Published online Nov 7, 2013. doi: 10.3748/wjg.v19.i41.7121
Revised: August 6, 2013
Accepted: August 20, 2013
Published online: November 7, 2013
AIM: To evaluate the suitability of reference genes in gastric tissue samples and cell lines.
METHODS: The suitability of genes ACTB, B2M, GAPDH, RPL29, and 18S rRNA was assessed in 21 matched pairs of neoplastic and adjacent non-neoplastic gastric tissues from patients with gastric adenocarcinoma, 27 normal gastric tissues from patients without cancer, and 4 cell lines using reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR). The ranking of the best single and combination of reference genes was determined by NormFinder, geNorm™, BestKeeper, and DataAssist™. In addition, GenEx software was used to determine the optimal number of reference genes. To validate the results, the mRNA expression of a target gene, DNMT1, was quantified using the different reference gene combinations suggested by the various software packages for normalization.
RESULTS: ACTB was the best reference gene for all gastric tissues, cell lines and all gastric tissues plus cell lines. GAPDH + B2M or ACTB + B2M was the best combination of reference genes for all the gastric tissues. On the other hand, ACTB + B2M was the best combination for all the cell lines tested and was also the best combination for analyses involving all the gastric tissues plus cell lines. According to the GenEx software, 2 or 3 genes were the optimal number of references genes for all the gastric tissues. The relative quantification of DNMT1 showed similar patterns when normalized by each combination of reference genes. The level of expression of DNMT1 in neoplastic, adjacent non-neoplastic and normal gastric tissues did not differ when these samples were normalized using GAPDH + B2M (P = 0.32), ACTB + B2M (P = 0.61), or GAPDH + B2M + ACTB (P = 0.44).
CONCLUSION: GAPDH + B2M or ACTB + B2M is the best combination of reference gene for all the gastric tissues, and ACTB + B2M is the best combination for the cell lines tested.
Core tip: Gene expression studies have revealed much about the molecular basis of gastric cancer. However, the normalization of expression data using reference genes without validation may undermine the results. In the present study, we evaluated the suitability of possible reference genes in gastric tissues and cell lines. To our knowledge, our study is the first to determine and validate reference genes for gastric samples in a Western population. In addition, the inclusion of normal gastric tissues from patients without cancer in determining the best reference genes is original in the literature.
- Citation: Wisnieski F, Calcagno DQ, Leal MF, Santos LCD, Gigek CO, Chen ES, Pontes TB, Assumpção PP, Assumpção MB, Demachki S, Burbano RR, Smith MAC. Reference genes for quantitative RT-PCR data in gastric tissues and cell lines. World J Gastroenterol 2013; 19(41): 7121-7128
- URL: https://www.wjgnet.com/1007-9327/full/v19/i41/7121.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i41.7121
Gastric cancer (GC) is the fourth most common cancer and the second leading cause of cancer-related death[1]. Although approximately 90% of gastric tumors are adenocarcinomas, the etiology and disease evolution may vary among populations, primary tumor location, histological subtypes of adenocarcinoma, and other variables. Among these factors, ethnicity can determine different levels of susceptibility and aggressiveness of gastric tumors[2,3]. An understanding of GC biology is important to identify cancer biomarkers, which may help in early diagnosis and in the development of new targets therapies and, therefore, contribute to reduce mortality or morbidity rates.
Although gene expression studies have revealed much about the molecular basis of GC, the detailed mechanisms remain unclear. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) is currently considered the gold standard for accurate, sensitive, and rapid measurements of gene expression[4-6]. However, to obtain reliable data, the gene expression levels must be normalized using two or more reference genes[7-9]. Ideally, reference genes should be stable, unregulated, and invariable under the conditions of the experiment[10,11]; therefore, a validation experiment for the evaluation of reference gene expression stability for each target tissue and disease is recommended[12,13]. To our knowledge, only one previous study has aimed to assess the best single and combination of reference genes for gastric adenocarcinoma and non-neoplastic samples in an East Asian population[14]. In contrast, there is no information about the stability of candidate reference genes in gastric samples from other populations.
In this study, we assessed the suitability of 5 possible reference genes in 21 matched pairs of neoplastic and non-neoplastic gastric tissues from patients with gastric adenocarcinoma and 27 normal gastric tissues from patients without cancer. We also included 4 cell lines in the analysis. The stability analysis was performed using 4 freely available software packages.
The ACP02 and ACP03 cell lines were established by our research group from primary gastric adenocarcinomas classified as diffuse and intestinal types, respectively[15]. The PG100 and MRC-5 cell lines were obtained from Rio de Janeiro Cell Bank, Brazil, and were established from a primary gastric adenocarcinoma and from normal human fibroblasts, respectively. All the cell lines were cultured at 37 °C in RPMI media 1640 (GIBCO®, Grand Island, NY) supplemented with 10% fetal bovine serum (GIBCO®, Grand Island, NY), and 0.02 mg/mL kanamycin (GIBCO®, Grand Island, NY).
Twenty-one matched pairs of neoplastic and adjacent non-neoplastic gastric tissues were obtained from patients with gastric adenocarcinoma who were subjected to gastric resection. Twenty-seven normal gastric tissues were obtained from patients subjected to routine endoscopic examination. Table 1 shows the clinicopathological features of the studied patients. All the gastric tissue samples were obtained from João de Barros Barreto University Hospital (HUJBB) in Pará State, Brazil, and were snap-frozen in liquid nitrogen and stored frozen until use. All patients had negative histories of exposure to either chemotherapy or radiotherapy before surgery, and there was no other co-occurrence of diagnosed cancers. Written informed consent with approval of the ethics committee of HUJBB was obtained from all patients prior to sample collection.
| Clinicopathological feature | Patients with gastric adenocarcinoma | Patients without gastric adenocarcinoma |
| Age (yr, mean ± SD) | 57 ± 15.6 | 49 ± 14.5 |
| Gender | ||
| Male | 15 (71) | 12 (44) |
| Female | 6 (29) | 15 (56) |
| Location | ||
| Cardia | 2 (9) | 0 (0) |
| Non-cardia | 19 (91) | 27 (100) |
| Histopathological type1 | ||
| Intestinal | 16 (76) | NA |
| Diffuse | 5 (24) | NA |
| Stage2 | ||
| Early | 4 (19) | NA |
| Advanced | 17 (81) | NA |
| Tumor Invasion | ||
| T1/T2 | 9 (43) | NA |
| T3/T4 | 12 (57) | NA |
| Lymph node metastasis | ||
| Absent | 3 (14) | NA |
| Present | 18 (86) | NA |
| Distant metastasis | ||
| Unknown/absent | 18 (86) | NA |
| Present | 3 (14) | NA |
Total RNA was extracted from the cell lines and tissue samples using the AllPrep DNA/RNA/Protein Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The concentration and quality of the extracted RNA were measured using a Nanodrop ND-1000 (Thermo Scientific, Wilmington, DE), and the integrity was determined by gel electrophoresis. The complementary DNA was synthesized using High-Capacity® cDNA Reverse Transcription (Life Technologies, Foster City, CA) following the manufacturer’s protocol.
The reaction to detect the expression range of the 5 candidate reference genes was performed in triplicate using TaqMan® inventoried Assays-on-Demand probes (Life Technologies, Foster City, CA) and the Applied Biosystems 7500 fast real-time PCR system. We also quantified the mRNA expression of a target gene, DNMT1, using the possible candidate genes for normalization. For this analysis, we evaluated 18 matched pairs of adjacent non-neoplastic and neoplastic gastric tissues from patients with gastric adenocarcinoma and 19 normal gastric tissues from patients without cancer. The analyzed genes, their respective TaqMan® assay identification and efficiencies are provided in Table 2. The relative quantification (RQ) of DNMT1 expression was calculated according to the Livak method[16]. A sample from a patient without cancer was designated as a calibrator.
| Symbol | Gene name (Assay ID1) | Location | Description | Efficiency |
| ACTB | β-Actin | 7p22 | Cytoskeletal structural protein | 109% |
| (Hs03023943_g1) | ||||
| B2M | β-2-Microglobulin | 15q21 | Beta-chain of major histocompatibility complex class I molecules | 104% |
| (Hs00984230_m1) | ||||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 12p13 | Oxidoreductase in glycolysis and gluconeogenesis | 102% |
| (Hs99999905_m1) | ||||
| RPL29 | Ribosomal protein L29 | 3p21 | Structural constituent of the ribosome | 100% |
| (Hs00426490_g1) | ||||
| 18S rRNA | 18S ribosomal RNA | 22p12 | Ribosome subunit | 110% |
| (Hs99999901_s1) | ||||
| DNMT1 | DNA (cytosine-5-)-methyltransferase 1 | 19p13 | Regulation of tissue-specific patterns of methylated cytosine residues | 98% |
| (Hs00945875_m1) |
We categorized the gastric tissues and cell lines into the following groups: (1) neoplastic tissues; (2) adjacent non-neoplastic tissues; (3) matched pairs of adjacent non-neoplastic and neoplastic gastric tissues; (4) normal tissues; ( 5) all gastric tissues; (6) cell lines; and (7) all gastric tissues plus cell lines. For the stability comparisons of the candidate reference genes, we used the software NormFinder version 20 (http://www.mdl.dk/publicationsnormfinder.htm)[17], geNorm™ (http://medgen.ugent.be/~jvdesomp/genorm/http://medgen.ugent.be/~jvdesomp/genorm/)[7], BestKeeper1 (http://www.gene-quantification.de/bestkeeper.html)[18], and DataAssist™ (http://www.lifetechnologies.com/us/en/home/technical-resources/software-downloads/dataassist-software.html) according to the recommendations of the authors. The software GenEx (http://genex.gene-quantification.info/) was used to determine the optimal number of reference genes by calculating the Accumulated Standard Deviation (Acc.S.D).
In the analysis using geNorm, the reference genes were ranked according to the expression stability value M (average pair-wise variation of a gene with all other tested candidate reference genes). Using NormFinder, the set of candidate reference genes was ranked according to their expression stability (combination of the intra- and intergroup variation). The ranking of the 5 reference genes by Bestkeeper was based on the standard deviation (SD) and coefficient of variance (CV) expressed as a percentage of the cycle threshold (Ct) level. Lastly, DataAssist provides a metric to measure reference gene stability based on the geNorm algorithm. Unlike all the other programs, DataAssist uses RQ to calculate the stability value of individual candidate reference genes. The two genes that showed the highest stability were considered the best combination of reference genes.
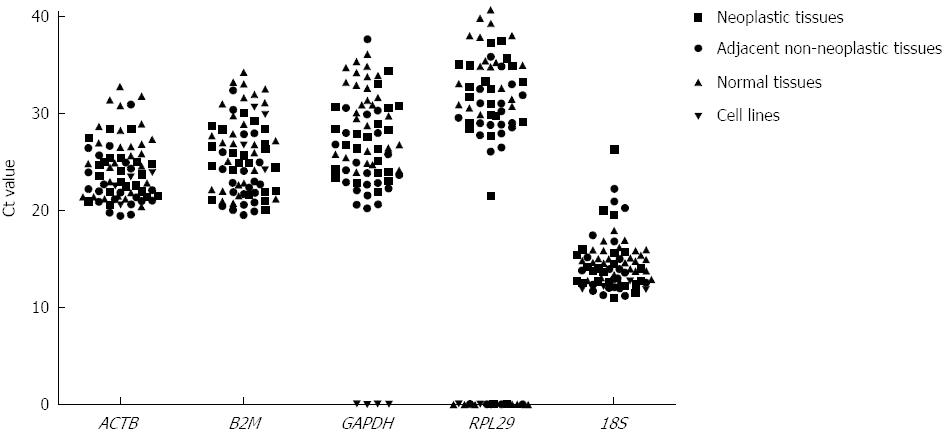
The expression levels of 5 candidate reference genes as the Ct value are shown in Figure 1. These genes displayed a wide range of expression levels. 18S rRNA showed the highest expression level in the gastric tissues and cell lines. In contrast, RPL29 showed the lowest expression level and did not amplify in 3 samples of neoplastic tissue, 2 samples of adjacent non-neoplastic tissue, and 9 samples of normal tissue. Similarly, RPL29 and GAPDH did not amplify in any of the 4 cell lines studied. Therefore, RPL29 was excluded from the ensuing analysis, and GAPDH was excluded from the set of candidate reference genes in the cell line analysis.
Table 3 demonstrates the stability value ranking of the single candidate reference genes calculated using the 4 different software packages. Although the various software packages suggested different reference genes, ACTB was the gene most cited as the best reference gene in the different gastric tissue categories, followed by GAPDH and B2M. ACTB was also the best reference gene in the cell line and all gastric tissues plus cell line categories.
| NormFinder | geNorm™ | BestKeeper | DataAssist™ | |||||
| Stability value1 | Ranking | M value1 | Ranking | Coefficient of variance1 | Ranking | Score1 | Ranking | |
| Neoplastic tissues | ||||||||
| 0.24 | ACTB | 0.07 | GAPDH | 9.78 | ACTB | 1.58 | ACTB | |
| 0.73 | B2M | 0.07 | B2M | 11.77 | B2M | 1.70 | B2M | |
| 0.99 | GAPDH | 0.08 | ACTB | 13.39 | GAPDH | 1.88 | GAPDH | |
| 1.41 | 18S rRNA | 0.12 | 18S rRNA | 17.14 | 18S rRNA | 2.25 | 18S rRNA | |
| Adjacent non-neoplastic tissues | ||||||||
| 0.61 | B2M | 0.05 | GAPDH | 7.80 | ACTB | 1.65 | B2M | |
| 0.65 | GAPDH | 0.05 | B2M | 9.61 | B2M | 1.77 | ACTB | |
| 0.65 | ACTB | 0.07 | ACTB | 10.52 | GAPDH | 1.81 | GAPDH | |
| 1.53 | 18S rRNA | 0.13 | 18S rRNA | 16.57 | 18S rRNA | 2.46 | 18S rRNA | |
| Matched pairs of adjacent non-neoplastic and neoplastic gastric tissues | ||||||||
| 0.14 | ACTB | 0.06 | GAPDH | 9.24 | ACTB | 1.83 | GAPDH | |
| 0.29 | B2M | 0.06 | B2M | 11.31 | B2M | 1.83 | ACTB | |
| 0.41 | GAPDH | 0.07 | ACTB | 12.52 | GAPDH | 2.00 | B2M | |
| 0.54 | 18S rRNA | 0.13 | 18S rRNA | 16.86 | 18S rRNA | 2.60 | 18S rRNA | |
| Normal gastric tissues | ||||||||
| 0.38 | GAPDH | 0.06 | GAPDH | 7.38 | 18S rRNA | 1.82 | GAPDH | |
| 0.41 | ACTB | 0.06 | ACTB | 11.08 | GAPDH | 2.24 | ACTB | |
| 1.07 | B2M | 0.07 | B2M | 11.46 | ACTB | 2.40 | B2M | |
| 2.24 | 18S rRNA | 0.11 | 18S rRNA | 13.34 | B2M | 3.91 | 18S rRNA | |
| All gastric tissues | ||||||||
| 0.14 | ACTB | 0.07 | GAPDH | 10.89 | ACTB | 2.04 | ACTB | |
| 0.36 | B2M | 0.07 | B2M | 13.12 | B2M | 2.09 | B2M | |
| 0.69 | GAPDH | 0.08 | ACTB | 13.24 | 18S rRNA | 2.12 | GAPDH | |
| 0.96 | 18S rRNA | 0.13 | 18S rRNA | 13.43 | GAPDH | 3.34 | 18S rRNA | |
| Cell lines | ||||||||
| 0.53 | ACTB | 0.06 | ACTB | 2.36 | 18S rRNA | 1.71 | ACTB | |
| 1.55 | B2M | 0.06 | B2M | 4.81 | ACTB | 2.45 | B2M | |
| 1.73 | 18S rRNA | 0.13 | 18S rRNA | 8.60 | B2M | 2.63 | 18S rRNA | |
| All gastric tissues + cell lines | ||||||||
| 0.45 | ACTB | 0.09 | ACTB | 10.67 | ACTB | 2.73 | B2M | |
| 1.11 | B2M | 0.09 | B2M | 13.02 | B2M | 3.32 | ACTB | |
| 1.19 | 18S rRNA | 0.16 | 18S rRNA | 13.41 | 18S rRNA | 3.38 | 18S rRNA | |
Table 4 shows the best combination of reference genes suggested by the 4 software packages. Overall, for the different gastric tissue categories, GAPDH + B2M were the genes more cited as the best combination of reference gene, followed by ACTB + B2M and GAPDH + ACTB. ACTB + B2M was also the best combination of reference genes suggested for the cell lines and all gastric tissues plus cell line categories.
| Neoplastic tissues | Adjacentnon-neoplastic tissues | Normal tissues | Matched pairs of adjacent nonneoplastic and neoplastic gastric tissues | All gastric tissues | Cell lines | All gastric tissues+cell lines |
| ACTB + B2M | GAPDH + B2M | GAPDH + ACTB | ACTB + B2M | ACTB + B2M | ACTB + B2M | ACTB + 18S rRNA |
| GAPDH + B2M | GAPDH + B2M | GAPDH + ACTB | GAPDH + B2M | GAPDH + B2M | ACTB + B2M | ACTB + B2M |
| GAPDH + B2M | GAPDH + B2M | GAPDH + B2M | GAPDH + B2M | GAPDH + B2M | ACTB + B2M | ACTB + B2M |
| ACTB + B2M | GAPDH + B2M | GAPDH + B2M | GAPDH + B2M | GAPDH + B2M | ACTB + B2M | ACTB + B2M |
Although the software indicated up to 2 genes as the best combination of reference genes, we also used GenEx software to determine the optimal number of reference genes. This software revealed that an Acc.S.D. of 0.03 was the lowest when 2 or 3 reference genes were used for both the matched pairs of adjacent non-neoplastic and neoplastic gastric tissues and all gastric tissues categories.
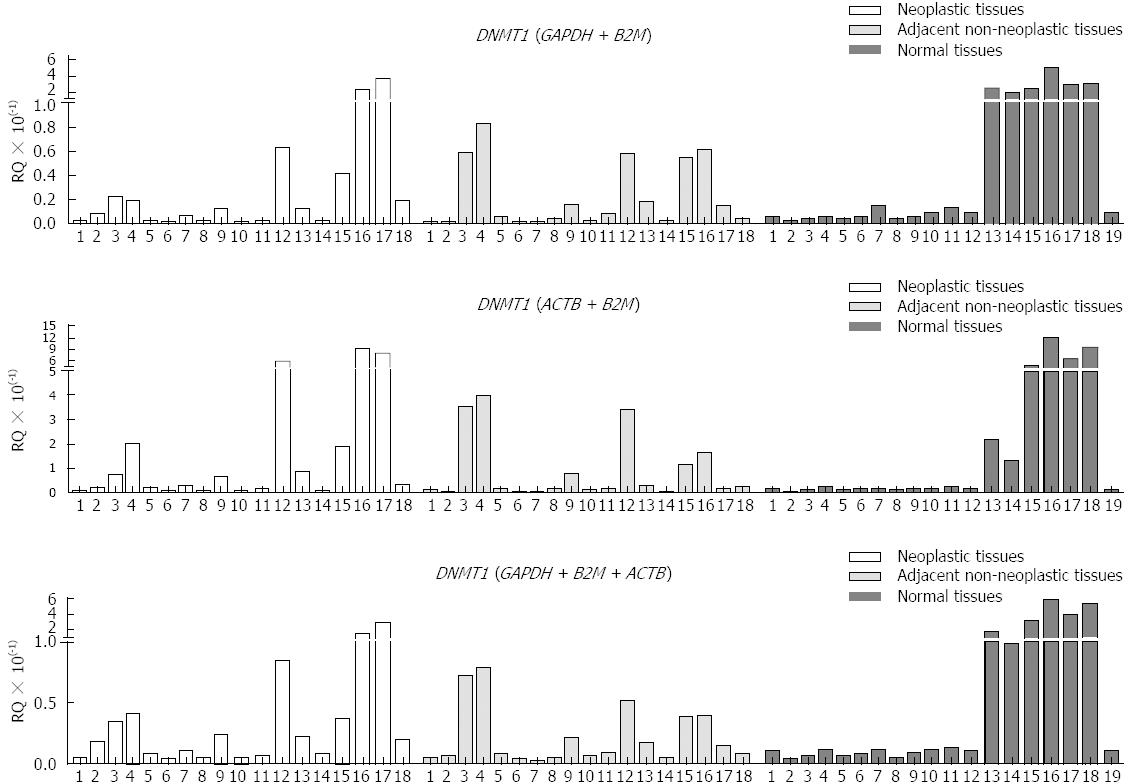
Because GAPDH + B2M and ACTB + B2M or GAPDH + B2M + ACTB were identified as the best combinations of references genes for gastric tissues, we evaluated the expression of DNMT1, as normalized by these combinations of reference genes. The RQ of DNMT1 normalized by each combination of reference genes showed similar patterns (Figure 2). The level of expression of DNMT1 in neoplastic, adjacent non-neoplastic and normal gastric tissues did not differ when these samples were normalized using GAPDH + B2M (P = 0.32, Kruskal-Wallis test), ACTB + B2M (P = 0.61, Kruskal-Wallis test), or GAPDH + B2M + ACTB (P = 0.44, Kruskal-Wallis test).
Reference genes have been described for RT-qPCR studies in several diseases and tissues[19-23]. However, with regard to gastric adenocarcinoma samples, there is only one previous study that evaluated the best single and combination of reference genes in an East Asian Population[14]. Because ethnicity can determine different levels of gene expression, it is important to determine the suitability of reference genes considering the population in addition to the disease type and target tissue. To our knowledge, our study is the first to determine and validate reference genes for gastric samples in a Western Population. The population in Pará State, Brazil, is composed of interethnic crosses between three main groups: European (mainly represented by Portuguese), Africans, and Amerindians[24]. In addition, to our knowledge, the study of normal gastric tissue from patients without gastric cancer and its inclusion in determining the best single and combination of reference genes is original in the literature.
The software packages NormFinder, geNorm™, BestKeeper, and DataAssist™ are statistical tools that aid in the selection of appropriate reference genes. Although these software packages differed in the suggestion of the best single and combination of reference genes, at least two programs agreed with the results for each group evaluated (Tables 3 and 4), emphasizing the importance of using more than one software to assess the best reference genes among a set of candidate genes. When considering all gastric tissues, our results showed that ACTB and GAPDH + B2M or ACTB + B2M were the best single and combination of reference genes, respectively. Despite the software packages indicating up to 2 genes as the best combination, the GenEx software revealed that 2 or 3 reference genes were necessary for gene normalization in all gastric tissue. When the expression of the target gene DNMT1 was evaluated using the 3 different combinations of reference genes for normalization (GAPDH + B2M, ACTB + B2M, and GAPDH + B2M + ACTB), no differences in DNMT1 expression were detected among the neoplastic, adjacent non-neoplastic, and normal tissues. These results validated the combination of reference genes suggested by the software, proving that combinations of 2 genes can be used and that it is not necessary to use 3 or more reference genes for all gastric tissues. ACTB and ACTB + B2M were the best single and combination of reference genes, respectively, for all the cell lines. Our results showed that ACTB + B2M was the best option under circumstances that require the use of the same combination of reference genes for all gastric tissues and cell lines. Although the measure of stability for 18S rRNA was within the range of acceptance when using Bestkeeper, it has repeatedly been documented that this is not a good reference gene because the regulation of its synthesis is not representative of mRNA levels[25-28].
Rho et al[14] proposed different reference genes for the study of gene expression in gastric tissues and cell lines, suggesting RPL29 and RPL29 + B2M and B2M and GAPDH + B2M as the best single and combination of reference genes, respectively. Interestingly, the genes suggested by Rho et al[14], RPL29 and GAPDH, did not amplify in our cell lines and in some tissue samples. The different methodologies applied can explain the different results. In the present study, we evaluated gene expression using commercially available TaqMan® assays, whereas Rho et al[14] evaluated gene expression using SYBR green and primers previously reported in the literature that can detect non-specific reaction products with variable sensitivity. In addition, it should be considered that samples obtained from different ethnicities could contribute to the different results of our group and Rho et al[14].
In conclusion, our suitability analysis suggested ACTB and GAPDH + B2M or ACTB + B2M as the best single and combination of reference genes for all gastric tissues, with ACTB and ACTB + B2M as the best single and combination of reference genes for all cell lines tested. When circumstances require the use of the same combination of reference genes for all gastric tissues and cell lines, our results showed that ACTB + B2M was the best option. The use of these genes for RT-qPCR data normalization may enhance the robustness of transcription level determination in gastric samples.
Gastric cancer is the fourth most common cancer worldwide, with high rates of mortality and morbidity. Reverse transcription quantitative polymerase chain reaction is currently considered the gold standard for the accurate, sensitive, and rapid measurement of gene expression. To obtain reliable data, a validation experiment to evaluate the best reference genes for the normalization of gene expression data is recommended for each target tissue and disease.
The etiology and disease evolution of gastric adenocarcinomas vary among patients due to several factors. Among them, ethnicity can determine different levels of gastric tumor susceptibility and aggressiveness. The understanding of gastric cancer biology is important to identify cancer biomarkers, which may help in the early diagnosis and development of new targets therapies and, therefore, contribute to reduce mortality and morbidity rates.
Only one previous study aimed to evaluate the best reference genes for gastric adenocarcinoma in an East Asian population. To their knowledge, the present study is the first to determine and validate reference genes for gastric samples in a Western population. In addition, the analysis of normal gastric tissue from patients without gastric cancer and its inclusion in determining the best reference genes is original in the literature.
The use of the combination of reference genes determined and validated in our study for reverse transcriptional quantitative polymerase chain reaction data normalization may enhance the robustness of transcription level determination in gastric samples.
Reference genes are internal controls used in reverse transcription quantitative polymerase chain reaction analysis to avoid the sample biases related to variability in the total RNA content, RNA stability, and enzymatic efficiency. Ideal reference genes should be stable, unregulated, and invariable under the conditions of the experiment.
The authors evaluated the suitability of five possible reference genes in matched pairs of non-neoplastic and neoplastic gastric tissues from patients with gastric adenocarcinoma and normal gastric tissues from patients without cancer. Four cell lines were also included in this analysis. The stability analysis was performed using four freely available software packages. This study validated GAPDH + B2M or ACTB + B2M as the best combination of reference genes for all gastric tissues. In addition, ACTB + B2M were suggested as the best combination of reference genes for cell lines. When circumstances require the use of the same combination of reference genes for all gastric tissues and cell lines, the ACTB + B2M combination was found to be the best option.
P- Reviewer: Guo JM S- Editor: Zhai HH L- Editor: A E- Editor: Liu XM
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (3)] |
| 2. | Crew KD, Neugut AI. Epidemiology of gastric cancer. World J Gastroenterol. 2006;12:354-362. [PubMed] [Cited in This Article: ] |
| 3. | Shah MA, Ajani JA. Gastric cancer--an enigmatic and heterogeneous disease. JAMA. 2010;303:1753-1754. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 4. | Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169-193. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2695] [Cited by in F6Publishing: 2537] [Article Influence: 105.7] [Reference Citation Analysis (0)] |
| 5. | Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR--a perspective. J Mol Endocrinol. 2005;34:597-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 885] [Cited by in F6Publishing: 807] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 6. | Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50:227-230. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 493] [Cited by in F6Publishing: 463] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 7. | Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. [PubMed] [Cited in This Article: ] |
| 8. | de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS One. 2007;2:e898. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 524] [Cited by in F6Publishing: 549] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 9. | Ho-Pun-Cheung A, Bascoul-Mollevi C, Assenat E, Bibeau F, Boissière-Michot F, Cellier D, Ychou M, Lopez-Crapez E. Validation of an appropriate reference gene for normalization of reverse transcription-quantitative polymerase chain reaction data from rectal cancer biopsies. Anal Biochem. 2009;388:348-350. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Li YL, Ye F, Hu Y, Lu WG, Xie X. Identification of suitable reference genes for gene expression studies of human serous ovarian cancer by real-time polymerase chain reaction. Anal Biochem. 2009;394:110-116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 80] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Ohl F, Jung M, Xu C, Stephan C, Rabien A, Burkhardt M, Nitsche A, Kristiansen G, Loening SA, Radonić A. Gene expression studies in prostate cancer tissue: which reference gene should be selected for normalization? J Mol Med (Berl). 2005;83:1014-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 161] [Cited by in F6Publishing: 178] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 12. | Bustin SA, Mueller R. Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci (Lond). 2005;109:365-379. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 310] [Cited by in F6Publishing: 309] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 13. | Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, Borghi L, Verbrugghe P, Kalaydjieva L, Bleuler S, Laule O. RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics. 2011;12:156. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 210] [Cited by in F6Publishing: 208] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 14. | Rho HW, Lee BC, Choi ES, Choi IJ, Lee YS, Goh SH. Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qPCR. BMC Cancer. 2010;10:240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 15. | Leal MF, Martins do Nascimento JL, da Silva CE, Vita Lamarão MF, Calcagno DQ, Khayat AS, Assumpção PP, Cabral IR, de Arruda Cardoso Smith M, Burbano RR. Establishment and conventional cytogenetic characterization of three gastric cancer cell lines. Cancer Genet Cytogenet. 2009;195:85-91. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117419] [Cited by in F6Publishing: 121468] [Article Influence: 5281.2] [Reference Citation Analysis (0)] |
| 17. | Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245-5250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5406] [Cited by in F6Publishing: 4894] [Article Influence: 244.7] [Reference Citation Analysis (0)] |
| 18. | Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3334] [Cited by in F6Publishing: 3306] [Article Influence: 165.3] [Reference Citation Analysis (0)] |
| 19. | Rubie C, Kempf K, Hans J, Su T, Tilton B, Georg T, Brittner B, Ludwig B, Schilling M. Housekeeping gene variability in normal and cancerous colorectal, pancreatic, esophageal, gastric and hepatic tissues. Mol Cell Probes. 2005;19:101-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 207] [Cited by in F6Publishing: 220] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 20. | Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ. Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer. 2008;8:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 79] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 21. | Fu J, Bian L, Zhao L, Dong Z, Gao X, Luan H, Sun Y, Song H. Identification of genes for normalization of quantitative real-time PCR data in ovarian tissues. Acta Biochim Biophys Sin (Shanghai). 2010;42:568-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Shen Y, Li Y, Ye F, Wang F, Lu W, Xie X. Identification of suitable reference genes for measurement of gene expression in human cervical tissues. Anal Biochem. 2010;405:224-229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 23. | Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med. 2012;126:943-952. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 111] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 24. | Batista dos Santos SE, Rodrigues JD, Ribeiro-dos-Santos AK, Zago MA. Differential contribution of indigenous men and women to the formation of an urban population in the Amazon region as revealed by mtDNA and Y-DNA. Am J Phys Anthropol. 1999;109:175-180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 25. | Solanas M, Moral R, Escrich E. Unsuitability of using ribosomal RNA as loading control for Northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal Biochem. 2001;288:99-102. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 109] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 26. | Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293-300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 421] [Cited by in F6Publishing: 449] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 27. | Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A. Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun. 2004;313:856-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1176] [Cited by in F6Publishing: 1146] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 28. | Mogal A, Abdulkadir SA. Effects of Histone Deacetylase Inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Mol Cell Probes. 2006;20:81-86. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 29. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] [Cited in This Article: ] |
| 30. | Washington K. 7th edition of the AJCC cancer staging manual: stomach. Ann Surg Oncol. 2010;17:3077-3079. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 702] [Cited by in F6Publishing: 801] [Article Influence: 61.6] [Reference Citation Analysis (0)] |