Published online Apr 7, 2015. doi: 10.3748/wjg.v21.i13.4006
Peer-review started: November 3, 2014
First decision: November 26, 2014
Revised: December 10, 2014
Accepted: January 8, 2015
Article in press: January 8, 2015
Published online: April 7, 2015
AIM: To determine the association between rapid viral response and IL28B, IL28RA, IL10RB and MxA polymorphisms in the Chinese Han population.
METHODS: The study cohort consisted of 238 chronic hepatitis C patients treated with interferon (IFN)-α-2b and ribavirin. Six single nucleotide polymorphisms were genotyped using the ABI TaqMan allelic discrimination assay. Biochemical indices were measured at baseline. Serum hepatitis C virus (HCV) RNA was detected at weeks 0, 4, 12 and 24 of therapy.
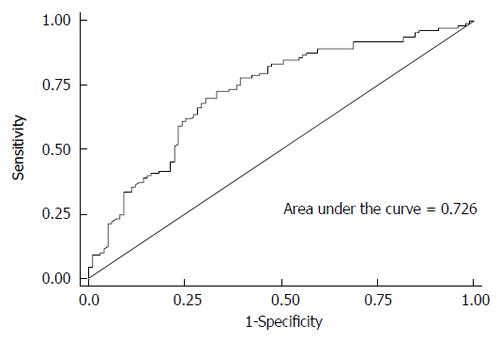
RESULTS: Only IL28B rs12980275 was associated with treatment response in the Chinese Han population. Patients carrying AG/GG genotypes had a reduced rapid viral response compared with patients carrying the AA genotype (additive model: adjusted OR = 0.43, 95%CI: 0.24-0.75). It took less time for patients with the AA genotype to achieve a viral load < 500 copies/mL (log-rank test, P = 0.004). In addition, the protective effect of genotype AA was independent of baseline viral load. HCV genotype, and baseline white blood cell count, α-fetoprotein and viral load might also help predict treatment response. The area under the receiver-operating characteristic curve was 0.726.
CONCLUSION: IL28B rs12980275 AA genotype is a strong predictor of positive response to IFN therapy in Chinese Han patients with hepatitis C.
Core tip: The association between IL28B rs12980275 and viral response to pegylated-interferon (IFN) plus ribavirin treatment has been observed in Japanese patients, but rarely in Chinese patients. Because pegylated-IFN is more expensive, non-pegylated instead of pegylated IFN-α is more commonly used for chronic hepatitis C treatment in Chinese primary hospitals. Therefore, the role of IFN-λ-related genes in the response to non-pegylated IFN-α treatment should be established to help guide clinical decisions and improve cost-effectiveness.
- Citation: Zhang YY, Chen HB, Xu Y, Huang P, Wang J, Zhang Y, Yu RB, Su J. Interferon-λ-related genes and therapeutic response in Chinese hepatitis C patients. World J Gastroenterol 2015; 21(13): 4006-4013
- URL: https://www.wjgnet.com/1007-9327/full/v21/i13/4006.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i13.4006
Hepatitis C virus (HCV) poses a serious global health problem due to its adverse clinical outcomes, such as cirrhosis and hepatocellular carcinoma. The estimated prevalence of HCV is 1%-1.9% in the general population of Mainland China, with 75%-80% of those chronically infected[1,2]. The treatment for chronic hepatitis C (CHC) consists of interferon (IFN) plus ribavirin (RBV) and protease inhibitors such as telaprevir and boceprevir. Sustained virological response (SVR), which refers to a negative HCV-RNA test 6 mo after cessation of therapy, is defined as a positive treatment response. Rapid viral response (RVR; negative HCV-RNA test 4 wk after treatment) is thought to be a powerful on-treatment predictor of SVR[3,4]. Patients who achieve RVR are more likely to achieve SVR. The treatment response likely depends on a complex host-virus interaction. Many studies have suggested a range of factors that are associated with RVR and SVR, including HCV genotype, viral load, liver function, and host immune status.
The influence of host gene polymorphisms has drawn attention in recent years. Genome-wide association studies (GWASs) have demonstrated that polymorphisms near the IL28B gene, which codes for IFN-λ3, affect the response of CHC to pegylated (PEG)-IFN-α/RBV therapy[5-7]. IFN-λ acts through binding to IL28RA and IL10RB genes, which subsequently activates the Janus kinase-signal transducer and activator of transcription pathway to up- or down-regulate hundreds of genes, such as MxA, OAS1 and PKR, and it is then involved in the immune response pathways[8]. IL10RB and IL28RA gene polymorphisms can predict the natural outcomes of HCV infection in the Chinese population[9]. According to our previous meta-analysis, MxA gene polymorphisms may also be associated with virological response to IFN in the Chinese population[10].
Given that the cost of PEG-IFN treatment is higher than non-PEG-IFN treatment, many patients in Chinese primary hospitals cannot afford PEG-IFN treatment. As a result, non-PEG IFN-α is more commonly used in the treatment of chronic hepatitis C. The previous GWASs were based on observations in Australian, European, African-American and Japanese, but not Chinese populations. Therefore, we aimed to establish pre-treatment predictors for response to non-PEG IFN-α/RBV in Chinese patients to help guide clinical decisions and improve cost-effectiveness. We investigated HCV kinetics during non-PEG IFN-α/RBV therapy, clarified the association of IL28B, IL10RB, IL28RA and MxA gene polymorphisms with RVR to non-PEG IFN-α-2b/RBV therapy, and determined the predictors of RVR in CHC.
Two hundred and fifty-six patients with CHC from Jurong Peoples’ Hospital, China were enrolled in this study, fulfilling the following criteria: (1) treatment naïve; (2) positive for HCV antibody (anti-HCV) and HCV RNA for > 6 mo; and (3) without hepatitis B virus (HBV) or HIV co-infection, or other liver diseases.
All patients were treated for 48 wk with non-PEG IFN-α-2b/RBV and treatment was discontinued according to standard guidelines[11]. Blood samples for biochemical analysis, SNP determination, and HCV genotyping were collected prior to antiviral therapy. HCV-RNA viral load was determined at weeks 0, 4, 12 and 24 of therapy.
Ethical approval was obtained from the participating hospital and the study was carried out in accordance with the guidelines of the International Conference on Harmonization for Good Clinical Practice[12]. All patients gave signed informed consent for DNA genotyping before enrollment.
Serum hepatitis B surface antigen and anti-HCV were measured using an ELISA (Beijing Wantai Biological Pharmacy Engineering Co. Ltd., Beijing, China). Serum HCV RNA and HCV genotype were determined by reverse-transcriptase polymerase chain reaction (TaKaRa Biotechnology, Dalian, China)[13,14].
IL28B rs12980275, IL28RA rs10903035 and rs11249006, MxA rs2071430 and rs17000900, and IL10RB rs2834167 were chosen for genotyping. These SNPs are possibly associated with treatment or natural clearance of HCV[5-7,9,10]. Genomic DNA was isolated from peripheral blood mononuclear cells using protease K digestion and phenol-chloroform purification according to a standard protocol[15]. Genotyping was performed using the ABI TaqMan allelic discrimination assay on the ABI 7900HT sequence Detection System (Applied Biosystems, San Diego, CA, United States)[16]. The primers used for genotyping are listed in Table 1.
| SNPs | Primer and probe (5’-3’) | |
| rs10903035 | Forward primer | TTGCCACCCTTGACCTCAG |
| Reverse primer | GAGGTTTTGTTTAGAGGGATCCAC | |
| Probe-FAM | TAGCAAACCACTCCTT | |
| Probe-HEX | TTAGCAAATCACTCCTT | |
| rs11249006 | Forward primer | AACTGGAAGGGAGAATGGGACT |
| Reverse primer | GTAACATGGCAGGAATCGGACT | |
| Probe-FAM | CCACAACAGTCAACCA | |
| Probe-HEX | CACAACGGTCAACCA | |
| rs2834167 | Forward primer | TACCACCTCCCGAAAATGTCA |
| Reverse primer | GGTGCGTTCCTGCCAATAGT | |
| Probe-FAM | TTCCCTTTGGCAAAAG | |
| Probe-HEX | TTCCCTTCGGCAAAA | |
| rs2071430 | Forward primer | CCGAGAACCTGCGTCTCC |
| Reverse primer | CGCGAAGAAATGAAACTCACAGAC | |
| Probe-FAM | CGTTTCTGCGCCCG | |
| Probe-HEX | CGTTTCTGCTCCCG | |
| rs17000900 | Forward primer | CCGAGAACCTGCGTCTCC |
| Reverse primer | CGCGAAGAAATGAAACTCACAGAC | |
| Probe-FAM | CAAGTGCTGCAGGTG | |
| Probe-HEX | CAAGTGCTGAAGGTG | |
| rs12980275 | Forward primer | TGAGGTGCTGAGAGAAGTCAAATT |
| Reverse primer | CGCTACCCCGGCAAATATT | |
| Probe-FAM | CTAGAAACGGACGTGTC | |
| Probe-HEX | CTAGAAACAGACGTGTCT |
The statistical methods were reviewed by Zhao Yang, Department of Epidemiology and Biostatistics, School of Public Health, Nanjing Medical University. The distribution of patient characteristics and clinical features at baseline between the RVR and non-RVR groups were analyzed by χ2 test. The association of genotypes with RVR were estimated by odds ratio (OR) and 95%CI using univariate and multivariate logistic regression analysis, with adjustment for sex, HCV genotype, HCV-RNA viral load at baseline, alanine aminotransferase (ALT), white blood cell (WBC) count, and α-fetoprotein (AFP). Receiver-operating characteristic (ROC) curves and areas under the curve (AUC) were calculated for the predictive model. Statistical significance between the genotypes and the time of first virus inhibition rates were analyzed using Kaplan-Meier curves and the log-rank test[17]. All statistical analyses were carried out using Stata version 10.0, and P < 0.05 in a two-sided test was considered statistically significant.
The baseline characteristics of the 256 enrolled patients are described in Table 2. Four patients withdrew because of intolerable side effects and 14 were lost to follow-up. A total of 238 patients were screened for analysis. After 4 wk of treatment, 133 patients achieved RVR (55.88%). Treatment response was not related to patient age, sex, or HCV genotype (P > 0.05). Among the tested biochemical indices, levels of ALT, WBCs and AFP differed between the RVR and non-RVR groups. Patients with high ALT/AFP and low WBC levels at baseline were more likely to achieve a worse treatment response.
| Variables | RVR(n = 133) | NRVR(n = 105) | P value |
| Age (yr) | |||
| ≤ 50 | 51 (38.35) | 40 (38.10) | |
| > 50 | 82 (61.65) | 65 (61.90) | 0.968 |
| Sex | |||
| Male | 41 (30.83) | 21 (20.00) | |
| Female | 92 (69.17) | 84 (80.00) | 0.059 |
| HCV genotype | |||
| 1a/1b | 93 (69.92) | 70 (66.67) | |
| 3 | 14 (10.53) | 5 (4.76) | |
| Mixed | 26 (19.55) | 30 (28.57) | 0.102 |
| ALT (U/L) | |||
| ≤ 40 | 64 (48.12) | 28 (26.67) | |
| (40-80] | 38 (28.57) | 42 (40.00) | |
| > 80 | 30 (22.56) | 35 (33.33) | 0.003 |
| ALB (g/L) | |||
| [40-55] | 99 (74.44) | 82 (78.10) | |
| < 40 | 33 (24.81) | 23 (21.90) | 0.577 |
| WBC (× 109/L) | |||
| [4-10] | 75 (56.39) | 73 (69.52) | |
| < 4 | 57 (42.86) | 31 (29.52) | 0.035 |
| > 10 | 1 (0.75) | 1 (0.95) | 0.985 |
| PLT (× 109/L) | |||
| [100-300] | 90 (67.67) | 68 (64.76) | |
| < 100 | 41 (30.83) | 37 (35.24) | 0.522 |
| AFP (ng/mL) | |||
| ≤ 7 | 95 (71.43) | 68 (64.76) | |
| > 7 | 19 (14.29) | 30 (28.57) | 0.016 |
| HGB (g/L) | |||
| [110-150] | 99 (74.44) | 89 (84.76) | |
| < 110 | 13 (9.77) | 8 (7.62) | |
| > 150 | 21 (15.79) | 8 (7.62) | 0.116 |
| Baseline RNA (lg, IU/mL) | 6.02 ± 0.97 | 6.19 ± 0.74 | 0.138 |
To examine the effects of IL28B rs12980275, IL28RA rs10903035 and rs11249006, MxA rs2071430 and rs17000900, and IL10RB rs2834167 on RVR, each SNP was analyzed in four genetic models (co-dominant, dominant, recessive, and additive). The results for all six SNPs are shown in Table 3. P values of all the adjusted factors were < 0.2 in the univariate analysis. Statistical significance in any model was considered to show a potential relationship with treatment response. As shown in Table 4, the distribution of two SNPs appeared to be associated with different treatment responses. In the co-dominant genetic model, mutant G allele of IL-28RA rs11249006 increased RVR (crude OR = 2.75, 95% CI: 1.07-7.07). However, there was no significant difference after adjusting for multiple variables (adjusted OR = 2.33, 95%CI: 0.84-6.48). Mutant G allele of IL28B rs12980275 was associated with decreased RVR in the co-dominant, dominant, and additive models. The adjusted OR was 0.11 (95%CI: 0.04-0.30), 0.19 (95%CI: 0.09-0.43), and 0.43 (95%CI: 0.24-0.75), respectively. The association of IL28B rs12980275 with RVR to IFN-α-2b/RBV therapy was still significant after Bonferroni correction. The results of genetic analyses suggested that IL28B rs12980275 is an indicator of response to IFN therapy.
| Genotype | RVR | NRVR | Crude OR (95%CI) | Adjusted OR (95%CI) |
| rs11249006 | n = 133 | n = 105 | ||
| AA | 51 (38.35) | 49 (46.67) | 1 | 1 |
| AG | 62 (46.62) | 49 (46.67) | 1.22 (0.71-2.09) | 0.86 (0.45-1.63) |
| GG | 20 (15.04) | 7 (6.67) | 2.75 (1.07-7.07) | 2.33 (0.84-6.48) |
| Dominant | 1.41 (0.84-2.36) | 1.06 (0.58-1.94) | ||
| Recessive | 2.48 (1.01-6.11) | 2.52 (0.96-6.64) | ||
| Additive | 1.47 (0.99-2.19) | 1.27 (0.82-1.98) | ||
| rs12980275 | n = 133 | n = 105 | ||
| AA | 118 (88.72) | 70 (66.67) | 1 | 1 |
| AG | 8 (6.02) | 28 (26.67) | 0.17 (0.07-0.39) | 0.11 (0.04-0.30) |
| GG | 7 (5.26) | 7 (6.67) | 0.59 (0.20-1.76) | 0.54 (0.17-1.74) |
| Dominant | 0.25 (0.13-0.50) | 0.19 (0.09-0.43) | ||
| Recessive | 0.78 (0.26-2.29) | 0.82 (0.26-2.56) | ||
| Additive | 0.46 (0.28-0.76) | 0.43 (0.24-0.75) | ||
| rs2834167 | n = 133 | n = 105 | ||
| AA | 37 (27.82) | 28 (26.67) | 1 | 1 |
| AG | 79 (59.40) | 62 (59.05) | 0.96 (0.53-1.74) | 0.90 (0.45-1.78) |
| GG | 17 (12.78) | 15 (14.29) | 0.86 (0.37-2.01) | 0.66 (0.23-1.88) |
| Dominant | 0.94 (0.53-1.68) | 0.85 (0.44-1.66) | ||
| Recessive | 0.88 (0.42-1.86) | 0.71 (0.28-1.83) | ||
| Additive | 0.93 (0.62-1.41) | 0.83 (0.51-1.36) | ||
| rs10903035 | n = 133 | n = 105 | ||
| AA | 43 (32.33) | 27 (25.71) | 1 | 1 |
| AG | 56 (42.11) | 48 (45.71) | 0.73 (0.40-1.36) | 0.68 (0.34-1.37) |
| GG | 34 (32.28) | 30 (28.57) | 0.71 (0.36-1.42) | 0.91 (0.41-2.02) |
| Dominant | 0.72 (0.41-1.28) | 0.76 (0.40-1.44) | ||
| Recessive | 0.86 (0.48-1.53) | 1.14 (0.58-2.25) | ||
| Additive | 0.84 (0.60-1.19) | 0.94 (0.63-1.40) | ||
| rs2071430 | n = 127 | n = 96 | ||
| GG | 63 (49.61) | 49 (51.04) | 1 | 1 |
| GT | 49 (38.58) | 40 (41.67) | 0.95 (0.54-1.67) | 0.94 (0.50-1.80) |
| TT | 15 (11.81) | 7 (7.29) | 1.67 (0.63-4.40) | 2.13 (0.66-6.85) |
| Dominant | 1.14 (0.68-1.91) | 1.26 (0.70-2.28) | ||
| Recessive | 1.78 (0.70-4.54) | 2.28 (0.73-7.08) | ||
| Additive | 1.15 (0.77-1.72) | 1.21 (0.76-1.94) | ||
| rs17000900 | n = 130 | n = 99 | ||
| AA | 97 (74.62) | 69 (69.70) | 1 | 1 |
| AG | 29 (22.31) | 27 (27.27) | 0.76 (0.42-1.40) | 0.79 (0.39-1.61) |
| GG | 4 (3.08) | 3 (3.03) | 0.95 (0.21-4.38) | 0.68 (0.13-3.70) |
| Dominant | 0.83 (0.46-1.47) | 0.83 (0.43-1.62) | ||
| Recessive | 1.05 (0.23-4.82) | 0.77 (0.14-4.16) | ||
| Additive | 0.84 (0.51-1.38) | 0.80 (0.45-1.42) |
| Genotype | RVR (n = 133) | NRVR (n = 105) | Crude OR (95%CI) | P value | Adjusted OR (95%CI) | P value |
| rs11249006 | ||||||
| AA | 51 (38.35) | 49 (46.67) | 1 | 1 | ||
| AG | 62 (46.62) | 49 (46.67) | 1.22 (0.71-2.09) | 0.048 | 0.86 (0.45-1.63) | 0.637 |
| GG | 20 (15.04) | 7 (6.67) | 2.75 (1.07-7.07) | 0.036 | 2.33 (0.84-6.48) | 0.104 |
| Dominant | 1.41 (0.84-2.36) | 0.197 | 1.06 (0.58-1.94) | 0.852 | ||
| Recessive | 2.48 (1.01-6.11) | 0.049 | 2.52 (0.96-6.64) | 0.061 | ||
| Additive | 1.47 (0.99-2.19) | 0.055 | 1.27 (0.82-1.98) | 0.282 | ||
| rs12980275 | ||||||
| AA | 118 (88.72) | 70 (66.67) | 1 | 1 | ||
| AG | 8 (6.02) | 28 (26.67) | 0.17 (0.07-0.39) | < 0.001 | 0.11 (0.04-0.30) | < 0.001 |
| GG | 7 (5.26) | 7 (6.67) | 0.59 (0.20-1.76) | 0.347 | 0.54 (0.17-1.74) | 0.301 |
| Dominant | 0.25 (0.13-0.50) | < 0.001 | 0.19 (0.09-0.43) | < 0.001 | ||
| Recessive | 0.78 (0.26-2.29) | 0.648 | 0.82 (0.26-2.56) | 0.736 | ||
| Additive | 0.46 (0.28-0.76) | 0.002 | 0.43 (0.24-0.75) | 0.003 |
Stepwise regression analysis showed that IL28B rs12980275, WBC count, AFP level, HCV genotype, and HCV-RNA viral load at baseline were independent predictors of RVR (Table 5). In addition, the ROC of these variables covered an AUC of 0.726 (Figure 1). The probability of RVR can be predicted using the following formula: log odds (RVR) = 4.13 + 0.67 × WBC (abnormal vs normal) - 0.98 × AFP (abnormal vs normal) -0.39 × HCV-genotype1-0.46 × log (base viral load) - 1.05 × rs12980275AG/GG.
| Variable | Coef | OR (95%CI) | P value |
| rs12980275 | -1.05 | 0.35 (0.20-0.62) | < 0.001 |
| WBC-group | 0.67 | 1.94 (1.08-3.50) | 0.027 |
| AFP-group | -0.98 | 0.38 (0.19-0.76) | 0.006 |
| HCV genotype | -0.39 | 0.67 (0.48-0.96) | 0.014 |
| Baseline RNA(lg) | -0.46 | 0.63 (0.44-0.91) | 0.027 |
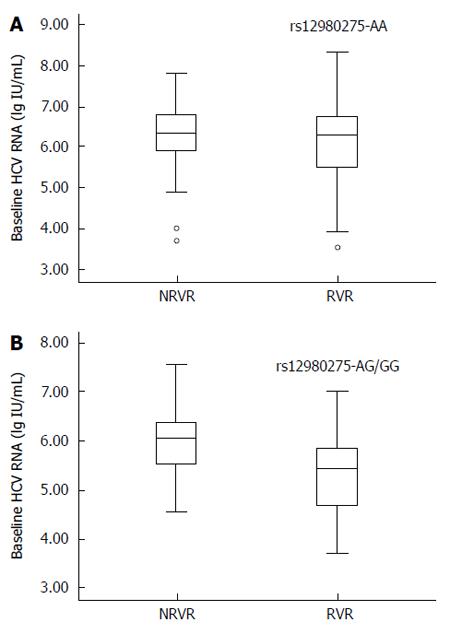
The predictive value of IL28B rs12980275 was further analyzed in stratified analyses. The treatment response in patients with HCV genotype AA was not affected by baseline HCV-RNA viral load. The mean HCV-RNA viral load (log value ± SD) in the non-RVR and RVR groups was 6.28 ± 0.75 lg(copies/mL) and 6.10 ± 0.95 lg(copies/mL), respectively (Figure 2A; P = 0.143). For patients carrying mutant G allele, lower baseline viral load was favored for RVR. The mean viral load in the non-RVR and RVR groups was 5.97 ± 0.67 lg(copies/mL) and 5.37 ± 1.01 lg(copies/mL), respectively (Figure 2B; P = 0.018).
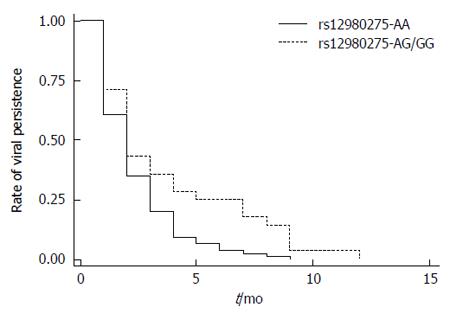
The Kaplan-Meier method and log-rank test were conducted to examine the association of IL28B rs12980275 (dominant model) with the time of initial virus inhibition (time of reaching HCV-RNA viral load < 500 copies/mL after therapy) in CHC patients. Figure 3 shows that the median time of initial inhibition response was 2 mo (95%CI: 1.72-2.28) for the rs12980275 AA group and 2 mo (95%CI: 1.36-2.64) for the AG/GG group. Although similar, the difference was significant (log-rank test, P = 0.004). Also the viral inhibition trends indicated that the inhibition rates were achieved faster in the AA group than in the AG/GG group.
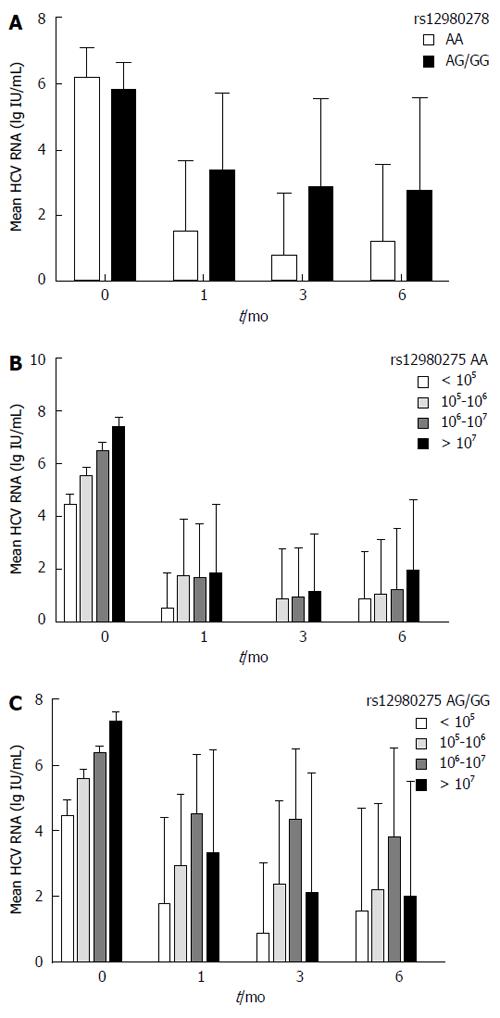
Patients carrying the IL28B rs12980275 AA genotype achieved a greater reduction in HCV-RNA viral load at 1 mo (B-1), 3 mo (B-3) and 6 mo (B-6) than those carrying the AG/GG genotype (B-1: 6.18 ± 0.87 vs 5.77 ± 0.82 log IU/mL, P = 0.003; B-3: 6.18 ± 0.90 vs 5.79 ± 0.80 log IU/mL, P = 0.01; B-6: 6.27 ± 0.86 vs 5.89 ± 0.79 log IU/mL, P = 0.021), respectively (Figure 4A). Considering the confounding effect of baseline HCV-RNA levels, patients were further divided into four groups (baseline HCV RNA < 105 IU/mL, 105-106 IU/mL, 106-107 IU/mL, and ≥ 107 IU/mL). IL28B rs12980275 AA carriers dropped to a similar viral load at 1 mo regardless of the baseline HCV-RNA levels (F = 2.11, P = 0.1) (Figure 4B). Meanwhile, the viral kinetics in the non-AA group were associated with baseline HCV-RNA levels (F = 17.64, P < 0.001). Viral load declined faster in patients with lower baseline level of virus (Figure 4C). The results of viral kinetics were consistent with the stratified analyses (Figure 2).
GWAS studies have identified IL28B rs12980275 as a strong SNP associated with HCV treatment in various populations[5-7,18]. Consistent with those studies, we also found that rs12980275 AA was a strong positive response predictor of non-PEG IFN-α/RBV treatment in the Chinese Han population. In addition, patients carrying the AA genotype were likely to achieve faster virological suppression compared with those carrying non-AA loci. The earliest difference among IL28B rs12979860 genotypes can occur at week 2[19]. The change in viral load seemed to vary in different rs12980275 genotypes (Figure 4). Unlike AG/GG genotypes, the protective effect of the AA genotype was not affected by baseline viral load. These results suggest that the AA genotype is a strong predictor of HCV treatment. The biological reason might be interpreted by another study of HBV infection. Serum IL28B level is higher in patients with the AA genotype and may reduce HBV viral load and liver inflammation[20]. However, the current study did not reveal a significant association between treatment response and polymorphisms in the selected downstream genes of IL28B.
Our results suggest that IL28B rs12980275 is the most important single predictor of RVR by the Random Forest Model (data not shown). In addition, including other viral and host factors, such as baseline viral load, HCV genotype, WBC count and AFP level, improved the accuracy of the predictive model (Figure 1). The predictive model in our study was similar to that in another Japanese study[21].
After 1 mo of treatment, 55.88% of the patients achieved RVR. Since 256 patients (92.02%) were infected with HCV genotype 1, this low rate of efficacy was understandable. The response rates to IFN therapy are usually higher among patients with HCV genotype 2/3, ranging from 75% to 94%, while patients with HCV genotype 1/4 have poorer response rates of about 50%[22,23]. The fact that HCV genotype 1 was the major strain in Jurong was consistent with a previous study[24].
In conclusion, our findings imply that the genetic variants of IL28B rs12980275 may play an important role in determining the response to non-PEG IFN-α-2b/RBV in the Chinese Han population.
We are indebted to the doctors and nurses from the Jurong Peoples’ Hospital for obtaining blood samples for this study.
Hepatitis C virus (HCV) poses a serious global health problem due to its adverse clinical outcomes, such as cirrhosis and hepatocellular carcinoma. Treatment for chronic hepatitis C consists of interferon (IFN) plus ribavirin (RBV) and protease inhibitors such as telaprevir and boceprevir. The treatment response likely depends on a complex host-virus interaction. The influence of host gene polymorphisms has attracted attention in recent years. Therefore, establishing calculable pre-treatment predictors for response to IFN-α/RBV in the Chinese population should guide clinical decisions and improve cost-effectiveness.
Many studies have suggested a range of factors associated with treatment response, including HCV genotype, viral load, liver function, and host immune status. Previous genome wide association studies (GWASs) have demonstrated that polymorphisms near the IL28B gene, which codes for IFN-λ3, affect the response to pegylated (PEG)-IFN-α/RBV in CHC.
Previous GWASs were based on observations in Australian, European, African-American, and Japanese, but not Chinese populations. In addition, because PEG-IFN is more expensive, non-PEG-IFN-α is more commonly used for chronic hepatitis C (CHC) in Chinese primary hospitals. The authors in their previous studies showed that IL10RB and IL28RA gene polymorphisms could predict the natural outcomes of HCV infection in the Chinese population. The present study aimed to clarify the association of IFN-λ-related genes with Rapid viral response to non-IFN-α-2b/RBV therapy in the Chinese Han population.
The results suggest that IL28B rs12980275 AA genotype is a strong predictor of positive response to IFN therapy in the Chinese Han population with CHC, and HCV genotype, baseline levels of white blood cells, α-fetoprotein, and viral load may help predict treatment response.
It is important to know new predictive factors in the treatment of this disease. The study is innovative in nature. The original study conducted on large groups of patients is very valuable.
P- Reviewer: Zielinski J S- Editor: Qi Y L- Editor: Webster JR E- Editor: Wang CH
| 1. | Sievert W, Altraif I, Razavi HA, Abdo A, Ahmed EA, Alomair A, Amarapurkar D, Chen CH, Dou X, El Khayat H. A systematic review of hepatitis C virus epidemiology in Asia, Australia and Egypt. Liver Int. 2011;31 Suppl 2:61-80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 385] [Cited by in F6Publishing: 417] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 2. | Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 589] [Cited by in F6Publishing: 598] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 3. | Gill U, Aziz H, Gill ML. Rapid virological response tailors the duration of treatment in hepatitis C virus genotype 3 patients treated with pegylated interferon alfa-2a and ribavirin in Pakistan. Int J Infect Dis. 2013;17:e1017-e1021. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 4. | Huang CI, Huang CF, Huang JF, Dai CY, Yeh ML, Hsieh MY, Lin ZY, Chen SC, Wang LY, Yu ML. Treatment efficacy of pegylated interferon plus ribavirin therapy in chronic hepatitis C patients with mixed genotype 1/2 infection. J Gastroenterol Hepatol. 2014;29:1012-1018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2776] [Cited by in F6Publishing: 2666] [Article Influence: 177.7] [Reference Citation Analysis (0)] |
| 6. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1505] [Cited by in F6Publishing: 1482] [Article Influence: 98.8] [Reference Citation Analysis (0)] |
| 7. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1747] [Cited by in F6Publishing: 1746] [Article Influence: 116.4] [Reference Citation Analysis (0)] |
| 8. | Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1416] [Cited by in F6Publishing: 1411] [Article Influence: 67.2] [Reference Citation Analysis (0)] |
| 9. | Cui Q, Zhang YX, Su J, Chen X, Ding K, Lei N, Liu Y, Li J, Zhang Y, Yu RB. Genetic variation in IL28RA is associated with the outcomes of HCV infection in a high-risk Chinese population. Infect Genet Evol. 2011;11:1682-1689. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Chen H, Zhang Y, Huang P, Xu Y, Wang J, Su J, Yu R. Host genetic variations are associated with virological response to interferon therapy of chronic HCV in Han Chinese patients. J Biomed Res. 2014;28:476-483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 11. | Chinese Society of Hepatology, Chinese Society of Infectious Diseases and Parasitic Diseases. Prevention guide of hepatitis C. Zhonghua Liuxingbing Xue Zazhi. 2004;25:7. [Cited in This Article: ] |
| 12. | Dixon JR. The International Conference on Harmonization Good Clinical Practice guideline. Qual Assur. 1998;6:65-74. [PubMed] [Cited in This Article: ] |
| 13. | Simmonds P, McOmish F, Yap PL, Chan SW, Lin CK, Dusheiko G, Saeed AA, Holmes EC. Sequence variability in the 5’ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661-668. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 302] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 14. | Choo QL, Richman KH, Han JH, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr PJ. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451-2455. [PubMed] [Cited in This Article: ] |
| 15. | Taniuchi S, Masuda M, Teraguchi M, Ikemoto Y, Komiyama Y, Takahashi H, Kino M, Kobayashi Y. Polymorphism of Fc gamma RIIa may affect the efficacy of gamma-globulin therapy in Kawasaki disease. J Clin Immunol. 2005;25:309-313. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1667] [Cited by in F6Publishing: 1635] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 17. | Goldman AI. The cure model and time confounded risk in the analysis of survival and other timed events. J Clin Epidemiol. 1991;44:1327-1340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Liu T, Sha K, Yang L, Wang Y, Zhang L, Liu X, Yang F. IL-28B polymorphisms correlated with treatment response in HCV-4 mono-infected patients: a meta-analysis. PLoS One. 2014;9:e91316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Rivero-Juárez A, Camacho Espejo A, Perez-Camacho I, Neukam K, Caruz A, Mira JA, Mesa P, García-Lázaro M, Torre-Cisneros J, Pineda JA. Association between the IL28B genotype and hepatitis C viral kinetics in the early days of treatment with pegylated interferon plus ribavirin in HIV/HCV co-infected patients with genotype 1 or 4. J Antimicrob Chemother. 2012;67:202-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | Li W, Jiang Y, Jin Q, Shi X, Jin J, Gao Y, Pan Y, Zhang H, Jiang J, Niu J. Expression and gene polymorphisms of interleukin 28B and hepatitis B virus infection in a Chinese Han population. Liver Int. 2011;31:1118-1126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Ochi H, Hayes CN, Abe H, Hayashida Y, Uchiyama T, Kamatani N, Nakamura Y, Chayama K. Toward the establishment of a prediction system for the personalized treatment of chronic hepatitis C. J Infect Dis. 2012;205:204-210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Yu ML, Chuang WL. Treatment of chronic hepatitis C in Asia: when East meets West. J Gastroenterol Hepatol. 2009;24:336-345. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 200] [Cited by in F6Publishing: 196] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 23. | Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335-1374. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2252] [Cited by in F6Publishing: 2192] [Article Influence: 146.1] [Reference Citation Analysis (1)] |
| 24. | Yue M, Gao CF, Wang JJ, Wang CJ, Feng L, Wang J, Yu RB, Peng ZH, Xue XX, Cai L. Toll-like receptor 7 variations are associated with the susceptibility to HCV infection among Chinese females. Infect Genet Evol. 2014;27:264-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |