59,60].
The inflammatory response, in addition to inducing oxidative DNA damage, can cause changes in cell turnover[61]. H. pylori infection is also responsible for inducing apoptosis, which stimulates gastric epithelial cell proliferation in response[62,63]. In addition, it has been shown that the proliferation of gastric cells is associated with the appearance of somatic mutations due to errors in replication and/or inappropriate DNA repair capacity[64,65]. Therefore, these changes in cell turnover could accelerate the progression to atrophic gastritis, thereby increasing the risk of developing gastric cancer[64,66]. Additionally, it has been shown that LPS is also able to interfere with the DNA repair machinery of intestinal cells, thus increasing the risk of permanent genotoxic effects[67].
The host-pathogen interactions are considered to be among the most plastic and dynamic systems in nature that lead to changes in epigenetic program. “Epigenetics”, as introduced by Conrad Waddington in 1946, is defined as a set of interactions between genes and the surrounding environment that determines the phenotype or physical traits in an organism. Currently, epigenetics is defined as heritable changes in gene expression that are not necessarily accompanied by changes in the DNA sequence[68]. To date, the epigenetic role of different gene expression patterns in determining the cellular phenotype has been broadly studied. These processes include DNA methylation, histone modifications that affect the chromatin structure and DNA silencing by noncoding RNAs (ncRNAs)[69-71]; ncRNA is not addressed in this review. The combination of these modifications has been called the “epigenetic code”, and because it is reversible, it has recently emerged as a promising area for cancer research. Studying the epigenetic pattern shared by different cancers provides exciting potential for powerful and more specific anticancer therapeutics.
DNA methylation is one of the most common epigenetic events in the human genome. This modification is heritable and reversible; therefore, it is an important therapeutic target[72]. DNA methylation is a covalent modification of nucleotides, and the most frequently methylated nucleotide in the human genome is a cytosine that is subsequently followed by a guanine in the DNA sequence, which constitutes a CpG dinucleotide. The cytosine is methylated in the C-5 position by a family of DNA methyltransferases (DNMTs) using the universal methyl donor S-adenosyl-L-methionine (SAM). It has been reported that 5-methylcytosines account for approximately 1% of the DNA bases in the human genome and affect 70%-80% of the CpG sites in a human somatic cell[73].
The genome of the cancer cell is globally hypomethylated in comparison with normal tissue. Hypomethylation is related to both chromosomal instability and the activation of proto-oncogenes[74]. Although the biological significance of this cancer-specific DNA hypomethylation has not been fully elucidated, animal studies have confirmed a causal connection between hypomethylation and tumor formation[75]. On the one hand, it has been shown that DNA hypomethylation may reactivate genes that are normally silenced by DNA methylation[76]. On the other hand, the de novo methylation of CpG islands in gene promoters is associated with the loss of expression of several cancer-related pathways, including BRCA1 (breast cancer 1, early onset), CDKN2A (cyclin-dependent kinase inhibitor 2A) and MLH1[72].
Currently, the importance of gene promoter methylation is well known in the development of cancer. Thus, as observed for other tumor tissues, the effects of DNA methylation on gastric carcinogenesis has been extensively studied. To date, aberrant DNA methylation has been described in more than 100 genes[77]. The hypomethylation of oncogenes and cancer-associated genes and the consequent gene activation have been associated with tumorigenesis and with progression and metastasis of gastric cancers[1,77]. In contrast, the hypermethylation of CpG islands, which results the in gene silencing of tumor-suppressor, pro-apoptotic and DNA repair genes, has been observed in relation to gastric carcinogenesis[1,77].
An aberrant methylation pattern was also associated with the presence of H. pylori infection. Maekita et al[78] evaluated the effect of H. pylori infection on the quantity of methylated DNA in noncancerous gastric mucosae and examined its association with gastric cancer risk. Those authors observed bacterial infection potently induces the methylation of multiple CpG islands in noncancerous gastric mucosa associated with the risk of gastric cancer in H. pylori-positive individuals. Kang et al[79] analyzed the methylation profiles of 27 cancer-related genes in samples from patients with gastric cancer and chronic gastritis. In their study, it was observed that the number of methylated genes was significantly higher in samples from patients with gastric cancer than in those from patients with chronic gastritis. It has been shown that the loss of RUNX3 expression was also associated with promoter methylation[80]. Additionally, the methylation of this gene is a risk factor for the carcinogenesis of chronic atrophic gastritis with H. pylori infection[81].
Aside from these examples, more than one hundred papers have been published associating H. pylori infection with an altered DNA methylation pattern and the risk of developing gastric cancer[1,77]. Thus, it is worth emphasizing the important role played by the H. pylori-induced inflammatory process on aberrant methylation[82,83]. It has been shown that IL-1β directly induces the promoter methylation of E-cadherin and is an important mediator of TGF-β1 promoter methylation[84-86]. In addition, it has been shown that the eradication of the bacteria decreases the level of methylation of several genes (CDH1, p16, APC, MLH1, and COX2) that are associated with carcinogenesis[87]. A study conducted in a Mongolian gerbil model indicated that 5-aza-dC treatment prevents the development of H. pylori-induced gastric cancer[88].
Histone proteins contribute to the maintenance and regulation of the dynamic chromatin structure, affecting the activation or inhibition of genes, accessibility to the DNA repair machinery and many other processes in the cell nucleus. Histones are hydrophilic, basic nuclear proteins that are subunits of the nucleosomes that constitute one of the structural core units of chromatin. Nucleosomes are histone octamers that consist of two copies of each of the four canonical histone isotypes (H2A, H2B, H3 and H4). DNA is wrapped around these octamers, which are located along the DNA with a spacing of 177-207 base pairs, in a pearls-on-a-string manner[89]. The N-terminal tails of histones extend outwards from the nucleosome and are the sites for regulatory covalent modifications, including acetylation, methylation, phosphorylation, ubiquitylation, ADP-ribosylation, crotonylation and glutarylation[90]. The packaging of chromatin defines the gene state: the euchromatin represents open and transcriptionally active regions, whereas the heterochromatin represents condensed regions with high levels of repetitive sequences[91]. Histone modifications directly affect the structure of chromatin and drive gene regulation with distinct functional activities. The architecture of chromatin can create binding sites for the recruitment of chromatin-modifying proteins and alter the stability of the interaction between DNA and histones[92]. However, these modification patterns are not static entities but dynamically changing and complex landscapes that evolve in a cell context-dependent fashion, are frequently misregulated in cancer and thus represent an interesting target for therapeutics.
The covalent histone modifications are controlled by enzymes that are able to add or remove different modifications in specialized domains that other enzymes recognize to activate or repress the gene expression. This is also known as the “histone code”, a key factor in the establishment and maintenance of epigenetic cellular memory. The best-characterized covalent histone modification is the acetylation of lysine residues that are regulated by histone acetyltransferases (HATs). This modification results in a neutralization of lysine charges, in turn weakening the interaction between histone and DNA with subsequent activation of transcription[93]. In contrast, histone deacetylases (HDACs) are enzymes that are responsible for erasing this mark and deacetylating the lysine residues, which results in a repressive state of chromatin and may thus directly influence cancer development by silencing tumor suppressor genes[94].
Furthermore, the lysine residues of histones can be methylated, and depending on the methylation level, gene expression is elevated or repressed. This epigenetic mark is catalyzed by a different protein complex, methyltransferase, which is dependent on a specific domain. H3K27me3 is a modification that has been found in many types of cancer and is regulated by Polycomb proteins, which recognize this modification through the chromodomain-containing protein CBX1 (chromobox homolog 1) and induce the compaction of chromatin, resulting in transcriptional repression[95-98]. To maintain a balance between histone methylation patterns, there are other enzymes (called KDMs) that remove the methyl groups from lysine residues and, as in the case of H3K27, upon its demethylation by KDM6A (UTX) and KDM6B (JMJD3), allow for active transcription[99].
The histone modifiers have a key role in the development and progression of gastric cancer. Thus, these post-translational alterations in chromatin are all suggested to be predictors for gastric cancer recurrence and survival[100]. There is evidence that the overexpression of phosphorylated histone H3S10 is an indicator of poor prognosis for gastric cancer[101] and that hypoacetylation of this histone in the p21(WAF1/CIP1) promoter reduces the expression of this gene in gastric cancer specimens[102]. Recent reports have shown that there is an induction of the histone H3K4 demethylase KDM1A (LSD1) in some gastric cancer cells associated with more aggressive behavior of these cells, whereas the HDAC SIRT1 (sirtuin 1), which is downregulated in gastric cancer, plays a tumor-suppressive role in gastric cancer development by inhibiting NF-κB signaling[103,104]. In addition, the H3K9/K36 demethylases KDM4B and JMJD1C, H3K27 methyltransferase EZH2, and histone lysine acetyltransferase KAT5 (TIP60) act as potential markers for the malignancy of gastric cancer because they are correlated with cell proliferation and lymph node metastasis[98,105].
To date, few studies are available concerning the effects of H. pylori infection on histone modifications. It has been shown in vitro that H. pylori causes the upregulation of p21WAF1 expression in both a gastric epithelial cell line and primary gastric cells. The increased p21WAF1 expression is associated with increased HDAC1 recruitment from the p21WAF1 promoter and hyperacetylation of histone H4[106]. In addition, it was demonstrated that H. pylori induces the dephosphorylation and deacetylation of histone H3 in gastric epithelial cells in a cagPAI-dependent manner. Such modifications are associated with changes in host gene expression, including the upregulation of c-Jun and down-regulation of hsp70[107,108].
The DNA repair machinery plays a key role in maintaining genomic stability by preventing the appearance of mutations. The DNA repair enzymes continuously monitor the chromosomes to correct any damage caused by exogenous and endogenous mutagens. The following DNA repair mechanisms respond to such DNA damage: (1) BER, which is essential for removing oxidized or chemically modified bases; (2) NER, which repairs pyrimidine dimers; (3) DNA MMR, which is required to correct any errors that occur during DNA replication (base-base errors, deletions and insertions); and (4) DSBR, which is vital for every living organism and acts at different stages of the cell cycle[109].
It is estimated that each human cell repairs approximately 10000-20000 DNA lesions per day[110]. To achieve this repair, enzymes involved in the BER system recognize damage at DNA bases and catalyze the excision and replacement of the damaged nucleotide[111]. This repair is initiated by the action of specific DNA glycosylases that recognize the DNA base damaged and cleave the N-glycosidic bond that links the DNA sugar-phosphate backbone[112]. The appearance of an abasic site (apurinic/apyrimidinic site or AP site) is then processed by an AP endonuclease (APE1 in human cells), which cleaves the 5’ phosphodiester bond to the AP site, thereby generating a DNA single strand break that contains a hydroxyl residue on the 3’ end and a phosphate on the deoxyribose 5’ end. This arrangement allows the DNA polymerase to incorporate a new nucleotide, and the DNA ligase (XRCC1-DNA complex ligase IIIα) then connects the terminal portion of the DNA[112,113].
The BER is involved in the repair of small changes in DNA bases, which may occur by both the short path, which is required for the removal of one nucleotide, and the long path, which removes from 2 to 13 nucleotides. The most common injuries removed by this system are an oxidized base, i.e., 8-oxodG, which can pair with either cytosine or adenine, resulting in transversions from G:C to T:A[114]. In addition, BER is involved in the removal of uracil in DNA formed by spontaneous cytosine deamination, which results in erroneous matching U:G[115]. A number of human pathologies, including cancer, result from oxidative DNA damage caused by endogenous and exogenous agents. In this sense, many epidemiological studies have investigated the association between common variants in BER genes and human cancer[116].
In addition, BER proteins may also play an important role in epigenetic regulation of gene expression. It has been recently demonstrated that BER proteins are necessary for both DNA methylation- and histone modification-mediated epigenetic regulation separate from its main function in maintaining genome stability[117]. Regarding the effects of promoter methylation controlling gene expression, it has been reported that some BER genes such as, MBD4, TGD, and OGG1, are significantly methylated in vitro[118-120] and in vivo[120].
Concurrent with the BER repair system, organisms have evolved a hierarchy of different pathways to handle several injuries caused to DNA to maintain genomic stability. Among the mechanisms of DNA repair, NER is the most versatile and can repair a large repertoire of chemically and structurally distinct injuries. There are more than 30 proteins that act in a sequential and concerted manner to remove DNA damage. During this process, the phosphodiester bonds 3’ and 5’ from the DNA damage are hydrolyzed by enzyme machinery called “excinuclease”. Then, a short oligonucleotide containing the lesion is removed, and the resulting gap is filled by a polymerase. In summary, this pathway consists of five steps: damage recognition, incision, excision, repair synthesis, and ligation[121]. In addition, NER can work together with transcription. Transcription-coupled repair ensures that the strand containing active genes are repaired with a higher priority than the rest of the genome, most likely because of RNA polymerase II (RNAPII), which acts as a sensor of injury[122]. Although NER confers protection against the accumulation of DNA lesions and maintains genome integrity, reducing NER activity may be beneficial for cancer patients who are undergoing chemotherapy to ensure the efficient action of the DNA damage-inducing drugs[123]. Additionally, it is well known that genetic defects in NER components cause xeroderma pigmentosum, an autosomal recessive disorder characterized by photosensitivity and predisposition to skin cancer[124].
It has been described that XPC, a NER gene, is highly methylated in different cell lines (Calu-1, H1355, and H441)[125]. Among bladder cancer patients, the XPC hypermethylation was related with lower mRNA levels[126]. In addition, the genes RAD23A and ERCC1, are also inactivated through promoter methylation in vitro[127,128].
The primary function of MMR is to eliminate base-base mismatches and insertion-deletion loops that arise as a consequence of DNA polymerase slippage during DNA replication[129]. The MMR system is composed of two protein complexes: the MutS, including protein MSH2 (mutS homolog 2), MSH3 (mutS homolog 3) and MSH6 (mutS homolog 6), and the MutL, comprising MLH1 (mutL homolog 1), PMS1 (postmeiotic increased segregation 1), PMS2 (postmeiotic segregation Increased 2) and MLH3 (mutL homolog 3). The operation of the MMR system requires that the protein complex MutL binds to MutSα (MSH2, and MSH6) or MutS-β (MSH2, and MSH3). It is believed that a deficiency in this repair system could be responsible for the accumulation of mutations[130,131]. Furthermore, the DNA repair enzyme MGMT (O-6-Methylguanine DNA methyltransferase) protects the DNA from mutations caused by alkylating agents, and the loss of MGMT expression can lead to the development of cancer[132].
MSI is one hallmark of DNA MMR deficiency that is involved in carcinogenesis. Microsatellites are short DNA sequence repeats that are scattered throughout the human genome. Errors in the DNA MMR mechanisms of tumor cells can result in the expansion or contraction of these repeated sequences and thus MSI. MSI was first described in 1993 in patients with hereditary nonpolyposis colorectal cancer (HNPCC), and from its discovery to date, it has been described in various types of cancer[8,133].
It has recently reported that trimethylated the histone modification H3K36 (H3K36me3) plays a critical role during initiation of MMR in vivo[134]. In this brilliant manuscript the authors showed that H3K36me3 interacts with and recruits MutSα to chromatin through the MSH6 PWWP domain. The abundance of H3K36me3 increases and reaches a plateau in late G1/early S, which correlates with the most critical need for MutSα on chromatin. In contrast, they found that the abundance of H3K36me3 decreases rapidly in late S and G2, when MMR is no longer relevant or helpful.
Additionally, it has been shown that methylation in the promoter region of MLH1 is related with a decreased activity of the gene in several types of cancer, such as: HNPCC[135], in sporadic endometrial carcinoma[136], gastric cancers[137], sporadic colorectal cancer[138], ovarian tumors[139], non-small cell lung cancer[140], oral squamous cell carcinoma[141], neck[142], and acute myeloid leukemia[143]. In summary several studies have been shown that MLH1 promoter methylation may have considerable importance in cancer development and as a prognostic factor. In addition, recent evidences reported that MSH2, MSH3, and MSH6 are also regulated by promoter methylation[144].
DNA double-strand breaks are critical injuries to the DNA molecule, which can result in cell death or a variety of genetic alterations such as deletions, LOH, translocations and chromosomal loss that are considered hallmarks of cancer development[145]. It is believed that such lesions may be attributed to the action of exogenous agents (such as ionizing radiation, chemotherapy drugs and infectious agents), endogenous agents (reactive oxygen species - ROS) and mechanical stress acting on chromosomes[146]. Two main strategies are employed for DSB repair: homologous recombination (HR) and non-homologous end-joining (NHEJ). It has been demonstrated that failures in any of the repair systems described above are important risk factors in carcinogenesis[147]. Additionally, several authors have shown that such changes could potentially lead to a disruption in the cell cycle and/or apoptosis[148,149].
HR initiates with extensive 5′ to 3′ end processing at broken ends. XRCC3 (X-ray repair Complementing defective repair in Chinese hamster cells 3) and RAD51 (RAD51 recombinase) are two important members of the HR repair pathway. The BRCA2-RAD51 complex is the central player for HR and catalyzes the homology search and strand exchange reaction, thereby allowing for the repair of the damaged region[150].
NHEJ starts in a stepwise manner, beginning with end processing by the MRE11/RAD50/NBS1 (MRN) complex and Ku70 and Ku80 subunits and resulting in the activation of ataxia telangiectasia mutated kinase (ATM), a member of the phosphatidylinositol 3-kinase-related kinase family. The association of ATM with the MRN complex leads to the activation of serine residue 1981 by phosphorylation, which results in the phosphorylation of the downstream targets involved in DNA repair and cell cycle checkpoints, including checkpoint kinase 2 (CHK2) and p53. Activated CHK2 can inhibit downstream targets, resulting in cell cycle arrest. ATM pathways also contribute to stabilization of the tumor suppressor protein p53 and lead to cell cycle arrest at the G1 phase[145].
Among epigenetic changes associated with DSBR, it has been shown that the DNA damage repair occurs in the context of chromatin. Among chromatin modifications linked to DSB response it is clear that phosphorylation of H2AX occurs following the break[151,152]. This phosphorylation occurs in a unique conserved SQE motif in the C-terminal tail at serine 139 (S139), so-called γ-H2AX[153]. Following phosphorylation the DNA repair and checkpoint proteins as well as the chromatin-remodeling complexes will form foci that colocalize with γ-H2AX[154,155]. It has been shown that the presence of γ-H2AX is not required for the initial signaling and recruitment of DNA repair factors. However, it is essential for their accumulation and retention at the break site, and subsequent amplification of the signal[154-156]. Additionally, methylation of histones H3K79 and H4K20 has also been shown to be important in the DSBR pathway[157].
Concerning the effects of DNA methylation on DSBR, it has been shown that the HR gene, BRCA1, was frequently methylated in several types of cancer such as breast cancer[158], ovarian cancer[159], gastric cancer[160], non-small cell lung cancer[161], uterine cancer[162], and bladder cancer[163]. In addition in has been reported that DNA methylation of XRCC5, a NHEJ gene, was found in patients with non-small cell lung cancer and squamous cell carcinoma[161]. High ATM methylation rate was also found in brain tumor patients[164].
As previously described, the cellular consequences of H. pylori infection produce a large number of different types of damage, such as 8-oxodG and AP sites, which in turn lead to single- and double-stranded breaks[165], DNA crosslinking and mutation[166-168]. Such alterations are removed predominantly by BER, which is critical for maintaining genome stability during the chronic inflammation that occurs during bacterial infection[169].
Studying BER-deficient cells, Meira et al[169] showed that H. pylori infection enhanced the inflammatory response and, as a consequence, increased the production of ROS and tumor-promoting cytokines. Moreover, the coculture of H. pylori in murine and human cancer cell lines increased the DSB levels[170]. Finally, a study showed that H. pylori infection induces the accumulation of AP sites in DNA that are further processed into DSBs, resulting in genomic instability and cellular transformation[171].
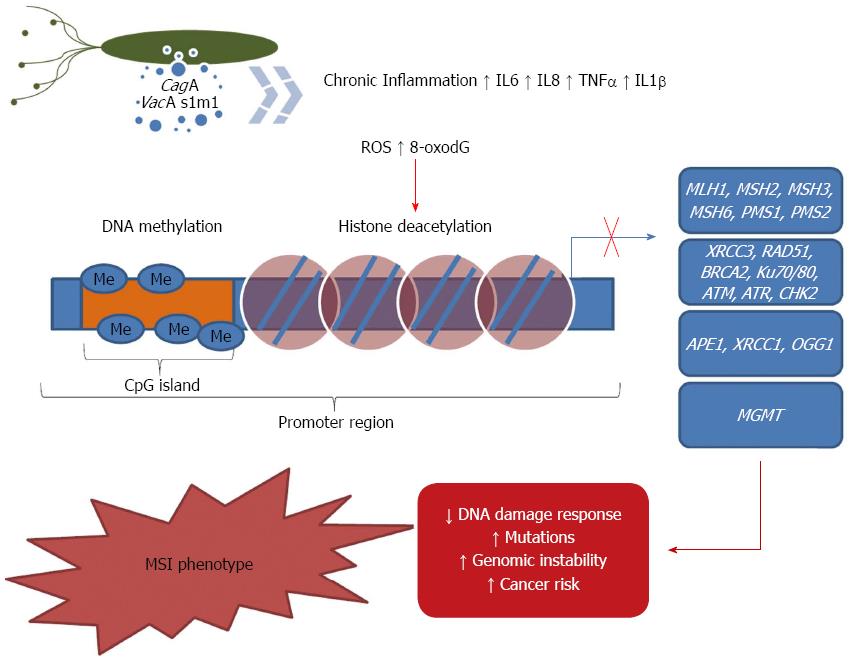
It has been proposed that the ROS generated by H. pylori infection, in either epithelial cell lines or cells isolated from mucosal biopsy samples, increases APE1 protein and mRNA levels, which indicates that the infected cells have a higher capacity to repair oxidative DNA damage[172]. Additionally, Futagami et al[173] observed that APE-1 expression is higher in gastric tissues from H. pylori-infected subjects compared with tissues from uninfected subjects. They also found that APE-1 is mainly localized in epithelial cells within gastric adenoma samples and in mesenchymal cells of gastric cancer tissues. The authors demonstrated that APE-1 expression in gastric cancer tissues with or without H. pylori was significantly reduced compared with that in H. pylori-infected gastric adenomas, whereas evidence of DNA damage did not differ between these neoplastic tissue types. These findings implicated that APE-1 plays a role in H. pylori-mediated human inflammatory and neoplastic gastric diseases. However, Machado et al[174] studied BER genes and detected a significant down-regulation of APE-1 in infected gastric cells, whereas no changes in OGG1 expression were observed. Because in BER AP sites are generated by OGG1 and then repaired by APE1, the authors postulated that an imbalance between the generation and repair of AP sites could be mutagenic though the generation of an excess of AP sites that could be converted into mutations by DNA polymerases or other repair enzymes (Figure 1).
XRCC1, another BER member, has been studied in gastric carcinogenesis. Wang et al[175] showed that the XRCC1 mRNA levels were lower in tumor tissues than in the corresponding adjacent non-tumorous tissues. Furthermore, the authors observed that methylation of the XRCC1 promoter was more frequent in tumor tissues, which indicated that methylation might contribute to the regulation of the transcriptional inactivation of XRCC1. Although XRCC1 repression may be involved in gastric carcinogenesis, there are no data available concerning the effects of H. pylori on epigenetic modulation of XRCC1. Similarly, further studies are necessary to understand the role of H. pylori infection on the epigenetic regulation of APE-1 and OGG1 (Figure 1).
MMR is one of the most important DNA repair pathways for maintaining genomic stability. MMR impairment results in the accumulation of mutations and an increased risk of MSI during replication. It has been shown that H. pylori infection is associated with the reduced efficiency of the DNA repair machinery, which favors the accumulation of mutations and genomic instability as well as gastric carcinogenesis[168,174,176,177]. Over the last several decades, there has been a growing body of evidence showing that genomic stability is affected by failures in MMR and that H. pylori is able to down-regulate the expression of several MMR effectors such as MLH1, MSH2, MSH6, PMS1 and PMS2, in vitro and in Big Blue transgenic mice[166,167,174,176]. Additionally, studies conducted in humans demonstrated that the infection reduced the MLH1 levels compared with uninfected individuals[178,179]
MLH1 down-regulation has been attributed to an increase in CpG methylation of its promoter region in vitro[176] and in vivo[87,179,180]. It has been reported that MLH1 promoter methylation occurs late in the progression of gastric carcinoma and that methylation depends partly on the persistence of the H. pylori infection[87,179]. Regarding the effects of other epigenetic mechanisms regulating MLH1 expression, the data from Fahrner et al[181] favor the idea that DNA hypermethylation, not a particular combination of histone modifications, is the dominant epigenetic mechanism involved in maintaining the silencing of MLH1.
Additionally, the presence of MSI in sporadic colorectal carcinomas has been significantly associated with the loss of MLH1 expression[182,183]. This phenomenon was associated with the hypermethylation of the MLH1 promoter, which is the underlying mechanism that causes MSI in gastric adenomas and early gastric cancers[179,184,185]. It is well known that H. pylori infection causes an increased rate of cell turnover in the gastric mucosa and thus overwhelms the DNA repair system. This process might allow for the accumulation of mutations that are consequent to H. pylori infection and other environmental risk factors[186].
The effects of methylation status on other MMR members (MSH2, MSH6 and PMS2) were evaluated previously in diffuse- and intestinal-type gastric cancer samples[187]. The data presented indicated that PMS2 methylation was associated with both diffuse- and intestinal-type cancer. Diffuse-type cancer was also significantly associated with MSH2 methylation, and MSH6 does not seem to be regulated by DNA promoter methylation. Although an association of MSH2 and PMS2 promoter methylation with gastric cancer was shown, the effects of H. pylori infection on their methylation has not been evaluated (Figure 1).
Recently, it was reported that the mRNA levels of MGMT, the gene product of which is required for the repair of O-6-methylguanine, were reduced in the gastric epithelium from patients with gastritis infected by H. pylori[188] and in gastric cancer patients[179,189]. These results were associated with an increased effect of H. pylori infection on MGMT CpG promoter methylation compared with uninfected patients[188]. Additionally, it has been suggested that the methylation of the MGMT promoter in H. pylori-infected patients is related to tumor progression[190,191]. The effects of histone modifications regulating MGMT expression on gastric carcinogenesis were also described. Meng et al[192], after treatment with 5-aza-2’-deoxycytidine and/or Trichostatin A, described histone H3K9 dimethylation, H3K4 dimethylation, H3K9 acetylation and DNA methylation working in combination to silence MGMT. However, the role of H. pylori infection in histone modifications regulating MGMT expression have not yet been evaluated (Figure 1).
The accumulation of DNA damage has been proposed to be a principal mechanism of infection, inflammation and cancer. The damaged DNA can be repaired through DSBR by either NHEJ or HR. ATM and ATR are critical molecules initiating the HR repair process, whereas Ku70/80 initiates the NHEJ DNA repair process. Therefore, the activation of both ATM/ATR and Ku70/80 is important in the DNA repair process[145,147,150].
It has been reported by Toller et al[170] that in vitro, H. pylori infection induces DSBs in a BabA adhesion-dependent manner. The authors also showed that damaged DNA triggers a damage signaling and repair response that involves the sequential ATM-dependent recruitment of 53BP1 and MDC1 and the phosphorylation of histone H2AX, a marker of DSB. In summary, they propose that H. pylori DSB induction contributes to the genetic instability and frequent chromosomal aberrations that are a hallmark of gastric cancer.
Subsequently, the results described by Hanada et al[193] demonstrated that ATM is activated in vivo and that this effect is related to H. pylori. They also showed that activated ATM and γ-H2AX, a marker of DSBs, are both present in H. pylori-infected human gastric epithelium. From these results, the authors concluded that it is likely that the ATM-dependent response occurs in response to H. pylori-induced DSBs to prevent or reduce chromosome aberrations. Accordingly, it has been shown that damaged DNA induces the induction of ATM, ATR, Ku proteins and cell cycle transition as well as the activation of p53 in H. pylori-infected tissues[194]. Studying the DNA-dependent protein kinase (DNA-PK), which is a serine/threonine kinase that consists of a 465-kDa catalytic subunit (DNA-PKcs) and the heterodimeric regulatory complex Ku [composed of a 70-kDa (Ku70) and an 86-kDa (Ku86) polypeptide], Lee et al[195] observed an increased expression in DNA-PKcs in H. pylori-associated gastritis, which may be associated with epithelial hyperproliferation or transcriptional changes. They also found that gastric cancers negative for DNA-PKcs are associated with an advanced stage, MSI phenotype; a high prevalence of lymph node metastasis; and poor patient survival.
A higher number of DSBs have been described in infections from H. pylori strains that are positive for the cagA virulence factor than from strains that are negative for it[193]. Additionally, they found that CagA inactivated RAD51, which suggested that higher levels of DSBs may be related in part to the reduced activity of DSB repair via HR[193]. In a complementary way, Bae et al[194] concluded that H. pylori-induced oxidative stress mediates a DNA damage response through NHEJ and HR repair processes, cell cycle arrest, and apoptosis in gastric mucosa of Mongolian gerbils (Figure 1).
It is well known that a major aspect of cellular response to DSBs occurs through specific interactions with chromatin structure and its modulation, which implicates highly dynamic post-translational modifications of histones that are critical for DNA damage recognition/signaling, repair of the lesion and release of cell cycle arrest. It has been reported that histone modification are disrupted in human cancers, implying that altered chromatin structure in tumor cells may impact DSB repair, increasing genomic instability and contributing to the progression of cancer[157,196]. Concerning the role of histone modifications regulating DSBR on gastric carcinogenesis, at the moment, there are no data available in the literature. Similarly, the putative effect of H. pylori infection in this mechanism has never been studied.
Over the last decade, the role of epigenetic alterations in gastric carcinogenesis has received greater attention. As described in this review, the disruption of epigenetic processes can lead to altered gene function and malignant cellular transformation. Considering the data expounded, it is clear that H. pylori plays a role in modulating the expression of BER, MMR and DSBR. Although aberrant epigenetic modifications are now believed to be essential players in DNA repair regulation, the epigenetic modulation of the DNA repair machinery in H. pylori-induced gastric carcinogenesis still requires further study.
P- Reviewer: Li YY, Slomiany BL S- Editor: Ma YJ L- Editor: A E- Editor: Ma S
| 1. | Kang C, Song JJ, Lee J, Kim MY. Epigenetics: an emerging player in gastric cancer. World J Gastroenterol. 2014;20:6433-6447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 51] [Cited by in F6Publishing: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 2. | Figueiredo C, Garcia-Gonzalez MA, Machado JC. Molecular pathogenesis of gastric cancer. Helicobacter. 2013;18 Suppl 1:28-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 3. | Lauren P. The Two Histological Main Types of Gastric Carcinoma: Diffuse and So-Called Intestinal-Type Carcinoma. An Attempt at a Histo-Clinical Classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] [Cited in This Article: ] |
| 4. | Hamilton JP, Meltzer SJ. A review of the genomics of gastric cancer. Clin Gastroenterol Hepatol. 2006;4:416-425. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 79] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Woolf CM, Isaacson EA. An analysis of 5 “stomach cancer families” in the state of Utah. Cancer. 1961;14:1005-1016. [PubMed] [Cited in This Article: ] |
| 6. | Kuniyasu H, Yasui W, Kitadai Y, Yokozaki H, Ito H, Tahara E. Frequent amplification of the c-met gene in scirrhous type stomach cancer. Biochem Biophys Res Commun. 1992;189:227-232. [PubMed] [Cited in This Article: ] |
| 7. | Hattori Y, Odagiri H, Nakatani H, Miyagawa K, Naito K, Sakamoto H, Katoh O, Yoshida T, Sugimura T, Terada M. K-sam, an amplified gene in stomach cancer, is a member of the heparin-binding growth factor receptor genes. Proc Natl Acad Sci USA. 1990;87:5983-5987. [PubMed] [Cited in This Article: ] |
| 8. | Smith MG, Hold GL, Tahara E, El-Omar EM. Cellular and molecular aspects of gastric cancer. World J Gastroenterol. 2006;12:2979-2990. [PubMed] [Cited in This Article: ] |
| 9. | Hibi K, Sakata M, Sakuraba K, Kitamura YH, Shirahata A, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G. Methylation of the DCC gene is lost in advanced gastric cancer. Anticancer Res. 2010;30:107-109. [PubMed] [Cited in This Article: ] |
| 10. | Yang TS, Yang XH, Chen X, Wang XD, Hua J, Zhou DL, Zhou B, Song ZS. MicroRNA-106b in cancer-associated fibroblasts from gastric cancer promotes cell migration and invasion by targeting PTEN. FEBS Lett. 2014;588:2162-2169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 11. | Wu YC, Lv P, Han J, Yu JL, Zhu X, Hong LL, Zhu WY, Yu QM, Wang XB, Li P. Enhanced serum methylated p16 DNAs is associated with the progression of gastric cancer. Int J Clin Exp Pathol. 2014;7:1553-1562. [PubMed] [Cited in This Article: ] |
| 12. | Bellini MF, Cadamuro AC, Succi M, Proença MA, Silva AE. Alterations of the TP53 gene in gastric and esophageal carcinogenesis. J Biomed Biotechnol. 2012;2012:891961. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 13. | Jang BG, Kim WH. Molecular pathology of gastric carcinoma. Pathobiology. 2011;78:302-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 94] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 14. | Aoyagi K, Kouhuji K, Miyagi M, Imaizumi T, Kizaki J, Isobe T, Shirouzu K. Expression of p27Kip1 protein in gastric carcinoma. Hepatogastroenterology. 2013;60:390-394. [PubMed] [Cited in This Article: ] |
| 15. | Velho S, Fernandes MS, Leite M, Figueiredo C, Seruca R. Causes and consequences of microsatellite instability in gastric carcinogenesis. World J Gastroenterol. 2014;20:16433-16442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 54] [Cited by in F6Publishing: 58] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 16. | Lee HJ, Jang YJ, Lee EJ, Kim JH, Park SS, Park SH, Kim CS, Mok YJ. The significance of mismatch repair genes in gastric cancer. J Cancer Res Ther. 2013;9:80-83. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Peek RM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1317] [Cited by in F6Publishing: 1298] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 18. | Sepulveda AR. Helicobacter, Inflammation, and Gastric Cancer. Curr Pathobiol Rep. 2013;1:9-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Infection with Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum. 1994;61:177-240. [PubMed] [Cited in This Article: ] |
| 20. | Figura N, Oderda G. Reflections on the first description of the presence of Helicobacter species in the stomach of mammals. Helicobacter. 1996;1:4-5. [PubMed] [Cited in This Article: ] |
| 21. | Marshall B. Helicobacter pylori: 20 years on. Clin Med. 2002;2:147-152. [PubMed] [Cited in This Article: ] |
| 22. | Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1:1273-1275. [PubMed] [Cited in This Article: ] |
| 23. | Peleteiro B, Bastos A, Ferro A, Lunet N. Prevalence of Helicobacter pylori infection worldwide: a systematic review of studies with national coverage. Dig Dis Sci. 2014;59:1698-1709. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 215] [Article Influence: 21.5] [Reference Citation Analysis (5)] |
| 24. | McColl KE. Helicobacter pylori 1988-1998. Eur J Gastroenterol Hepatol. 1999;11:13-16. [PubMed] [Cited in This Article: ] |
| 25. | Perez-Perez GI, Rothenbacher D, Brenner H. Epidemiology of Helicobacter pylori infection. Helicobacter. 2004;9 Suppl 1:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 156] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 26. | Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449-490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1446] [Cited by in F6Publishing: 1410] [Article Influence: 78.3] [Reference Citation Analysis (1)] |
| 27. | Bruce MG, Maaroos HI. Epidemiology of Helicobacter pylori infection. Helicobacter. 2008;13 Suppl 1:1-6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 100] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 28. | Nouraie M, Latifi-Navid S, Rezvan H, Radmard AR, Maghsudlu M, Zaer-Rezaii H, Amini S, Siavoshi F, Malekzadeh R. Childhood hygienic practice and family education status determine the prevalence of Helicobacter pylori infection in Iran. Helicobacter. 2009;14:40-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Queiroz DM, Carneiro JG, Braga-Neto MB, Fialho AB, Fialho AM, Goncalves MH, Rocha GA, Rocha AM, Braga LL. Natural history of Helicobacter pylori infection in childhood: eight-year follow-up cohort study in an urban community in northeast of Brazil. Helicobacter. 2012;17:23-29. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Fujimoto Y, Furusyo N, Toyoda K, Takeoka H, Sawayama Y, Hayashi J. Intrafamilial transmission of Helicobacter pylori among the population of endemic areas in Japan. Helicobacter. 2007;12:170-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 31. | Quigley EM, Turnberg LA. pH of the microclimate lining human gastric and duodenal mucosa in vivo. Studies in control subjects and in duodenal ulcer patients. Gastroenterology. 1987;92:1876-1884. [PubMed] [Cited in This Article: ] |
| 32. | Schreiber S, Konradt M, Groll C, Scheid P, Hanauer G, Werling HO, Josenhans C, Suerbaum S. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc Natl Acad Sci USA. 2004;101:5024-5029. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 223] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 33. | Amieva MR, Salama NR, Tompkins LS, Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol. 2002;4:677-690. [PubMed] [Cited in This Article: ] |
| 34. | Eaton KA, Krakowka S. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect Immun. 1994;62:3604-3607. [PubMed] [Cited in This Article: ] |
| 35. | Eaton KA, Krakowka S. Avirulent, urease-deficient Helicobacter pylori colonizes gastric epithelial explants ex vivo. Scand J Gastroenterol. 1995;30:434-437. [PubMed] [Cited in This Article: ] |
| 36. | Roesler BM, Rabelo-Gonçalves EM, Zeitune JM. Virulence Factors of Helicobacter pylori: A Review. Clin Med Insights Gastroenterol. 2014;7:9-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Herrera V, Parsonnet J. Helicobacter pylori and gastric adenocarcinoma. Clin Microbiol Infect. 2009;15:971-976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 172] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 38. | Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2635] [Cited by in F6Publishing: 2543] [Article Influence: 94.2] [Reference Citation Analysis (0)] |
| 39. | Kuck D, Kolmerer B, Iking-Konert C, Krammer PH, Stremmel W, Rudi J. Vacuolating cytotoxin of Helicobacter pylori induces apoptosis in the human gastric epithelial cell line AGS. Infect Immun. 2001;69:5080-5087. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Amieva MR, El-Omar EM. Host-bacterial interactions in Helicobacter pylori infection. Gastroenterology. 2008;134:306-323. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 379] [Cited by in F6Publishing: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 41. | Slomiany BL, Slomiany A. Involvement of p38 MAPK-dependent activator protein (AP-1) activation in modulation of gastric mucosal inflammatory responses to Helicobacter pylori by ghrelin. Inflammopharmacology. 2013;21:67-78. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 42. | Asaka M, Sugiyama T, Nobuta A, Kato M, Takeda H, Graham DY. Atrophic gastritis and intestinal metaplasia in Japan: results of a large multicenter study. Helicobacter. 2001;6:294-299. [PubMed] [Cited in This Article: ] |
| 43. | Correa P. Helicobacter pylori as a pathogen and carcinogen. J Physiol Pharmacol. 1997;48 Suppl 4:19-24. [PubMed] [Cited in This Article: ] |
| 44. | Bertuccio P, Chatenoud L, Levi F, Praud D, Ferlay J, Negri E, Malvezzi M, La Vecchia C. Recent patterns in gastric cancer: a global overview. Int J Cancer. 2009;125:666-673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 475] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 45. | Robinson K, Argent RH, Atherton JC. The inflammatory and immune response to Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21:237-259. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 119] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 46. | Qadri Q, Rasool R, Gulzar GM, Naqash S, Shah ZA. H. pylori infection, inflammation and gastric cancer. J Gastrointest Cancer. 2014;45:126-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 47. | Chiba T, Marusawa H, Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143:550-563. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 274] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 48. | Cadamuro AC, Rossi AF, Maniezzo NM, Silva AE. Helicobacter pylori infection: host immune response, implications on gene expression and microRNAs. World J Gastroenterol. 2014;20:1424-1437. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 74] [Cited by in F6Publishing: 72] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 49. | Meyer-ter-Vehn T, Covacci A, Kist M, Pahl HL. Helicobacter pylori activates mitogen-activated protein kinase cascades and induces expression of the proto-oncogenes c-fos and c-jun. J Biol Chem. 2000;275:16064-16072. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 203] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 50. | Sepulveda AR, Tao H, Carloni E, Sepulveda J, Graham DY, Peterson LE. Screening of gene expression profiles in gastric epithelial cells induced by Helicobacter pylori using microarray analysis. Aliment Pharmacol Ther. 2002;16 Suppl 2:145-157. [PubMed] [Cited in This Article: ] |
| 51. | Keates S, Keates AC, Warny M, Peek RM, Murray PG, Kelly CP. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag- Helicobacter pylori. J Immunol. 1999;163:5552-5559. [PubMed] [Cited in This Article: ] |
| 52. | Baik SC, Youn HS, Chung MH, Lee WK, Cho MJ, Ko GH, Park CK, Kasai H, Rhee KH. Increased oxidative DNA damage in Helicobacter pylori-infected human gastric mucosa. Cancer Res. 1996;56:1279-1282. [PubMed] [Cited in This Article: ] |
| 53. | Farinati F, Cardin R, Cassaro M, Bortolami M, Nitti D, Tieppo C, Zaninotto G, Rugge M. Helicobacter pylori, inflammation, oxidative damage and gastric cancer: a morphological, biological and molecular pathway. Eur J Cancer Prev. 2008;17:195-200. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 58] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 54. | Farinati F, Cardin R, Degan P, Rugge M, Mario FD, Bonvicini P, Naccarato R. Oxidative DNA damage accumulation in gastric carcinogenesis. Gut. 1998;42:351-356. [PubMed] [Cited in This Article: ] |
| 55. | Ladeira MS, Rodrigues MA, Salvadori DM, Queiroz DM, Freire-Maia DV. DNA damage in patients infected by Helicobacter pylori. Cancer Epidemiol Biomarkers Prev. 2004;13:631-637. [PubMed] [Cited in This Article: ] |
| 56. | Ladeira MS, Bueno RC, Dos Santos BF, Pinto CL, Prado RP, Silveira MG, Rodrigues MA, Bartchewsky W, Pedrazzoli J, Ribeiro ML. Relationship among oxidative DNA damage, gastric mucosal density and the relevance of cagA, vacA and iceA genotypes of Helicobacter pylori. Dig Dis Sci. 2008;53:248-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 57. | Ladeira MS, Rodrigues MA, Salvadori DM, Neto PP, Achilles P, Lerco MM, Rodrigues PA, Gonçalves I, Queiroz DM, Freire-Maia DV. Relationships between cagA, vacA, and iceA genotypes of Helicobacter pylori and DNA damage in the gastric mucosa. Environ Mol Mutagen. 2004;44:91-98. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 58. | Obst B, Wagner S, Sewing KF, Beil W. Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis. 2000;21:1111-1115. [PubMed] [Cited in This Article: ] |
| 59. | Fu S, Ramanujam KS, Wong A, Fantry GT, Drachenberg CB, James SP, Meltzer SJ, Wilson KT. Increased expression and cellular localization of inducible nitric oxide synthase and cyclooxygenase 2 in Helicobacter pylori gastritis. Gastroenterology. 1999;116:1319-1329. [PubMed] [Cited in This Article: ] |
| 60. | Grisham MB, Jourd’heuil D, Wink DA. Review article: chronic inflammation and reactive oxygen and nitrogen metabolism--implications in DNA damage and mutagenesis. Aliment Pharmacol Ther. 2000;14 Suppl 1:3-9. [PubMed] [Cited in This Article: ] |
| 61. | Israel DA, Peek RM. pathogenesis of Helicobacter pylori-induced gastric inflammation. Aliment Pharmacol Ther. 2001;15:1271-1290. [PubMed] [Cited in This Article: ] |
| 62. | Konturek PC, Pierzchalski P, Konturek SJ, Meixner H, Faller G, Kirchner T, Hahn EG. Helicobacter pylori induces apoptosis in gastric mucosa through an upregulation of Bax expression in humans. Scand J Gastroenterol. 1999;34:375-383. [PubMed] [Cited in This Article: ] |
| 63. | Bartchewsky W, Martini MR, Squassoni AC, Alvarez MC, Ladeira MS, Salvatore DM, Trevisan MA, Pedrazzoli J, Ribeiro ML. Effects of Helicobacter pylori infection on the expressions of Bax and Bcl-2 in patients with chronic gastritis and gastric cancer. Dig Dis Sci. 2010;55:111-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 64. | Cahill RJ, Xia H, Kilgallen C, Beattie S, Hamilton H, O’Morain C. Effect of eradication of Helicobacter pylori infection on gastric epithelial cell proliferation. Dig Dis Sci. 1995;40:1627-1631. [PubMed] [Cited in This Article: ] |
| 65. | Lynch DA, Mapstone NP, Clarke AM, Sobala GM, Jackson P, Morrison L, Dixon MF, Quirke P, Axon AT. Cell proliferation in Helicobacter pylori associated gastritis and the effect of eradication therapy. Gut. 1995;36:346-350. [PubMed] [Cited in This Article: ] |
| 66. | Björkholm B, Falk P, Engstrand L, Nyrén O. Helicobacter pylori: resurrection of the cancer link. J Intern Med. 2003;253:102-119. [PubMed] [Cited in This Article: ] |
| 67. | Cavallo P, Cianciulli A, Mitolo V, Panaro MA. Lipopolysaccharide (LPS) of helicobacter modulates cellular DNA repair systems in intestinal cells. Clin Exp Med. 2011;11:171-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 68. | Waddington CH. The epigenotype. 1942. Int J Epidemiol. 2012;41:10-13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 530] [Cited by in F6Publishing: 497] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 69. | Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465-476. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2134] [Cited by in F6Publishing: 2053] [Article Influence: 128.3] [Reference Citation Analysis (0)] |
| 70. | Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3359] [Cited by in F6Publishing: 3483] [Article Influence: 267.9] [Reference Citation Analysis (0)] |
| 71. | Hannon GJ, Rivas FV, Murchison EP, Steitz JA. The expanding universe of noncoding RNAs. Cold Spring Harb Symp Quant Biol. 2006;71:551-564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Virani S, Colacino JA, Kim JH, Rozek LS. Cancer epigenetics: a brief review. ILAR J. 2012;53:359-369. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 71] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 73. | Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709-2721. [PubMed] [Cited in This Article: ] |
| 74. | Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400-5413. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1124] [Cited by in F6Publishing: 1058] [Article Influence: 48.1] [Reference Citation Analysis (0)] |
| 75. | Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489-492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1147] [Cited by in F6Publishing: 1070] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 76. | Zendman AJ, Ruiter DJ, Van Muijen GN. Cancer/testis-associated genes: identification, expression profile, and putative function. J Cell Physiol. 2003;194:272-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 198] [Cited by in F6Publishing: 194] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 77. | Nakamura J, Tanaka T, Kitajima Y, Noshiro H, Miyazaki K. Methylation-mediated gene silencing as biomarkers of gastric cancer: a review. World J Gastroenterol. 2014;20:11991-12006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 34] [Cited by in F6Publishing: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 78. | Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989-995. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 474] [Cited by in F6Publishing: 460] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 79. | Kang GH, Lee S, Cho NY, Gandamihardja T, Long TI, Weisenberger DJ, Campan M, Laird PW. DNA methylation profiles of gastric carcinoma characterized by quantitative DNA methylation analysis. Lab Invest. 2008;88:161-170. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 80. | Kitajima Y, Ohtaka K, Mitsuno M, Tanaka M, Sato S, Nakafusa Y, Miyazaki K. Helicobacter pylori infection is an independent risk factor for Runx3 methylation in gastric cancer. Oncol Rep. 2008;19:197-202. [PubMed] [Cited in This Article: ] |
| 81. | Lu XX, Yu JL, Ying LS, Han J, Wang S, Yu QM, Wang XB, Fang XH, Ling ZQ. Stepwise cumulation of RUNX3 methylation mediated by Helicobacter pylori infection contributes to gastric carcinoma progression. Cancer. 2012;118:5507-5517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 82. | Niwa T, Tsukamoto T, Toyoda T, Mori A, Tanaka H, Maekita T, Ichinose M, Tatematsu M, Ushijima T. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70:1430-1440. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 289] [Cited by in F6Publishing: 305] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 83. | Hur K, Niwa T, Toyoda T, Tsukamoto T, Tatematsu M, Yang HK, Ushijima T. Insufficient role of cell proliferation in aberrant DNA methylation induction and involvement of specific types of inflammation. Carcinogenesis. 2011;32:35-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Qian X, Huang C, Cho CH, Hui WM, Rashid A, Chan AO. E-cadherin promoter hypermethylation induced by interleukin-1beta treatment or H. pylori infection in human gastric cancer cell lines. Cancer Lett. 2008;263:107-113. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 85. | Wang YQ, Li YM, Li X, Liu T, Liu XK, Zhang JQ, Guo JW, Guo LY, Qiao L. Hypermethylation of TGF-β1 gene promoter in gastric cancer. World J Gastroenterol. 2013;19:5557-5564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 18] [Cited by in F6Publishing: 16] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 86. | Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter. 1998;3:241-253. [PubMed] [Cited in This Article: ] |
| 87. | Perri F, Cotugno R, Piepoli A, Merla A, Quitadamo M, Gentile A, Pilotto A, Annese V, Andriulli A. Aberrant DNA methylation in non-neoplastic gastric mucosa of H. Pylori infected patients and effect of eradication. Am J Gastroenterol. 2007;102:1361-1371. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 88. | Niwa T, Toyoda T, Tsukamoto T, Mori A, Tatematsu M, Ushijima T. Prevention of Helicobacter pylori-induced gastric cancers in gerbils by a DNA demethylating agent. Cancer Prev Res (Phila). 2013;6:263-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 89. | Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251-260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6680] [Cited by in F6Publishing: 6347] [Article Influence: 235.1] [Reference Citation Analysis (0)] |
| 90. | Kim JH, Choi YK, Kwon HJ, Yang HK, Choi JH, Kim DY. Downregulation of gelsolin and retinoic acid receptor beta expression in gastric cancer tissues through histone deacetylase 1. J Gastroenterol Hepatol. 2004;19:218-224. [PubMed] [Cited in This Article: ] |
| 91. | Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 162] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 92. | Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559-599. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 613] [Cited by in F6Publishing: 598] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 93. | Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261-1270. [PubMed] [Cited in This Article: ] |
| 94. | Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1425] [Cited by in F6Publishing: 1473] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 95. | Kaustov L, Ouyang H, Amaya M, Lemak A, Nady N, Duan S, Wasney GA, Li Z, Vedadi M, Schapira M. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 96. | Benard A, Goossens-Beumer IJ, van Hoesel AQ, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ. Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and histone modification H3K27me3 in colorectal cancer. PLoS One. 2014;9:e108265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 97. | Healey MA, Hu R, Beck AH, Collins LC, Schnitt SJ, Tamimi RM, Hazra A. Association of H3K9me3 and H3K27me3 repressive histone marks with breast cancer subtypes in the Nurses’ Health Study. Breast Cancer Res Treat. 2014;147:639-651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 33] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 98. | He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu DZ, Luo GY, Zhu SL, Xie D. Prognostic significance of overexpression of EZH2 and H3k27me3 proteins in gastric cancer. Asian Pac J Cancer Prev. 2012;13:3173-3178. [PubMed] [Cited in This Article: ] |
| 99. | Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 741] [Cited by in F6Publishing: 817] [Article Influence: 74.3] [Reference Citation Analysis (0)] |
| 100. | Park YS, Jin MY, Kim YJ, Yook JH, Kim BS, Jang SJ. The global histone modification pattern correlates with cancer recurrence and overall survival in gastric adenocarcinoma. Ann Surg Oncol. 2008;15:1968-1976. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 127] [Cited by in F6Publishing: 137] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 101. | Takahashi H, Murai Y, Tsuneyama K, Nomoto K, Okada E, Fujita H, Takano Y. Overexpression of phosphorylated histone H3 is an indicator of poor prognosis in gastric adenocarcinoma patients. Appl Immunohistochem Mol Morphol. 2006;14:296-302. [PubMed] [Cited in This Article: ] |
| 102. | Mitani Y, Oue N, Hamai Y, Aung PP, Matsumura S, Nakayama H, Kamata N, Yasui W. Histone H3 acetylation is associated with reduced p21(WAF1/CIP1) expression by gastric carcinoma. J Pathol. 2005;205:65-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 103. | Zheng YC, Duan YC, Ma JL, Xu RM, Zi X, Lv WL, Wang MM, Ye XW, Zhu S, Mobley D. Triazole-dithiocarbamate based selective lysine specific demethylase 1 (LSD1) inactivators inhibit gastric cancer cell growth, invasion, and migration. J Med Chem. 2013;56:8543-8560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 170] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 104. | Yang Q, Wang B, Gao W, Huang S, Liu Z, Li W, Jia J. SIRT1 is downregulated in gastric cancer and leads to G1-phase arrest via NF-κB/Cyclin D1 signaling. Mol Cancer Res. 2013;11:1497-1507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 105. | Sakuraba K, Yokomizo K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Hibi K. TIP60 as a potential marker for the malignancy of gastric cancer. Anticancer Res. 2011;31:77-79. [PubMed] [Cited in This Article: ] |
| 106. | Xia G, Schneider-Stock R, Diestel A, Habold C, Krueger S, Roessner A, Naumann M, Lendeckel U. Helicobacter pylori regulates p21(WAF1) by histone H4 acetylation. Biochem Biophys Res Commun. 2008;369:526-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 107. | Fehri LF, Rechner C, Janssen S, Mak TN, Holland C, Bartfeld S, Brüggemann H, Meyer TF. Helicobacter pylori-induced modification of the histone H3 phosphorylation status in gastric epithelial cells reflects its impact on cell cycle regulation. Epigenetics. 2009;4:577-586. [PubMed] [Cited in This Article: ] |
| 108. | Ding SZ, Goldberg JB, Hatakeyama M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010;6:851-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 109. | Shin A, Lee KM, Ahn B, Park CG, Noh SK, Park DY, Ahn SH, Yoo KY, Kang D. Genotype-phenotype relationship between DNA repair gene genetic polymorphisms and DNA repair capacity. Asian Pac J Cancer Prev. 2008;9:501-505. [PubMed] [Cited in This Article: ] |
| 110. | Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709-715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3896] [Cited by in F6Publishing: 3591] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 111. | Dianov GL, Thybo T, Dianova II, Lipinski LJ, Bohr VA. Single nucleotide patch base excision repair is the major pathway for removal of thymine glycol from DNA in human cell extracts. J Biol Chem. 2000;275:11809-11813. [PubMed] [Cited in This Article: ] |
| 112. | Dianov GL, Hübscher U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 2013;41:3483-3490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 113. | Nash RA, Caldecott KW, Barnes DE, Lindahl T. XRCC1 protein interacts with one of two distinct forms of DNA ligase III. Biochemistry. 1997;36:5207-5211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 179] [Cited by in F6Publishing: 191] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 114. | Peterson CL, Côté J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602-616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 227] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 115. | Jagannathan I, Cole HA, Hayes JJ. Base excision repair in nucleosome substrates. Chromosome Res. 2006;14:27-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 116. | Karahalil B, Bohr VA, Wilson DM. Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk. Hum Exp Toxicol. 2012;31:981-1005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 90] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 117. | Li J, Braganza A, Sobol RW. Base excision repair facilitates a functional relationship between Guanine oxidation and histone demethylation. Antioxid Redox Signal. 2013;18:2429-2443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 118. | Peng B, Hurt EM, Hodge DR, Thomas SB, Farrar WL. DNA hypermethylation and partial gene silencing of human thymine- DNA glycosylase in multiple myeloma cell lines. Epigenetics. 2006;1:138-145. [PubMed] [Cited in This Article: ] |
| 119. | Howard JH, Frolov A, Tzeng CW, Stewart A, Midzak A, Majmundar A, Godwin A, Heslin M, Bellacosa A, Arnoletti JP. Epigenetic downregulation of the DNA repair gene MED1/MBD4 in colorectal and ovarian cancer. Cancer Biol Ther. 2009;8:94-100. [PubMed] [Cited in This Article: ] |
| 120. | Guan H, Ji M, Hou P, Liu Z, Wang C, Shan Z, Teng W, Xing M. Hypermethylation of the DNA mismatch repair gene hMLH1 and its association with lymph node metastasis and T1799A BRAF mutation in patients with papillary thyroid cancer. Cancer. 2008;113:247-255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 121. | Nouspikel T. DNA repair in mammalian cells : Nucleotide excision repair: variations on versatility. Cell Mol Life Sci. 2009;66:994-1009. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 239] [Cited by in F6Publishing: 204] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 122. | Deaconescu AM, Artsimovitch I, Grigorieff N. Interplay of DNA repair with transcription: from structures to mechanisms. Trends Biochem Sci. 2012;37:543-552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 123. | Liu L, Lee J, Zhou P. Navigating the nucleotide excision repair threshold. J Cell Physiol. 2010;224:585-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 124. | Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J Rare Dis. 2011;6:70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 239] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 125. | Wu YH, Tsai Chang JH, Cheng YW, Wu TC, Chen CY, Lee H. Xeroderma pigmentosum group C gene expression is predominantly regulated by promoter hypermethylation and contributes to p53 mutation in lung cancers. Oncogene. 2007;26:4761-4773. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 126. | Yang J, Xu Z, Li J, Zhang R, Zhang G, Ji H, Song B, Chen Z. XPC epigenetic silence coupled with p53 alteration has a significant impact on bladder cancer outcome. J Urol. 2010;184:336-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 127. | Peng B, Hodge DR, Thomas SB, Cherry JM, Munroe DJ, Pompeia C, Xiao W, Farrar WL. Epigenetic silencing of the human nucleotide excision repair gene, hHR23B, in interleukin-6-responsive multiple myeloma KAS-6/1 cells. J Biol Chem. 2005;280:4182-4187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 128. | Chen HY, Shao CJ, Chen FR, Kwan AL, Chen ZP. Role of ERCC1 promoter hypermethylation in drug resistance to cisplatin in human gliomas. Int J Cancer. 2010;126:1944-1954. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 129. | Jiricny J, Nyström-Lahti M. Mismatch repair defects in cancer. Curr Opin Genet Dev. 2000;10:157-161. [PubMed] [Cited in This Article: ] |
| 130. | Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89-96. [PubMed] [Cited in This Article: ] |
| 131. | Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, Collins FS. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24:27-35. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 217] [Cited by in F6Publishing: 238] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 132. | Iwakuma T, Shiraishi A, Fukuhara M, Kawate H, Sekiguchi M. Organization and expression of the mouse gene for DNA repair methyltransferase. DNA Cell Biol. 1996;15:863-872. [PubMed] [Cited in This Article: ] |
| 133. | Halling KC, Harper J, Moskaluk CA, Thibodeau SN, Petroni GR, Yustein AS, Tosi P, Minacci C, Roviello F, Piva P. Origin of microsatellite instability in gastric cancer. Am J Pathol. 1999;155:205-211. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 116] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 134. | Li F, Mao G, Tong D, Huang J, Gu L, Yang W, Li GM. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590-600. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 429] [Cited by in F6Publishing: 426] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 135. | Arends MJ. Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol. 2013;21:97-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 81] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 136. | Esteller M, Levine R, Baylin SB, Ellenson LH, Herman JG. MLH1 promoter hypermethylation is associated with the microsatellite instability phenotype in sporadic endometrial carcinomas. Oncogene. 1998;17:2413-2417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 347] [Cited by in F6Publishing: 317] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 137. | Yamamoto H, Imai K. Microsatellite instability: an update. Arch Toxicol. 2015;89:899-921. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 138. | Li X, Yao X, Wang Y, Hu F, Wang F, Jiang L, Liu Y, Wang D, Sun G, Zhao Y. MLH1 promoter methylation frequency in colorectal cancer patients and related clinicopathological and molecular features. PLoS One. 2013;8:e59064. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 139. | Murphy MA, Wentzensen N. Frequency of mismatch repair deficiency in ovarian cancer: a systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int J Cancer. 2011;129:1914-1922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 90] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 140. | Wang YC, Lu YP, Tseng RC, Lin RK, Chang JW, Chen JT, Shih CM, Chen CY. Inactivation of hMLH1 and hMSH2 by promoter methylation in primary non-small cell lung tumors and matched sputum samples. J Clin Invest. 2003;111:887-895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 113] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 141. | Czerninski R, Krichevsky S, Ashhab Y, Gazit D, Patel V, Ben-Yehuda D. Promoter hypermethylation of mismatch repair genes, hMLH1 and hMSH2 in oral squamous cell carcinoma. Oral Dis. 2009;15:206-213. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 142. | Steinmann K, Sandner A, Schagdarsurengin U, Dammann RH. Frequent promoter hypermethylation of tumor-related genes in head and neck squamous cell carcinoma. Oncol Rep. 2009;22:1519-1526. [PubMed] [Cited in This Article: ] |
| 143. | Seedhouse CH, Das-Gupta EP, Russell NH. Methylation of the hMLH1 promoter and its association with microsatellite instability in acute myeloid leukemia. Leukemia. 2003;17:83-88. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 144. | Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3:51-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 129] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 145. | Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134-147. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 906] [Cited by in F6Publishing: 908] [Article Influence: 56.8] [Reference Citation Analysis (0)] |
| 146. | Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686-690. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 399] [Cited by in F6Publishing: 389] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 147. | Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553-1558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 486] [Cited by in F6Publishing: 453] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 148. | Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH. DNA damage response and transcription. DNA Repair (Amst). 2011;10:743-750. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 149. | Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J. MicroRNAs, the DNA damage response and cancer. Mutat Res. 2011;717:54-66. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 67] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 150. | Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296-3307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 273] [Cited by in F6Publishing: 285] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 151. | Redon CE, Nakamura AJ, Zhang YW, Ji JJ, Bonner WM, Kinders RJ, Parchment RE, Doroshow JH, Pommier Y. Histone gammaH2AX and poly(ADP-ribose) as clinical pharmacodynamic biomarkers. Clin Cancer Res. 2010;16:4532-4542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 202] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 152. | Thiriet C, Hayes JJ. Chromatin in need of a fix: phosphorylation of H2AX connects chromatin to DNA repair. Mol Cell. 2005;18:617-622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 153. | Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858-5868. [PubMed] [Cited in This Article: ] |
| 154. | Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922-927. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1067] [Cited by in F6Publishing: 1006] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 155. | Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675-679. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 728] [Cited by in F6Publishing: 784] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 156. | Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc Natl Acad Sci USA. 2002;99:8173-8178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 407] [Cited by in F6Publishing: 403] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 157. | Rossetto D, Truman AW, Kron SJ, Côté J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin Cancer Res. 2010;16:4543-4552. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 158. | Karsli-Ceppioglu S, Dagdemir A, Judes G, Ngollo M, Penault-Llorca F, Pajon A, Bignon YJ, Bernard-Gallon D. Epigenetic mechanisms of breast cancer: an update of the current knowledge. Epigenomics. 2014;6:651-664. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 159. | Koukoura O, Spandidos DA, Daponte A, Sifakis S. DNA methylation profiles in ovarian cancer: implication in diagnosis and therapy (Review). Mol Med Rep. 2014;10:3-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 160. | Bernal C, Vargas M, Ossandón F, Santibáñez E, Urrutia J, Luengo V, Zavala LF, Backhouse C, Palma M, Argandoña J. DNA methylation profile in diffuse type gastric cancer: evidence for hypermethylation of the BRCA1 promoter region in early-onset gastric carcinogenesis. Biol Res. 2008;41:303-315. [PubMed] [Cited in This Article: ] |
| 161. | Lee MN, Tseng RC, Hsu HS, Chen JY, Tzao C, Ho WL, Wang YC. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res. 2007;13:832-838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 162. | Xing D, Scangas G, Nitta M, He L, Xu X, Ioffe YJ, Aspuria PJ, Hedvat CY, Anderson ML, Oliva E. A role for BRCA1 in uterine leiomyosarcoma. Cancer Res. 2009;69:8231-8235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 163. | Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13:7296-7304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 156] [Cited by in F6Publishing: 168] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 164. | Mehdipour P, Karami F, Javan F, Mehrazin M. Linking ATM Promoter Methylation to Cell Cycle Protein Expression in Brain Tumor Patients: Cellular Molecular Triangle Correlation in ATM Territory. Mol Neurobiol. 2015;52:293-302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 165. | Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195-1214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2156] [Cited by in F6Publishing: 2077] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 166. | Kim JJ, Tao H, Carloni E, Leung WK, Graham DY, Sepulveda AR. Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology. 2002;123:542-553. [PubMed] [Cited in This Article: ] |
| 167. | Park DI, Park SH, Kim SH, Kim JW, Cho YK, Kim HJ, Sohn CI, Jeon WK, Kim BI, Cho EY. Effect of Helicobacter pylori infection on the expression of DNA mismatch repair protein. Helicobacter. 2005;10:179-184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 168. | Touati E, Michel V, Thiberge JM, Wuscher N, Huerre M, Labigne A. Chronic Helicobacter pylori infections induce gastric mutations in mice. Gastroenterology. 2003;124:1408-1419. [PubMed] [Cited in This Article: ] |
| 169. | Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest. 2008;118:2516-2525. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 259] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 170. | Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci USA. 2011;108:14944-14949. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 230] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 171. | Kidane D, Murphy DL, Sweasy JB. Accumulation of abasic sites induces genomic instability in normal human gastric epithelial cells during Helicobacter pylori infection. Oncogenesis. 2014;3:e128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 172. | Ding SZ, O’Hara AM, Denning TL, Dirden-Kramer B, Mifflin RC, Reyes VE, Ryan KA, Elliott SN, Izumi T, Boldogh I. Helicobacter pylori and H2O2 increase AP endonuclease-1/redox factor-1 expression in human gastric epithelial cells. Gastroenterology. 2004;127:845-858. [PubMed] [Cited in This Article: ] |
| 173. | Futagami S, Hiratsuka T, Shindo T, Horie A, Hamamoto T, Suzuki K, Kusunoki M, Miyake K, Gudis K, Crowe SE. Expression of apurinic/apyrimidinic endonuclease-1 (APE-1) in H. pylori-associated gastritis, gastric adenoma, and gastric cancer. Helicobacter. 2008;13:209-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 174. | Machado AM, Figueiredo C, Touati E, Máximo V, Sousa S, Michel V, Carneiro F, Nielsen FC, Seruca R, Rasmussen LJ. Helicobacter pylori infection induces genetic instability of nuclear and mitochondrial DNA in gastric cells. Clin Cancer Res. 2009;15:2995-3002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 175. | Wang P, Tang JT, Peng YS, Chen XY, Zhang YJ, Fang JY. XRCC1 downregulated through promoter hypermethylation is involved in human gastric carcinogenesis. J Dig Dis. 2010;11:343-351. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 176. | Yao Y, Tao H, Park DI, Sepulveda JL, Sepulveda AR. Demonstration and characterization of mutations induced by Helicobacter pylori organisms in gastric epithelial cells. Helicobacter. 2006;11:272-286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 177. | Machado AM, Desler C, Bøggild S, Strickertsson JA, Friis-Hansen L, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection affects mitochondrial function and DNA repair, thus, mediating genetic instability in gastric cells. Mech Ageing Dev. 2013;134:460-466. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 178. | Mirzaee V, Molaei M, Shalmani HM, Zali MR. Helicobacter pylori infection and expression of DNA mismatch repair proteins. World J Gastroenterol. 2008;14:6717-6721. [PubMed] [Cited in This Article: ] |
| 179. | Alvarez MC, Santos JC, Maniezzo N, Ladeira MS, da Silva AL, Scaletsky IC, Pedrazzoli J, Ribeiro ML. MGMT and MLH1 methylation in Helicobacter pylori-infected children and adults. World J Gastroenterol. 2013;19:3043-3051. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 180. | Ferrasi AC, Pinheiro NA, Rabenhorst SH, Caballero OL, Rodrigues MA, de Carvalho F, Leite CV, Ferreira MV, Barros MA, Pardini MI. Helicobacter pylori and EBV in gastric carcinomas: methylation status and microsatellite instability. World J Gastroenterol. 2010;16:312-319. [PubMed] [Cited in This Article: ] |
| 181. | Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62:7213-7218. [PubMed] [Cited in This Article: ] |
| 182. | Edmonston TB, Cuesta KH, Burkholder S, Barusevicius A, Rose D, Kovatich AJ, Boman B, Fry R, Fishel R, Palazzo JP. Colorectal carcinomas with high microsatellite instability: defining a distinct immunologic and molecular entity with respect to prognostic markers. Hum Pathol. 2000;31:1506-1514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 63] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 183. | Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261-268. [PubMed] [Cited in This Article: ] |
| 184. | Baek MJ, Kang H, Kim SE, Park JH, Lee JS, Paik YK, Kim H. Expression of hMLH1 is inactivated in the gastric adenomas with enhanced microsatellite instability. Br J Cancer. 2001;85:1147-1152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 185. | Fleisher AS, Esteller M, Tamura G, Rashid A, Stine OC, Yin J, Zou TT, Abraham JM, Kong D, Nishizuka S. Hypermethylation of the hMLH1 gene promoter is associated with microsatellite instability in early human gastric neoplasia. Oncogene. 2001;20:329-335. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 186. | Gologan A, Graham DY, Sepulveda AR. Molecular markers in Helicobacter pylori-associated gastric carcinogenesis. Clin Lab Med. 2005;25:197-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 187. | Moura Lima E, Ferreira Leal M, Cardoso Smith Mde A, Rodríguez Burbano R, Pimentel de Assumpção P, Bello MJ, Rey JA, Ferreira de Lima F, Casartelli C. DNA mismatch repair gene methylation in gastric cancer in individuals from northern Brazil. Biocell. 2008;32:237-243. [PubMed] [Cited in This Article: ] |
| 188. | Sepulveda AR, Yao Y, Yan W, Park DI, Kim JJ, Gooding W, Abudayyeh S, Graham DY. CpG methylation and reduced expression of O6-methylguanine DNA methyltransferase is associated with Helicobacter pylori infection. Gastroenterology. 2010;138:1836-1844. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 189. | Bartchewsky W, Martini MR, Squassoni AC, Alvarez MC, Ladeira MS, Salvatore DM, Trevisan MA, Pedrazzoli J, Ribeiro ML. Influence of Helicobacter pylori infection on the expression of MLH1 and MGMT in patients with chronic gastritis and gastric cancer. Eur J Clin Microbiol Infect Dis. 2009;28:591-597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 190. | Zou XP, Zhang B, Zhang XQ, Chen M, Cao J, Liu WJ. Promoter hypermethylation of multiple genes in early gastric adenocarcinoma and precancerous lesions. Hum Pathol. 2009;40:1534-1542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 191. | Schneider BG, Peng DF, Camargo MC, Piazuelo MB, Sicinschi LA, Mera R, Romero-Gallo J, Delgado AG, Bravo LE, Wilson KT. Promoter DNA hypermethylation in gastric biopsies from subjects at high and low risk for gastric cancer. Int J Cancer. 2010;127:2588-2597. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 192. | Meng CF, Zhu XJ, Peng G, Dai DQ. Role of histone modifications and DNA methylation in the regulation of O6-methylguanine-DNA methyltransferase gene expression in human stomach cancer cells. Cancer Invest. 2010;28:331-339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 193. | Hanada K, Uchida T, Tsukamoto Y, Watada M, Yamaguchi N, Yamamoto K, Shiota S, Moriyama M, Graham DY, Yamaoka Y. Helicobacter pylori infection introduces DNA double-strand breaks in host cells. Infect Immun. 2014;82:4182-4189. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 79] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 194. | Bae M, Lim JW, Kim H. Oxidative DNA Damage Response in Helicobacter pylori-Infected Mongolian Gerbils. J Cancer Prev. 2013;18:271-275. [PubMed] [Cited in This Article: ] |
| 195. | Lee HS, Choe G, Park KU, Park do J, Yang HK, Lee BL, Kim WH. Altered expression of DNA-dependent protein kinase catalytic subunit (DNA-PKcs) during gastric carcinogenesis and its clinical implications on gastric cancer. Int J Oncol. 2007;31:859-866. [PubMed] [Cited in This Article: ] |
| 196. | Lorat Y, Schanz S, Schuler N, Wennemuth G, Rübe C, Rübe CE. Beyond repair foci: DNA double-strand break repair in euchromatic and heterochromatic compartments analyzed by transmission electron microscopy. PLoS One. 2012;7:e38165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |