Published online Aug 7, 2012. doi: 10.3748/wjg.v18.i29.3896
Revised: March 6, 2011
Accepted: May 12, 2012
Published online: August 7, 2012
AIM: To analyze the epidermal growth factor receptor pathway substrate 8 (EPS8) expression status and role in colorectal carcinogenesis given that EPS8 has a conserved actin barbed-end capping function that is required for proper maturation in intestinal cells.
METHODS: We studied 8 colon cancer cell lines and 58 colorectal tumors (19 adenomas and 39 carcinomas). We performed expression microarray analysis of colon cancer cell lines followed by loss of heterozygosity (LOH) analysis and immunohistochemistry for EPS8 expression in colon tumors. Subsequently, we performed mutation analysis by direct sequencing and methylation analysis by bisulfite sequencing and methylation-specific polymerase chain reaction assays.
RESULTS: Expression microarray analysis of colon cancer cell lines showed overexpression of EPS8 transcript in all lines but RKO. Genome wide loss of heterozygosity (LOH) analysis of colon tumors, showed considerable LOH at the EPS8 gene locus. Immunohistochemically, EPS8 was constitutively expressed in normal colonic mucosa with a dot-like supranuclear localization with accentuation at the luminal surface supporting its proposed role in epithelial maturation. Nineteen colon tumors (4 adenoma, 15 carcinoma) out of 51 (37%) showed strikingly tumor specific EPS8 protein loss. Of the remaining tumors, 5/51 (2 adenoma, and 3 carcinoma, 10%) showed marked overexpression, while 27/51 tumors (53%) showed retained expression. Mutation analysis revealed a missense mutation (c.794C>T, p.R265C) in exon 8 in RKO. The EPS8 promoter was also methylated in RKO, but there was no significant methylation in other cell lines or carcinoma specimens.
CONCLUSION: The loss of EPS8 expression in colorectal adenomas and carcinomas suggests that down regulation of this gene contributes to the development of a subset of colorectal cancers, a finding which could have applications in diagnosis and treatment.
- Citation: Abdel-Rahman WM, Ruosaari S, Knuutila S, Peltomäki P. Differential roles of EPS8 in carcinogenesis: Loss of protein expression in a subset of colorectal carcinoma and adenoma. World J Gastroenterol 2012; 18(29): 3896-3903
- URL: https://www.wjgnet.com/1007-9327/full/v18/i29/3896.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i29.3896
Epidermal growth factor receptor pathway substrate 8 (EPS8) is a 97-kDa protein that is tyrosine phosphorylated following stimulation of receptor tyrosine kinases (RTK)[1]. EPS8 plays a role in signal transduction from RTK and PI3K[2,3] leading to Rac-mediated actin remodeling, ruffle formation and cell motility[4]. In Caenorhabditis elegans (C. elegans), eps-8 knockdown animals were zygotic lethal due to major defects in the gut, and the isoform EPS-8A was shown to be required for proper apical morphogenesis in the intestinal cells. This phenotype was correlated with an actin barbed-end capping activity, which is present in the C terminus of the EPS-8A isoform and is required for coordinately terminated elongation of the microvillar actin bundle core[5]. This function of EPS8 protein is conserved throughout evolution[6].
EPS8 was recently shown to be overexpressed in advanced stage human cancers including colon cancer cell lines and specimens[7]. Our expression microarray analysis of colon cancer cell lines confirmed this overexpression. Interestingly, there was a strikingly low level of EPS8 in RKO, a colon cancer cell line with a marked lack of constitutive β-catenin regulated transcription[8], which prompted us to conduct a comprehensive immunohistochemical, genetic, and epigenetic analysis of EPS8 alterations in colorectal cell lines and patient specimens.
We studied 8 colon cancer cell lines (RKO, HCA7, KM12, LoVo, DLD1, HCT116, SW48, LIM1215) and 58 colorectal tumors (19 adenomas and 39 carcinomas) of which 21 tumors (4 adenomas and 17 carcinomas) belong to a well characterized series of familial colon cancer type X (FCC-X). 15 adenomas and 22 carcinomas were sporadic. Clinicopathological characteristics of these cohorts are available in our previous publications[9-11]. The FCC-X originated from 19 cancer families clinically indistinguishable from Lynch syndrome (hereditary non-polyposis colon cancer), but screening negative for the known predisposing genes by multiple techniques[12]. We identified distinct molecular features in these tumors including high frequency of genomically stable carcinomas with membranous β-catenin, however, the predisposing defects in these families remain elusive[9]. The sporadic colorectal tumors were selected from a larger cohort with the aim to include equal numbers of tumors with membranous vs nuclear β-catenin.
Fresh frozen and/or paraffin derived specimens of tumor and matching normal tissues were collected from pathology departments of different hospitals and used for immunohistochemical analysis and DNA extraction according to standard protocols. All human specimens were obtained after informed consent and approvals from the appropriate institutional review boards of the Helsinki University Central Hospital.
Analyses were performed using HG-U133 Plus 2.0 array (Affymetrix, Santa Clara, CA, United States). The protocols for HG-U133 Plus 2.0 arrays were as described by the manufacturer (Affymetrix, Santa Clara, CA, United States). Briefly, total RNA was extracted from cell lines by RNeasy (Qiagen, Valencia, CA, United States). An aliquot of each RNA sample was run on a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, United States) to visualize and quantify the degree of RNA integrity. Double-stranded cDNA was synthesized from 5 μg of total RNA using the GeneChip One-Cycle cDNA synthesis kit, followed by cleanup with the GeneChip Sample Cleanup Module, in vitro transcription (IVT) and Biotin labeling reaction using the GeneChip IVT Labeling kit, and clean-up and quantification of the biotin-labeled cRNA yield by spectrophotometric analysis. All kits were from Affymetrix. Fragmentation of the 8 μg cRNA and hybridizations to test chips and the HG-U133 Plus 2.0 array were carried out according to Affymetrix protocols, and microarrays were processed by the Affymetrix Fluidics Station 450 and scanned with an Affymetrix GeneChip Scanner 7G. Captured images were analyzed using Microarray Suite version 5.0 algorithm (Affymetrix). All quality control criteria recommended by Affymetrix were observed in the “Test” chips and sample chips.
The hybridization data were pre-processed using Robust Multi-array Average (RMA[13]), designed to enhance the comparability of expression measures between separate arrays. RMA pre-processing produces a single expression measure for each probe set in the Affymetrix array which can be readily used in subsequent analyses. As duplicate arrays were available for each cell line, the median of the two RMA values was used as the expression value. Gene assignments of the probes were extracted from the Affymetrix annotation files and genes with ambiguous information about the physical location were excluded from the analysis.
For the EPS8 loss of heterozygosity (LOH) analysis we chose two microsatellite markers spanning the EPS8 gene locus at Ensembl cytogenetic band 12p12.3 and surrounding the gene from both directions (http://www.ensembl.org). The physical distances between loci in mega-bases according to Ensembl are given in parentheses: pter D12S1580 - (2.4 Mb) - EPS8 - (0.4 Mb) - D12S1728 qter. The polymerase chain reaction (PCR) amplification primers were from Généthon Microsatellite Maps at http://www.genlink.wustl.edu/genethon_frame. The forward primers were fluorescently labeled with carboxyfluorescein and PCR fragments were run on the ABI3730 sequencer/genotyper and results analyzed using GeneMapper v3 software (Applied Biosystems, Forster City, CA, United States) as described previously[9]. A sample was scored as showing LOH, if one of the alleles had decreased 40% or more, and borderline LOH or allelic imbalance, if the decrease was 25%-39% for one allele.
Microsatellite instability status was determined using the Bethesda panel of 5 microsatellite markers and additional markers as described[9,14,15]. Tumors with two or more unstable markers were considered to have high-degree microsatellite instability (MSI-H), while those with one unstable marker had low-degree microsatellite instability (MSI-L) and those with no unstable markers were microsatellite stable (MSS). MSI-H cancers were mostly excluded from this study cohort to enable the LOH study.
Four-micrometer sections from formalin-fixed paraffin-embedded tissues were mounted on silanized slides (Dako, Glostrup, Denmark) and air-dried overnight at 37 °C. After de-waxing and re-hydration in distilled water, sections were subject to heat-induced target retrieval in 1 mmol/L ethylenediaminetetraacetic acid buffer pH 8.0 for 5 min at 750 W followed by 5 min at 450 W in a microwave oven. After cooling, the slides were washed in Tris-buffered saline pH 7.2 and subsequent staining steps were performed manually with the Dako EnVision+ System, Peroxidase (DAB), according to the manufacturer’s instructions (Dako, Glostrup, Denmark). In addition, after blocking endogenous peroxidase activity, and prior to incubation with the primary antibody, the sections were incubated with 10% normal (non-immune) goat serum (Dako, Glostrup, Denmark) for 30 min. The primary antibodies were purified rabbit polyclonal anti EPS8 Antibody (C-terminal, clone RB4006, Abgent, San Diego, CA, United States) and purified mouse monoclonal anti-β-catenin antibody (clone 14, BD Transduction Laboratories, Erembodegem, Belgium). Paired tumor and normal mucosa were in the same section and the normal tissues were used as an internal reference for evaluation of staining results. β-catenin immunohistochemical staining for identification of its sub-cellular localization and the interpretation of results were performed as described[9]. β-catenin expression was considered aberrant if there was nuclear staining of more than 10% or cytoplasmic staining of more than 50% of tumor cells (not observed in the matching normal tissue). For approximately half of the tumors, β-catenin data were available from our earlier studies[9], while for the rest, these results were generated in the present investigation.
All coding exons of the EPS8 gene were examined by direct sequencing. The primer sequences and PCR conditions are given in Table 1. EPS8 sequences were compared to that of GenBank accession number RefSeq NC_000012.10, and exon information was from Ensembl ENST00000389337. DNA mutation numbering is based on cDNA sequence where +1 corresponds to the A of the ATG translation initiation codon and the initiation codon is codon 1. Sequence changes reported here were present in sequence tracing from both the forward and reverse direction and were reproducibly found in 2 independent PCR products from cases of interest.
| Primer | Sequence | Primer for sequence | Tm | PCR fragment (bp) |
| EPS8ex1F | tcctggcagcaacacatatt | F | 59 | 227 |
| EPS8ex1R | ccaaatcaaattcccccaaa | 62 | ||
| EPS8ex2F | aacccaacacaatgaccttttt | 60 | 251 | |
| EPS8ex2R | tcactgcctcattccaaaca | R | 60 | |
| EPS8ex3F | gagatagccacatgataccaaca | 59 | 195 | |
| EPS8ex3R | tgttcctcaagggtcactctaaa | F | 60 | |
| EPS8ex4F | tctttttcctttttgccaat | 56 | 280 | |
| EPS8ex4R | ttccatccattttcaaacaatc | R | 58 | |
| EPS8ex5F | gattgtttgaaaatggatggaa | F | 59 | 261 |
| EPS8ex5R | aaagctcccagacaatctgc | 59 | ||
| EPS8ex6F | tcagacaaggaacaatcccttt | F | 60 | 251 |
| EPS8ex6R | tttttctaactctttggggaaaaa | 60 | ||
| EPS8ex7F | agtaccacaagttgagttaattgat | F | 55 | 264 |
| EPS8ex7R | tcccaacccaaagtaagtgttc | 60 | ||
| EPS8ex8F | ggcaaatggtcctccttttt | F | 60 | 206 |
| EPS8ex8R | ccagtgatctaaaggcgactc | 59 | ||
| EPS8ex9F | tgggctgcttccttttctaa | F | 60 | 280 |
| EPS8ex9R | ctggagatctaaccaggcatt | 58 | ||
| EPS8ex10F | cctctcctctcgcttatttca | F | 58 | 234 |
| EPS8ex10R | cacacccccacaaaatctat | 57 | ||
| EPS8ex11F | gaccgtcccctctgtgtcta | F | 60 | 229 |
| EPS8ex11R | ccagacagacacttggggtta | 60 | ||
| EPS8ex12F | cttgtttttgccatgggttt | F | 60 | 265 |
| EPS8ex12R | aaggcattataggtggtaaatgct | 59 | ||
| EPS8ex13F | tatgccttcattccctcctg | F | 60 | 297 |
| EPS8ex13R | tgaataaaatgagaacttgcaatca | 60 | ||
| EPS8ex14F | tgacctgagtgctgattcaaa | F | 59 | 274 |
| EPS8ex14R | gacactgtcacctctgttagcac | 59 | ||
| EPS8ex15F | ctttaggaagagctagcagaat | 54 | 250 | |
| EPS8ex15R | aatacttttgaaggaaagtttagttat | R | 54 | |
| EPS8ex16F | gggaacttctttcgtagaatgg | F | 59 | 267 |
| EPS8ex16R | aagagtgataacttcgtaaatgtgt | 56 | ||
| EPS8ex17F | aaagtataatttgttttcctagcc | F | 55 | 315 |
| EPS8ex17R | tgcctccctgggaaacttac | 61 | ||
| EPS8ex18F | ggggttctagagagggtgatgt | F | 61 | 292 |
| EPS8ex18R | tgttgtacacacagaattgcaaag | 60 | ||
| EPS8ex19F | tttctcctttgtttgtaggcaat | 59 | 256 | |
| EPS8ex19R | aatagttgtttccagagctttcaa | R | 59 | |
| EPS8ex20F | gcagcctgcacaagtcagta | F | 60 | 203 |
| EPS8ex20R | aatgccaaaaacaatggagtt | 59 |
To search for CpG islands in the EPS8 promoter, the EMBOSS CpG Plot program was used with default definitions (http://www.ebi.ac.uk/emboss). Two adjacent CpG islands were identified, together spanning 750 bp within and upstream of the untranslated exon number 1. This area was divided into two overlapping segments to screen cell lines and normal lymphocytes for methylation by bisulfite sequencing. The primers for the distal region were, forward, 5’-gggagatttttagggatttgatgg-3’ and reverse, 5’-ccaaattatcaaaaccacaatcaaaatc-3’, and for the proximal region (closest to and in part including the untranslated exon 1), forward, 5’-ttagttagttttgttagggtatttttgg-3’ and reverse, 5’-ctaactactacctataaaatctaaaacc-3’. Only the distal region showed any evidence of methylation, which is why we focused on this region when designing methylation-specific PCR (MSP) assays for the studies of patient specimens.
MSP[11] was performed to separately amplify either methylated or unmethylated alleles from the distal region of the EPS8 promoter (see above). Two alternative pairs of primers (MF1 + MR1, or MF3 + MR3) were used for the methylated reaction, and primers UF1 + UR1 for the unmethylated reaction. The primer sequences were: MF1, 5’-tggttattagatgcgttttgtttgtc-3’, MR1, 5’-gtataaaaaacttcgcccccgacg-3’; MF3, 5’-ggtgttgtaatttgagcgtttttttc-3’, MR3, 5’-aacgtataaaaaacttcgcccccg-3’; and UF1, 5’-ttggttattagatgtgttttgtttgtt-3’, UR1 5’-ccaacaaaaataaacaccccaaaca-3’. DNA (1 μg) was modified with sodium bisulfite treatment (CpGenome DNA Modification kit, Chemicon) and subjected to MSP. MSP was performed in a volume of 25 μL containing 24 ng of bisulfite-modified template per reaction with HotStarTaq DNA polymerase (Qiagen). Cycling conditions were according to the manufacturers’ standard cycling protocol for HotStarTaq DNA polymerase, with 35 cycles. Annealing temperatures were 58 °C for the methylated reaction MF1 + MR1, 64 °C for the methylated reaction MF3 + MR3, and 58 °C for the unmethylated reaction. MSP products were run through 2%-3% agarose gel, stained with ethidium bromide, and visualized with ultraviolet transillumination. All sodium bisulfite modifications and MSP runs were repeated at least twice. A negative control without template was included in each MSP run.
RKO is a special colon cancer cell line as it lacks constitutive β-catenin regulated transcription compared to other colon cancer cell lines[8]. To detect genes that show the most remarkable differential expression in the RKO cell line compared to all the other cell lines (HCA7, KM12, LoVo, DLD1, HCT116, SW48, and LIM1215, each being mismatch repair-deficient like RKO), maximum deviance in signal between RKO and the remaining lines was calculated for each Affymetrix probe. When identifying putative over-expressed genes, the deviance was defined as the difference between the signal in RKO and the maximum signal in the other cell lines. In the case of under-expressed genes, the deviance was defined as the difference between the signal in RKO and the minimum signal in the other cell lines. The Affymetrix probe “202609_at” corresponding to the EPS8 gene showed remarkable reduction in the RKO cell line when compared to the other cell lines. The signal detected in RKO was 118, whereas the other cell lines showed signals of 1454, 1361, 3792, 429, 683, 758 and 1804 (Log2 ratio-3.53).
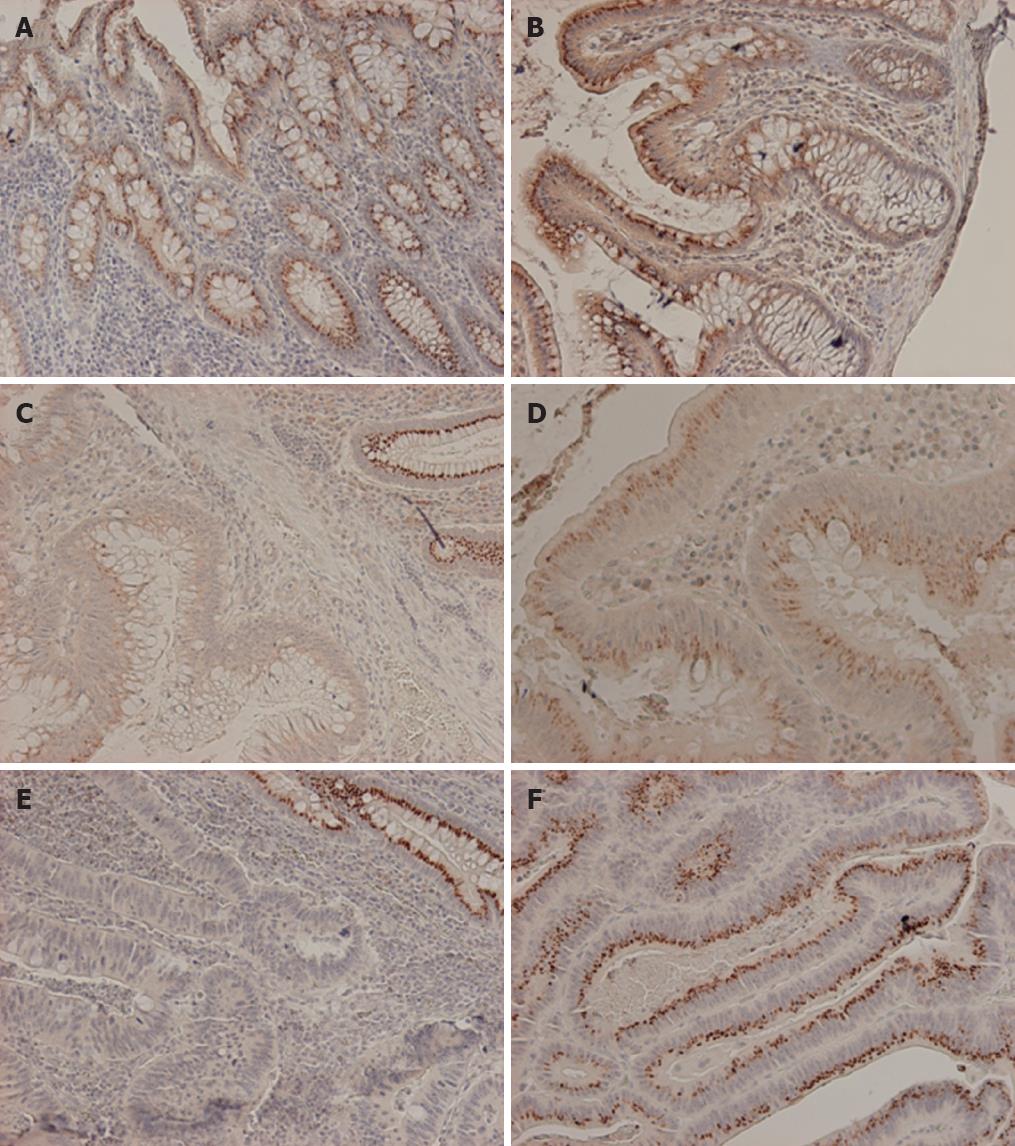
Most patient samples were MSS apart from 5/43 (12%) which showed the MSI phenotype. By immunohistochemical analysis of clinical specimens, normal colonic mucosa showed dot-like supranuclear cytoplasmic expression pattern of EPS8 protein (Figure 1A). In some cases we noticed a gradient of expression with more intense staining at the luminal surface that faded away towards the intestinal crypts (Figure 1B). Colorectal adenomas and carcinomas showed three patterns of expression compared to their matching normal mucosae. 19 (4 adenoma, 15 carcinoma) out of 51 tumors (37%) showed tumor specific EPS8 protein loss, 5 (2 adenoma, and 3 carcinoma) out of 51 (10%) showed marked overexpression, while 27/51 tumors (53%) showed retained expression comparable to what was observed in the matching normal mucosae (Table 2, Figure 1C-F). However, there was no significant correlation between EPS8 expression pattern and β-catenin sub-cellular localization (in contrast to the finding in the RKO cell line), or tumor stage and location within the colon.
As possible mechanisms underlying expression changes, LOH, mutation, and promoter methylation were evaluated. We report LOH at EPS8 locus if, at least, one of the two markers D12S1580 and D12S1728 showed a clear cut LOH (40% or more reduction) while borderline-LOH (ratio reduction ranging from (25%-39%) at one marker only was ignored. Overall, 20/51 (39%) tumors showed EPS8 locus LOH with similar frequencies in adenomas (40%) and carcinomas (37%-41%) (Table 2). LOH was observed in 6/16 (38%) of informative tumors with absent EPS8 protein vs 9/39 (23%) of cases with retained or elevated protein expression (P = 0.53).
All coding exons and flanking intronic regions of the EPS8 gene were examined by direct sequencing. The focus of this analysis was the cell line RKO and tumors with loss of EPS8 expression and/or LOH. We also included one of the control cell lines (HCA7 because of other special features[16]) We identified only one tumor specific missense mutation c.794C>T (p.R265C) in exon 8 in the RKO cell line. Since the matching normal tissue for the RKO cell line was not available, we further analyzed more than 100 normal DNA samples and none of them showed this change. To our knowledge, this nucleotide change is also not reported in any sequence or single nucleotide polymorphism (SNP) database. The nature of the amino acid change suggests that this is not likely to be a SNP since Arginine (R) is a positively charged, large polar amino acid that mostly prefers to substitute for the other positively charged amino acid Lysine, although in some circumstances it will also tolerate a change to other polar amino acids, but substitution with the small amino acid cysteine (C) is not tolerated in any cellular location[17]. We tested this particular substitution using the SIFT program (http://blocks.fhcrc.org/sift/SIFT.html) that sorts intolerant from tolerant amino acid substitutions based on evolutionary conservation, and cysteine substitution was regarded as intolerant.
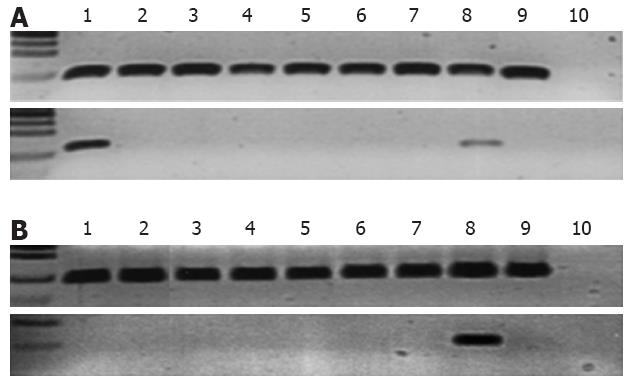
Regarding EPS8 methylation analysis, our very first observation was that the well-established human lymphoblastoid cell line TK6 was methylated in the distal region of the EPS8 promoter, which indicated that the promoter was sensitive to methylation in general (Figure 2A). EPS8 promoter methylation was examined by bisulfite sequencing in all cancer cell lines. These included cell lines in which the MLH1 promoter was known to be methylated (RKO, KM12, HCA7) as well as cell lines with unmethylated MLH1 promoter (HCT15, HCT116, LoVo, LIM1215)[11]. Only RKO was methylated (Figure 2A). Encouraged by EPS8 methylation in RKO, we designed MSP reactions to investigate EPS8 methylation status in patient specimens of colorectal cancer. We focused on those cases that had no LOH at chromosome 12 markers (including cases that were uninformative for LOH), yet EPS8 protein was reduced or lost by immunohistochemistry, suggesting that there had to be alternative mechanisms for inactivation. There was no methylation in any of the seven tumor specimens analyzed, including five MSS tumors and two with MSI (Figure 2B). Given the lack of methylation in these samples of perhaps the highest interest, we did not extend these analyses to additional specimens.
Our data shed light on the role of EPS8 in tumorigenesis in several important respects. EPS8 is involved in actin dynamics through its actin barbed-end capping activity and its ability to modulate Rac activity. Accordingly, EPS8 is crucial for the formation of actin networks that support cellular structures such as lamellipodia, filopodia, stress fibers and focal adhesions[18]. It appears that this is the most significant function of EPS8 in carcinogenesis also, since it did not colocalize with epidermal growth factor receptors but colocalized with F-actin in circular ruffles and at the leading edge of pancreatic cancer cells[19]. The data presented here support an important role of EPS8 in maturation and differentiation of the normal human colonic mucosa since normal colonic mucosa showed strong constitutive supranuclear cytoplasmic expression of EPS8 with increasing intensity towards the luminal surface away from the crypt base. These data are consistent with the well established role of EPS8 in the maturation of intestinal epithelium in C. elegans[5] and the previously described expression pattern in pancreatic ductal cells[19]. Potential roles of EPS8 in normal colonic epithelium might include the migration of proliferating cells from the bases of the crypt to the colonic luminal surface and/or stabilization of cell-cell junctions, as EPS8 was shown to be involved in cell-cell junction stability in fibroblasts[20], and EPS8 knockdown impaired actin cell-cell junction in confluent pancreatic cancer cells[19].
Regarding colon carcinoma, we noticed high levels of EPS8 mRNA in all cell lines except RKO. However, immunohistochemical analysis of EPS8 protein in uncultured tumor biopsies showed that only around 10% of uncultured patient biopsies showed protein overexpression. This discrepancy between the cell line mRNA approach and patient biopsies’ protein expression was consistent with the observation in pancreatic ductal adenocarcinomas[19] and may be explained by the apparent need of the cell lines to over-express motility and invasion markers. We are currently undertaking studies to explore the role of miRNA in posttranscriptional regulation of EPS8 protein expression. However, studies showed good correlation between mRNA and protein levels within the same tumor model[21].
