Published online Jun 21, 2013. doi: 10.3748/wjg.v19.i23.3562
Revised: March 5, 2013
Accepted: March 22, 2013
Published online: June 21, 2013
AIM: To determine the features of Enterococcus that contribute to the development and maintenance of the inflammatory process in patients with inflammatory bowel disease (IBD).
METHODS: Multiplex polymerase chain reaction (PCR) was applied to assess the presence of genes that encode virulence factors [surface aggregating protein (asa1), gelatinase (gelE), cytolysin (cylA), extracellular surface protein (esp) and hyaluronidase (hyl)] in the genomic DNA of 28 strains of Enterococcus isolated from the intestinal tissues of children with IBD (n = 16) and of children without IBD (controls; n = 12). Additionally, strains with confirmed presence of the gelE gene were tested by PCR for the presence of quorum sensing genes (fsrA, fsrB, fsrC) that control the gelatinase production. Gelatinase activity was tested on agar plates containing 1.6% gelatin. We also analysed the ability of Enterococcus strains to release and decompose hydrogen peroxide (using Analytical Merckoquant peroxide test strips) and tested their ability to adhere to Caco-2 human gut epithelium cells and form biofilms in vitro.
RESULTS: A comparison of the genomes of Enterococcus strains isolated from the inflamed mucosa of patients with IBD with those of the control group showed statistically significant differences in the frequency of the asa1 gene and the gelE gene. Furthermore, the cumulative occurrence of different virulence genes in the genome of a single strain of Enterococcus isolated from the IBD patient group is greater than in a strain from the control group, although no significant difference was found. Statistically significant differences in the decomposition of hydrogen peroxide and adherence to the Caco-2 epithelial cell line between the strains from the patient group and control group were demonstrated. The results also showed that profuse biofilm production was more frequent among Enterococcus strains isolated from children with IBD than in control strains.
CONCLUSION: Enterococcus strains that adhere strongly to the intestinal epithelium, form biofilms and possess antioxidant defence mechanisms seem to have the greatest influence on the inflammatory process.
Core tip: In this research we have attempted to show which features make Enterococcus strains contributing to the development and maintenance of the inflammatory process in patients with inflammatory bowel disease. The outcome of this research may have an impact on better understanding of the pathomechanisms of this disease, as its etiology is not fully known. The study results suggest that Enterococcus strains which adhere strongly to the intestinal epithelium, form biofilm as well as possess the enzymatic mechanisms protecting them against the effects of reactive oxygen species, seem to have the highest chances to survive and influence the inflammatory process.
-
Citation: Golińska E, Tomusiak A, Gosiewski T, Więcek G, Machul A, Mikołajczyk D, Bulanda M, Heczko PB, Strus M. Virulence factors of
Enterococcus strains isolated from patients with inflammatory bowel disease. World J Gastroenterol 2013; 19(23): 3562-3572 - URL: https://www.wjgnet.com/1007-9327/full/v19/i23/3562.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i23.3562
Inflammatory bowel disease (IBD) refers to two disease entities: ulcerative colitis (UC) and Crohn’s disease (CD). UC is a chronic inflammatory process of the mucous membranes of the rectum and colon. Inflammatory changes in the form of recurrent surface ulcerations are located along the whole length of the large intestine. The course of UC course ranges from mild disease activity, lasting for years, to severe disease that can result in death after only a few weeks. CD is a chronic inflammatory process that can affect any part of the alimentary system, from the oral cavity to the rectum.
The aetiology of IBD is not yet fully understood[1]. Based on numerous clinical observations and results of extensive in vitro and in vivo experiments, it is currently thought that this disease develops as a result of the concurrence of three factors: genetic predisposition, disorders of the immune system and the influence of the environmental factors. Among the environmental factors, changes in the composition of the bacterial flora that colonise the human alimentary tract are considered to be of great influence[2]. Consequently, a great deal of attention is currently devoted to the study of the bacterial species that constitute the normal alimentary tract flora compared to those found in the tract of the IBD patients.
From 2004 to 2006, research on the qualitative and quantitative changes in the microflora of the alimentary tracts of a group of children who had been diagnosed with CD or UC for the first time was performed in our department[3,4]. No single aetiological factor that could be responsible for the exacerbation of the progressing inflammatory process was confirmed based on the obtained results.
However, the analysis of the quantitative changes in the group of children with CD identified a significant increase in the population of cocci (including Streptococcus and Enterococcus) and bacteria belonging to the genus Lactobacillus in the inflammatorily changed sites with a simultaneous decrease in the number of strictly anaerobic bacteria, particularly those belonging to the genus Bifidobacterium.
All of the above-mentioned bacteria are important members of the commensal flora of the human colon. Therefore, we must consider the mechanisms by which these bacteria may contribute to the development and/or maintenance of the inflammatory process in patients suffering from IBD. The virulence potential of the genus Enterococcus has been established in many publications, and it is this genus contributes to infections such as peritonitis, bloodstream and urinary tract infections and endocarditis[5,6]. Their potential role in the pathomechanisms leading to IBD has also been highlighted by research utilising IL-10 gene knockout mice to show that Enterococcus faecalis (E. faecalis) can induce IBD[7].
Recent studies on the pathogenicity of enterococci indicate that the genomes of strains that are able to cause tissue damage and inflammation contain a pathogenicity island that encodes aggregation substance (AS), gelatinase, extracellular surface proteins (Esp), cytolysin, hyaluronidase and other proteins[8,9]. Enterococci that express AS were found to resist phagocytosis significantly better than an isogenic AS-negative strain by inhibiting the respiratory burst of macrophages[10]. Gelatinase, a protease produced by enterococci, is capable of hydrolysing gelatin, collagen, casein, haemoglobin and other peptides[9]. The Esp enhance biofilm formation in E. faecalis[11]. Cytolysin produced by the enterococci is lethal for a broad range of prokaryotic and eukaryotic cells[12]. Hyaluronidase is mainly a degradative enzyme that is associated with tissue damage[13].
Little attention has been devoted to the ability of enterococci to release hydrogen peroxide into the extracellular space[14]. Pursuant to the results obtained previously by our group, select members of several genera, including Streptococcus, Enterococcus and Lactobacillus, are, under aerobic conditions, able to produce amounts of hydrogen peroxide comparable to those released by cells of the immune system during the oxidative burst[15]. This additional source of hydrogen peroxide could help sustain, or even exacerbate, gut inflammation[16]. Notably, certain Enterococcus strains can defend themselves against the surplus of reactive oxygen species (ROS) by producing anti-oxidative enzymes to increase their chances of survival in unfavourable conditions[17].
Thus, the main objective of this work was to compare the occurrence of genes encoding selected virulence factors [surface aggregating protein (asa1), gelatinase (gelE), cytolysin (cylA), extracellular surface protein (esp) and hyaluronidase (hyl)] in the genomes of Enterococcus strains isolated from patients suffering from IBD with the occurrence of these genes in the genomes of strains derived from the control group subjects. Additionally, strains confirmed positive for the gelE gene were tested for the presence of the quorum sensing genes fsrA-C that regulate gelatinase production, and gelatinase activity was tested on agar plates containing 1.6% gelatin. The adherence of the Enterococcus strains to Caco-2 epithelial cells and their ability to form biofilms were also tested. Furthermore, the ability of strains isolated from inflamed or non-inflamed gut mucosa to release and decompose extracellular hydrogen peroxide was assessed.
Enterococcus strains were isolated from colon biopsies of 34 children who were diagnosed with IBD for the first time and 24 patients from the control group comprised of children with functional bowel disorders that were hospitalised in the same hospital during the same period of time. Neither the IBD patients nor the participants in the control group received antibiotics during the two weeks prior to the study. The diagnosis of CD or UC was based on endoscopic, histopathological, immunological and radiological criteria[3,4]. The biopsies were taken from both groups of patients during colonoscopy procedures carried out at the Clinic of Paediatrics, Gastroenterology and Nutrition of the Polish-American Institute of Paediatrics of the Jagiellonian University Medical College in Cracow pursuant to the approval of the Bioethical Committee (No. KBET/236/B/2002). The patients were prepared for the colonoscopy using routine washing procedures described by Gosiewski et al[4]. During the colonoscopy, the location and intensity of the lesions was assessed, and tissue fragments were obtained for histopathological and microbiological examinations. The biopsy samples were transported to the microbiological laboratory in Schaedler broth (SAB, Difco, United States, + 10% glycerol) at -20 °C.
In the laboratory, the frozen samples were thawed, weighed and homogenised in 1 mL of SAB and quantitatively analysed for the main cultivable bacterial constituents of the colon microflora using differential media in aerobic and anaerobic conditions[3,4]. These steps were performed aseptically in an anaerobic chamber (MACS - MG 500 Work Station, DW Scientific, Shipley, United Kingdom) in N2 (85%) + H2 (10%) + CO2 (5%) atmosphere. The homogenised samples were serially diluted with SAB and 100 μL aliquots plated on the following media: McConkey Agar (Oxoid, Basingstoke, United Kingdom) for Enterobacteriaceae, Columbia Blood Agar (Difco, Lawrence, Kansas, United States) with 5% sheep blood for streptococci, Enterococcosel agar (BBL, BD, Franklin Lakes, United States) for enterococci, MRS agar (Oxoid) for lactobacilli and other lactic acid bacteria, BL agar for bifidobacteria and Wilkins-Chalgren agar base (Difco) with supplements for Bacteroides.
The morphology of the grown colonies was analysed using a magnifying glass, and several colonies of each morphological type were subcultured on appropriate aerobic and anaerobic media and subsequently Gram-stained. After further incubation and culture purity checks, the phenotypic identification was conducted with the use of commercial identification systems (API 20E, API20A, APIStaph, APIStrep, bioMerieux, Marcy l’Etoile, France; BBL Crystal ID System, BD, Franklin Lakes, United States). The identification was confirmed by species-specific multiplex polymerase chain reaction (PCR) as described by Jackson et al[18].
The selected strains were preserved at -80 °C on glass beads in BBL Nutrient Broth with the addition of 15% glycerol.
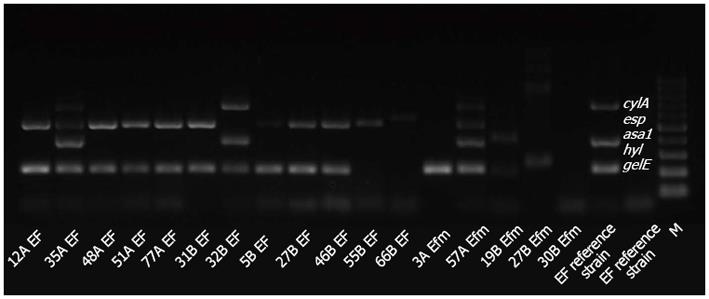
To detect presence of genes encoding selected virulence factors (asa1, gelE, cylA, esp, hyl) in the genomes of the tested strains, multiplex PCR was applied according to the methods of Vankerckhoven[19].
To investigate the presence of quorum sensing genes (fsrA, fsrB, fsrC) in the genomes of all the strains positive for the gelE gene, PCR was applied pursuant to Qin et al[20].
The reference strain E. faecalis ATTC29212 containing the gene fsrC was used as the positive control. The negative control was a reference strain of E. faecium (ATTC 35667) that did not possess any of the examined genes.
The product sizes for fsrA, fsrB and fsrC were 484 bp, 574 bp and 580 bp, respectively (BLAST).
The production of gelatinase in Enterococcus strains was detected by the method described by Steck et al[6]. Supernatants from overnight cultures were spotted onto tryptic soy agar supplemented with 0.5 g/L L-cysteine and 1.6% Difco gelatine. The zone of clearance was measured after 24 h of incubation.
The ability of Enterococcus strains to adhere to Caco-2 human gut epithelium cells (ATCC HTB-37) was assessed using an in vitro assay according to Górska-Frączek[21]. Caco-2 cells were cultured for 24 h in a 12-well flat bottom tissue culture plate at a density of 1 × 106 cells/mL (Iwaki, Japan) in Eagle’s 1959 medium (MEM) with L-glutamine and NaHCO3 (IITD, Wrocław, Poland) containing 5% foetal calf serum (Sigma-Aldrich Chemie, Germany) and antibiotics (penicillin 100 UI/mL, streptomycin 100 UI/mL, neomycin 200 μg/mL) (Sigma Aldrich Chemie, Germany) and were washed twice with PBS. Overnight cultures of bacteria were diluted with MRS+MEM to a concentration of about 108 CFU/mL. The Caco-2 cells were inoculated with the bacterial cultures. After incubating at 37 °C for 30 min, wells were washed twice with PBS to release unbound bacteria. Then, the cells were fixed with 3.7% formaldehyde for 1 h, washed twice with PBS and stained with crystal violet stain (Merck, Germany). The degree of adhesion was evaluated using a semiquantitative scoring system ranging from (-) to (+++). The adherent Enterococcus cells were counted in 20 randomly selected microscopic fields, as we have previously described[22].
Fifteen strains of enterococci collected from children with IBD and 10 strains from the control children were examined for biofilm formation in sterile plastic 96-well plates with an adherent surface (Greiner Bio-One, United States). Bacterial cells were grown in 10 mL of trypticase soy broth (TSB, Difco) at 37 °C for 24 h in aerobic conditions. The culture was then centrifuged (2000 r/min; 10 min) and washed with 10 mL of saline. A suspension of each strain (1 × 105 CFU/mL) was prepared by serial dilution of bacteria in saline using MacFarland’s scale, and then 20 μL of the prepared suspension was added to a well followed by 180 μL of sterile TSB. The final concentration of bacteria was 1 × 104 CFU/mL. The plates were centrifuged for 10 min at 2000 r/min to sediment the bacteria onto the bottom of each well and were then incubated for 48 h (37 °C, aerobic conditions). The biofilm quantity (total mass of bacterial polysaccharides) was measured using Congo red dye according to a modified procedure described by Reuter et al[23]. Briefly, at two different time points (0 and 48 h), the plates were centrifuged, the culture medium was gently removed from the wells and 200 μL of 0.1% Congo red solution was immediately added. The plates were left at room temperature for 30 min and then washed twice with buffered saline to remove unbound dye. The absorbance was measured at a wavelength of 492 nm using a spectrophotometer (Awareness Technology, Inc.). All measurements were performed in triplicate, and the mean values ± SD were calculated. The degree of biofilm production by the tested cocci was arbitrarily categorised as either highly positive (A492≥ 0.81) or weakly positive (A492≤ 0.8).
Analytical Merckoquant peroxide test strips (Merck, NJ, United States) were used to measure H2O2 production by the tested strains on a detection scale between 0 and 100 mg/L. The tested bacteria were suspended in 2 mL of TSB broth (TSB) (Difco, Kansas, United States) and cultured at 37 °C in aerobic conditions. The measurements of H2O2 were performed after 4 and 24 h according to the procedure provided by the manufacturer. The mean density of bacterial growth at 4 h was approximately 3 × 106 CFU/mL and increased to 1 × 107 CFU/mL after 24 h. Uninoculated TSB broth was used as a negative control. The amounts of H2O2 are presented as mmol/L.
The bacterial strains were cultured in 10 mL of TSB for 24 h at 37 °C in aerobic conditions. Their final density was approximately 0.5 on the McFarland scale. After 24 h, chemically pure hydrogen peroxide (Sigma-Aldrich, United States) was added to each test culture at a final concentration of 2 mmol/L. The culture was incubated under the same conditions for 4 h, and the amount of hydrogen peroxide remaining in the test tube was then determined using Analytical Merckoquant peroxide test strips (Merck, NJ, United States). The resulting amount of H2O2 was converted from mg/L into mmol/L. Then, the number of bacterial cells in each culture was determined using the viable count method. These levels were comparable for all strains and equalled approximately 8 × 106 CFU/mL. The negative control was sterile TSB containing 2 mmol/L H2O2.
The statistical analysis was focused on comparison of probabilities of the analyzed genes presence between the group of the bacteria isolated from IBD children and this from control group. Such comparison was done using frequency χ2 test. If the given frequencies didn’t fulfill the assumption of the test it’s less strong equivalents: likelihood ratio of Fisher’s exact test were used. The value P < 0.05 was regarded as the threshold for statistical significance. All calculations were performed using JMP 7.0.2 (SAS, United States) software package.
In total, 48 strains belonging to the genus Enterococcus were isolated. Of these, 34 strains originated from children diagnosed with IBD, including 21 from children diagnosed with CD and 13 from children with UC. In this group, 16 (47%) strains belonged to E. faecalis, 10 (29%) to E. avium and 8 (24%) to E. faecium. In the control group, 5 (36%) out of 14 strains were identified as E. faecalis, 4 (29%) as E. avium, 3 (21%) as E. faecium and 2 (14%) as E. durans (Table 1).
| Number of isolates | Strain number | Species (API 20 Strep/Multiplex PCR) | Disease entity/Patient number |
| 1 | 3A | Enterococcus faecium | CD/1 |
| 2 | 3B | Enterococcus faecium | |
| 3 | 10A | Enterococcus avium | CD/2 |
| 4 | 12A/1 | Enterococcus faecalis | |
| 5 | 12A/2 | Enterococcus avium | CD/3 |
| 6 | 12B | Enterococcus faecium | |
| 7 | 25A | Enterococcus faecium | CD/4 |
| 8 | 25B | Enterococcus avium | |
| 9 | 31B | Enterococcus faecalis | CD/5 |
| 10 | 42A/1 | Enterococcus faecalis | CD/6 |
| 11 | 42A/2 | Enterococcus faecium | |
| 12 | 51A/1 | Enterococcus faecalis | CD/7 |
| 13 | 51A/2 | Enterococcus avium | |
| 14 | 57A | Enterococcus faecium | CD/8 |
| 15 | 8A | Enterococcus faecalis | CD/9 |
| 16 | 8B/1 | Enterococcus faecalis | |
| 17 | 8B/2 | Enterococcus avium | |
| 18 | 19B | Enterococcus faecium | CD/10 |
| 19 | 22B | Enterococcus faecalis | CD/11 |
| 20 | 69A | Enterococcus faecalis | CD/12 |
| 21 | 79A | Enterococcus faecalis | CD/13 |
| 22 | 32B | Enterococcus faecalis | UC/1 |
| 23 | 35A | Enterococcus faecalis | UC/2 |
| 24 | 40A | Enterococcus avium | UC/3 |
| 25 | 40B | Enterococcus avium | |
| 26 | 48A | Enterococcus faecalis | UC/4 |
| 27 | 58A | Enterococcus faecalis | UC/5 |
| 28 | 77A | Enterococcus faecalis | UC/6 |
| 29 | 33B | Enterococcus avium | UC/7 |
| 30 | 29B | Enterococcus avium | UC/8 |
| 31 | 34A/1 | Enterococcus faecium | UC/9 |
| 32 | 34A/2 | Enterococcus avium | |
| 33 | 64B | Enterococcus faecalis | UC/10 |
| 34 | 65A | Enterococcus faecalis | UC/11 |
| 35 | 5B | Enterococcus faecalis | Control/1 |
| 36 | 7B | Enterococcus faecium | Control/2 |
| 37 | 15B/1 | Enterococcus avium | Control/3 |
| 38 | 15B/2 | Enterococcus durans | Control/4 |
| 39 | 16B/1 | Enterococcus avium | Control/5 |
| 40 | 16B/2 | Enterococcus durans | |
| 41 | 27B/1 | Enterococcus faecalis | Control/6 |
| 42 | 27B/2 | Enterococcus faecium | |
| 43 | 27B/3 | Enterococcus avium | |
| 44 | 30B/1 | Enterococcus faecium | Control/7 |
| 45 | 30B/2 | Enterococcus avium | |
| 46 | 46B | Enterococcus faecalis | Control/8 |
| 47 | 55B | Enterococcus faecalis | Control/9 |
| 48 | 66B | Enterococcus faecalis | Control/10 |
For further studies, 28 Enterococcus strains were randomly selected, including 16 strains from patients with IBD (n = 9 CD, n = 7 UC) and 12 strains from the control group (Tables 2, 3 and 4). Molecular typing of all Enterococcus isolates using PFGE procedure (not described here) was performed before selection to eliminate redundant pulsotypes. No clustering within individual species or across the entire set of strains was noted.
| Source | Strains | Detection of genes | Gelatinase activity on 16% gelatin plates | |||
| gelE | fsrA | fsrB | fsrC | |||
| Crohn's disease | Enterococcus faecalis 12A/1 | + | - | - | + | - |
| Enterococcus faecalis 31B | + | - | + | + | - | |
| Enterococcus faecalis 51A/1 | + | - | - | + | - | |
| Enterococcus faecium 3A | + | + | + | + | + | |
| Enterococcus faecium 57A | + | - | - | - | - | |
| Enterococcus faecium 19B | + | + | - | - | - | |
| Enterococcus avium 51A/2 | - | - | - | - | - | |
| Enterococcus avium 25B | + | - | - | + | - | |
| Enterococcus avium 10A | + | - | - | - | - | |
| Ulcerative colitis | Enterococcus faecalis 35A | + | - | - | - | - |
| Enterococcus faecalis 48A | + | - | - | + | - | |
| Enterococcus faecalis 77A | + | - | - | + | - | |
| Enterococcus faecalis 32B | + | + | + | + | + | |
| Enterococcus avium 40A | + | - | - | - | - | |
| Enterococcus avium 34A/2 | - | - | - | - | - | |
| Enterococcus avium 40B | + | - | - | - | - | |
| Control | Enterococcus faecalis 5B | + | - | - | + | |
| Enterococcus faecalis 27B/1 | + | + | + | + | + | |
| Enterococcus faecalis 46B | + | - | + | - | - | |
| Enterococcus faecalis 55B | - | - | - | - | - | |
| Enterococcus faecalis 66B | - | - | - | - | - | |
| Enterococcus faecium 27B/2 | + | - | - | - | - | |
| Enterococcus faecium 30B | - | - | - | - | - | |
| Enterococcus avium 15B/1 | - | - | - | - | - | |
| Enterococcus avium 16B/1 | - | - | - | - | - | |
| Enterococcus avium 27B/3 | + | - | - | - | - | |
| Enterococcus durans 15B/2 | - | - | - | - | - | |
| Enterococcus durans 16B/2 | - | - | - | - | - | |
| ATCC control | Enterococcus faecalis 29212 | + | - | - | + | - |
| Enterococcus faecium 35667 | - | - | - | - | - | |
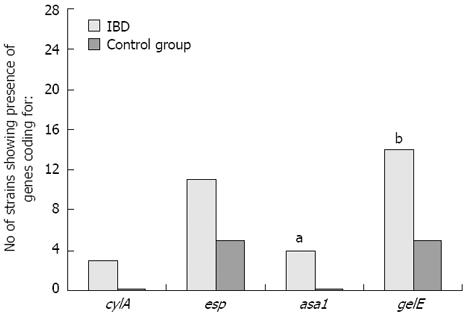
While identifying genes coding for selected virulence factors (asa1, gelE, cylA, esp, hyl) in the genomes of the Enterococcus strains isolated from the inflamed mucosa of IBD patients and from control group patients, statistically significant differences in the frequencies of the asa1 gene, which encodes the surface aggregating proteins, and the gelE gene (P = 0.0091), which encodes the gelatinase that is responsible for decomposing collagen and elastin, were confirmed (Tables 2 and 3; Figures 1 and 2).
| Source | Strains | asa1 | Adherence | Biofilm production (after 48 h) A492 nm± SD |
| Crohn's disease | Enterococcus faecalis 12A/1 | - | +++ | 0.995 ± 0.351 |
| Enterococcus faecalis 31B | - | +++ | 1.302 ± 0.585 | |
| Enterococcus faecalis 51A/1 | - | ++ | 1.083 ± 0.413 | |
| Enterococcus faecium 3A | - | - | 1.286 ± 0.088 | |
| Enterococcus faecium 57A | + | +++ | 0.698 ± 0.066 | |
| Enterococcus faecium 19B | + | +++ | 1.087 ± 0.368 | |
| Enterococcus avium 51A/2 | - | - | 2.101 ± 0.424 | |
| Enterococcus avium 25B | - | - | 2.474 ± 0.683 | |
| Enterococcus avium 10A | - | - | 1.423 ± 0.284 | |
| Ulcerative colitis | Enterococcus faecalis 35A | + | +++ | 1.528 ± 0.326 |
| Enterococcus faecalis 48A | - | +++ | 0.767 ± 0.582 | |
| Enterococcus faecalis 77A | - | ++ | 0.822 ± 0.196 | |
| Enterococcus faecalis 32B | + | ++ | 0.557 ± 0.344 | |
| Enterococcus avium 40A | - | +++ | 0.859 ± 0.152 | |
| Enterococcus avium 34A/2 | - | - | ||
| Enterococcus avium 40B | - | - | 0.282 ± 0.078 | |
| Control | Enterococcus faecalis 5B | - | 0.512 ± 0.065 | |
| Enterococcus faecalis 27B/1 | - | - | 0.934 ± 0.103 | |
| Enterococcus faecalis 46B | - | ++ | 0.507 ± 0.095 | |
| Enterococcus faecalis 55B | - | - | 0.408 ± 0.242 | |
| Enterococcus faecalis 66B | - | - | 0.172 ± 0.031 | |
| Enterococcus faecium 27B/2 | - | 0.917 ± 0.269 | ||
| Enterococcus faecium 30B | - | - | 0.359 ± 0.087 | |
| Enterococcus avium 15B/1 | - | - | ||
| Enterococcus avium 16B/1 | - | - | 0.305 ± 0.104 | |
| Enterococcus avium 27B/3 | - | - | 0.657 ± 0.182 | |
| Enterococcus durans 15B/2 | - | - | ||
| Enterococcus durans 16B/2 | - | - | 0.315 ± 0.246 | |
| ATCC control | Enterococcus faecalis 29212 | + | +++ | 0.877 ± 0.221 |
| Enterococcus faecium 35667 | - | ++ | 0.722 ± 0.371 |
The cylA gene, which encodes cytolysin, was detected in 2 strains of E. faecalis and 1 strain of Enterococcus faecium isolated from the inflamed sites. This gene was not detected in the control strains. The obtained results were at the limit of statistical significance (P = 0.0569). The esp gene encoding the extracellular surface protein was present in 11 strains isolated from patients with IBD and in 5 strains isolated from the control group. None of the 28 strains examined, including strains from both research groups, contained the hyl gene that encodes hyaluronidase. No statistically significant differences were observed in the prevalence of esp or hyl between the two patient groups (Figures 1 and 2).
The cumulative occurrence of different virulence genes in the genome of a single strain of Enterococcus isolated from patients suffering from IBD was greater than that of a strain isolated from the control group (mean of 2.6 vs 2.0, respectively), but no significant difference was found. For instance, the strains E. faecalis 35A and E. faecium 57A were isolated from patients with IBD and possessed as many as 4 virulence genes (cylA, esp, asa1 and gelE), but strains containing all four of these genes were not observed in Enterococcus isolates from the control group.
Further analysis was performed on strains in which the presence of the gelE gene was confirmed (n = 19). Based on the results of a PCR assay for the presence of the regulator genes fsrA-C, statistically significant differences between the two groups were noted for the occurrence of fsrC. This gene was confirmed in 8 Enterococcus strains isolated from the study group, but it was detected in only 2 strains from the control group (P = 0.0195). Furthermore, the presence of fsrA was confirmed in 3 strains from the study group and in 1 Enterococcus strain from the control group. The fsrB gene was detected in the genome of 3 strains isolated from the IBD children and in 2 strains from the control group children. These results are shown in Table 2.
Enterococci containing the gene encoding gelatinase also underwent an in vitro test in which the activity of gelatinase on agar plates containing 1.6% gelatin was examined. Zones of clearance were demonstrated only on plates inoculated with cultures of strains that had all three regulator genes (fsrA, B and C): 2 Enterococcus strains from the studied group and 1 strain from the control group. No statistically significant differences were shown between these two groups. The results are shown in Table 2.
While testing the ability of the enterococci to adhere to Caco-2 cells, statistically significant differences were demonstrated between the bacteria isolated from IBD children compared to controls. Ten of the 24 enterococcal strains from the IBD group strongly adhered to the Caco-2 cells (P = 0.0238). Only 1 strain from the control group adhered to the epithelial cell line used in the study. It is important to note that adherence was demonstrated by all strains having the asa1 gene, which encodes the surface aggregating protein and allows for increased adherence of the bacteria to the host’s tissues. The results of this study are shown in Table 3. However, 6 strains from the control group and 1 strain from the study group with no confirmed presence of asa1 gene also adhered to the cells. Therefore, there are other virulence factors that increase bacterial adherence to host tissues.
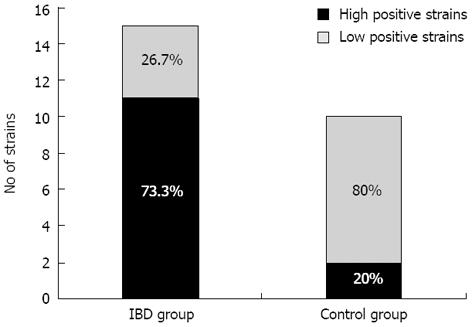
All of the tested Enterococcus strains were able to form biofilms structure within 48 h. However, the results demonstrated that profuse biofilm production was more frequent among strains isolated from children with IBD than in control strains. Among the enterococcal strains isolated from the studied group, 11 (73.3%) were abundant biofilm producers (high positive; A492≥ 0.81), and 4 (26.7%) were weak biofilm producers (low positive; A492≤ 0.8). Of the 10 strains isolated from the control group, 2 (20%) were classified as abundant biofilm producers and 8 (80%) were weak producers. The difference between the IBD and control groups was not statistically significant. The results of the biofilm formation analysis are shown in Table 3 and Figure 3.
While analysing the ability of the strains to produce extracellular hydrogen peroxide, we observed that only E. faecium and E. avium species released hydrogen peroxide in amounts equal to or higher than 0.3 mmol/L. In total, 5 of the 16 Enterococcus strains isolated from patients with IBD and 3 of the 12 strains isolated from the control group were able to produce extracellular hydrogen peroxide, but this difference was not statistically significant (Table 4).
| Source | Strains | H2O2 production (mmol/L) after 4 h | H2O2 production (mmol/L) after 24 h |
| Crohn's disease | Enterococcus faecalis 12A/1 | 0 | 0 |
| Enterococcus faecalis 31B | 0 | 0 | |
| Enterococcus faecalis 51A/1 | 0 | 0 | |
| Enterococcus faecium 3A | 0 | 0 | |
| Enterococcus faecium 57A | 0 | 0 | |
| Enterococcus faecium 19B | 0 | 0.3 | |
| Enterococcus avium 51A/2 | 0 | 0.3 | |
| Enterococcus avium 25B | 0 | 0.3 | |
| Enterococcus avium 10A | 0.3 | 1 | |
| Ulcerative colitis | Enterococcus faecalis 35A | 0 | 0 |
| Enterococcus faecalis 48A | 0 | 0 | |
| Enterococcus faecalis 77A | 0 | 0 | |
| Enterococcus faecalis 32B | 0 | 0 | |
| Enterococcus avium 40A | 0 | 0 | |
| Enterococcus avium 34A/2 | 0 | 0.3 | |
| Enterococcus avium 40B | 0 | 0 | |
| Control | Enterococcus faecalis 5B | 0 | 0 |
| Enterococcus faecalis 27B/1 | 0 | 0 | |
| Enterococcus faecalis 46B | 0 | 0 | |
| Enterococcus faecalis 55B | 0 | 0 | |
| Enterococcus faecalis 66B | 0 | 0 | |
| Enterococcus faecium 27B/2 | 0 | 0.3 | |
| Enterococcus faecium 30B | 0 | 0 | |
| Enterococcus avium 15B/1 | 0.3 | 1 | |
| Enterococcus avium 16B/1 | 0 | 0 | |
| Enterococcus avium 27B/3 | 0 | 0.3 | |
| Enterococcus durans 15B/2 | 0 | 0 | |
| Enterococcus durans 16B/2 | 0 | 0 |
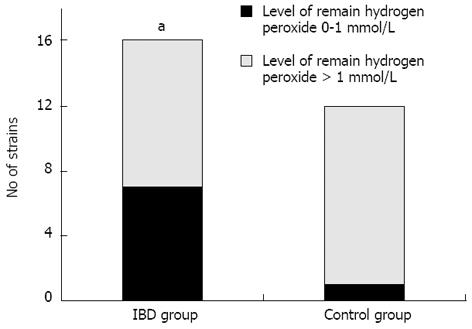
On the other hand, significant differences in the ability of the strains to decompose H2O2 were observed upon examination of the 28 strains of Enterococcus isolated from both groups. Two subgroups could be distinguished based on the amount of decomposed hydrogen peroxide. The first group contained enterococci that decomposed 2 mmol/L of hydrogen peroxide to a level of 0-1 mmol/L in 4 h. The second group comprised strains that decomposed hydrogen peroxide to a level higher than 1 mmol/L. The difference in hydrogen peroxide decomposition ability was statistically significant between the Enterococcus strains isolated from patients with IBD and the strains isolated from control group patients (P = 0.04; Figure 4).
Recently, an important role has been ascribed to the quantitative changes in the composition of the intestinal flora during inflammation[15,24]. These changes are likely related to the alterations in structure and function of the gut epithelium under inflammatory conditions that affect oxygenation, acidity and the functions of many enzymes and may lead to the preferential selection of microbial strains that are able to adapt to and/or interact with the inflamed epithelium.
In this work, Enterococcus strains were chosen for further studies because of their potential to be highly virulent. We sought to detect the differences between the virulence potential of the Enterococcus strains that colonised the intestines of children with IBD and control strains isolated from children without signs of gut inflammation.
The virulence of Enterococci can be linked to the presence of specific virulence factors encoded by specific genes, including cytolytic toxin, extracellular surface protein and AS, serine protease, gelatinase, cell wall adhesins, collagen-binding proteins and capsular polysaccharides[19,25].
In the presented study, statistically significant differences were demonstrated for the occurrence of the asa1 gene in the population of Enterococcus strains isolated from gut biopsies taken from patients with IBD in comparison to those cultured from those of the control group without gut inflammation. The asa1 gene is responsible for bacterial aggregation on the surface of the host tissues (particularly enterocytes, neutrophil granulocytes and epithelial cells of the urinary tract), and it also enables conjugation between bacteria due to increased hydrophobicity of the cell wall surface[26]. Consequently, strains that possess the asa1 gene can develop large aggregations of bacterial cells during an infection and may simultaneously increase the bacterial population[27]. This property is likely to be important in the pathomechanism underlying IBD because during episodes of diarrhoea, one of the main symptoms of IBD, the majority of the alimentary tract flora is removed with the intestinal contents. Enterococci that express the asa1 gene are able to strongly adhere to intestinal surfaces lacking a mucous layer and are able to form biofilms for protection against unfavourable environmental factors.
Furthermore, experiments using a rat model of endocarditis have shown that enterococci equipped with the asa1 gene are better able to survive inside stimulated immune cells, indicating that they may possess enzymatic mechanisms that protect them against the influence of ROS secreted by macrophages during the respiratory burst[10].
The gelE gene was more frequently found in the genomes of enterococci isolated from the group of children with IBD. However, the presence of the gelE gene alone is not sufficient for the production of gelatinase protein by bacteria. Therefore, we also detected the quorum sensing genes that regulate gelatinase production. Gelatinase is a zinc-dependent metalloproteinase that is capable of hydrolysing gelatin, collagen, casein, haemoglobin and human endothelin[28]. Its activity is one of the main causes of the pathological changes in the host’s body upon infection with E. faecalis[25]. It is therefore possible that colonisation of the alimentary tract of patients with IBD with Enterococcus strains possessing the gelE gene can weaken the tight junction protein connections between the epithelial cells lining the intestinal walls of the host, thus leading to the disruption of the mucous barrier[6].
Cytolysin, encoded by the cylA gene, contributes to another pathomechanism of IBD that causes the excessive lysis of erythrocytes, leading to the uncontrolled release of large amounts of haemoglobin and subsequently heme and iron ions that influence the populations of Enterobacteriaceae, which are able to acquire iron[29]. Therefore, the increased number of enterococci that are able to decompose haemoglobin may be related to increases in the E. coli population observed in patients with IBD[30].
Another Enterococcus virulence factor examined in this study was the esp gene, which encodes the extracellular surface protein (Esp). The esp gene was present in 11 strains of enterococci from the group of children with the IBD and in 5 strains from the control group. This finding may indicate that enterococci found in biopsy samples from ulcerations were able to adhere to and colonise the intestinal tissues more easily because Esp promotes primary attachment and biofilm formation in E. faecalis, as proved demonstrated by Toledo-Arana et al[31].
It should be noted here that our studies on the selected virulence factors were based only on the detection of the specified genes that encode these factors, and more information could be obtained from the direct detection of these factors in bacteria using enterocytes in in vitro systems or in animal experiments because the expression of the studied genes may be specifically altered in inflamed tissues[32].
In the alimentary tract of patients suffering from IBD, there can be a local accumulation of large amounts of ROS released by stimulated phagocytic cells, such as macrophages and neutrophils, due to the progressing acute and subsequently chronic inflammatory process. Additionally, some species of Lactobacillus, Streptococcus and Enterococcus that colonise the human intestinal tract are able to produce substantial amounts of superoxide (O2-) and hydrogen peroxide (H2O2) that are comparable to those released by immune cells[33,34]. However, as has been shown by Al-Mushrif and Jones, the oxygen concentration present in the specific ecological niche regulates the bacterial production of hydrogen peroxide[35]. As we have previously demonstrated and discussed elsewhere (Strus et al[15]), the oxygen tension in inflamed tissues during the course of IBD enables the propagation of facultative aerobes such as Enterococcus spp. and the production of considerable amounts of hydrogen peroxide and other metabolites. Selected Enterococcus strains belonging to the species E. avium and E. faecium isolated from patients with IBD and children from the control group were able to produce extracellular hydrogen peroxide in our studies. This result may indicate that the production of hydrogen peroxide is due to certain Enterococcus species rather than the inflammatory conditions at the site of their isolation.
Bacteria belonging to the genera Streptococcus, Lactobacillus and Enterococcus occur in increased numbers in the alimentary tracts patients with IBD and can produce significant amounts of H2O2 that can consequently limit the population of anaerobic flora, stimulate the cells of the immune system to release pro-inflammatory cytokines and stimulate the apoptosis of intestinal epithelial cells deprived of their protective mucous layer[36]. Denning et al[16] showed that hydrogen peroxide concentrations ranging from 0.5 up to 2 mmol/L were able to induce apoptosis in the cells of the abdominal lining and intestinal epithelium; similar amounts of H2O2 were produced in vitro by the Enterococcus strains examined in this study. These observations support the studies by Kruidenier et al[37] and underline the importance of oxidative stress in the pathogenesis of IBD.
During the inflammatory process, the intestinal epithelial cells are exposed to the effects of ROS. If this is a short-term process, no significant damage or disruption of host cell function are observed due to the production of enzymes that inactivate ROS, such as catalase and peroxidase, and DNA repair mechanisms. A significant risk for the intestinal epithelium occurs when ROS are generated at variable concentrations for a longer period of time. Then, not only can the host cells be damaged, but the qualitative and quantitative composition of the specific bacterial species that comprise the commensal flora changes as well[35].
In this work, we showed that enterococci isolated from patients with IBD were able to decompose hydrogen peroxide considerably faster than those isolated from the control group. This may indicate that these strains survived the selective pressures of the inflammatory process. It is likely that only bacterial species possessing a system capable of enzymatic deactivation of ROS were able to survive and subsequently increase their populations on the surface of the damaged intestinal epithelial cells in patients with IBD. Until recently, bacteria belonging to the genera Enterococcus, Lactobacillus and Pediococcus were regarded as catalase negative. However, it appears that these bacteria have a very efficient system of hydrogen peroxide deactivation based on manganese-containing catalase or haeme-dependent catalase[38].
In this study, we present data that suggest that processes in the alimentary tract of patients with IBD (most probably based on inflammatory background of this disease) may lead to the selection of certain bacterial species or strains from the constituents of the commensal flora. Our observations have been supported by recent studies on microbiota from biopsies taken from patients with UC and controls performed by Fite et al[39], who detected statistically significant differences in the bacterial populations of the UC mucosa and in the control group that varied over the study period. High clinical activity indices and sigmoidoscopy scores were associated with enterobacteria, desulfovibrios, Type E Clostridium perfringens and E. faecalis. We believe that in case of Enterococcus, the strains that adhere strongly to the intestinal epithelium, build three-dimensional biofilm structures and possess enzymatic mechanisms to protect against the effects of ROS produced by the immune cells and other bacterial species have the highest chances of survival.
Inflammatory bowel disease (IBD) is a chronic inflammation of all or part of the digestive tract. The aetiology of IBD is not fully known, but much attention is currently focused on the role of bacterial flora in the development and/or maintenance of the inflammatory process in patients suffering from this disease. Recent studies indicate that bacteria belonging to the genus Enterococcus play an important role in the pathomechanisms underlying IBD based on their virulence potential.
Changes in the composition of the bacterial flora colonising the human alimentary tract are considered influential in the maintenance of the inflammatory process in patients with IBD. The virulence potential the enterococci may be particularly important. It will be interesting to identify other factors that allow enterococci to develop and maintain the inflammatory process while increasing their chances of survival in unfavourable conditions.
Recent studies have shown that no single aetiological factor could be responsible for the exacerbation of the inflammatory process in IBD. However, an analysis of the quantitative changes in the intestinal microflora of patients with Crohn’s disease (CD) demonstrated a significant increase in the population of cocci (including Enterococcus and Streptococcus) at the sites of inflammation. In the present study, authors observed that virulence genes, such as asa1 and gelE, occurred more frequently in the genomes of Enterococcus strains isolated from patients with IBD than in strains isolated from healthy patients. Moreover, authors demonstrated statistically significant differences in the ability of these strains to decompose hydrogen peroxide. Enterococci isolated from patients with IBD decomposed hydrogen peroxide considerably faster than those isolated from the control group. This result indicates that only select bacterial species with an efficient system of enzymatic deactivation of reactive oxygen species (ROS) are able to survive and subsequently increase their populations on the surface of the damaged intestinal epithelial cells.
The results of this study suggest that Enterococcus strains that adhere strongly to the intestinal epithelium, form biofilms and possess enzymatic mechanisms that protect against the effects of ROS have the highest chances surviving and influencing the inflammatory process. Their research improves our understanding of the pathomechanisms underlying IBD.
An interesting publication in which authors determine the features of Enterococcus that contribute to the development and maintenance of the inflammatory process in patients with IBD. The results are interesting and suggest that Enterococcus strains that adhere strongly to the intestinal epithelium, form biofilms and possess antioxidant defence mechanisms seem to have the greatest influence on the inflammatory process.
P- Reviewer Hays J S- Editor Wen LL L- Editor A E- Editor Zhang DN
| 1. | Blumberg RS, Strober W. Prospects for research in inflammatory bowel disease. JAMA. 2001;285:643-647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 2. | Sands BE. Inflammatory bowel disease: past, present, and future. J Gastroenterol. 2007;42:16-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in F6Publishing: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 3. | Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sładek M, Pieczarkowski S, Adamski P, Kochan P. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15:5287-5294. [PubMed] [Cited in This Article: ] |
| 4. | Gosiewski T, Strus M, Fyderek K, Kowalska-Duplaga K, Wedrychowicz A, Jedynak-Wasowicz U, Sladek M, Pieczarkowski S, Adamski P, Heczko PB. Horizontal distribution of the fecal microbiota in adolescents with inflammatory bowel disease. J Pediatr Gastroenterol Nutr. 2012;54:20-27. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 5. | Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. 2009;155:1749-1757. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 657] [Cited by in F6Publishing: 651] [Article Influence: 43.4] [Reference Citation Analysis (0)] |
| 6. | Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. 2011;141:959-971. [PubMed] [DOI] [Cited in This Article: ] |
| 7. | Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253-2257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 232] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 8. | Shankar N, Baghdayan AS, Gilmore MS. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature. 2002;417:746-750. [PubMed] [Cited in This Article: ] |
| 9. | Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of enterococcus: an emerging nosocomial pathogen. Indian J Med Microbiol. 2009;27:301-305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 57] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 10. | Süssmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun. 2000;68:4900-4906. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 121] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 11. | Tendolkar PM, Baghdayan AS, Shankar N. The N-terminal domain of enterococcal surface protein, Esp, is sufficient for Esp-mediated biofilm enhancement in Enterococcus faecalis. J Bacteriol. 2005;187:6213-6222. [PubMed] [Cited in This Article: ] |
| 12. | Coburn PS, Gilmore MS. The Enterococcus faecalis cytolysin: a novel toxin active against eukaryotic and prokaryotic cells. Cell Microbiol. 2003;5:661-669. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 109] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Kayaoglu G, Ørstavik D. Virulence factors of Enterococcus faecalis: relationship to endodontic disease. Crit Rev Oral Biol Med. 2004;15:308-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 249] [Cited by in F6Publishing: 272] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 14. | Tendolkar PM, Baghdayan AS, Shankar N. Pathogenic enterococci: new developments in the 21st century. Cell Mol Life Sci. 2003;60:2622-2636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 105] [Cited by in F6Publishing: 98] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 15. | Strus M, Gosiewski T, Fyderek K, Wedrychowicz A, Kowalska-Duplaga K, Kochan P, Adamski P, Heczko PB. A role of hydrogen peroxide producing commensal bacteria present in colon of adolescents with inflammatory bowel disease in perpetuation of the inflammatory process. J Physiol Pharmacol. 2009;60 Suppl 6:49-54. [PubMed] [Cited in This Article: ] |
| 16. | Denning TL, Takaishi H, Crowe SE, Boldogh I, Jevnikar A, Ernst PB. Oxidative stress induces the expression of Fas and Fas ligand and apoptosis in murine intestinal epithelial cells. Free Radic Biol Med. 2002;33:1641-1650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 17. | Pugh SY, Knowles CJ. Synthesis of catalase by “Streptococcus faecalis subsp. zymogenes”. Arch Microbiol. 1983;136:60-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Jackson CR, Fedorka-Cray PJ, Barrett JB. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J Clin Microbiol. 2004;42:3558-3565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 303] [Cited by in F6Publishing: 291] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 19. | Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, Jabes D, Goossens H. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473-4479. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 410] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 20. | Qin X, Singh KV, Weinstock GM, Murray BE. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect Immun. 2000;68:2579-2586. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 249] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 21. | Górska-Frączek S, Sandström C, Kenne L, Rybka J, Strus M, Heczko P, Gamian A. Structural studies of the exopolysaccharide consisting of a nonasaccharide repeating unit isolated from Lactobacillus rhamnosus KL37B. Carbohydr Res. 2011;346:2926-2932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Strus M, Kukla G, Rurańska-Smutnicka D, Przondo-Mordarska A, Heczko P. [Surface properties of Lactobacillus strains. II. Adherence to tissue culture surfaces]. Med Dosw Mikrobiol. 2001;53:253-258. [PubMed] [Cited in This Article: ] |
| 23. | Reuter M, Mallett A, Pearson BM, van Vliet AH. Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl Environ Microbiol. 2010;76:2122-2128. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 24. | Loh G, Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes. 2012;3:544-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 25. | Gilmore MS, Coburn S, Nallapareddy SR, Murray BE. Enterococcal Virulence. The Enterococci. Pathogenesis, Molecular Biology and Antibiotic Resistance. Washington: ASM PRESS 2002; 301-354. [Cited in This Article: ] |
| 26. | Clewell DB, Flannagan SE. The conjugative transposons of gram positive bacteria. Bacterial Conjugation. New York: Plenum Press 1993; 369-393. [Cited in This Article: ] |
| 27. | Kreft B, Marre R, Schramm U, Wirth R. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect Immun. 1992;60:25-30. [PubMed] [Cited in This Article: ] |
| 28. | Mäkinen PL, Mäkinen KK. The Enterococcus faecalis extracellular metalloendopeptidase (EC 3.4.24.30; coccolysin) inactivates human endothelin at bonds involving hydrophobic amino acid residues. Biochem Biophys Res Commun. 1994;200:981-985. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 29. | Andrews SC, Robinson AK, Rodríguez-Quiñones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215-237. [PubMed] [Cited in This Article: ] |
| 30. | Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380-3389. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 595] [Cited by in F6Publishing: 628] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 31. | Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, Amorena B, Leiva J, Penadés JR, Lasa I. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538-4545. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 392] [Cited by in F6Publishing: 401] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 32. | Hanin A, Sava I, Bao Y, Huebner J, Hartke A, Auffray Y, Sauvageot N. Screening of in vivo activated genes in Enterococcus faecalis during insect and mouse infections and growth in urine. PLoS One. 2010;5:e11879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 33. | Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529-536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 292] [Cited by in F6Publishing: 288] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 34. | Nathan C. Points of control in inflammation. Nature. 2002;420:846-852. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1820] [Cited by in F6Publishing: 1759] [Article Influence: 80.0] [Reference Citation Analysis (0)] |
| 35. | Al-Mushrif S, Jones BM. A study of the prevalence of hydrogen peroxide generating Lactobacilli in bacterial vaginosis: the determination of H2O2 concentrations generated, in vitro , by isolated strains and the levels found in vaginal secretions of women with and without infection. J Obstet Gynaecol. 1998;18:63-67. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 36. | Rezaie A, Parker RD, Abdollahi M. Oxidative stress and pathogenesis of inflammatory bowel disease: an epiphenomenon or the cause? Dig Dis Sci. 2007;52:2015-2021. [PubMed] [Cited in This Article: ] |
| 37. | Kruidenier L, Kuiper I, Lamers CB, Verspaget HW. Intestinal oxidative damage in inflammatory bowel disease: semi-quantification, localization, and association with mucosal antioxidants. J Pathol. 2003;201:28-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 262] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 38. | Abriouel H, Herrmann A, Stärke J, Yousif NM, Wijaya A, Tauscher B, Holzapfel W, Franz CM. Cloning and heterologous expression of hematin-dependent catalase produced by Lactobacillus plantarum CNRZ 1228. Appl Environ Microbiol. 2004;70:603-606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 42] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 39. | Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT, Macfarlane GT. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51:849-856. [PubMed] [Cited in This Article: ] |