Published online Aug 21, 2012. doi: 10.3748/wjg.v18.i31.4156
Revised: April 23, 2012
Accepted: April 27, 2012
Published online: August 21, 2012
AIM: To screen for genes related to metabotropic receptors that might be involved in the development of chronic hepatitis.
METHODS: Assessment of 20 genes associated with metabotropic receptors was performed in liver specimens obtained by punch biopsy from 12 patients with autoimmune and chronic hepatitis type B and C. For this purpose, a microarray with low integrity grade and with oligonucleotide DNA probes complementary to target transcripts was used. Evaluation of gene expression was performed in relation to transcript level, correlation between samples and grouping of clinical parameters used in chronic hepatitis assessment. Clinical markers of chronic hepatitis included alanine and aspartate aminotransferase, γ-glutamyltranspeptidase, alkaline phosphatase and cholinesterase activity, levels of iron ions, total cholesterol, triglycerides, albumin, glucose, hemoglobin, platelets, histological analysis of inflammatory and necrotic status, fibrosis according to METAVIR score, steatosis, as well as anthropometric body mass index, waist/hip index, percentage of adipose tissue and liver size in ultrasound examination. Gender, age, concomitant diseases and drugs were also taken into account. Validation of oligonucleotide microarray gene expression results was done with the use of quantitative real-time polymerase chain reaction (qRT-PCR).
RESULTS: The highest (0.002 < P < 0.046) expression among genes encoding main components of metabotropic receptor pathways, such as the α subunit of G-coupled protein, phosphoinositol-dependent protein kinase or arrestin was comparable to that of angiotensinogen synthesized in the liver. Carcinogenesis suppressor genes, such as chemokine ligand 4, transcription factor early growth response protein 1 and lysophosphatidic acid receptor, were characterized by the lowest expression (0.002 < P < 0.046), while the factor potentially triggering hepatic cancer, transcription factor JUN-B, had a 20-fold higher expression. The correlation between expression of genes of protein kinases PDPK1, phosphoinositide 3-kinase and protein kinase A (Spearman’s coefficient range: 0.762-0.769) confirmed a functional link between these enzymes. Gender (P = 0.0046) and inflammation severity, measured by alanine aminotransferase activity (P = 0.035), were characterized by diverse metabotropic receptor gene expression patterns. The Pearson’s coefficient ranging from -0.35 to 0.99 from the results of qRT-PCR and microarray indicated that qRT-PCR had certain limitations as a validation tool for oligonucleotide microarray studies.
CONCLUSION: A microarray-based analysis of hepatocyte metabotropic G-protein-related gene expression can reveal the molecular basis of chronic hepatitis.
- Citation: Cieśla A, Kuśmider M, Faron-Górecka A, Dziedzicka-Wasylewska M, Bociąga-Jasik M, Owczarek D, Ciećko-Michalska I, Cibor D, Mach T. Intrahepatic expression of genes related to metabotropic receptors in chronic hepatitis. World J Gastroenterol 2012; 18(31): 4156-4161
- URL: https://www.wjgnet.com/1007-9327/full/v18/i31/4156.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i31.4156
The natural course of chronic viral hepatitis is affected by progression of fibrosis and the risk of hepatocellular carcinoma development[1]. Current data indicate that intracellular signaling disturbances have an impact on progression of inflammation and fibrosis, as well as carcinogenesis, in the course of chronic hepatitis.
G-protein-coupled receptors (GPCRs) are a family of cell surface receptors which receive, integrate and enhance the majority of extracellular signals. After stimulation with different signals, GPCRs activate amplifying enzymatic cascades, regulatory proteins and ion channels. This activation regulates cellular responses, including growth, proliferation, and cell survival.
In the present study, using microarray DNA analysis, we attempt to define genes related to metabotropic receptors associated with progression of chronic hepatitis.
DNA technology with genomic profiling and cluster analysis allows determination of the role of genes in the pathogenesis of liver injury[2]. We assessed the activity of 20 genes encoding metabotropic receptors, some of which have been documented to have probable significance in the progression of chronic hepatitis.
The assessment of expression of genes of the main component of GPCR, such as the G-protein α subunit (GNAS), 3-phosphoinositide-dependent protein kinase-1 (PDPK1), phosphoinositide 3-kinase (PIK3CG), protein kinase A (AKT1) and arrestin β (ARRB2) was performed. We determined transcription factor JUN-B, ETS-domain protein (ELK4), early growth response protein 1 (EGR1) activated as the result of GPCR stimulation, angiotensinogen (AGT), which is a GPCR-ligand, dual-specificity protein phosphatases 14 (DUSP14), which is responsible for dephosphorylation of kinase products, calcitonin receptor proteins (CALCR), thyrotropin receptor (TSHR), colony stimulating factor-3 (CSF3), sphingosine-1-phosphate receptor 1 (EDG1), and lysophosphatidic acid receptor (EDG2) associated with G-protein. The selection of EDG2, EGR1, JUN-B, chemokine (C-C motif) ligand 4 (CCL4), ELK4 genes was based on their association with cellular proliferation, differentiation and apoptosis[3-6]. Additionally, the selected group represented genes involved in regulation of inflammatory response and liver fibrosis, which included genes for interleukin 1β (IL-1β) and its receptors IL1 type I (IL1R1), IL1 type II (IL1R2), CALCR, EDG1, CCL4, AGT and adhesion molecules vascular cell adhesion molecule 1 (VCAM1)[7-14].
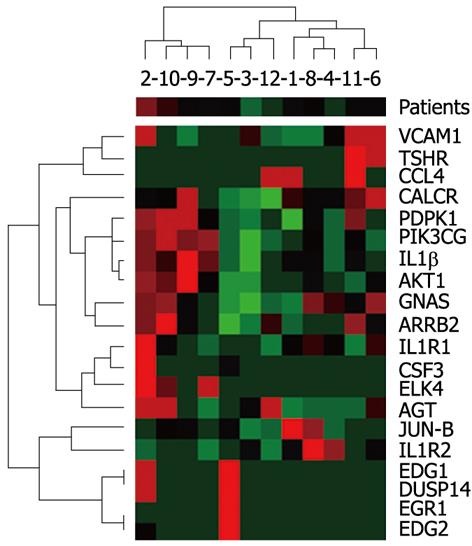
In the group of 12 patients (7 men, 5 women; age 36 ± 10.8 years) with chronic hepatitis type B (2 patients) and C (8 patients) and autoimmune hepatitis (2 patients), according to clinical indications, a liver biopsy was performed by the Menghini technique. From the obtained liver sections, a sample of 2-3 mm in length was frozen at -75 °C until the analysis of mRNA of 20 selected genes was performed (gene list Figure 1). During the histopathological investigation of the biopsies, the degree of inflammation and fibrosis was assessed by the METAVIR score and steatosis by the steatosis scoring system.
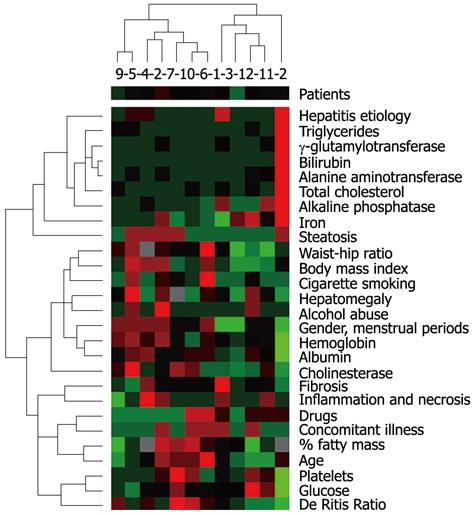
On the day of liver biopsy, the activity of serum alanine aminotransferase (ALT/GPT Cobas, Roche Diagnostics, Mannheim, Germany), aspartate aminotransferase (ASPAT/GOT Cobas), γ-glutamyltransferase (GGT Cobas), alkaline phosphatase (ALP Cobas), cholinesterase (CHE Cobas), the level of iron (Fe Cobas), total cholesterol (CHOD-PAP Cobas), triglycerides (TG Cobas), albumin (ALB plus Cobas), glucose (GLU Cobas), hemoglobin and platelet (Sysmex XE-2100, Sysmex Europe GMBH, Norderstedt, Germany) were assessed. Anthropometric measurements, including body mass index, waist/hip index and the percentage of fatty mass was assessed by skin fold thickness. The size of the liver was measured during ultrasound examination of the abdomen. Patient characteristics included the presence of concomitant illnesses, drug history, as well as the use of such substances as alcohol and cigarettes (Figure 2). In women, the menstrual cycle phase and menopause were taken into account. Biochemical, histological and anthropometric parameters are presented in Figure 2. The gene expression studies were performed using microarray (with low integration level), with DNA oligonucleotides complementary to the investigated transcripts. Total RNA was isolated from liver samples using the TRI Reagent (Sigma-Aldrich, St. Louis, United States), and then purified using the RNeasy MiniElute spin columns with a DNA eliminator (Qiagen, Hilden, Germany). The quantity of isolated RNA was assessed by a spectrophotometer (NanoDrop, Wilmington, United States), and subsequently its degradation level was measured by a capillary electrophoresis system (Experion, Bio-Rad, Hercules, United States). For further analysis only samples without evidence of RNA degradation were qualified (RQI > 8.5 Rna Quality Index). Subsequently, based on the obtained RNA, probes were synthesized according to the manufacturer’s instructions (SABiosciences, Frederick, United States), and then hybridized to a microarray. We used the Oligo GEArray Human GPCR Signaling Pathway Finder Microarray (OHS-071) supplied by SABioscences. Detection of the array probes is achieved based on chemiluminescence, using the FujiLAS System (FujiFilm, Tokyo, Japan). The resulting images (the signal density) were quantified using the OligoAnalyser (SABiosciences). The obtained results describing the relative levels of gene expression (with respect to the reference gene) were further examined.
Diversification in gene expression was assessed by agglomerative hierarchical clustering methods. Using the Spearman’s rank correlation coefficient for activity of the investigated genes, we searched for pairs of objects and then for clusters with the smallest distance (Figure 1). An analogous classification was carried out using biochemical, histological and anthropometric parameters (Figure 2). Determination of a direct correlation between gene expression and clinical features was done based on agglomerative hierarchical clustering of both the investigated indicators of chronic hepatitis. The genotype-phenotype distinction was analyzed using Fisher’s exact test, to determine the difference between clustering of patients achieved on the basis of a dendrogram of clinical signs and a dendrogram of gene expression (Figures 1 and 2). Differentiation of clinical parameters between groups of patients with the biggest difference in gene expression was performed by the Mann-Whitney U test and Fisher’s exact test. In the present study, these were patients number 2, 10, 9, 7 vs the rest (Figure 1). The Wilcoxon signed-rank test was used to assess differential expression between the selected genes, to determine the three groups of genes with the highest, moderate, and lowest activity (Figure 3).
The results of the microarray experiment were verified by means of quantitative real-time polymerase chain reaction (qRT-PCR) for IL1B, VCAM1, PIK3CG, AGT, PDPK1, GNAS, JUN-B, EDG2, CCL4, EGR1, IL1R1, IL1R2, CALCR, AKT1 and ARRB2. Total RNA acquired from the tissues of interest was reverse-transcribed using the High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, United States) for the 2-step qRT-PCR assays. The qRT-PCR was performed on a Chromo 4 (Bio-Rad), using the ABI SYBR Green master mix (Applied Biosystems). Primer sets against sequences of genes are indicated by microarray. They are commercially available at OriGene (Rockville, United States), but were additionally checked for specificity with National Center for Biotechnology Information (NCBI) The Basic Local Alignment Search Tool (BLAST). The optimum annealing temperature for each primer set was determined prior to the analysis of experimental samples. Following amplification, dissociation curves were analyzed for each reaction. A sample volume of 20 μL was used for all assays which contained a 1X final concentration of SYBR green PCR master mix, 100 nmol gene specific primers, and 1 μL of template. The assays were run using the following protocol: 95 °C for 10 min, 95 °C for 40 s, gene specific annealing temperature (58-62 °C) for 60 s for 40 cycles, followed by a gradual increase in temperature from 55 °C to 95 °C during the dissociation stage.
Following amplification, the instrument software was used to set the baseline and threshold for each reaction, as well as to determine the reaction efficiency. A cycle threshold (Ct) was assigned at the beginning of the logarithmic phase of PCR amplification and the difference in the Ct values [corrected for reaction efficiency: Ct = Ct × log(efficiency)/log(2)] of the housekeeping genes [mean Ct of glucuronidase (GUS) and beta-actin (BAKT)] and the gene of interest were used to determine the relative expression of the gene in each sample. Relative expression levels were then calculated as fold changes to the housekeeping genes, where each PCR cycle represented a 2-fold change.
The data of PCR and microarray experiments were then correlated using Pearson’s coefficient.
The study was carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association.
Using the agglomerative hierarchical clustering methods we found statistically significant correlations between the EDG1, DUSP14, EGR1 and EDG2 genes and PDPK1, PIK3CG, AKT1, as well as clustering, though not statistically significant, for GNAS and ARRB2 (Figure 1). Spearman’s rank correlation coefficients were 0.762 for PDPK1 and AKT1, 0.769 (P < 0.05) for PDPK1 and PIK3CG, 0.769 (P < 0.05) for AKT1 and PIK3CG, and 0.699 (P < 0.05) for GNAS and ARRB2. In case of EDG1, DUSP14, EGR1 and EDG2, in 10 out of 12 patients there was no transcriptional activity. Except for the transcriptional factor JUN-B and the stage of fibrosis r = 0.667 (P < 0.05), we did not observe any correlation between the activity of the selected genes and the investigated clinical factors. Assessing patient distribution based on dendrograms of the investigated genotype and phenotype, a difference on the border of statistical significance was found in Fisher’s exact test (P = 0.08). The patients with the highest diversity in gene expression (Figure 1) showed a statistically significant difference in gender and ALT activity (P = 0.0046 and P = 0.035, respectively). Based on expression activity, the genes were divided into a group with high activity, which included GNAS, PDPK1, AGT and ARRB2 and statistically differed (0.002 < P < 0.046) from the genes with moderate and low expression. The group of genes with moderate activity of AKT1, PIK3CG, IL1β, VCAM1, JUN-B and CALCR also showed a statistically significant difference (0.002 < P < 0.041) compared with genes which were classified as belonging to the group with low expression (Figure 3).
For the following genes: JUN-B, EDG2, CCL4 and EGR1, estimated with the use of microarrays and qRT-PCR, there was a strong positive correlation, with Pearson’s coefficient within the range of 0.61-0.99. For other genes, IL1B, VCAM1, PIK3CG, AGT, PDPK1, and GNAS, the correlation was weakly positive, with Pearson’s coefficient ranging from 0.07 to 0.43; and for IL1R1, IL1R2, CALCR, AKT1, and ARRB2 the correlation of expression estimated with the two methods was weakly negative (Pearson’s coefficient ranging from -0.1 to -0.35).
With the exception of the correlation between JUN-B and fibrosis stage, we did not reveal any direct relationship between expression of genes related to metabotropic receptors in hepatocytes and anthropometric, histological and biochemical parameters that are commonly used for monitoring progression of chronic hepatitis. In assessing the connection between gene expression and the investigated clinical features, we found an impact of gender and concentration of ALT in the serum on changes in gene expression. The effect of these two factors was detected as the result of the analysis of not any single, but rather all the investigated genes. In the probability test, it was gender which better determined changes in gene activity rather than ALT. In chronic hepatitis C and B, gender is the factor which defines the course and prognosis of the disease[1]. Changes in gene expression resulting from gender differences can influence the progression of liver disease.
In the present study, among different markers of inflammation and fibrosis in the liver parenchyma, only the ALT level differentiated the gene activity. Among the investigated genes, there were important mediators of inflammation, including IL-1β, with their receptors, as well as chemokines and adhesion factors. Also protein products of such genes as EDG1, EDG2, CALCR are involved in the induction of the inflammatory response[9,15]. In contrast to ALT, a small histological difference used to assess the inflammatory process in the liver, classified in the majority of cases as score 1 or 2 according to METAVIR, were not useful in assessing gene expression.
The reported values of genes expression and their statistical diversity allow distinction between three groups of genes with high, moderate and low activity. Because there is no direct correlation between gene activity and clinical markers of chronic hepatitis, it is difficult to determine if the observed gene expression is induced or constitutional.
In the group of genes with the highest activity there were genes of the main metabotropic receptor proteins, GNAS, PDPK1 and ARRB2. Their high transcriptional activity is related to their essential role in the metabotropic receptor system. GNAS expression, despite individual differences, showed the highest correlation with ARRB2 among the investigated genes. Proportional to the level of stimulation of GPCR, ARRB2 triggers the mechanism of receptor internalization, which is an adaptation to overstimulation[16]. Also mRNA encoding AGT was characterized by high expression. This observation remains in accordance with current literature and is associated with the liver being the main site of AGT synthesis[17]. Because AGT is incorporated only indirectly by the renin-angiotensin system in the progression of liver fibrosis[18,19], its gene activity did not correlate with the histological assessment of this process.
In the group of genes with moderate activity, there were genes which encode products involved in the pathogenesis of liver injury. Disturbances of PIK3CG described in chronic hepatitis as caused by NS5 protein of hepatitis C virus and HBx protein of hepatitis B virus destabilize and damage hepatocytes[20,21]. However, in the present study, expression of PIK3CG did not correlate with the markers of liver injury.
As there were no cases with advanced fibrosis among the investigated patients, we did not observe any increased expression of VCAM1, which is characteristic only for liver cirrhosis[22].
The correlation of expression of phosphatidylinositol kinases observed in the study confirms the functional association of the investigated enzymes. A significantly higher expression of PDPK1 compared with PIK3CG is due to the amplification of PDPK1 during phosphorylation catalyzed by PIK3CG. PDPK1 in turn activates AKT1 correspondingly; therefore, the 3-fold higher activity of PDPK1 is noteworthy.
Among the investigated phosphatases that dephosphorylate active kinases, DUSP14 was measured. However, this phosphatase, which is not functionally associated with the presently studied kinases, showed low expression values and was not correlated with the expression of kinases. Interestingly, a statistically significant correlation was determined between the stage of fibrosis and expression of transcriptional factor JUN-B. This factor, in the malfunctioning of the liver, is responsible for reprogramming of hepatocyte genes to the phase of cell proliferation[23,24]. Its increase is frequently observed in liver injury and hepatic carcinogenesis[25].
The higher expression of this factor linked to carcinogenesis correlated with a low activity of the genes CCL4, EGR1, EDG2 involved in the induction of apoptosis and suppression of neoplasia[26].
The results obtained in the present study indicate that qRT-PCR has certain limitations as a validation tool for oligonucleotide microarray studies. Only four genes have shown a similar expression pattern between results obtained with the use of both techniques. A weak positive and negative correlation observed for other genes might result from potential pitfalls inherent in both approaches, and might be a source of errors encountered while employing each method.
However, the range of differences in the correlation coefficient observed in the present study remains within the range described in the literature, from -0.48 to 0.94[27,28]. The results obtained in this study reflect the debate over which methods produce the most accurate measurements of gene expression.
In conclusion, gender and inflammation activity, as determined by ALT level, were associated with a more diverse pattern of metabotropic receptor gene expression. The highest gene expression was observed for mRNA of the main components of the metabotropic receptor pathway, such as GNAS, PDPK1, ARRB2, and correlated with mRNA of angiotensinogen synthesized in the liver. The correlation of expression of protein kinases PDPK1, PIK3CG and AKT1 points to a functional association of these enzymes. The genes suppressing carcinogenesis, CCL4, EGR, EDG2, were characterized by the lowest expression levels among the investigated genes. On the other hand, JUN-B, a factor potentially involved in the development of hepatocellular cancer, was characterized by a 20-fold higher level of expression.
Metabotropic G protein-coupled receptors activate various signaling pathways, which trigger multiple sub-cellular reactions. Microarray-based analysis of expression of hepatocyte genes related to metabotropic receptors can reveal the molecular basis of liver diseases.
The natural course of chronic viral hepatitis is associated with progression of fibrosis and the risk of hepatocellular carcinoma development. Current data indicate that intracellular signaling disturbances have an impact on progression of inflammation and fibrosis as well as carcinogenesis in the course of chronic hepatitis.
The highest gene expression was in the mRNAs of the main components of the metabotropic receptor pathways, such as the α subunit of G-coupled protein, phosphoinositol-dependent protein kinase (PDPK1) and arrestin β and correlated with the mRNA for angiotensinogen synthesized in liver. Carcinogenesis suppressor genes such as chemokine CCL4, transcription factor EGR1 and lysophosphatidic acid receptor were characterized by the lowest expression, while the factor potentially triggering hepatic cancer, JUN-B, had 20-fold higher expression. Comparable expression of genes encoding protein kinases PDPK1, phosphoinositide 3-kinase and protein kinase A confirms a functional link between these enzymes. Gender and inflammation severity, measured by alanine aminotransferase activity, were characterized by different expression patterns of genes related to metabotropic receptors.
Results of the presented work enables better delineation of mechanisms governing the course of chronic hepatitis and form the basis for future investigations.
G-protein-coupled receptors are a family of the cell surface receptors, which receive, integrate and enhance most of the extracellular signals implicated in cell growth, proliferation, and survival. Microarray DNA technology with genomic profiling and cluster analysis allows determination of the role of genes in the pathogenesis of liver injury.
This is a good descriptive study in which authors screen for genes related to metabotropic receptors family that might be involved in the development of chronic hepatitis. The results are interesting and suggest that a microarray-based analysis of hepatocyte metabotropic G protein-related gene expression can reveal the molecular basis of chronic hepatitis.
Peer reviewer: Dr. Shashi Bala, Department of Medicine, Umass Medical School, 364 Plantation Street, LRB270l, Worcester, MA 01605, United States
S-Editor Gou SX L-Editor Cant MR E-Editor Zhang DN
| 1. | Poynard T, Bedossa P, Opolon P. Natural history of liver fibrosis progression in patients with chronic hepatitis C. The OBSVIRC, METAVIR, CLINIVIR, and DOSVIRC groups. Lancet. 1997;349:825-832. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2199] [Cited by in F6Publishing: 2083] [Article Influence: 77.1] [Reference Citation Analysis (0)] |
| 2. | Furuta K, Sato S, Yamauchi T, Ozawa T, Harada M, Kakumu S. Intrahepatic gene expression profiles in chronic hepatitis B and autoimmune liver disease. J Gastroenterol. 2008;43:866-874. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 3. | van Meeteren LA, Moolenaar WH. Regulation and biological activities of the autotaxin-LPA axis. Prog Lipid Res. 2007;46:145-160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 280] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 4. | Cooper AB, Wu J, Lu D, Maluccio MA. Is autotaxin (ENPP2) the link between hepatitis C and hepatocellular cancer? J Gastrointest Surg. 2007;11:1628-1634; discussion 1634-1635. [PubMed] [Cited in This Article: ] |
| 5. | Martínez Martínez CM, Hernández Pando R. [Chemokines, a new family of cytokines in inflammatory cell recruitment]. Rev Invest Clin. 1999;51:255-268. [PubMed] [Cited in This Article: ] |
| 6. | Goto T, Kato N, Yoshida H, Otsuka M, Moriyama M, Shiratori Y, Koike K, Matsumura M, Omata M. Synergistic activation of the serum response element-dependent pathway by hepatitis B virus x protein and large-isoform hepatitis delta antigen. J Infect Dis. 2003;187:820-828. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 7. | Hay DL, Christopoulos G, Christopoulos A, Poyner DR, Sexton PM. Pharmacological discrimination of calcitonin receptor: receptor activity-modifying protein complexes. Mol Pharmacol. 2005;67:1655-1665. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 146] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Zeremski M, Petrovic LM, Talal AH. The role of chemokines as inflammatory mediators in chronic hepatitis C virus infection. J Viral Hepat. 2007;14:675-687. [PubMed] [Cited in This Article: ] |
| 9. | Kroeger I, Erhardt A, Abt D, Fischer M, Biburger M, Rau T, Neuhuber WL, Tiegs G. The neuropeptide calcitonin gene-related peptide (CGRP) prevents inflammatory liver injury in mice. J Hepatol. 2009;51:342-353. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Haruta I, Tokushige K, Komatsu T, Ikeda I, Yamauchi K, Hayashi N. Clinical implication of vascular cell adhesion molecule-1 and very late activation antigen-4 interaction, and matrix metalloproteinase-2 production in patients with liver disease. Can J Gastroenterol. 1999;13:721-727. [PubMed] [Cited in This Article: ] |
| 11. | Knittel T, Dinter C, Kobold D, Neubauer K, Mehde M, Eichhorst S, Ramadori G. Expression and regulation of cell adhesion molecules by hepatic stellate cells (HSC) of rat liver: involvement of HSC in recruitment of inflammatory cells during hepatic tissue repair. Am J Pathol. 1999;154:153-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Watanabe Y, Morita M, Ikematsu N, Akaike T. Tumor necrosis factor alpha and interleukin-1 beta but not interferon gamma induce vascular cell adhesion molecule-1 expression on primary cultured murine hepatocytes. Biochem Biophys Res Commun. 1995;209:335-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 13. | García-Monzón C, Sánchez-Madrid F, García-Buey L, García-Arroyo A, García-Sánchez A, Moreno-Otero R. Vascular adhesion molecule expression in viral chronic hepatitis: evidence of neoangiogenesis in portal tracts. Gastroenterology. 1995;108:231-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 96] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Proost P, Wuyts A, van Damme J. The role of chemokines in inflammation. Int J Clin Lab Res. 1996;26:211-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 139] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 15. | Kaneko T, Murakami T, Kawana H, Takahashi M, Yasue T, Kobayashi E. Sphingosine-1-phosphate receptor agonists suppress concanavalin A-induced hepatic injury in mice. Biochem Biophys Res Commun. 2006;345:85-92. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Klaasse EC, Ijzerman AP, de Grip WJ, Beukers MW. Internalization and desensitization of adenosine receptors. Purinergic Signal. 2008;4:21-37. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Takahashi D, Tamura K, Ushikubo T, Moriya A, Yokoyama N, Nyui N, Chiba E, Hibi K, Ishigami T, Yabana M. Relationship between hepatic angiotensinogen mRNA expression and plasma angiotensinogen in patients with chronic hepatitis. Life Sci. 1997;60:1623-1633. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 18. | Forrest EH, Thorburn D, Spence E, Oien KA, Inglis G, Smith CA, McCruden EA, Fox R, Mills PR. Polymorphisms of the renin-angiotensin system and the severity of fibrosis in chronic hepatitis C virus infection. J Viral Hepat. 2005;12:519-524. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Xiao F, Wei H, Song S, Li G, Song C. Polymorphisms in the promoter region of the angiotensinogen gene are associated with liver cirrhosis in patients with chronic hepatitis B. J Gastroenterol Hepatol. 2006;21:1488-1491. [PubMed] [Cited in This Article: ] |
| 20. | Wang ZL, Wu XH, Song LF, Wang YS, Hu XH, Luo YF, Chen ZZ, Ke J, Peng XD, He CM. Phosphoinositide 3-kinase gamma inhibitor ameliorates concanavalin A-induced hepatic injury in mice. Biochem Biophys Res Commun. 2009;386:569-574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Mukherji A, Janbandhu VC, Kumar V. HBx protein modulates PI3K/Akt pathway to overcome genotoxic stress-induced destabilization of cyclin D1 and arrest of cell cycle. Indian J Biochem Biophys. 2009;46:37-44. [PubMed] [Cited in This Article: ] |
| 22. | Bruno CM, Sciacca C, Cilio D, Bertino G, Marchese AE, Politi G, Chinnici L. Circulating adhesion molecules in patients with virus-related chronic diseases of the liver. World J Gastroenterol. 2005;11:4566-4569. [PubMed] [Cited in This Article: ] |
| 23. | Beauchamp RD, Papaconstantinou J, Henderson AM, Sheng HM, Townsend CM, Thompson JC. Activation of hepatic proliferation-associated transcription factors by lipopolysaccharide. Surgery. 1994;116:367-376; discussion 376-7. [PubMed] [Cited in This Article: ] |
| 24. | Morello D, Lavenu A, Babinet C. Differential regulation and expression of jun, c-fos and c-myc proto-oncogenes during mouse liver regeneration and after inhibition of protein synthesis. Oncogene. 1990;5:1511-1519. [PubMed] [Cited in This Article: ] |
| 25. | Liao DZ, Blanck A, Gustafsson JA, Hällström IP. Expression of the c-jun, jun-B, ets-2 and liver regeneration factor-1 (LRF-1) genes during promotion and progression of rat liver carcinogenesis in the resistant hepatocyte model. Cancer Lett. 1996;100:215-221. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 26. | Baek SJ, Wilson LC, Hsi LC, Eling TE. Troglitazone, a peroxisome proliferator-activated receptor gamma (PPAR gamma ) ligand, selectively induces the early growth response-1 gene independently of PPAR gamma. A novel mechanism for its anti-tumorigenic activity. J Biol Chem. 2003;278:5845-5853. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 153] [Cited by in F6Publishing: 155] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 27. | Etienne W, Meyer MH, Peppers J, Meyer RA. Comparison of mRNA gene expression by RT-PCR and DNA microarray. Biotechniques. 2004;36:618-20, 622, 624-6. [PubMed] [Cited in This Article: ] |
| 28. | Beckman KB, Lee KY, Golden T, Melov S. Gene expression profiling in mitochondrial disease: assessment of microarray accuracy by high-throughput Q-PCR. Mitochondrion. 2004;4:453-470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 26] [Article Influence: 1.3] [Reference Citation Analysis (0)] |