The remarkable finding in this work was loss of EPS8 protein expression in subsets of colon adenoma and carcinoma. This finding is intriguing given the large number of published reports on EPS8 upregulation in different types of cancers, including those of the colon[7,19,22-24]. We also noted this upregulation at the levels of mRNA and protein expression in some tumors as discussed above. A careful analysis of the published reports shows, however, that most cases of EPS8 upregulation were characteristic of advanced stage and metastatic cancers[7,19,23,24]. The published literature suggests that EPS8 is most likely to be upregulated at the stage of metastasis. This hypothesis is best highlighted by the finding of EPS8 upregulation in the metastatic cell line SW620 as compared to its primary colon cancer cell line SW480[7]. These two cell lines are a well established model that have been used to study the markers associated with metastasis in colon carcinomas[25,26]. Similarly, the metastatic HN12 cells expressed high levels of EPS8 compared to its primary squamous cell carcinoma-derived cell line HN4[22]. In pancreatic cancer, cell lines from primary tumors had low levels of EPS8 mRNA expression; cell lines from pancreatic cancer metastases had medium levels of EPS8 mRNA expression; and a cell line derived from malignant ascites (AsPC-1) had high levels of EPS8 mRNA expression[19]. These data could explain the apparent lack of mutation in our study, particularly in those tumors with loss of EPS8 protein which should leave a space for upregulation and overexpression at later stages; since reversion mutations are known to be extremely rare. In this regard, epigenetic and other regulatory mechanisms that could be easily reversed would be a preferable mode for controlling this gene expression status. Consistent with this, we noted the susceptibility of the EPS8 promoter to methylation in the lymphoblastoid TK6 cells (Figure 2A) and its methylation in the RKO cell line associated with EPS8 mRNA underexpression. This is in agreement with RKO being a prototype of CpG island methylator phenotype that is usually observed in combination with MSI tumors[27]. We, however, did not observe promoter methylation in the primary tumors considered to have the highest a priori likelihood for methylation, suggesting that EPS8 inactivation in these tumors occurred by other, as yet unknown mechanisms.
In conclusion, we report EPS8 loss of expression in colorectal adenomas and carcinomas and propose that EPS8 downregulation plays a role in the development of these tumors.
Esa Perkiö is thanked for help in methylation analyses, Saila Saarinen for expert technical assistance throughout this work and Sanna Heino and Tiina Wirtanen for technical advice through the microarray experiment.
Epidermal growth factor receptor pathway substrate 8 (EPS8) is a 97-kDa protein that is required for intestinal cell maturation. EPS8 was recently shown to be overexpressed in advanced stage human cancers including colon cancer cells. In this study, the authors analyzed EPS8 status in colorectal cancers.
This work applies multiple approaches to gain insight into the expression status of EPS8 in colorectal cancer cell lines and primary tumors. Furthermore, it sheds light on the possible mechanisms of the observed expression alterations.
The remarkable finding in this work was loss of EPS8 protein expression in colorectal adenoma and carcinoma. This finding is intriguing given the previously published reports on EPS8 upregulation in different types of cancers, including those of the colon. Thus, the results show, for the first time, that EPS8 downregulation plays a role in the development of subsets of colorectal tumors.
The current findings could have applications in diagnosis and treatment of a subset of colon tumors. The observed expression differences of EPS8 here raise a note of caution about generalization of the previously reported findings of EPS8 overexpression in some tumors and re-emphasize the significance of personalized medicine in the treatment of cancer patients.
It is an interesting study worth to be considered.
Peer reviewer: Dr. Jan Mollenhauer, PhD, Professor, Head of Molecular Oncology, Institute for Medical Biology, University of Southern Denmark, Winsloewparken 25, 5000 Odense C, Denmark
S- Editor Cheng JX L- Editor Webster JR E- Editor Zheng XM
| 1. | Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong WT, Di Fiore PP. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799-3808. [PubMed] [Cited in This Article: ] |
| 2. | Scita G, Tenca P, Areces LB, Tocchetti A, Frittoli E, Giardina G, Ponzanelli I, Sini P, Innocenti M, Di Fiore PP. An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J Cell Biol. 2001;154:1031-1044. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 109] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | Innocenti M, Tenca P, Frittoli E, Faretta M, Tocchetti A, Di Fiore PP, Scita G. Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J Cell Biol. 2002;156:125-136. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 147] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 4. | Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegård M, Betsholtz C, Di Fiore PP. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290-293. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 266] [Cited by in F6Publishing: 268] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 5. | Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier MF, Scita G, Baumeister R, Di Fiore PP. A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol. 2004;6:1173-1179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 6. | Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6:1180-1188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 160] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 7. | Maa MC, Lee JC, Chen YJ, Chen YJ, Lee YC, Wang ST, Huang CC, Chow NH, Leu TH. Eps8 facilitates cellular growth and motility of colon cancer cells by increasing the expression and activity of focal adhesion kinase. J Biol Chem. 2007;282:19399-19409. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 56] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | da Costa LT, He TC, Yu J, Sparks AB, Morin PJ, Polyak K, Laken S, Vogelstein B, Kinzler KW. CDX2 is mutated in a colorectal cancer with normal APC/beta-catenin signaling. Oncogene. 1999;18:5010-5014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 98] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Abdel-Rahman WM, Ollikainen M, Kariola R, Järvinen HJ, Mecklin JP, Nyström-Lahti M, Knuutila S, Peltomäki P. Comprehensive characterization of HNPCC-related colorectal cancers reveals striking molecular features in families with no germline mismatch repair gene mutations. Oncogene. 2005;24:1542-1551. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 65] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Abdel-Rahman WM, Kalinina J, Shoman S, Eissa S, Ollikainen M, Elomaa O, Eliseenkova AV, Bützow R, Mohammadi M, Peltomäki P. Somatic FGF9 mutations in colorectal and endometrial carcinomas associated with membranous beta-catenin. Hum Mutat. 2008;29:390-397. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Joensuu EI, Abdel-Rahman WM, Ollikainen M, Ruosaari S, Knuutila S, Peltomäki P. Epigenetic signatures of familial cancer are characteristic of tumor type and family category. Cancer Res. 2008;68:4597-4605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Renkonen E, Zhang Y, Lohi H, Salovaara R, Abdel-Rahman WM, Nilbert M, Aittomaki K, Jarvinen HJ, Mecklin JP, Lindblom A. Altered expression of MLH1, MSH2, and MSH6 in predisposition to hereditary nonpolyposis colorectal cancer. J Clin Oncol. 2003;21:3629-3637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 74] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 13. | Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3769] [Cited by in F6Publishing: 3860] [Article Influence: 183.8] [Reference Citation Analysis (0)] |
| 14. | Kuismanen SA, Moisio AL, Schweizer P, Truninger K, Salovaara R, Arola J, Butzow R, Jiricny J, Nyström-Lahti M, Peltomäki P. Endometrial and colorectal tumors from patients with hereditary nonpolyposis colon cancer display different patterns of microsatellite instability. Am J Pathol. 2002;160:1953-1958. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 84] [Cited by in F6Publishing: 88] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Ollikainen M, Abdel-Rahman WM, Moisio AL, Lindroos A, Kariola R, Järvelä I, Pöyhönen M, Butzow R, Peltomäki P. Molecular analysis of familial endometrial carcinoma: a manifestation of hereditary nonpolyposis colorectal cancer or a separate syndrome? J Clin Oncol. 2005;23:4609-4616. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 101] [Cited by in F6Publishing: 95] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Abdel-Rahman WM, Lohi H, Knuutila S, Peltomäki P. Restoring mismatch repair does not stop the formation of reciprocal translocations in the colon cancer cell line HCA7 but further destabilizes chromosome number. Oncogene. 2005;24:706-713. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 17. | Betts MJ, Russell RB. Amino acid properties and consequences of substitutions. Bioinformatics for Geneticists. New York: Wiley 2003; 289-316. [DOI] [Cited in This Article: ] |
| 18. | Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5:635-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 234] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 19. | Welsch T, Endlich K, Giese T, Büchler MW, Schmidt J. Eps8 is increased in pancreatic cancer and required for dynamic actin-based cell protrusions and intercellular cytoskeletal organization. Cancer Lett. 2007;255:205-218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Matoskova B, Wong WT, Salcini AE, Pelicci PG, Di Fiore PP. Constitutive phosphorylation of eps8 in tumor cell lines: relevance to malignant transformation. Mol Cell Biol. 1995;15:3805-3812. [PubMed] [Cited in This Article: ] |
| 21. | Xu M, Shorts-Cary L, Knox AJ, Kleinsmidt-DeMasters B, Lillehei K, Wierman ME. Epidermal growth factor receptor pathway substrate 8 is overexpressed in human pituitary tumors: role in proliferation and survival. Endocrinology. 2009;150:2064-2071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Yao J, Weremowicz S, Feng B, Gentleman RC, Marks JR, Gelman R, Brennan C, Polyak K. Combined cDNA array comparative genomic hybridization and serial analysis of gene expression analysis of breast tumor progression. Cancer Res. 2006;66:4065-4078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 137] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 23. | Wang H, Patel V, Miyazaki H, Gutkind JS, Yeudall WA. Role for EPS8 in squamous carcinogenesis. Carcinogenesis. 2009;30:165-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Chen YJ, Shen MR, Chen YJ, Maa MC, Leu TH. Eps8 decreases chemosensitivity and affects survival of cervical cancer patients. Mol Cancer Ther. 2008;7:1376-1385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Gagos S, Hopwood VL, Iliopoulos D, Kostakis A, Karayannakos P, Yatzides H, Skalkeas GD, Pathak S. Chromosomal markers associated with metastasis in two colon cancer cell lines established from the same patient. Anticancer Res. 1995;15:369-378. [PubMed] [Cited in This Article: ] |
| 26. | Abdel-Rahman WM, Katsura K, Rens W, Gorman PA, Sheer D, Bicknell D, Bodmer WF, Arends MJ, Wyllie AH, Edwards PA. Spectral karyotyping suggests additional subsets of colorectal cancers characterized by pattern of chromosome rearrangement. Proc Natl Acad Sci USA. 2001;98:2538-2543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 130] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 27. | Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci USA. 1998;95:8698-8702. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 461] [Cited by in F6Publishing: 489] [Article Influence: 18.8] [Reference Citation Analysis (0)] |