Published online May 28, 2014. doi: 10.3748/wjg.v20.i20.6345
Revised: December 15, 2013
Accepted: January 14, 2014
Published online: May 28, 2014
AIM: To clarify the associations between G-protein beta polypeptide 3 (GNB3) C825T polymorphism and risk of the irritable bowel syndrome (IBS) by a meta-analysis.
METHODS: We searched relevant studies in PubMed, EMBASE, CNKI, Google Scholar, Ovid and Cochrane library prior to October 2013. The strengths of the associations between GNB3 C825T polymorphism and IBS risk were estimated by odds ratios (ORs) with 95% confidence interval (CIs).
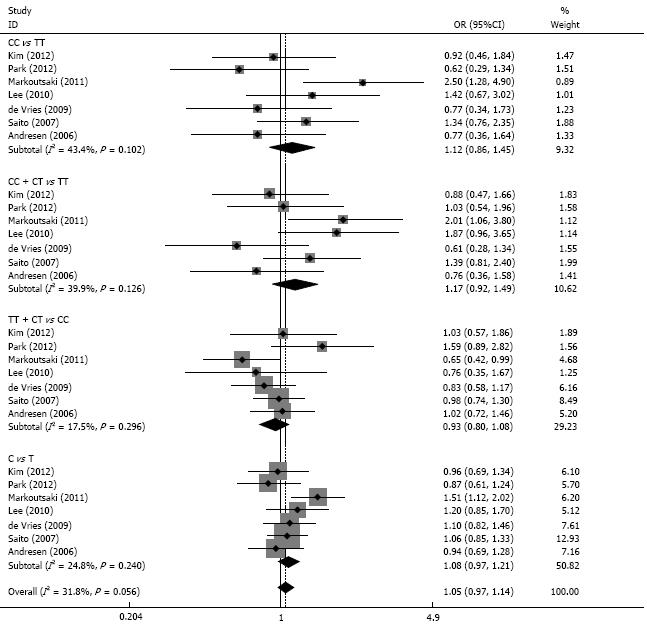
RESULTS: We identified seven case-control studies with 1085 IBS cases and 1695 controls for the analysis. We found no significantly associations of GNB3 C825T polymorphism with IBS risk in the overall population (CC vs TT, OR = 1.12, 95%CI: 0.86-1.45; CC + CT vs TT, OR = 1.17, 95%CI: 0.92-1.49; TT + CT vs CC, OR = 0.93, 95%CI: 0.80-1.08; C vs T, OR = 1.08, 95%CI: 0.97-1.21). Subgroup analysis did not reveal significant associations either in Asian population or Caucasian population. The pooled results of four studies fail to show associations of GNB3 C825T polymorphism with subtypes of IBS (constipation-dominant type, diarrhea-dominant type and mixed type).
CONCLUSION: The present study suggests no associations of GNB3 C825T polymorphism with IBS risk.
Core tip: G-protein beta polypeptide 3 (GNB3) polymorphisms have been identified as an independent risk factor causally associated with functional gastrointestinal disorder and receive extensive interest. However, their association with irritable bowel syndrome (IBS) remains controversial. In the present paper, the authors combined the data from seven case-control studies with 1085 IBS cases and 1695 controls, and found that there were associations between GNB3 C825T polymorphism and IBS risk in the overall population; subgroup analysis did not reveal significant associations either in Asian population or Caucasian population. In regard to the subtypes of IBS (constipation-dominant type, diarrhea-dominant type and mixed type), no associations were found either.
- Citation: Pan ZG, Xiao C, Su DX. No association of G-protein beta polypeptide 3 polymorphism with irritable bowel syndrome: Evidence from a meta-analysis. World J Gastroenterol 2014; 20(20): 6345-6352
- URL: https://www.wjgnet.com/1007-9327/full/v20/i20/6345.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i20.6345
Irritable bowel syndrome (IBS) is a functional gastrointestinal disorder (FGID) of multifactorial origin. Population-based studies show that the prevalence and annual incidence of IBS range from 10%-20% and 1%-2%, respectively[1]. Several factors are associated with IBS, including abnormal gastrointestinal motor function, visceral hypersensitivity, psychological distress (including depression, anxiety, and abuse), immune dysregulation, dietary factors, and family history of IBS[2]. In addition, evidence indicating that IBS cases cluster in families[3] and that identical twins have a greater chance of developing IBS than dizygotic twins[4] suggests that genetic factors also contribute to the pathogenesis of IBS. Accordingly, the genes encoding several proteins, such as serotonin (5-hydroxytryptamine, 5-HT), interleukin (IL)-10, and tumor necrosis factor (TNF)-α, have been proven to be associated with the risk of IBS[5,6].
G-protein was also recently indicated to be another emerging candidate gene for the risk of IBS[5]. G-protein, which comprises an α, a β, and a γ subunit, integrates signals between receptors and effector proteins. The G-protein β polypeptide 3 (GNB3) gene is implicated in the pathogenesis of FGID[7,8] and IBS[6,9,10]. The C825T polymorphism is located in the GNB3 gene encoding the β3 subunit of heterotrimeric G-proteins[11,12]. Furthermore, the T allele of GNB3 C825T is associated with alternative splicing of the gene and the formation of a truncated but functionally active splice variant[13]. Although several studies have investigated the association between GNB3 C825T polymorphism and IBS risk, the results are inconsistent[5,10,14,15]. Such inconsistencies are generally due to ethnic differences or variations in sample size. Therefore, the present meta-analysis aimed to clarify the associations between GNB3 C825T polymorphism and IBS risk.
In order to find all studies investigating the associations between GNB3 polymorphism and IBS risk, we searched the Cochrane Clinical Trials Database, Medline, EMBASE and the Chinese National Knowledge Infrastructure, Google Scholar, the Ovid Library, and conference abstracts prior to October 2013 using the following search terms: “irritable bowel syndrome,”“G-protein beta3 subunit,” and “polymorphism.” The search was not limited by language or publication status. Potentially relevant articles were screened by at least 2 independent reviewers, and disagreements were resolved by discussion or input from a third reviewer if required.
The inclusion criteria were as follows: (1) a study examining the association between GNB3 polymorphism and IBS risk; (2) case-control design; and (3) sufficient information of genotype frequency. In cases of studies with the overlapping data, the most recent one was selected.
The exclusion criteria were as follows: (1) non-case-control design; and (2) genotype frequency or allele frequency not adequately reported/data unable to be obtained by contacting the authors.
Two investigators independently extracted data from the identified publications, including the first author’s name, year of publication, source of publication, IBS diagnostic criteria, genotyping method, numbers of cases and controls, genotype frequency, and allele frequency. Discrepancies in data extraction were resolved by repeating the study review and discussing the results.
Hardy-Weinberg equilibrium (HWE) was applied to the control population to evaluate the data quality, and HWE analysis was carried out using the χ2 test. The respective associations under the allele contrast, codominant, dominant, and recessive models were analyzed. The strengths of the associations between GNB3 polymorphism and IBS risk were estimated according to odds ratios (ORs) with 95% confidence intervals (CIs). The heterogeneity among studies was assessed in a meta-analysis using the Cochran Q test. The inconsistency index I2 was also calculated to quantify heterogeneity[16]. A fixed-effects model (P > 0.05) was used to pool the results if the result of the heterogeneity test was not significant; otherwise, a random-effects model was selected (P < 0.05). Sensitivity analysis was performed by excluding studies that deviated from HWE[17]. Subgroup analyses were carried out with respect to ethnicity and IBS subtype. Publication bias was examined by using the Begg’s test and Egger’s test. All statistical tests were 2-sided and the level of significance was set at P < 0.05. STATA version 11.2 (Stata Corporation, College Station, TX, United States) was used for all analyses.
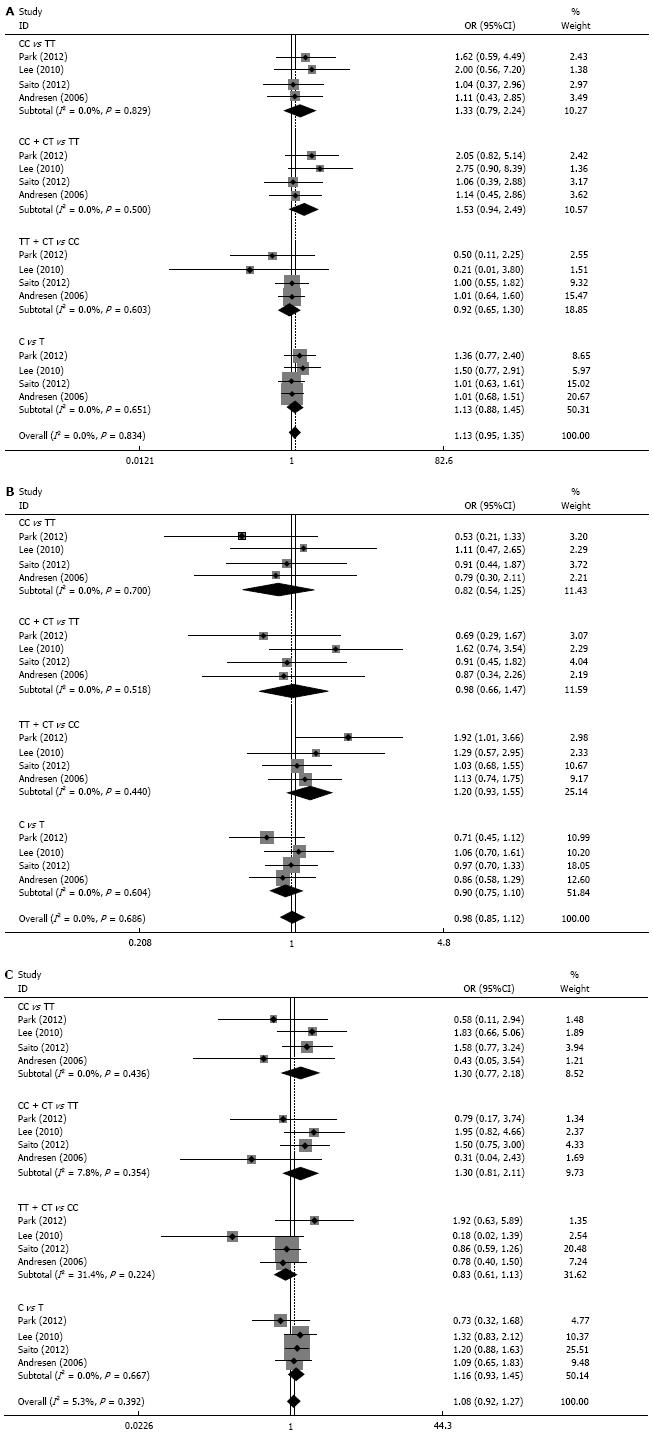
Seven case-control studies[5,6,9,10,14,15,18] with a total of 1085 cases and 1695 controls were finally included in the present meta-analysis. Four of them[10,14,18] specifically investigated the associations between GNB3 C825T polymorphisms and 3 IBS subtypes including the constipation-dominant type (IBS-C), diarrhea-dominant type (IBS-D), and mixed (IBS-M).
Four studies[5,6,10,15] used the Rome III criteria to select IBS patients, while the other 3[9,14,18] used the Rome II criteria. The subjects from 4 studies[6,9,14,18] were Caucasian, while those in the other 3 studies[5,10,15] were Asian. The characteristics of the 7 studies are shown in Table 1.
| Ref. | Year | Country | IBS/controls | Diagnosis criteria | HWE of control |
| Kim et al[15] | 2012 | South Korea | 60/434 | Rome III | 0.850 |
| Park et al[10] | 2012 | South Korea | 72/148 | Rome III | 0.411 |
| Saito et al[18] | 2012 | United States | 385/262 | Rome II | 0.154 |
| Lee et al[5] | 2010 | South Korea | 94/88 | Rome III | 0.010 |
| Markoutsaki et al[6] | 2011 | Greece | 124/238 | Rome III | 0.825 |
| de Vries et al[9] | 2009 | Netherlands | 136/373 | Rome II | 0.099 |
| Andresen et al[14] | 2006 | United States | 214/152 | Rome II | 0.514 |
There were no significant associations between GNB3 C825T and IBS risk among the 4 models (CC vs TT, OR = 1.12, 95%CI: 0.86-1.45; CC+CT vs TT, OR = 1.17, 95%CI: 0.92-1.49; TT+CT vs CC, OR = 0.93, 95% CI: 0.80-1.08; C vs T, OR = 1.08, 95%CI: 0.97-1.21). There was no significant heterogeneity among the 4 models (P > 0.05 for all). The results of the sensitivity analysis were similar to the overall results after excluding 1 study in which the controls deviated from HWE (Figure 1, Table 2). The publication bias test showed little publication bias among the 4 models (P > 0.05 for all).
| GNbeta3 C825T | Pvalue | OR (95%CI) | I2 | Pheterogeneity | Begg’s test | Egger’s test |
| CC vs TT | 0.392 | 1.12 (0.86-1.45) | 43.4 | 0.102 | 0.352 | 0.133 |
| Caucasian | 0.177 | 1.26 (0.90-1.76) | 57.8 | 0.068 | ||
| Asian | 0.750 | 0.93 (0.61-1.42) | 11.7 | 0.322 | ||
| CC + CT vs TT | 0.205 | 1.17 (0.92-1.49) | 39.9 | 0.126 | 0.308 | 0.230 |
| Caucasian | 0.364 | 1.16 (0.84-1.60) | 58.0 | 0.067 | ||
| Asian | 0.374 | 1.18 (0.82-1.70) | 29.6 | 0.241 | ||
| TT + CT vs CC | 0.347 | 0.93 (0.80-1.08) | 17.5 | 0.296 | 1.000 | 0.710 |
| Caucasian | 0.169 | 0.89 (0.75-1.05) | 10.3 | 0.342 | ||
| Asian | 0.456 | 1.14 (0.80-1.65) | 16.0 | 0.304 | ||
| C vs T | 0.157 | 1.08 (0.97-1.21) | 24.8 | 0.240 | 0.756 | 0.548 |
| Caucasian | 0.089 | 1.13 (0.98-1.29) | 43.7 | 0.149 | ||
| Asian | 0.982 | 1.00 (0.82-1.22) | 0.0 | 0.417 |
The studies were divided into subgroups according Caucasian and Asian ethnicity. However, there were no associations among the 4 models of GNB3 C825T polymorphism and IBS risk (all P > 0.05). Next, the 3 subtypes of IBS (i.e., IBS-C, IBS-D, and IBS-M) were analyzed from the data of 4 studies. However, there were no associations between GNB3 C825T polymorphism and the risk of IBS-C, IBS-D, or IBS-M (Figure 2).
The results of the present study indicate that GNB3 C825T polymorphism is not associated with IBS risk regardless of Caucasian or Asian ethnicity. Pooling the data of 4 studies also revealed no associations between GNB3 C825T polymorphism and the 3 subtypes of IBS. The present results are of particular clinical significance because of the inconsistency regarding the associations between GNB3 C825T polymorphism and IBS risk.
G-protein regulates the functions of ion channels and protein kinases, and is essential for stimulus-response coupling in the intracellular system. It acts as the main mediator of signal transport into the cellular system[19]. The GNB3 gene encodes the β3 subunit of heterotrimeric G-proteins and can affect intracellular signal transduction and biological activity. The T allele of GNB3 C825T is associated with enhanced G-protein activation, and the CC genotype results in reduced G-protein translation[20]. In addition, patients with the CC genotype may have decreased immune response to infection and thus require a prolonged recovery period from any infection[11]; this may predispose such individuals to visceral hypersensitivity. The T allele is associated with depression[21], lymphocyte chemotaxis[22], and hypertension[23]. The C allele, which is predictive of diminished G-protein activation, is associated with functional dyspepsia[24]. Furthermore, reports from the USA indicate that meal-unrelated dyspepsia is associated with both the homozygous T and C genotypes[25]. Perturbed gut sensory and motor function, autonomic nervous system dysfunction, and psychiatric disturbances are common causes of IBS; all of these factors can alter intracellular signal transduction[26]. About 80% of all known membrane receptors linked to intracellular effector systems are coupled to G-proteins[27]. Accordingly, changes in G-proteins may lead to functional changes in intracellular signal transduction. Therefore, the GNB3 gene is likely involved in the generation of common pathophysiologic mechanisms of IBS. To date, several studies have investigated the association between GNB3 C825T polymorphism and IBS. However, current findings regarding these associations are controversial and therefore require further study.
Ethnicity is generally one of the causes of discrepancies in the associations between gene polymorphisms and disease risk. In the case of GNB3 C825T polymorphism, the T allele frequency is reported to be higher in Asian populations (42%-53%) than Caucasian populations (27%-42%)[13,28]. Furthermore, inconsistent results regarding the associations between GNB3 C825T polymorphism and IBS risk have been reported in studies with either Caucasian or Asian subjects. Two studies[14,18] involving American populations do not show associations between GNB3 C825T polymorphism and IBS risk. However, 2 other studies[6,9] involving Europeans support such associations. Likewise, Kim et al[15] do not show that GNB3 C825T polymorphism is associated with susceptibility to IBS in a Korean population, whereas 2 other Korean studies[5,10] show that the T allele is associated with IBS risk. In the present study, neither the overall results with the whole population nor the subgroup analysis by Caucasian and Asian ethnicity indicates associations between GNB3 C825T polymorphism and IBS risk. Thus, the results indicate that GNB3 C825T polymorphism may not contribute to the development of IBS.
Because the present results do not appear to support the notion that the disparity is due to ethnic differences, we further analyzed the associations between GNB3 C825T polymorphism and IBS subtypes. There are 4 subtypes of IBS: IBS-C, IBS-D, IBS-M, and unclassified IBS. The prevalence of IBS subtypes varies in the literature[29-31]. Early studies report the association of GNB3 C825T polymorphism with varying susceptibility to IBS subtypes. Lee et al[5] report that the T allele is associated with IBS-C. Park et al[10] found that the CC genotype is common in IBS-D while the TT genotype is common in IBS-C. However, 2 studies[14,18] failed to reveal associations between GNB3 C825T polymorphism and 3 IBS subtypes. In the present study, although the pooled results did not indicate associations with IBS subtypes, it remains difficult to conclude the null association between GNB3 C825T polymorphism and IBS subtypes. We did not analyze the associations with respect to ethnicity, because only 4 studies were included. Thus, additional studies that include data regarding IBS subtypes may produce different results.
To date, no genome-wide association study or comprehensive quantitative assessment has been conducted to determine the genetic association with IBS risk. The present study is the first meta-analysis to clarify the association between GNB3 C825T polymorphism and IBS risk. The large sample size of the present study enhances statistical power to detect more stable associations and provides more reliable estimates than any single previous study. Moreover, associations were assessed by testing 4 different genetic models to obtain more robust and accurate estimations. Furthermore, the low heterogeneity and publication bias demonstrate the precision and robustness of the present results.
Despite its strengths, this study has some limitations that should be mentioned. First, although the present meta-analysis contained a large sample, there were few studies in the subgroup analysis with respect to ethnicity and IBS subtype. Second, the diagnostic criteria for IBS are not uniform; the studies in the meta-analysis employed either the Rome II or III criteria, which inevitably caused selection bias. Third, all Asian subjects were Korean. Therefore, the genetic susceptibility to IBS with respect to GNB3 C825T polymorphism in other Asian ethnicities remains unknown. Fourth, other factors such as gene-environment and gene-gene interactions were not adjusted in the present meta-analysis because of a lack of information; this further confounds the reliability of the present results. Therefore, the present results should be interpreted cautiously.
In summary, the results of the present meta-analysis indicate that there are no associations between GNB3 C825T polymorphism and IBS risk. However, because of the limited available studies, further studies taking ethnicity and IBS subtypes into account are warranted.
The GNB3 gene encodes the β3 subunit of heterotrimeric G-proteins and is able to affect intracellular signal transduction and biological activity. The common causes of irritable bowel syndrome (IBS), such as disturbed gut sensory or motor function, dysfunction of autonomic nervous system and psychiatric disturbance, are all able to alter the intracellular signal transduction.
The current findings of the associations between GNB3 C825T polymorphism and IBS are controversial. The authors conducted a meta-analysis to clarify these associations.
This is the first meta-analysis to investigate the association between IL-10 polymorphisms and IBS risk. The authors showed no associations of GNB3 C825T polymorphism with IBS risk, regardless of Asian or Caucasian ethnicity, and no associations were found in subtypes of IBS.
This study furthers the understanding of the association of GNB3 C825T polymorphism and IBS risk.
This is a good meta-analysis in which authors analyze the associations between GNB3 C825T polymorphism and the risk of IBS. The results are interesting and suggest that no associations were found between GNB3 C825T polymorphism and IBS risk.
P- Reviewers: Aigner F, Rolle U, Tang WF S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Wang CH
| 1. | Drossman DA, Morris CB, Hu Y, Toner BB, Diamant N, Leserman J, Shetzline M, Dalton C, Bangdiwala SI. A prospective assessment of bowel habit in irritable bowel syndrome in women: defining an alternator. Gastroenterology. 2005;128:580-589. [PubMed] [Cited in This Article: ] |
| 2. | van Tilburg MA, Whitehead WE. New paradigm for studying genetic contributions to irritable bowel syndrome. Dig Dis Sci. 2012;57:2484-2486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 3. | Locke GR, Zinsmeister AR, Talley NJ, Fett SL, Melton LJ. Familial association in adults with functional gastrointestinal disorders. Mayo Clin Proc. 2000;75:907-912. [PubMed] [Cited in This Article: ] |
| 4. | Bengtson MB, Rønning T, Vatn MH, Harris JR. Irritable bowel syndrome in twins: genes and environment. Gut. 2006;55:1754-1759. [PubMed] [Cited in This Article: ] |
| 5. | Lee HJ, Lee SY, Choi JE, Kim JH, Sung IK, Park HS, Jin CJ. G protein beta3 subunit, interleukin-10, and tumor necrosis factor-alpha gene polymorphisms in Koreans with irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:758-763. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Markoutsaki T, Karantanos T, Gazouli M, Anagnou NP, Ladas SD, Karamanolis DG. Serotonin transporter and G protein beta 3 subunit gene polymorphisms in Greeks with irritable bowel syndrome. Dig Dis Sci. 2011;56:3276-3280. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Park H. Functional gastrointestinal disorders and overlap syndrome in Korea. J Gastroenterol Hepatol. 2011;26 Suppl 3:12-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 8. | Tahara T, Arisawa T, Shibata T, Wang F, Nakamura M, Sakata M, Hirata I, Nakano H. Homozygous 825T allele of the GNB3 protein influences the susceptibility of Japanese to dyspepsia. Dig Dis Sci. 2008;53:642-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 9. | de Vries DR, ter Linde JJ, van Herwaarden MA, Smout AJ, Samsom M. Gastroesophageal reflux disease is associated with the C825T polymorphism in the G-protein beta3 subunit gene (GNB3). Am J Gastroenterol. 2009;104:281-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 10. | Park CS, Uhm JH. Polymorphisms of the Serotonin Transporter Gene and G-Protein β3 Subunit Gene in Korean Children with Irritable Bowel Syndrome and Functional Dyspepsia. Gut Liver. 2012;6:223-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Baumgart D, Naber C, Haude M, Oldenburg O, Erbel R, Heusch G, Siffert W. G protein beta3 subunit 825T allele and enhanced coronary vasoconstriction on alpha(2)-adrenoceptor activation. Circ Res. 1999;85:965-969. [PubMed] [Cited in This Article: ] |
| 12. | Rosskopf D, Manthey I, Habich C, Kielbik M, Eisenhardt A, Nikula C, Urban M, Kohnen S, Graf E, Ravens U. Identification and characterization of G beta 3s2, a novel splice variant of the G-protein beta 3 subunit. Biochem J. 2003;371:223-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Rosskopf D, Manthey I, Siffert W. Identification and ethnic distribution of major haplotypes in the gene GNB3 encoding the G-protein beta3 subunit. Pharmacogenetics. 2002;12:209-220. [PubMed] [Cited in This Article: ] |
| 14. | Andresen V, Camilleri M, Kim HJ, Stephens DA, Carlson PJ, Talley NJ, Saito YA, Urrutia R, Zinsmeister AR. Is there an association between GNbeta3-C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985-1994. [PubMed] [Cited in This Article: ] |
| 15. | Kim HG, Lee KJ, Lim SG, Jung JY, Cho SW. G-Protein Beta3 Subunit C825T Polymorphism in Patients With Overlap Syndrome of Functional Dyspepsia and Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2012;18:205-210. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39087] [Cited by in F6Publishing: 41743] [Article Influence: 1987.8] [Reference Citation Analysis (1)] |
| 17. | Zintzaras E, Rodopoulou P, Sakellaridis N. Variants of the arachidonate 5-lipoxygenase-activating protein (ALOX5AP) gene and risk of stroke: a HuGE gene-disease association review and meta-analysis. Am J Epidemiol. 2009;169:523-532. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Saito YA, Larson JJ, Atkinson EJ, Ryu E, Almazar AE, Petersen GM, Talley NJ. The role of 5-HTT LPR and GNβ3 825C > T polymorphisms and gene-environment interactions in irritable bowel syndrome (IBS). Dig Dis Sci. 2012;57:2650-2657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 19. | Kleuss C, Scherübl H, Hescheler J, Schultz G, Wittig B. Different beta-subunits determine G-protein interaction with transmembrane receptors. Nature. 1992;358:424-426. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 343] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 20. | Siffert W, Rosskopf D, Siffert G, Busch S, Moritz A, Erbel R, Sharma AM, Ritz E, Wichmann HE, Jakobs KH. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18:45-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 555] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 21. | Zill P, Baghai TC, Zwanzger P, Schüle C, Minov C, Riedel M, Neumeier K, Rupprecht R, Bondy B. Evidence for an association between a G-protein beta3-gene variant with depression and response to antidepressant treatment. Neuroreport. 2000;11:1893-1897. [PubMed] [Cited in This Article: ] |
| 22. | Virchow S, Ansorge N, Rübben H, Siffert G, Siffert W. Enhanced fMLP-stimulated chemotaxis in human neutrophils from individuals carrying the G protein beta3 subunit 825 T-allele. FEBS Lett. 1998;436:155-158. [PubMed] [Cited in This Article: ] |
| 23. | Siffert W. G-protein beta3 subunit 825T allele and hypertension. Curr Hypertens Rep. 2003;5:47-53. [PubMed] [Cited in This Article: ] |
| 24. | Holtmann G, Siffert W, Haag S, Mueller N, Langkafel M, Senf W, Zotz R, Talley NJ. G-protein beta 3 subunit 825 CC genotype is associated with unexplained (functional) dyspepsia. Gastroenterology. 2004;126:971-979. [PubMed] [Cited in This Article: ] |
| 25. | Camilleri CE, Carlson PJ, Camilleri M, Castillo EJ, Locke GR, Geno DM, Stephens DA, Zinsmeister AR, Urrutia R. A study of candidate genotypes associated with dyspepsia in a U.S. community. Am J Gastroenterol. 2006;101:581-592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Corsetti M, Caenepeel P, Fischler B, Janssens J, Tack J. Impact of coexisting irritable bowel syndrome on symptoms and pathophysiological mechanisms in functional dyspepsia. Am J Gastroenterol. 2004;99:1152-1159. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 115] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Lindemann M, Virchow S, Ramann F, Barsegian V, Kreuzfelder E, Siffert W, Müller N, Grosse-Wilde H. The G protein beta3 subunit 825T allele is a genetic marker for enhanced T cell response. FEBS Lett. 2001;495:82-86. [PubMed] [Cited in This Article: ] |
| 28. | Hauner H, Meier M, Jöckel KH, Frey UH, Siffert W. Prediction of successful weight reduction under sibutramine therapy through genotyping of the G-protein beta3 subunit gene (GNB3) C825T polymorphism. Pharmacogenetics. 2003;13:453-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 29. | Guilera M, Balboa A, Mearin F. Bowel habit subtypes and temporal patterns in irritable bowel syndrome: systematic review. Am J Gastroenterol. 2005;100:1174-1184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 101] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 30. | Halder SL, Locke GR, Schleck CD, Zinsmeister AR, Melton LJ, Talley NJ. Natural history of functional gastrointestinal disorders: a 12-year longitudinal population-based study. Gastroenterology. 2007;133:799-807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 204] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 31. | Penny KI, Smith GD, Ramsay D, Steinke DT, Kinnear M, Penman ID. An examination of subgroup classification in irritable bowel syndrome patients over time: a prospective study. Int J Nurs Stud. 2008;45:1715-1720. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |