Published online Aug 14, 2013. doi: 10.3748/wjg.v19.i30.4867
Revised: June 4, 2013
Accepted: July 4, 2013
Published online: August 14, 2013
Hepatitis B virus (HBV) infection is a global public health problem that causes persistent liver diseases such as chronic hepatitis, cirrhosis, and hepatocellular carcinoma. A large amount of people die annually from HBV infection. However, the pathogenesises of the HBV-related diseases are ill defined and the therapeutic strategies for the diseases are less than optimum. The recently discovered microRNAs (miRNAs) are tiny noncoding RNAs that regulate gene expression primarily at the post-transcriptional level by binding to mRNAs. miRNAs contribute to a variety of physiological and pathological processes. A number of miRNAs have been found to play a pivotal role in the host-virus interaction including host-HBV interaction. Numerous studies have indicated that HBV infection could change the cellular miRNA expression patterns and different stages of HBV associated disease have displayed distinctive miRNA profiles. Furthermore, the differential expressed miRNAs have been found involved in the progression of HBV-related diseases, for instance some miRNAs are involved in liver tumorigenesis and tumor metastasis. Studies have also shown that the circulating miRNA in serum or plasma might be a very useful biomarker for the diagnosis and prognosis of HBV-related diseases. In addition, miRNA-based therapy strategies have attracted increasing attention, indicating a promising future in the treatment of HBV-related diseases.
Core tip: The cellular microRNAs (miRNAs) involved in host-hepatitis B virus (HBV) interaction and each stage of HBV-related disease show distinctive miRNA expression profiles at the tissue or serum level indicating that miRNAs have marked potential in detecting or treating of HBV infection.
- Citation: Wei YF, Cui GY, Ye P, Chen JN, Diao HY. MicroRNAs may solve the mystery of chronic hepatitis B virus infection. World J Gastroenterol 2013; 19(30): 4867-4876
- URL: https://www.wjgnet.com/1007-9327/full/v19/i30/4867.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i30.4867
The hepatitis B virus (HBV) is a hepadnavirus that causes persistent liver diseases and have a major effect on global public health[1,2]. HBV, discovered in 1966[3], is transmitted among humans by contact with the blood, semen or vaginal fluid of an infected person. Approximately, one third of the world’s population have infected HBV, and more than 350 million people have developed chronic HBV infection[4-6]. The severity of HBV-related disease varies widely, from a self-limited infection to acute hepatitis and from asymptomatic chronic infection to cirrhosis and hepatocellular carcinoma[7,8]. The factors affecting the prognosis of HBV infection have not been determined. miRNAs was discovered recently and researchers have determined that it plays a pivotal role in host-virus interactions[9-11]. By using the functions of miRNA, we may explain the mechanism of chronic HBV infection and discover novel biomarkers as well as new therapies for HBV associated diseases.
Since numerous researches discovered that RNA does more than simply serves an intermediary function in “central dogma”[12], the door to a brand new world of RNA had been opened. The genomes of organisms produce two types of RNA, and mRNAs belong to the first type which can be used as translation templates. Besides, genomes manufacture a variety of noncoding RNAs, including the components of the machinery of gene expression and regulatory RNAs[13]. MicroRNAs (miRNAs) are noncoding RNAs, and their mature forms are approximately 22 nucleotides (nt) in length. When these RNAs were initially described in Caenorhabditis elegans (C. elegans)[14], they were hypothesized to be peculiar to nematodes[12,13]. Subsequent work revealed that miRNAs are common tiny nucleic acid molecules that can be found in plants[15], animals[16] and other organisms[17]. To date, the record of miRNAs has increased significantly. MiRbase 19, released in August of 2012, increased the numbers of recognized hairpin and mature miRNAs to 21264 and 25141, respectively[18-23]. In human, while the expression profiles of some miRNAs in different cells or tissues are similar, other miRNAs may exhibit temporal or tissue-specific patterns[24,25], suggesting that miRNA may be involved in numerous physiological or pathological processes[26].
The biogenesis and action mechanism of these tiny but potential molecules had been detailed described[24,25,27]. Briefly, they are not born so small, in other words, they have some larger progenitors. The processing of the mature miRNA ancestors (primary and precursor miRNAs) is closely related to RNA polymerase II (pol II), Drosha, the GTP-dependent Ran/Exportin 5 complex, and the Dicer enzyme[24-32]. Generally, by binding to the 3’ untraslated regions of their target mRNAs, miRNAs can serve as gene expression regulators, fine-tune the expression primarily at the post-transcriptional level and play critical roles in a variety of physiological and pathological processes, including antiviral defense, developmental timing, cell apoptosis, cell proliferation, tumor generation and so on[24,30,33-39]. One computational prediction indicated that more than 30% of animal genes may be subject to regulation by miRNAs, which emphasizes the importance of miRNA-mediated gene regulation[40,41].
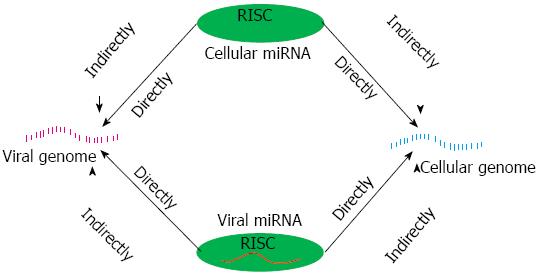
Viruses are generally harmful to human, in order to protect our health, the battle between virus and host break out shortly after infection initiated. In this war, a large amount of reports have indicated that cellular miRNAs serve a key role in protecting the host. However, we may be disappointed at the truth that viruses can use miRNAs as their weapons to fight the host. Remarkably, some features of miRNAs ensure their effectiveness as virally encoded regulators of host and viral gene expression: they are small, lack of immunogenicity and functional flexibility[42]. To facilitate an understanding of the intricacies of host-virus cross-talk mediated by miRNAs, we designed an illustration (Figure 1) base on the review of Scaria et al[9] In the interaction between virus and host, miRNAs can be divided into cellular miRNAs and viral miRNAs. To cellular miRNAs, their expression profiles changed at the infected state and the abnormal miRNAs often closely relate to the viral life cycle as well as host disorder. To viral miRNAs, they can evolved to regulate both viral and cellular gene expression[42].
Studies have noted that miRNA-mediated gene regulation involve in diverse biological processes in the mammalian system, including cellular miRNAs influence viral reproduction and pathogenesis[42,43]. Sometimes, viruses may exploit celluar miRNAs to facilitate certain steps of their life cycle, a living example is hepatitis C virus (HCV) use miR-122, a liver-specific cellular miRNA, to enhance its replication of itself by targeting the viral 5’ non-coding region[34,44]. Another study showed that miR-122 knockdown reduced the HCV load in infected chimpanzees[45] and the interferon-mediated down-regulation of miR-122 that contributes to antiviral effects[46]. In contrast, miR-122 serve as an antiviral role in HBV life cycle. For instance, Qiu et al[47] found that the miR-122 over-expression inhibited HBV expression, whereas the depletion of endogenous miR-122 resulted in increased production of HBV in transfected cells. Their subsequent study suggested that the miR-122 inhibitor also caused an increase in cellular heme oxygenase-1, which can decrease HBV covalently closed circular DNA (cccDNA) levels both in vitro and in vivo by reducing the stability of the HBV core protein[48]. A recent study by Wang et al[1], indicated that miR-122 expression in the liver was significantly down-regulated in patients with HBV infection compared with healthy controls. Depletion of endogenous miR-122 and over-expression of miR-122 led to enhanced HBV replication and inhibited viral production, respectively. Cyclin G1 was identified as an miR-122 target that specifically interacted with p53, resulting in the specific binding of p53 to the HBV enhancer elements and simultaneous abrogation of the p53-mediated inhibition of HBV transcription. Ji et al[49] found that miR-122 was significantly up-regulated in HBV-infected patients and could inhibit HBV replication in Huh7 and HepG2 cells. Overall, to HCV and HBV, miR-122 can promote and inhibit viral replication respectively. In other words, cellular miRNAs can influence viral lifecycles by accelerative or suppressive mechanisms.
Studies have reported the involvement of cellular miRNAs in numerous host-virus interactions. HIV-1 can use cellular miRNAs to repress the expression of viral proteins and evade the host immune system respose[11,50]. The replication of primate foamy virus can be inhibited by cellular miR-32[43]. miR-24 and miR-93 were responsible for the increased vesicular stomatitis virus replication in variant Dicer1d/d allele mice[51]. The above instances indicate the diversity of miRNA activity and indicate that host-derived miRNAs are essential for the host-virus interactions.
A number of the miRNAs that participate in the interaction between host and virus are viral. Pfeffer et al[52] initially discovered the existence viral miRNAs in the Epstein-Barr virus (EBV). Analogous to cellular miRNAs, viral miRNAs have multifaceted functions[42], that generally benefit the virus in maintaining its replication, latency and evasion of the host immune system[11]. Barth et al[53] showed that miR-BART2 down-regulates the viral DNA polymerase BALF5, inhibiting the transition from latent to lytic viral replication in EBV. Analogously, miR-BART-1p, miR-BART16 and miR-BART17-5p have been found to repress the translation of latency-associated membrane protein LMP-1 mRNA[11,54]. Additional examples of viral miRNAs that regulate viral gene expression are found in HCMV, SV40, MDV, HIV-1 and other viruses[11].
Although numerous miRNA-produced viruses have been identified, the HBV-encoded miRNAs have not been confirmed experimentally but have been suggested by computation[55,56]. This discrepancy may be the result of the limitations of current technology and HBV-derived miRNAs could be found in the future.
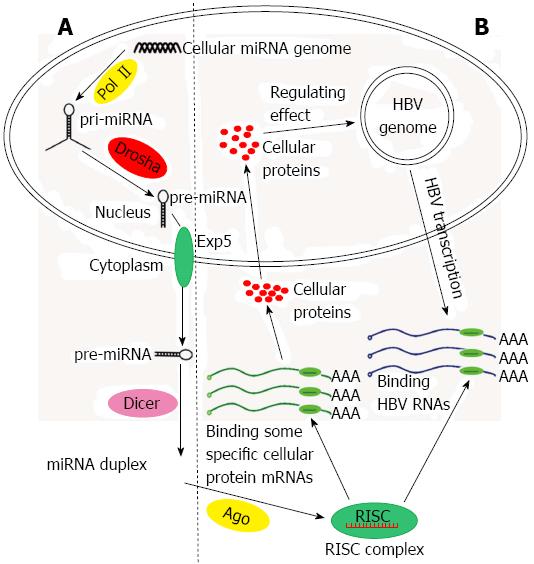
A number of cases of host-virus interaction at the miRNA level have been mentioned above. To emphasize the role of miRNAs in HBV infection, we intend to report additional details about the interaction between miRNAs and HBV (Figure 2).
Understanding the mechanisms of miRNAs influence HBV infection requires the knowledge that HBV is a noncytopathic virus that replicates preferentially in the hepatocytes. cccDNA which serves as a template for transcription of all viral RNA is synthetized And after HBV DNA enters the hepatocyte nucleus. The HBV genome is 3.2 kb in length and contains four overlapping open reading frames. It can transcribe viral pregenomic RNA that reverses transcription to synthesize the viral DNA genome and encode the hepatitis B virus surface antigen (HBsAg), hepatitis B virus core protein, viral reverse DNA polymerase (Pol) and X protein. Two enhancers, I and II, have been shown to function as two master regulators of the four viral promoters[57-59]. Although the viral miRNAs encoded by HBV have not been verified[56], there are cellular miRNAs capable of inhibiting or stimulating HBV viral replication and gene expression. In addition, the products of HBV can alter the miRNA expression profiles.
A study of Zhang et al[60], in attempt to determine whether host-encoded miRNAs affect HBV replication, antisense oligonucleotides of 328 identified human miRNAs were orderly transfected into HepG22.2.15 cells. The expression level of HBsAg, hepatitis B e antigen and cell proliferation were detected by enzyme-linked immunosorbent assay and methyltestosterone assay. Compared to the experimental controls, miR-199a-3p and miR-210 efficiently reduced the HBsAg expression without affecting HepG2 2.2.15 cell proliferation. Furthermore, they used the bioinformatics method to analyze six miRNAs, and the outcome suggested a putative binding site for miR-199a-3p in the HBsAg coding region and a binding site for miR-210 in the HBV pre-S1 region, respectively. Potenza et al[61] used MiRanda to analyze the HBV genome and found seven sites that were potential targets for human liver miRNAs. Their subsequent validation test found that hsa-miR-125a-5p interferes with the HBV translation and down-regulation of the expression of the surface antigen. These findings indicate that cellular miRNAs can alter HBV gene expression by targeting to HBV transcripts.
Cellular miRNAs can affect viral translation and change viral replication. In addition to the instance of the miR-122 inhibition of HBV replication, there are other cases about host miRNAs altering HBV replication. A study by Hu et al[33] suggested that miR-141 suppressed HBV replication by reducing HBV promoter activities through the down-regulation of peroxisome proliferator- activated receptor alpha. DNA hypermethylation might be closely related to the suppression of HBV cccDNA transcription[56], and miR-152 might be a factor involved in the regulation of the methylation of HBV cccDNA[62,63]. Zhang et al[64] revealed that cellular miRNAs do not consistently inhibit HBV replication. Collectively, miRNAs can directly or indirectly alter HBV replication.
A recently study by Wei et al[65] showed that the hepatitis B virus x (HBx) protein expression was found to have a significant inverse correlation with miR-101 expression in HBx-expressing HepG2 cells compared to control HepG2 cells. Ren et al[66] found that Drosha (a regulator of the biogenesis of miRNAs) mRNA and protein expression were down-regulated in cells expressing the HBV genome, and that the mechanism was related to a reduction in the activity of the Drosha gene promoter. By using RNA interference to knockdown the HBX gene, the expression of Drosha was significantly restored. Their date showed that HBV could inhibit Drosha expression by inhibiting the promoter activity and in turn, leading to an alteration of the host miRNA profiles[66]. These studies suggested that HBV infection can alter the miRNA expression profiles.
The consequences of HBV infection are diverse and can be ranged from asymptomatic chronic infection to cirrhosis and hepatocellular carcinoma. Numerous studies have detected that cellular miRNAs could influence the lifecycle of HBV and HBV could change the miRNA expression profiles, reversely. Taking these factors into consideration, the miRNA profiles may change along with the severity of HBV associated disease. So, we concentrated on the miRNAs expression patterns and their potential role in HBV associated chronic hepatitis, cirrhosis and HCC in the following contents.
The miRNA profiles of chronic hepatitis B (CHB) from numerous studies are controversial and complicated. On the one hand, a series of study indicated that the miRNA expression patterns of CHB are particular at the tissue or serum level[1,67-69]. For instance, a study of Ura et al[68] suggested that the miRNA expression profiles in chronic hepatitis B were different from those in the healthy controls and those in HBV-associated HCC, and hepatitis C. To the contrary, appliying massively parallel signature sequencing to conduct an in-depth analysis of the miRNomes in normal human, hepatitis and HCC liver tissues, Hou et al[70] found that, except for in HCC, the known miRNAs exhibited a similar distribution in each library based on classification of the transcripts permillion degrees.
A well-known trilogy of hepatitis B is that chronic hepatitis B progresses into liver cirrhosis and HCC. An increasing number of studies have focused on the expression patterns of miRNAs during the cirrhotic stage to uncover their function in the progression of hepatitis B and to seek novel therapies for cirrhosis. Roderburg et al[71] investigated the role of miRNAs in liver fibrosis by carbon tetrachloride and bile duct ligation models of liver fibrosis. Fibrosis-inducing injuries cause the abnormal expression of many miRNAs. All three members of the miR-29 family were significantly down-regulated under the disposes of these models. To correlate these findings with HBV in human, they measured the miRNA profiles of human liver samples, and found miR-29 family members were down-regulated in the fibrotic/cirrhotic tissues compared with the non-fibrotic tissues. In conclusion, miR-29 family members were down-regulation both in mouse models and in human fibrotic livers. Hepatic stellate cells (HSCs) play a key role in liver fibrosis[72,73]. Roderburg group’s further study revealed that miR-29b was down-regulated in HSCs, upon exposure to fibrotic stress. On a cellular level, miR-29 down-regulation in murine HSC cells was mediated by transforming growth factor (TGF)-β as well as inflammatory signaling and nuclear factor κB (NF-κB). Forced expression of miR-29b in murine HSCs can result in the repression of collagen expression[71].
Additional studies report on miRNA regulation in the progression of liver fibrosis. Compared with quiescent HSCs, Lakner et al[73] verified that miR-19b was a regulator of TGF-β signaling in activated HSCs, it play an inhibitory effect in HSC-mediated fibrogenesis. Another study suggested that liver fibrosis could cause the down-regulation of miR-150 and miR-194 in HSC, and that their over expressions could repress HSC activation.
Chronic hepatitis B is closely relate to HCC. In recent years, numerous studies that focused on the miRNA profiling in HBV-related HCC identified a number of deregulated miRNAs which are critical for the generation of HCC. Gao et al[74] isolated miRNAs from formalin fixed paraffin embedded dysplastic nodules (DNs), small HCCs, and their corresponding nontumorous livers. They investigated the expression changes of seven cancerrelated miRNAs, which have been reported to be frequently deregulated in human cancers and might play a role in liver carcinogenesis. They frequently observed the down-regulation of miR-145 and miR-199b as well as the up-regulation of miR-244 in premalignant DNs, moreover these alterations persisted throughout the HCC development. By restoring miR-145 in both HepG2 and Hep3B HCC cells, they found that cell proliferation, cell migration and cell invasion were significantly inhibited. What’s more, an anti-miR-145 inhibitor could impair these inhibitory functions of miR-145. This study suggested that miRNA deregulation was an early event and may accumulate throughout the generation of HBV-associated HCC[74]. A study from Hou et al[70] identified miR-199a/b-3p which consistently decreased in HCC, and its decrement significantly correlates with poor survival of HCC patients. Huang et al[63] suggested that miR-152 was aberrantly expressed and involved in the regulation of the abnormal DNA methylation status in HBV-related HCC.
Interestingly, one miRNA was found to be up-regulated and contribute to enhancing HBV-related HCC metastasis by repressing the expression of fibronectin[75]. Zhang et al[75] reported that the levels of miR-143 were significantly increased in p21-HBx transgenic mice and HCC patients with metastatic HBV-HCC. Furthermore, they found that the over-expression of miR-143 was transcribed by NF-κB and facilitates the invasive and metastatic behavior of liver tumor cell. In an athymic nude mouse model, they found that high levels of miR-143 administered by intratumoral administration could remarkably promote HCC metastasis. And they used p21-HBx transgenic mice to show in vivo that local liver metastasis and distant lung metastasis were significantly inhibited by blocking miR-143. What’s more, fibronectin type III domain containing 3B was identified in vivo and in vitro as the target of miR-143[75].
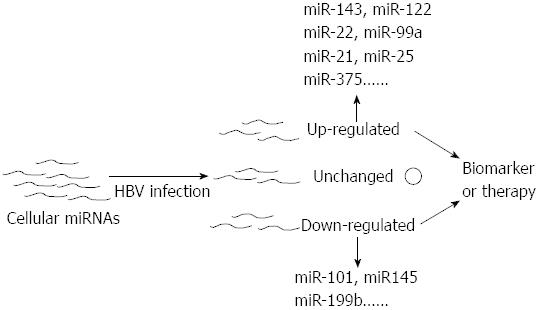
The expression profiles of miRNAs in different stages of HBV-associated diseases are always inconsistent. Moreover, a portion of miRNAs are closely related to the stage of this liver disease and often play a crucial role in their progression[68,69,71,73-75]. Therefore miRNAs can serve as the role of biomarker in the diagnosis of HBV-related disease (Figure 3). Studies have reported that miRNAs could be stably detected in plasma and serum[76-78]. Chen et al[77] demonstrated that miRNAs could be found in the plasma and serum of humans and that their levels in serum were stable, reproducible, and consistent among individuals of the same species. In their study, Solexa was employed to sequence all of the serum miRNAs of healthy Chinese subjects and to identify specific expression patterns of serum miRNAs for lung cancer, colorectal cancer, and diabetes. They validated two non-small cell lung cancer-specific serum miRNAs in an independent trail using quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays. These results showed the existence of human serum miRNAs and suggested that these miRNAs contain fingerprints for diverse diseases[77]. Hence, assaying miRNA profiles could become a novel approach for detecting HBV-related diseases.
More powerful biomarkers are needed to compensate for the defects of the existing diagnostic means for detecting HBV-related liver injury and HCC[69,79-83]. In blood samples, alanine aminotransferase (ALT) and aspartate aminotransferase (AST) are the most widely used enzymatic indicators for liver damage[84]. But enhanced ALT and AST activities have been detected in some other clinical disorders[81,82]. In clinical practice, these two markers are not always consistent with histomorphological alterations[80]. One of the reasons for the high mortality in HBV-related HCC is that the tumors are frequently identified after metastasis at a stage in which curative resection is no longer feasible[69]. We rely on radiology imaging methods such as ultrasonography, computed tomography, and magnetic resonance imaging to find a liver mass to diagnose HCC. These methods can not diagnose small lesions accurately[69]. The most commonly used serum HCC markers, α-fetoprotein (AFP), has insufficient sensitivity and specificity[69,85,86].
Collectively, there is an urgent need for novel strategies in the detection of HBV-related disease, and miRNAs could become a novel and powerful biomarker.
Several recent reports suggested that miRNAs could be used as an indicator of liver injury and HBV infection[49,67,84,87]. Zhang et al[84] selected and validated miRNA biomarkers using an extensive set of plasma samples from patients with HBV infection, patients with skeletal, and healthy controls. Combining these experimental results with their further investigation in liver injury mouse models, these authors reported that the plasma miR-122 concentration presented a disease severity-dependent change in the patients and mouse models that earlier than the alteration in aminotransferase activity. Their findings suggested that miR-122 had potential as a blood marker for liver injury including HBV associated injury[84]. Waidmann et al[67] investigated the relationship between miR-122 and HBV infection, suggested that the serum levels of miR-122 can discriminate between HBV infected patients and healthy controls. Ji et al[49] found that the numbers of circulating miRNAs increased with the symptom severity of HBV infected patients and that the expression of miR-122 was significantly up-regulated in these patients.
miRNAs other than miR-122 had been reported to get the ability of indicating HBV infection. Hayes et al[87] found a number of disease-specific serum miRNAs of HBV infection, including miR-122, miR-22, and miR-99a which were up-regulated at least 1.5-fold in the serum of HBV-infected patients.
Early diagnosis of HCC plays a vital role in reducing mortality, but the existing strategies are not effective. A number of miRNAs had been found to have the potential to become the diagnostic and prognostic markers of HCC[69,88-94].
Tomimaru et al[69] measured the plasma miR-21 levels of different subjects including HCC patients and chronic hepatitis patients. In their study, plasma miR-21 was significantly reduced after a curative resection in HCC, and the level in the HCC subjects was significantly higher than the levels in the patients with chronic hepatitis and healthy controls. These authors found that miR-21 could differentiate HCC from healthy controls with high sensitivity and specificity. In theory, miR-21 was superior to AFP in the diagnosis of HCC. In a study with several phases, Qi et al[93] found that the serum miR-122 level was significantly higher in HCC patients compared to healthy controls and post-operative subjects. Their findings indicate that miR-122 might serve as a novel biomarker for the detection of HCC in healthy subjects but is not useful for the detection of HCC in patients with chronic HBV infections[93].
Li et al[92], employed Solexa sequencing to screen and qRT-PCR to validate miRNAs in serum samples. Thirteen miRNAs were found that could accurately distinguish not only HBV cases from healthy and HCV individuals, but also HBV-postive HCC subjects from healthy and HBV subjects. Additionally, in a comparison of miRNA expression in the serum of HCC subjects and healthy controls, six miRNAs were found to be significantly elevated in the samples from HCC. Three miRNAs (miR-25, miR-375, and let-7f) can be used to separate HCC cases from healthy controls. In the prediction of HCC, miR-375 had an ROC of 0.96 (specificity: 96%; sensitivity: 100%).
Although the outcomes of these studies are not uniform, the data have shown that miRNAs are promising for detecting HCC or HBV-positive HCC. A number of reports have indicated that the expression of miRNAs could anticipate the prognosis of HCC[89,91]. Using Kaplan-Meier estimates and the log-rank test, Li et al[91] showed that high expression of has-miR-125b was related to good survival and a subsequent transfection assay showed that forced expression of miR-125b in the HCC cell line could perceptibly repress the cell growth and phosphorylation of Akt. Budhu et al[89] created a unique 20-miRNA metastasis signature that could significantly predict HCC tissues with venous metastases from metastasis-free solitary tumors with a 10-fold cross-validation. In the corresponding noncancerous liver tissues they could not identify significant miRNAs. A survival risk prediction analysis revealed that the majority of metastasis-related miRNAs were related to survival. Their additional validation experiments revealed that the 20-miRNA tumor signature could serve as a survival and relapse predictor of HCC[89].
Although miRNAs have significant potential, a number of problems remain. Too many miRNAs have been identified to be practically applied for routine clinical use, and the accuracy of the miRNA signatures has not been adequately evaluated[95]. These factors may result in inaccuracy or incorrect diagnosis and prediction outcomes.
The closely relationship between miRNAs and HBV-related diseases offers an opportunity to use miRNAs or antagomir in the treatment of these diseases (Figure 3). The feasibility of this method has been demonstrated[96-99]. Grimm et al[100] showed that anti-HBV shRNAs might cause serious toxicity in vivo. Although a miRNA-based strategy is promising, its therapeutic application must be dependent on rigorously demonstrated safety, efficient delivery to target tissues and optimization shRNAs dosing and sequencing[100,101]. To obtain an optimal solution for a miRNA-based strategy, Keck et al[102] produced improved HBV RNAi triggers, Ely et al[103] designed pri-miRNA expression cassettes and linear DNA sequences that expressed antiviral micro-RNA shuttles[104], and Xiangji et al[105] developed a lentiviral miRNA-based system. Improved miRNA-based therapeutic methods could successfully inhibit HBV replication or expression. A promising miRNA-based HBV therapy method has not been well established but could be designed successfully in the future.
