Published online May 14, 2012. doi: 10.3748/wjg.v18.i18.2245
Revised: December 6, 2011
Accepted: March 10, 2012
Published online: May 14, 2012
AIM: To determine antibiotic resistance of Helicobacter pylori (H. pylori) in Pakistan and its correlation with host and pathogen associated factors.
METHODS: A total of 178 strains of H. pylori were isolated from gastric biopsies of dyspeptic patients. Susceptibility patterns against first and second-line antibiotics were determined and trends of resistance were analyzed in relation to the sampling period, gastric conditions and cagA gene carriage. The effect of cagA gene on the acquisition of resistance was investigated by mutant selection assay.
RESULTS: The observations showed that monoresistant strains were prevalent with rates of 89% for metronidazole, 36% for clarithromycin, 37% for amoxicillin, 18.5% for ofloxacin and 12% for tetracycline. Furthermore, clarithromycin resistance was on the rise from 2005 to 2008 (32% vs 38%, P = 0.004) and it is significantly observed in non ulcerative dyspeptic patients compared to gastritis, gastric ulcer and duodenal ulcer cases (53% vs 20%, 18% and 19%, P = 0.000). On the contrary, metronidazole and ofloxacin resistance were more common in gastritis and gastric ulcer cases. Distribution analysis and frequencies of resistant mutants in vitro correlated with the absence of cagA gene with metronidazole and ofloxacin resistance.
CONCLUSION: The study confirms the alarming levels of antibiotic resistance associated with the degree of gastric inflammation and cagA gene carriage in H. pylori strains.
-
Citation: Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and
cagA gene correlation: A looming crisis ofHelicobacter pylori . World J Gastroenterol 2012; 18(18): 2245-2252 - URL: https://www.wjgnet.com/1007-9327/full/v18/i18/2245.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i18.2245
Helicobacter pylori (H. pylori) is among the most widespread infectious agents because of its high colonization rate and persistent nature in its host’s stomach. Mostly the colonization is silent but overt damage of gastric mucosa occurs in certain cases leading to the development of gastritis, duodenal ulcer (DU), gastric ulcer (GU), gastric cancer (GC) and mucosa associated lymphoid tissue lymphoma. It is believed that CagA toxin is responsible for the underlying virulence mechanism because of its ability to translocate into gastric epithelial cells. In host cells, it binds with cellular SRC homology 2 domain-containing tyrosine phosphatase (SHP-2) protein and subsequently damage of gastric mucosa.
Triple therapy constituting the combination of a proton pump inhibitor or bismuth citrate and two antibiotics such as amoxicillin (AML), clarithromycin (CLR) or metronidazole (MTZ) is the internationally recommended first-line regime to eradicate H. pylori in symptomatic patients[1,2]. However, the combination might be delivered concomitantly, sequentially or in the form of a traditional straight course for 10-15 d to attain the highest cure rate[3]. In case of treatment failure, several other antibiotics such as fluoroquinolones and tetracycline (TE) are used as secondary options[3,4]. The increased rates of CLR and MTZ resistance have further compounded the already challenging treatment strategy within the harsh acidic environment of the stomach. Patient compliance has declined to less than 80% due to resistance against one of the antibiotics used in the first-line regime[5,6].
Pakistan is among the countries with high prevalence of H. pylori infection. The organism is not only associated with severe clinical outcomes[7] but is also carried by the healthy population. Empirical treatment has always been in practice without examining whether it matches in vitro antibiotics susceptibility testing (AST) or not. As a result, patient compliance has diminished up to 70%-75% in the last decade[8,9]. As most of studies from Pakistan are based on the outcome of therapy, information is scanty on the status of AST profiles of local isolates. Previous attempts, some of which are based on alternate AST methods, provide limited information[10,11], while no data is available on AST profiles for second-line options.
The paucity of information on the issue and the key role it plays in controlling H. pylori infection led us to conduct the present investigation that not only provides a detailed AST profile of local H. pylori isolates against first- and second-line regimes but also analyzes their distribution in various groups of patients. The study also contributes toward the better understanding of the role of cagA gene in the evolution of resistance.
A total of 178 H. pylori strains isolated from gastroduodenal biopsies were included in this study. Biopsy samples were taken from symptomatic patients (n = 450) who underwent gastroduodenal endoscopy at Medical Unit II, Civil Hospital, Dow University of Health Sciences, Karachi, from March 2005 to November 2008. They were grouped as non ulcerative dyspepsia (NUD), gastritis, GU and DU on the basis of endoscopic findings. Patients with previous treatment history for H. pylori infection and/ or GC were excluded. Samples were collected in 20% sterile glucose solution, transported in ice, and processed within two hours of collection. The study was conducted upon approval from the ethical review board of the University of Karachi, Pakistan.
Genomic DNAs were extracted from crushed tissue samples by SDS-PK method[12] (Khan, 2006 No. 354). Molecular diagnosis for the presence of H. pylori was conducted by polymerase chain reaction (PCR) targeting the 16SrRNA gene as described previously[13]. Samples that were found positive with the 16SrRNA gene of H. pylori were further examined for the presence of the cagA gene by PCR using primers designed for the entire 3’ repeat conserved region. Amplification was performed at 35 cycles of 95 °C for 1 min, 52 °C for 1 min, and 72 °C for 1 min with a final extension of 7 min at 72 °C[14]. A segment of human β-globulin gene was amplified as the internal control.
For the isolation of H. pylori, biopsy samples were crushed with the help of a sterile disposable tissue homogenizer and inoculated on Columbia Blood Agar (Oxoid, United Kingdom) containing 7% laked horse blood (Oxoid, United Kingdom) and H. pylori selective supplement Dent (Oxoid, United Kingdom). Plates were incubated at 37 °C for 5 d under microaerophilic conditions using a Campygen gas generating kit (Oxoid, United Kingdom). Suspicious small dew drop colonies were subjected for morphological and biochemical identification.
For further confirmation, genomic DNA was extracted and subjected to PCR analysis for the 16SrRNA gene of H. pylori per the above-mentioned protocol.
Susceptibility patterns of H. pylori isolates was determined against a battery of antibiotics including MTZ, AML, CLR, DA, TE, ofloxacin (OFX) and erythromycin (E). Various concentrations of antibiotics were added to Mueller-Hinton agar containing 5% old sheep blood (SB-MHA). Bacterial suspensions were prepared in sterile phosphate buffered saline (PBS) with density equivalent to 3 McFarland’s turbidity standard. Ten microliters of each strain was spotted on the plates and incubated for 72 h under microaerophilic conditions. The lowest concentration of antibiotic able to inhibit visible bacterial growth was considered as minimum inhibitory concentrations (MIC). Results were interpreted according to standard criteria[15].
To determine the role of cagA gene in the emergence of resistance, two cagA+ and two cagA- strains were subjected to mutant selection assay. A total of 1.2 × 106-1.5 × 106 colony forming units (CFU) of bacterial strains suspended in PBS was spread over SB-MHA plates containing varying concentrations of antibiotics and incubated for 72 h at 37 °C under microaerophilic conditions. Frequency of resistant mutants was determined as the CFU of each strain grown on antibiotic supplemented plates divided by the starting inocula[16].
Statistical analyses were performed by PASW statistics 18 (SPSS Inc., Chicago, IL, United States). Pearson’s χ2 test was applied to compare categorical data. Linear regression was applied to correlate the frequency of resistance with cagA gene. A P value of < 0.05 was considered statistically significant.
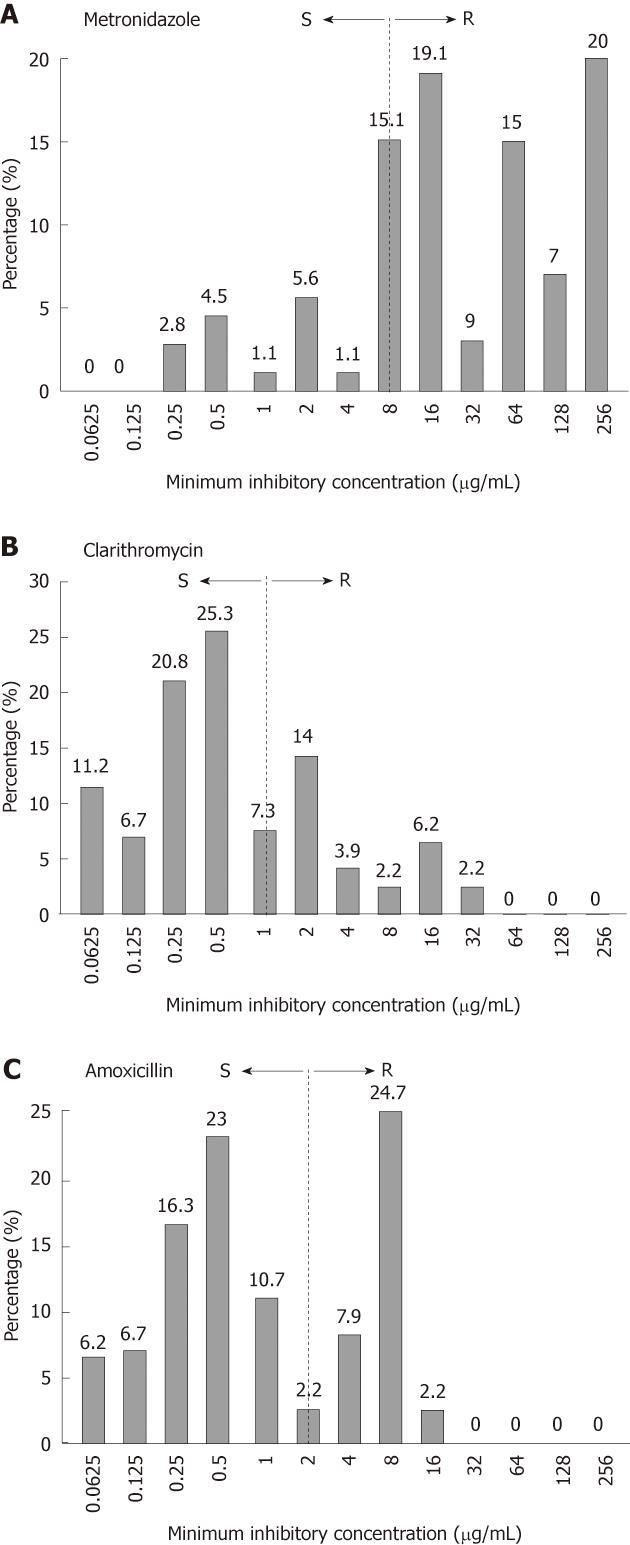
Out of 450 dyspeptic patients, 201 (45%) were found positive for H. pylori by PCR and 178 (40%) by culture. AST profile of 178 H. pylori strains revealed high levels of resistance against the first-line regime. A total of 149 (84%) were found to be resistant (MIC ≥ 8 μg/mL) to MTZ. As shown in Figure 1A, 34 (19%) strains had MIC 16 μg/mL, 16 (9%) had 32 μg/mL, 27 (15%) had 64 μg/mL, 12 (7%) had 128 μg/mL and 35 (20%) had MIC 256 μg/mL whereas 27 (15%) isolates were inhibited at boarder-line concentration 8 μg/mL. In case of CLR, 64 (36%) strains showed resistance with MICs ≥ 1 μg/mL. The highest MIC 32 μg/mL was observed only in 4 (2.2%) isolates while 25 (14%) had MIC 2 μg/mL (Figure 1B). Sixty six (37%) strains showed resistance to AML with MICs ≥ 2 μg/mL. Surprisingly, 44 (24.7%) strains exhibited MIC 8 μg/mL (Figure 1C).
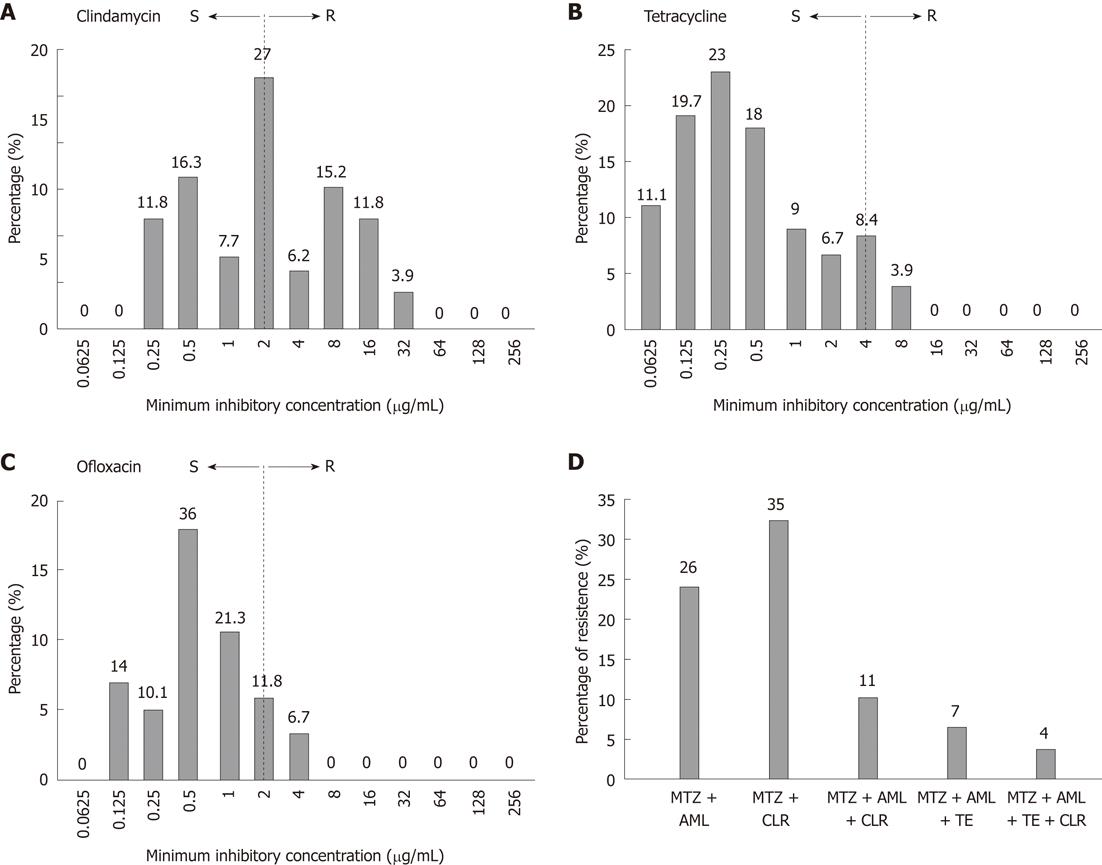
Emergence of resistance against first-line therapy led us to investigate the susceptibility patterns of antibiotics usually given as second-line treatment options, such as TE, fluoroquinolones and clindamycin (DA). Data analysis shows an evenly distributed pattern of DA activity with MICs ranging from 0.25 to 32 μg/mL (Figure 2A). Resistance to TE was found in 21 (12%) strains. MIC 4 μg/mL was observed in 15 (8.4%) strains and 8 μg/mL in 7(3.9%) strains (Figure 2B). As shown in Figure 2C, a total of 33 (18.5%) strains were resistant to OFX. MIC of OFX was 2 μg/mL and 4 μg/mL for 21 (11.8%) and 12 (6.7%) strains respectively. We further analyzed the rate of multidrug resistance (MDR) in our studied population. A total of 46 (26%) of the isolates were resistant to two antibiotics i.e. MTZ and AML whereas 62 (35%) were resistant to MTZ and CLR. MDR isolates who were resistant to all first-line antibiotics (R-phenotype; MTZrCLRrAMLr) were 20 (11%). Of these 7 (4%) were also resistant to tetracycline (R-phenotype; MTZrCLRrAMLrTEr) as shown in Figure 2D.
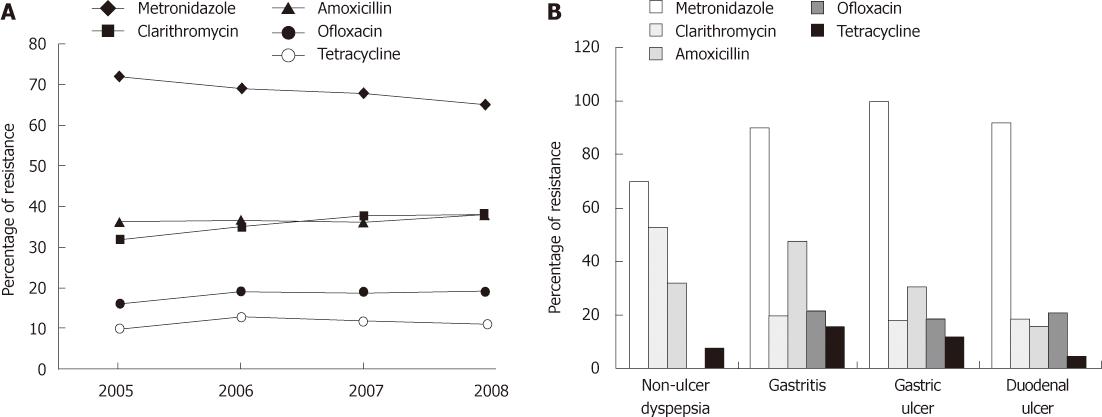
We next examined whether the proportion of strains showing antibiotic resistance were increased over time or not. For the purpose, the strains were divided into four groups; 2005 (n = 24), 2006 (n = 55), 2007 (n = 73) and 2008 (n =26) according to the year of sample collection. In general, a significantly progressive trend was only observed in CLR resistance from 32% (n = 8) in 2005 to 35% (n = 19) in 2006, 37.5% (n = 27) and 38% (n = 19) in 2008 (P = 0.004). On the contrary, MTZ resistant strains went down by year from 72% (n = 17) in 2005 to 69% (n = 38) in 2006, 68% (n = 50) in 2007 and 65% (n = 17) in 2008. However AML and OFX resistance remained steady with the rates of approximately 37% and 19% throughout the study period whereas overall resistance rates of TE were 10% (n = 2) in 2005, 13% (n = 7) in 2006, 12% (n = 9) in 2007 and 11% (n = 3) in 2008 (Figure 3A).
Comparative analysis was also performed according to the endoscopic findings of each patient from which H. pylori strains were isolated. Out of 178, a total of 25 strains were isolated from NUD cases, 89 gastritis cases, 26 GU cases and 38 from DU cases. Our findings indicate that AML and OFX resistance rates were more common in gastritis patients at 48% (n = 43) (P = 0.005) and 22% (n = 20) of cases (P = 0.08) respectively. In contrast, respective resistance rates of AML and OFX were 31% (n = 8) and 19% (n = 5) in GU and 16% (n = 6) and 21% (n = 8) in DU patients while no OFX resistant strain was found in NUD group. In case of MTZ, resistance rate was significantly higher among the patients with damaged mucosa such as gastritis (90%), GU (100%) and DU (92%) compared with NUD cases (70%) (P = 0.001). CLR resistance was observed in 53% (n = 13) of NUD, 20% (n = 18) gastritis, 18% (n = 4) GU and 19% (n = 5) of DU cases (P = 0.000) as shown in Figure 3B.
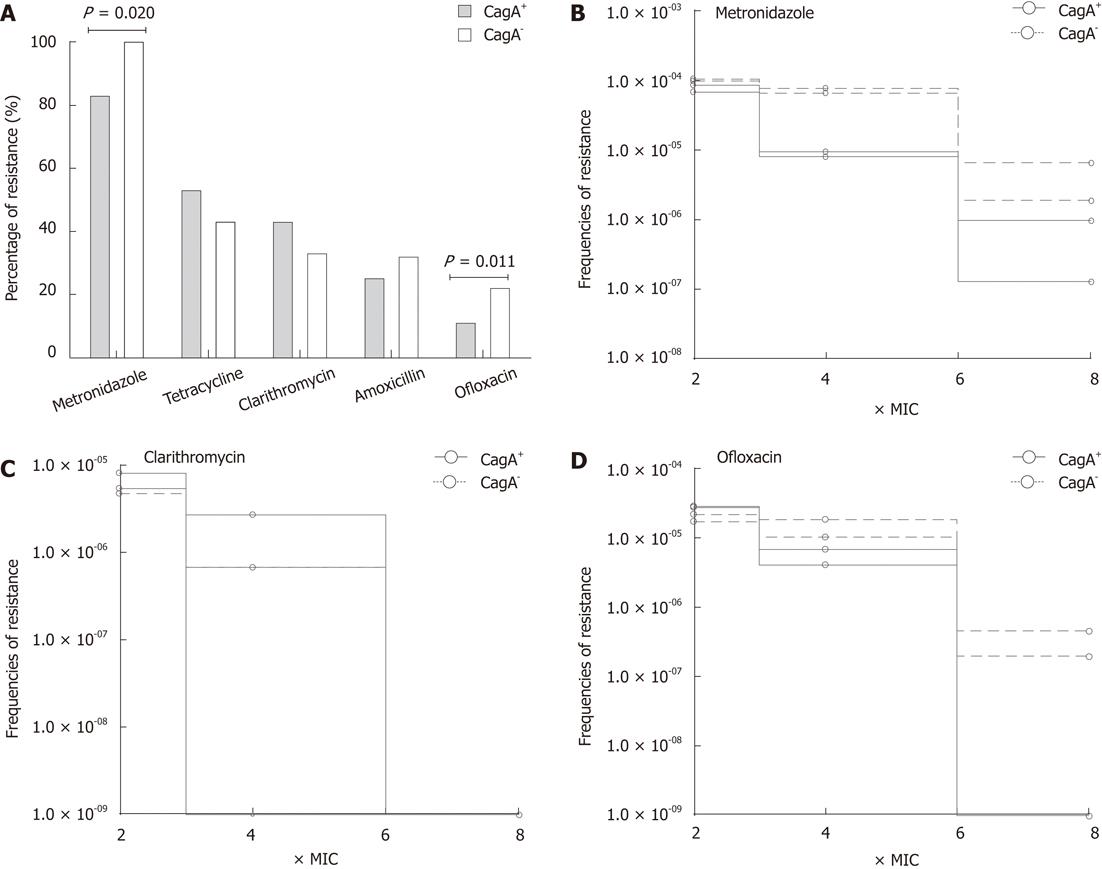
The cagA genotypes of H. pylori usually correlate with the severity of disease; therefore, we determined the relationship of cagA gene with the susceptibility profile in local isolates. In this study 83 out of 178 (47%) H. pylori strains carried the cagA gene. The percentage of cagA+ strains was 49% (n = 44) among patients with gastritis, 69% (n =18) GU and 60% (n = 23) DU whereas only 8% (n = 2) of the strains isolated from NUD cases carried this gene. Due to the low frequency of CagA+ strains in NUD cases, further analysis was only based only on a total of 153 strains which were isolated from gastritis, GU and DU cases. Of these, 81 (53%) were cagA+ and 72 (47%) were cagA-. Analysis of the drug resistance indicates the lower prevalence of OFX (11%, n = 9), MTZ (83%, n = 67) and AML (25%, n = 20) resistance in cagA+ strains compared with 22% (n = 16), 100% (n = 72) and 32% (n = 23) in cagA- strains respectively (Figure 4A). In contrast, CLR resistance was more prevalent in cagA+ strains (43%, n = 35) than in cagA- (33%, n = 24). The analysis indicates a possible link between cagA gene and the development of drug resistance.
To determine whether the rate of acquisition of antibiotic resistance varies between cagA+ and cagA- strains, we exposed selected strains to increasing concentrations of MTZ, OFX and CLR. Bacterial growth was monitored at each concentration of antibiotics and frequency of resistant mutants was determined as the CFU of H. pylori strain divided by the starting inocula. We observed that cagA- strains were able to mutate more frequently under the selective pressure of MTZ since they were able maintain their frequencies even after the exposure of 6 × MIC of MTZ (r2 0.9966, P = 0.0374). In contrast, more than 1 log10 decrease in bacterial growth was observed in cagA+ strains with increasing concentrations of MTZ (Figure 4B). Similarly cagA- strains maintained their frequency in the increasing concentration of OFX (r2 = 0.9966, P = 0.0374) whereas a sharp decline was observed in the development of resistant mutants of cagA+ strains (Figure 4D). However, no significant difference was observed in the case of clarithromycin (Figure 4C).
H. pylori is often neglected for antimicrobial susceptibility testing because of its complex growth requirement and low recovery rate by bacterial culture. Increasing reports of treatment failure necessitate surveillance studies to analyze the trend of drug resistance especially in developing countries where MDR is quite common in other bacterial species. To determine the trend of antibiotic resistance in Pakistan, we conducted a 4-year longitudinal study comprised of 178 H. pylori strains. AST profile revealed high levels of resistance against the first-line regime including MTZ (84%), CLR (36%) and AML (37%). Our results and those from recently published papers from other countries show comparable prevalence rates; for example 33% resistance to AML was observed in the United States[17] whereas MTZ and CLR resistance rates were 31% and 33% in Ireland[18], 61% and 26% in France[19], 48% and 28% in Saudi Arabia[20], 80% and 45% in India[21], and 77% and 15% in Bangladesh respectively[22]. Moreover the trends of CLR and MTZ resistance were also in agreement with previously published reports[6,23]. In contrast with our observations, available data indicate a low occurrence of such strains in southeast Asian countries such as Malaysia and Taiwan where H. pylori is endemic[6,24]. However, the distinct genotypic nature of Southeast Asian strains provides a possible explanation for the differences in resistance profiles compared with rest of the world, including Pakistan.
The global analysis of clinical data clearly indicates that drug resistance to AML, MTZ and CLR has a central role in poor patient compliance to “gold standard” triple therapy to H. pylori infection, especially in the case of CLR if the point mutations in peptidyltransferase of 23S rRNA gene are responsible to phenotypic behavior[25]. Therefore, Maastricht III consensus guidelines proposed not to provide CLR based empirical therapy if primary resistance rates are more than 15%-20% in the respective territory[4]. The present study clearly indicates the upward trend in the primary resistance to CLR in our population with an average of 36% in mono-resistance and 22% in MDR (R-phenotype; MTZrCLRrAMLr) strains which provides an possible reason for the poor patient compliance (up to 70%-75%) with CLR based therapies in Pakistan as reported earlier.[8,9] Although MDR strains were equally present in our studied population when compared with the rates in other countries[26,27], they were less prevalent than mono-resistant strains.
Fluroquinolones such as ofloxacin or ciprofloxacin and tetracycline are usually considered as second-line therapy for H. pylori infection. In this study, the prevalence of TE resistance was comparable to that of other countries, however resistance to OFX was at a higher level than that seen other countries[18,28]. Mutations in the gyrA gene that are responsible for fluoroquinolone resistance have been directly linked with the failure of H. pylori eradication[29] therefore the higher rate of OFX resistance is alarming. These antibiotics are generally used to treat gastrointestinal infections in Pakistan; consequently, the resistance occurs in other Gram negative bacteria such as Salmonella, Shigella and Escherichia coli[30] and therefore the transmission of resistance in H. pylori can be anticipated. To combat the situation, broad-spectrum fluoroquinolones such as levofloxacin have been introduced; however, the development of resistance and intense side effects hamper its wide use despite its better compliance rate[31].
Genotypic differences of H. pylori directly influence the pathogenesis of infection. Such effects have been widely evidenced with the cagA gene carriage however the exact mechanism remains elusive. In this study, the differential prevalence of MTZ and OFX resistance in cagA+ and cagA- strains clearly indicate the absence of cagA gene contributes in the acquisition of resistance which was further evidenced by the differential frequencies of resistant mutants developed with the increasing amount of each antibiotic as previously observed by Taneike et al[16] previously. The underlying phenomenon is usually explained by the ability of cagA+ strains to cause intense inflammation which might increase the availability of antibiotics at the site of infection and eventually lead to better eradication of infection. In other words, it describes no direct role of cagA gene in antibiotic resistance. However, undermining the hypothesis, we observed that drug sensitive strains were more prevalent in NUD cases, despite of the absence of cagA gene, compared to those patients with damaged gastric mucosa. Taken together, the present study suggests that cagA gene and the degree of tissue damage might be two independent factors that affect the drug susceptibility of H. pylori.
In summary, we observed that the magnitude of drug resistance in H. pylori strains is alarming in Pakistan. The degrees of gastric inflammation and bacterial genotypes are independently implicated in the development of resistance. The study reaffirms the need for both the continuous surveillance for drug resistance and the development of effective prevention and treatment strategies at national and regional levels.
We thank Nikki Kelvin for technical editing of the manuscript. We also acknowledge the staff of the Endoscopy Laboratory of Medical Unit II, Civil hospital and the Department of Microbiology, University of Karachi, for their cooperation.
Eradication of Helicobacter pylori (H. pylori) is directly associated with symptomatic relief in the patients of gastroduodenal diseases. However, the failure of combination therapy containing two antibiotics and a proton pump inhibitor often results because of antibiotic resistance.
Pattern of antibiotic resistance in H. pylori varies in different settings. However it is yet to determine that how host and pathogenic factors affect the prevalence of resistance.
This study indicates the alarming level of antibiotic resistance among Pakistani strains of H. pylori especially the magnitude of clarithromycin resistance is on rise and more commonly observed in non ulcerative dyspeptic patients. We further describe that cagA gene carriage and the degree of gastric inflammation are two independent factors affecting the metronizadole and ofloxacin resistance in H. pylori.
It is important to conduct continuous surveillance of antibiotic resistance in H. pylori. This study helps to comprehend antibiotic resistance pattern in H. pylori that facilitate to developing effective treatment strategy in different groups of patients.
Non ulcerative dyspepsia (NUD) is defined as presence of upper gastrointestinal tract symptoms such as stomachache, indigestion and vomiting in patients who did not have damaged gastric mucosa.
Overall, the study was well carried out and generally well written. However there are a few areas that needs further clarification mainly in the results section.
Peer reviewer: Cuong D Tran, PhD, Research Fellow, Affiliate Lecturer, University of Adelaide, Gastroenterology Unit, Children, Youth and Women’s Health Service, 72 King William Rd, North Adelaide, SA 5006, Australia
S- Editor Shi ZF L- Editor A E- Editor Xiong L
| 1. | Chey WD, Wong BC. American College of Gastroenterology guideline on the management of Helicobacter pylori infection. Am J Gastroenterol. 2007;102:1808-1825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 819] [Cited by in F6Publishing: 792] [Article Influence: 46.6] [Reference Citation Analysis (1)] |
| 2. | Asaka M, Kato M, Takahashi S, Fukuda Y, Sugiyama T, Ota H, Uemura N, Murakami K, Satoh K, Sugano K. Guidelines for the management of Helicobacter pylori infection in Japan: 2009 revised edition. Helicobacter. 2010;15:1-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 284] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 3. | Graham DY, Shiotani A. New concepts of resistance in the treatment of Helicobacter pylori infections. Nat Clin Pract Gastroenterol Hepatol. 2008;5:321-331. [PubMed] [Cited in This Article: ] |
| 4. | Malfertheiner P, Megraud F, O'Morain C, Bazzoli F, El-Omar E, Graham D, Hunt R, Rokkas T, Vakil N, Kuipers EJ. Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report. Gut. 2007;56:772-781. [PubMed] [Cited in This Article: ] |
| 5. | Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280-322. [PubMed] [Cited in This Article: ] |
| 6. | Poon SK, Lai CH, Chang CS, Lin WY, Chang YC, Wang HJ, Lin PH, Lin HJ, Wang WC. Prevalence of antimicrobial resistance in Helicobacter pylori isolates in Taiwan in relation to consumption of antimicrobial agents. Int J Antimicrob Agents. 2009;34:162-165. [PubMed] [Cited in This Article: ] |
| 7. | Shah SZA, Meron AS. Helicobacter pylori Infection in Cirrhotic Patients with Upper Gastrointestinal Bleeding. World Appl Sci J. 2010;8:137-140. [Cited in This Article: ] |
| 8. | Abbas Z, Yakoob J, Abid S, Jafri W, Islam M, Azam Z, Hilal I. Furazolidone, co-amoxiclav, colloidal bismuth subcitrate, and esomeprazole for patients who failed to eradicate Helicobacter pylori with triple therapy. Dig Dis Sci. 2009;54:1953-1957. [PubMed] [Cited in This Article: ] |
| 9. | Khokhar N. One-week therapy with omeprazole, clarithromycin and amoxicillin for eradication of Helicobacter pylori infection. J Coll Physicians Surg Pak. 2002;12:338-340. [Cited in This Article: ] |
| 10. | Mirza IA, Mirza SH, Ali AM. Antimicrobial susceptibility pattern of H. pylori in isolates from Northern Pakistan. Int J Pathol. 2007;5:18-20. [Cited in This Article: ] |
| 11. | Yakoob J, Abid S, Abbas Z, Jafri SN. Antibiotic susceptibility patterns of Helicobacter pylori and triple therapy in a high-prevalence area. Br J Biomed Sci. 2010;67:197-201. [PubMed] [Cited in This Article: ] |
| 12. | Khan S, Rai MA, Khanani MR, Khan MN, Ali SH. HIV-1 subtype A infection in a community of intravenous drug users in Pakistan. BMC Infect Dis. 2006;6:164. [PubMed] [Cited in This Article: ] |
| 13. | Chisholm SA, Owen RJ, Teare EL, Saverymuttu S. PCR-based diagnosis of Helicobacter pylori infection and real-time determination of clarithromycin resistance directly from human gastric biopsy samples. J Clin Microbiol. 2001;39:1217-1220. [PubMed] [Cited in This Article: ] |
| 14. | Yamaoka Y, Osato MS, Sepulveda AR, Gutierrez O, Figura N, Kim JG, Kodama T, Kashima K, Graham DY. Molecular epidemiology of Helicobacter pylori: separation of H. pylori from East Asian and non-Asian countries. Epidemiol Infect. 2000;124:91-96. [PubMed] [Cited in This Article: ] |
| 15. | Yamaoka Y; CLSI. Performance standards for antimicrobial susceptibility testing. Wayne, PA. : Clinical and Laboratory Standards Institute 2009; . [Cited in This Article: ] |
| 16. | Taneike I, Nami A, O'Connor A, Fitzgerald N, Murphy P, Qasim A, O'Connor H, O'Morain C. Analysis of drug resistance and virulence-factor genotype of Irish Helicobacter pylori strains: is there any relationship between resistance to metronidazole and cagA status? Aliment Pharmacol Ther. 2009;30:784-790. [PubMed] [Cited in This Article: ] |
| 17. | Qureshi NN, Morikis D, Schiller NL. Contribution of specific amino acid changes in penicillin binding protein 1 to amoxicillin resistance in clinical Helicobacter pylori isolates. Antimicrob Agents Chemother. 2011;55:101-109. [PubMed] [Cited in This Article: ] |
| 18. | O'connor A, Taneike I, Nami A, Fitzgerald N, Murphy P, Ryan B, O'connor H, Qasim A, Breslin N, O'moráin C. Helicobacter pylori resistance to metronidazole and clarithromycin in Ireland. Eur J Gastroenterol Hepatol. 2010;22:1123-1127. [PubMed] [Cited in This Article: ] |
| 19. | Raymond J, Lamarque D, Kalach N, Chaussade S, Burucoa C. High level of antimicrobial resistance in French Helicobacter pylori isolates. Helicobacter. 2010;15:21-27. [PubMed] [Cited in This Article: ] |
| 20. | Momenah AM, Asghar AH. Prevalence and antibiotic resistance among helicobacter pylori clinical isolates from main Hospitals in the Western Region of Saudi Arabia. Pak J Med Sci. 2008;24:100-103. [Cited in This Article: ] |
| 21. | Thyagarajan SP, Ray P, Das BK, Ayyagari A, Khan AA, Dharmalingam S, Rao UA, Rajasambandam P, Ramathilagam B, Bhasin D. Geographical difference in antimicrobial resistance pattern of Helicobacter pylori clinical isolates from Indian patients: Multicentric study. J Gastroenterol Hepatol. 2003;18:1373-1378. [PubMed] [Cited in This Article: ] |
| 22. | Nahar S, Mukhopadhyay AK, Khan R, Ahmad MM, Datta S, Chattopadhyay S, Dhar SC, Sarker SA, Engstrand L, Berg DE. Antimicrobial susceptibility of Helicobacter pylori strains isolated in Bangladesh. J Clin Microbiol. 2004;42:4856-4858. [PubMed] [Cited in This Article: ] |
| 23. | Rimbara E, Noguchi N, Tanabe M, Kawai T, Matsumoto Y, Sasatsu M. Susceptibilities to clarithromycin, amoxycillin and metronidazole of Helicobacter pylori isolates from the antrum and corpus in Tokyo, Japan, 1995-2001. Clin Microbiol Infect. 2005;11:307-311. [PubMed] [Cited in This Article: ] |
| 24. | Ahmad N, Zakaria WR, Mohamed R. Analysis of antibiotic susceptibility patterns of Helicobacter pylori isolates from Malaysia. Helicobacter. 2011;16:47-51. [PubMed] [Cited in This Article: ] |
| 25. | De Francesco V, Zullo A, Ierardi E, Giorgio F, Perna F, Hassan C, Morini S, Panella C, Vaira D. Phenotypic and genotypic Helicobacter pylori clarithromycin resistance and therapeutic outcome: benefits and limits. J Antimicrob Chemother. 2010;65:327-332. [PubMed] [Cited in This Article: ] |
| 26. | Sun QJ, Liang X, Zheng Q, Gu WQ, Liu WZ, Xiao SD, Lu H. Resistance of Helicobacter pylori to antibiotics from 2000 to 2009 in Shanghai. World J Gastroenterol. 2010;16:5118-5121. [PubMed] [Cited in This Article: ] |
| 27. | Torres J, Camorlinga-Ponce M, Pérez-Pérez G, Madrazo-De la Garza A, Dehesa M, González-Valencia G, Muñoz O. Increasing multidrug resistance in Helicobacter pylori strains isolated from children and adults in Mexico. J Clin Microbiol. 2001;39:2677-2680. [PubMed] [Cited in This Article: ] |
| 28. | Toledo H, López-Solís R. Tetracycline resistance in Chilean clinical isolates of Helicobacter pylori. J Antimicrob Chemother. 2010;65:470-473. [PubMed] [Cited in This Article: ] |
| 29. | Liou JM, Chang CY, Sheng WH, Wang YC, Chen MJ, Lee YC, Hung HW, Chian H, Chang SC, Wu MS. Genotypic resistance in Helicobacter pylori strains correlates with susceptibility test and treatment outcomes after levofloxacin- and clarithromycin-based therapies. Antimicrob Agents Chemother. 2011;55:1123-1129. [PubMed] [Cited in This Article: ] |
| 30. | Khan E, Jabeen K, Ejaz M, Siddiqui J, Shezad MF, Zafar A. Trends in antimicrobial resistance in Shigella species in Karachi, Pakistan. J Infect Dev Ctries. 2009;3:798-802. [PubMed] [Cited in This Article: ] |
| 31. | Kuo CH, Hu HM, Kuo FC, Hsu PI, Chen A, Yu FJ, Tsai PY, Wu IC, Wang SW, Li CJ. Efficacy of levofloxacin-based rescue therapy for Helicobacter pylori infection after standard triple therapy: a randomized controlled trial. J Antimicrob Chemother. 2009;63:1017-1024. [PubMed] [Cited in This Article: ] |