In this review, we limited our focus to the role of miRNAs in host-virus interactions, especially in host-HBV interactions. HBV infection is a global issue, but the pathogenesises and therapies of HBV-related diseases are not well defined. In the years since miRNA was discovered in C. elegans and subsequent studies revealed that miRNA are involved in many physiological and pathological processes in humans, scientists have observed that miRNAs played a key role in viral diseases and could serve a guardian or aggressor role. Regarding to HBV infection, cellular miRNAs were found to influence HBV translation and replication and HBV was found to change expression profiles of cellular miRNA. This finding led to the possibilities of miRNAs serving as biomarkers and of miRNAs or antagomirs serving as therapeutic tools in HBV-related diseases (Figure 3). Studies have indicated that the blood or tissue samples from the different stages of HBV-related disease presented distinctive miRNA expression patterns and that miRNA-based therapy is feasible.
Although many experimental studies have confirmed the capacity of miRNAs or antagomirs to detect or treat HBV-related diseases, adequate evaluation of their accuracy, efficacy, and cost-effectiveness is required. Further research into the relationship between miRNAs and chronic HBV infection may increase the understanding of hepatitis B virus infection and miRNAs could become accurate biomarkers and powerful therapy tools.
P- Reviewers Anand BS, Utama A, Zhang Y S- Editor Gou SX L- Editor A E- Editor Li JY
| 1. | Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730-741. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in F6Publishing: 201] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 2. | Maddrey WC. Hepatitis B: an important public health issue. J Med Virol. 2000;61:362-366. [PubMed] [Cited in This Article: ] |
| 3. | Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733-1745. [PubMed] [Cited in This Article: ] |
| 4. | Zuckerman JN, Zuckerman AJ. Current topics in hepatitis B. J Infect. 2000;41:130-136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 74] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 5. | Lavanchy D. Worldwide epidemiology of HBV infection, disease burden, and vaccine prevention. J Clin Virol. 2005;34 Suppl 1:S1-S3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 334] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 6. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 596] [Cited by in F6Publishing: 595] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 7. | Bläckberg J, Kidd-Ljunggren K. Occult hepatitis B virus after acute self-limited infection persisting for 30 years without sequence variation. J Hepatol. 2000;33:992-997. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 120] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 8. | Chu CM. Natural history of chronic hepatitis B virus infection in adults with emphasis on the occurrence of cirrhosis and hepatocellular carcinoma. J Gastroenterol Hepatol. 2000;15 Suppl:E25-E30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 126] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Scaria V, Hariharan M, Pillai B, Maiti S, Brahmachari SK. Host-virus genome interactions: macro roles for microRNAs. Cell Microbiol. 2007;9:2784-2794. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3:68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 157] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 11. | Ghosh Z, Mallick B, Chakrabarti J. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009;37:1035-1048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 12. | Dennis C. The brave new world of RNA. Nature. 2002;418:122-124. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 55] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 13. | Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823-826. [PubMed] [Cited in This Article: ] |
| 14. | Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673-676. [PubMed] [Cited in This Article: ] |
| 15. | Llave C, Kasschau KD, Rector MA, Carrington JC. Endogenous and silencing-associated small RNAs in plants. Plant Cell. 2002;14:1605-1619. [PubMed] [Cited in This Article: ] |
| 16. | Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336-338. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1353] [Cited by in F6Publishing: 1241] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 17. | Papenfort K, Vogel J. Regulatory RNA in bacterial pathogens. Cell Host Microbe. 2010;8:116-127. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 247] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 18. | Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152-D157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2680] [Cited by in F6Publishing: 2696] [Article Influence: 192.6] [Reference Citation Analysis (0)] |
| 19. | Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154-D158. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2784] [Cited by in F6Publishing: 3132] [Article Influence: 184.2] [Reference Citation Analysis (0)] |
| 20. | Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140-D144. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3282] [Cited by in F6Publishing: 3401] [Article Influence: 188.9] [Reference Citation Analysis (0)] |
| 21. | Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109-D111. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1612] [Cited by in F6Publishing: 1594] [Article Influence: 79.7] [Reference Citation Analysis (0)] |
| 22. | Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M. A uniform system for microRNA annotation. RNA. 2003;9:277-279. [PubMed] [Cited in This Article: ] |
| 23. | Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20:3186-3190. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 971] [Cited by in F6Publishing: 873] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 24. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] [Cited in This Article: ] |
| 25. | Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38 Suppl:S25-S30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 321] [Cited by in F6Publishing: 340] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 26. | Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 261] [Cited by in F6Publishing: 274] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 27. | Cullen BR. Derivation and function of small interfering RNAs and microRNAs. Virus Res. 2004;102:3-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 54] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 28. | Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597-610. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3165] [Cited by in F6Publishing: 3434] [Article Influence: 245.3] [Reference Citation Analysis (0)] |
| 29. | He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4964] [Cited by in F6Publishing: 5118] [Article Influence: 255.9] [Reference Citation Analysis (0)] |
| 30. | Baulcombe D. RNA silencing in plants. Nature. 2004;431:356-363. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1827] [Cited by in F6Publishing: 1534] [Article Influence: 76.7] [Reference Citation Analysis (0)] |
| 31. | Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjärvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol. 2005;79:9556-9565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 252] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 32. | Sano M, Kato Y, Taira K. Sequence-specific interference by small RNAs derived from adenovirus VAI RNA. FEBS Lett. 2006;580:1553-1564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 33. | Hu W, Wang X, Ding X, Li Y, Zhang X, Xie P, Yang J, Wang S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One. 2012;7:e34165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 34. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1993] [Cited by in F6Publishing: 1929] [Article Influence: 101.5] [Reference Citation Analysis (0)] |
| 35. | Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119-129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 130] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 36. | Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 709] [Cited by in F6Publishing: 649] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 37. | Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513-520. [PubMed] [Cited in This Article: ] |
| 38. | Ambros V. The functions of animal microRNAs. Nature. 2004;431:350-355. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7919] [Cited by in F6Publishing: 8282] [Article Influence: 414.1] [Reference Citation Analysis (0)] |
| 39. | Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151-1164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 306] [Cited by in F6Publishing: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 40. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8847] [Cited by in F6Publishing: 9072] [Article Influence: 477.5] [Reference Citation Analysis (0)] |
| 41. | Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3534] [Cited by in F6Publishing: 3551] [Article Influence: 186.9] [Reference Citation Analysis (0)] |
| 42. | Gottwein E, Cullen BR. Viral and cellular microRNAs as determinants of viral pathogenesis and immunity. Cell Host Microbe. 2008;3:375-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 317] [Cited by in F6Publishing: 342] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 43. | Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557-560. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 686] [Cited by in F6Publishing: 686] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 44. | Roberts AP, Jopling CL. Targeting viral infection by microRNA inhibition. Genome Biol. 2010;11:201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 45. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1284] [Cited by in F6Publishing: 1262] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 46. | Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 686] [Cited by in F6Publishing: 727] [Article Influence: 42.8] [Reference Citation Analysis (0)] |
| 47. | Qiu L, Fan H, Jin W, Zhao B, Wang Y, Ju Y, Chen L, Chen Y, Duan Z, Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem Biophys Res Commun. 2010;398:771-777. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 128] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 48. | Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hösel M, Schirmacher P, Tiegs G. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology. 2007;133:1156-1165. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 49. | Ji F, Yang B, Peng X, Ding H, You H, Tien P. Circulating microRNAs in hepatitis B virus-infected patients. J Viral Hepat. 2011;18:e242-e251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 118] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 50. | Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med. 2007;13:1241-1247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 583] [Cited by in F6Publishing: 612] [Article Influence: 36.0] [Reference Citation Analysis (0)] |
| 51. | Otsuka M, Jing Q, Georgel P, New L, Chen J, Mols J, Kang YJ, Jiang Z, Du X, Cook R. Hypersusceptibility to vesicular stomatitis virus infection in Dicer1-deficient mice is due to impaired miR24 and miR93 expression. Immunity. 2007;27:123-134. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 274] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 52. | Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C. Identification of virus-encoded microRNAs. Science. 2004;304:734-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1199] [Cited by in F6Publishing: 1176] [Article Influence: 58.8] [Reference Citation Analysis (0)] |
| 53. | Barth S, Pfuhl T, Mamiani A, Ehses C, Roemer K, Kremmer E, Jäker C, Höck J, Meister G, Grässer FA. Epstein-Barr virus-encoded microRNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36:666-675. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 259] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 54. | Lo AK, To KF, Lo KW, Lung RW, Hui JW, Liao G, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci USA. 2007;104:16164-16169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 266] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 55. | Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124-126. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 56. | Liu WH, Yeh SH, Chen PJ. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim Biophys Acta. 2011;1809:678-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 57. | Wang SH, Yeh SH, Lin WH, Wang HY, Chen DS, Chen PJ. Identification of androgen response elements in the enhancer I of hepatitis B virus: a mechanism for sex disparity in chronic hepatitis B. Hepatology. 2009;50:1392-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 58. | Buhlmann S, Racek T, Schwarz A, Schaefer S, Pützer BM. Molecular mechanism of p73-mediated regulation of hepatitis B virus core promoter/enhancer II: implications for hepatocarcinogenesis. J Mol Biol. 2008;378:20-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Lee GH, Wasser S, Lim SG. Hepatitis B pregenomic RNA splicing--the products, the regulatory mechanisms and its biological significance. Virus Res. 2008;136:1-7. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 21] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 173] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 61. | Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157-5163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 136] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 62. | Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51:881-890. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 63. | Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 268] [Cited by in F6Publishing: 287] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 64. | Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476-1485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 154] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 65. | Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 66. | Ren M, Qin D, Li K, Qu J, Wang L, Wang Z, Huang A, Tang H. Correlation between hepatitis B virus protein and microRNA processor Drosha in cells expressing HBV. Antiviral Res. 2012;94:225-231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 67. | Waidmann O, Bihrer V, Pleli T, Farnik H, Berger A, Zeuzem S, Kronenberger B, Piiper A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J Viral Hepat. 2012;19:e58-e65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 102] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 68. | Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 297] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 69. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I, Umeshita K. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 259] [Cited by in F6Publishing: 271] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 70. | Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232-243. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 542] [Cited by in F6Publishing: 570] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 71. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 604] [Cited by in F6Publishing: 629] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 72. | Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600-1609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 194] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 73. | Lakner AM, Steuerwald NM, Walling TL, Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL, Schrum LW. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300-310. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 164] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 74. | Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO. Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol. 2011;54:1177-1184. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 75. | Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 280] [Cited by in F6Publishing: 303] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 76. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5705] [Cited by in F6Publishing: 5849] [Article Influence: 324.9] [Reference Citation Analysis (0)] |
| 77. | Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3218] [Cited by in F6Publishing: 3383] [Article Influence: 211.4] [Reference Citation Analysis (0)] |
| 78. | Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513-10518. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5636] [Cited by in F6Publishing: 6046] [Article Influence: 377.9] [Reference Citation Analysis (0)] |
| 79. | Shabaneh Al-Tamimi HA, McDonald R. Elevated alanine aminotransferase levels associated with polymyositis: can this be due to muscle injury. J Clin Rheumatol. 2008;14:363-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 80. | Ozer J, Ratner M, Shaw M, Bailey W, Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 736] [Cited by in F6Publishing: 767] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 81. | Nathwani RA, Pais S, Reynolds TB, Kaplowitz N. Serum alanine aminotransferase in skeletal muscle diseases. Hepatology. 2005;41:380-382. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 236] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 82. | Kim WR, Flamm SL, Di Bisceglie AM, Bodenheimer HC. Serum activity of alanine aminotransferase (ALT) as an indicator of health and disease. Hepatology. 2008;47:1363-1370. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 521] [Cited by in F6Publishing: 530] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 83. | Amacher DE, Adler R, Herath A, Townsend RR. Use of proteomic methods to identify serum biomarkers associated with rat liver toxicity or hypertrophy. Clin Chem. 2005;51:1796-1803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, Fei M, Sun S. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830-1838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 307] [Cited by in F6Publishing: 314] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 85. | Marrero JA, Lok AS. Newer markers for hepatocellular carcinoma. Gastroenterology. 2004;127:S113-S119. [PubMed] [Cited in This Article: ] |
| 86. | Okuda H, Nakanishi T, Takatsu K, Saito A, Hayashi N, Takasaki K, Takenami K, Yamamoto M, Nakano M. Serum levels of des-gamma-carboxy prothrombin measured using the revised enzyme immunoassay kit with increased sensitivity in relation to clinicopathologic features of solitary hepatocellular carcinoma. Cancer. 2000;88:544-549. [PubMed] [Cited in This Article: ] |
| 87. | Hayes CN, Akamatsu S, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PLoS One. 2012;7:e47490. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 88. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 430] [Cited by in F6Publishing: 469] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 89. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 543] [Cited by in F6Publishing: 594] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 90. | Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419-427. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 387] [Cited by in F6Publishing: 414] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 91. | Li W, Xie L, He X, Li J, Tu K, Wei L, Wu J, Guo Y, Ma X, Zhang P. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616-1622. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 239] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 92. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 349] [Cited by in F6Publishing: 365] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 93. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 234] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 94. | Chen CJ, Lee MH. Early diagnosis of hepatocellular carcinoma by multiple microRNAs: validity, efficacy, and cost-effectiveness. J Clin Oncol. 2011;29:4745-4747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 95. | Cai G, Liu Y. Diagnosing advanced versus early-stage hepatocellular carcinoma. J Clin Oncol. 2012;30:2167; author reply 2168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 96. | Gao YF, Yu L, Wei W, Li JB, Luo QL, Shen JL. Inhibition of hepatitis B virus gene expression and replication by artificial microRNA. World J Gastroenterol. 2008;14:4684-4689. [PubMed] [Cited in This Article: ] |
| 97. | Ebert G, Poeck H, Lucifora J, Baschuk N, Esser K, Esposito I, Hartmann G, Protzer U. 5’ Triphosphorylated small interfering RNAs control replication of hepatitis B virus and induce an interferon response in human liver cells and mice. Gastroenterology. 2011;141:696-706, 706.e1-3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 98. | McCaffrey AP. RNA interference inhibitors of hepatitis B virus. Ann N Y Acad Sci. 2009;1175:15-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 99. | Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578-585. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 294] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 100. | Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1240] [Cited by in F6Publishing: 1305] [Article Influence: 72.5] [Reference Citation Analysis (0)] |
| 101. | Arbuthnot P. MicroRNA-like antivirals. Biochim Biophys Acta. 2011;1809:746-755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 102. | Keck K, Volper EM, Spengler RM, Long DD, Chan CY, Ding Y, McCaffrey AP. Rational design leads to more potent RNA interference against hepatitis B virus: factors effecting silencing efficiency. Mol Ther. 2009;17:538-547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 103. | Ely A, Naidoo T, Mufamadi S, Crowther C, Arbuthnot P. Expressed anti-HBV primary microRNA shuttles inhibit viral replication efficiently in vitro and in vivo. Mol Ther. 2008;16:1105-1112. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 104. | Chattopadhyay S, Ely A, Bloom K, Weinberg MS, Arbuthnot P. Inhibition of hepatitis B virus replication with linear DNA sequences expressing antiviral micro-RNA shuttles. Biochem Biophys Res Commun. 2009;389:484-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 105. | Xiangji L, Feng X, Qingbao C, Weifeng T, Xiaoqing J, Baihe Z, Feng S, Hongyang W, Mengchao W. Knockdown of HBV surface antigen gene expression by a lentiviral microRNA-based system inhibits HBV replication and HCC growth. J Viral Hepat. 2011;18:653-660. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |