Published online Nov 21, 2009. doi: 10.3748/wjg.15.5377
Revised: September 16, 2009
Accepted: September 23, 2009
Published online: November 21, 2009
Genome-wide association studies (GWAS) have been applied to various gastrointestinal and liver diseases in recent years. A large number of susceptibility genes and key biological pathways in disease development have been identified. So far, studies in inflammatory bowel diseases, and in particular Crohn’s disease, have been especially successful in defining new susceptibility loci using the GWAS design. The identification of associations related to autophagy as well as several genes involved in immunological response will be important to future research on Crohn’s disease. In this review, key methodological aspects of GWAS, the importance of proper cohort collection, genotyping issues and statistical methods are summarized. Ways of addressing the shortcomings of the GWAS design, when it comes to rare variants, are also discussed. For each of the relevant conditions, findings from the various GWAS are summarized with a focus on the affected biological systems.
- Citation: Melum E, Franke A, Karlsen TH. Genome-wide association studies - A summary for the clinical gastroenterologist. World J Gastroenterol 2009; 15(43): 5377-5396
- URL: https://www.wjgnet.com/1007-9327/full/v15/i43/5377.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.5377
The genetic epidemiology of gastrointestinal diseases has been hard to unravel, despite the fact that many of the diseases have a high sibling recurrence risk pointing to genetic risk factors[1,2]. However, over the last three years, with the recent advent of genome-wide association studies (GWAS), a wealth of susceptibility loci have been discovered. Several hundred GWAS have been reported to this end, with thousands of reports on novel disease genes and loci. Interestingly, inflammatory bowel disease (IBD) has been leading the race[3-17]. The genes and polymorphisms identified have revealed several biological pathways by which gastroenterological diseases develop and may eventually be treated. Although most of the associations are weak in a statistical sense [odds ratios (OR) of risk variants in the range of 1.1-1.2][10,18], they point to loci involved in biological systems worth investigating further with other methodologies. This current review includes studies published or available early online up to August 20, 2009.
The most important milestone in the development of GWAS has clearly been the HapMap project which involved genotyping of 1 million single nucleotide polymorphisms (SNPs) of the human genome in the first phase[19] and 3.1 million in the second phase[20]. Since the frequency of genetic variants varies between different geographical regions, genotyping was performed in populations from Nigeria (Yoruba), Japan, China and the United States [residents with ancestry from Northern and Western Europe, collected in 1980 by the Centre d’Etude du Polymorphisme Humain (CEPH) and used for other human genetic maps]. The HapMap website (http://www.hapmap.org) reports on the frequencies of the SNPs and their correlation through linkage disequilibrium (LD). The HapMap resource has become an invaluable tool for genetic research[21]. By taking into account the information regarding LD in HapMap, companies have been able to select SNPs for the most recent genome-wide genotyping arrays that provide information on more than 90% of the genetic variation in HapMap. In addition to SNPs selected via LD, modern genotyping arrays also contain markers with a high likelihood of biological relevance (e.g. non-synonymous SNPs leading to an amino acid change in the encoded protein), as well as probes designed to detect other types of genetic variation (e.g. deletions, insertions and duplications of DNA segments). Once completed, the currently ongoing 1000 genomes project (http://www.1000genomes.org/), which aims at sequencing 1000 individuals, will further add to the publicly available catalogues of genetic variants and aid in the design of novel genotyping products.
The GWAS design contrasts with the traditional hypothesis-driven studies of biomedical research. Initial criticism with regard to this design has been replaced by appraisal as the scientific community has realized that hypothesis-free data mining performed in a systematic manner[22] represents a powerful tool to pin-point biological systems and generate hypotheses for further research.
Table 1 summarizes the different parts in the study design that make up a GWAS. In the following paragraphs, detailed points are discussed regarding the GWAS study design, both for the researcher planning to embark on a GWAS and the casual reader trying to interpret the findings of an existing study.
| Sample panel building |
| Cases and healthy controls of same ethnicity (for power estimates see figure 2) |
| Enrichment with early onset cases and/or familial cases |
| Keep variability in phenotype at a minimum |
| Establish replication cohort(s) after the same principles. Other, yet similar, ethnicities may be included, although matched healthy controls should be collected |
| Genotyping |
| Sample preparation (DNA extraction, calibration) |
| Genotyping chip (cost vs number of samples) |
| Genetic coverage |
| Initial quality control |
| Exclude samples failing platform-specific QC measures |
| Exclude samples with low call-rate |
| Exclude SNPs with a low genotyping rate |
| Exclude SNPs with a low minor allele frequency and those grossly out of Hardy-Weinberg equilibrium (e.g. P < 10-4) |
| Statistical analysis |
| Imputation of non-genotyped SNPs using HapMap as the reference |
| Single-point association analysis, if needed include covariates of interest in the present study (e.g. gender, sex, smoking, imputation uncertainties etc.) |
| Manually inspect cluster plots for highly significant SNPs that should be followed-up |
| Select 1-2 SNPs from each associated locus to take forward in replication |
| Replication |
| Genotype (preferentially independent technology) in a panel of cases and healthy controls that are properly sized to detect effects in the same range as seen in the discovery panel |
| Follow-up experiments |
| Highly depends on results, i.e. nature of genetic finding, and normally not part of the GWAS design |
To maximize the power to detect even low-effect risk variants, the study panels of a GWAS should preferably contain DNA from as many patients and healthy controls as possible. However, this ideal is limited by several pragmatic factors such as the logistics of identifying and classifying patients, recruitment formalities and financial constraints. Given the significant investment, with a considerably higher price per sample than for genotyping of a few markers, great care should be taken in the sample collection process. The “lowest hanging fruits” with the highest risk effects were easily identified in the first round of GWAS and further studies in the same phenotypes will require larger study panels to identify the genes associated with a more modest risk.
The phenotype of the cases used in the study should be very homogeneous. For instance, for IBD it would be advisable to only include patients for whom long-lasting follow-up data is available in order to ensure the correct diagnosis was made (e.g. for IBD, 10% of the patients change diagnosis during the first year of the disease course[23]). Although a perfect phenotype description is the aim, a small misascertainment seems to only slightly reduce power[24]. If the alternative is not to do a study at all, slight uncertainties in phenotyping can probably be accepted and other aspects for increasing power applied instead.
To enrich the sample collection with risk alleles and thereby increase power, familiar cases[25], which are believed to have the respective disease due to a larger contribution of genetic factors, can be used. The risk estimates achieved in such a cohort should not be used to judge the risk effect of the allele in the general population, both due to enrichment of the associated alleles in these types of panels and preferential selection of the most associated markers (the so called ‘winner’s curse’)[26]. Therefore, replication of findings and final effect size calculations need to be performed in separate study panels sampled without bias.
In rare diseases or in meta-analyses (see separate section on this subject below), case-control cohorts recruited in different countries and even continents[18,27] might be necessary to make the study panels large enough to perform sensible genome-wide association analyses. The main challenge that arises when several case-control cohorts are combined is differences in allele frequencies between populations; a problem that is particularly pronounced in African populations[28], but also needs to be taken into account when combining study panels of European descent[29]. Recently, analytical tools utilizing genome-wide SNP data for correction of population heterogeneity/stratification (e.g. the EIGENSTRAT software[30] and similar approaches[31]) have become available to generate non-inflated population stratification-corrected test statistics.
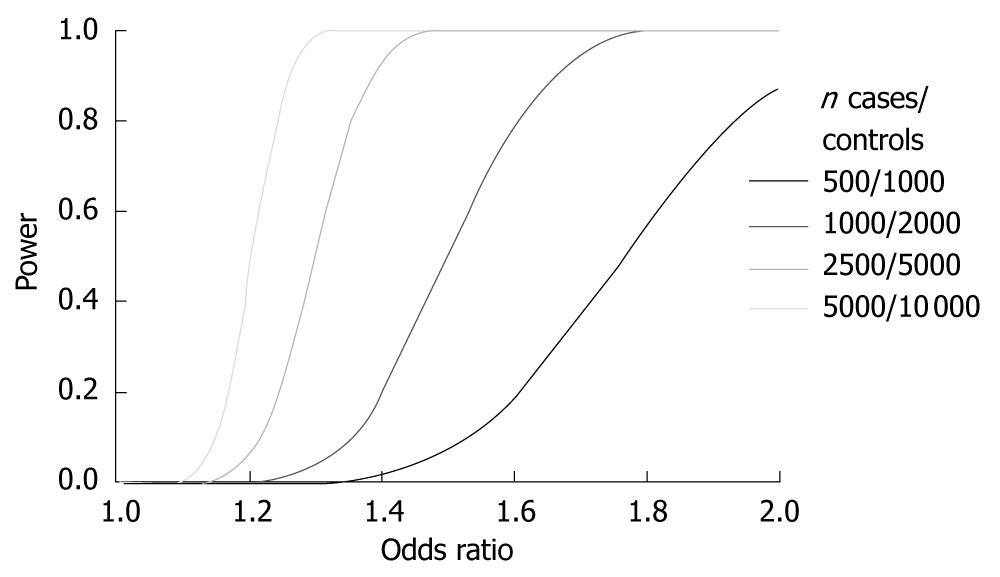
Healthy control data can be shared between groups working in different diseases, thereby reducing the cost of genotyping. Increasing the control-to-case ratio increases the power. This was recently done to an extreme degree in an Icelandic study with 37 196 controls and 192 cases[32]. It is important to keep in mind that the gain in power is minute when increasing the control-to-case ratio above 4. For example, the power at a P-value of 5 × 10-7 in a study including 1000 cases increases from 77% to 87% for an allele with a frequency of 0.2 and an OR of 1.4, by increasing the number of controls from 4000 to 10 000.
The high density genotyping chips now available have the potential to assay up to 1 million markers (Affymetrix SNP 6.0 and Illumina 1M). In essence, the chips consist of dense arrays of specific labeled probes for given DNA sequences that will emit a light signal in the case of hybridization (binding of a matching DNA sequence in the investigated sample).
Since the price is lower, it has recently been suggested that the most cost-effective way to perform a GWAS is to continue using the older and cheaper arrays with medium density (300 000-500 000 SNPs) and then computationally determine untyped SNPs in the remainder of the genome by means of the HapMap reference (so called imputation)[33]. An even cheaper approach is to employ DNA-pooling[34]. This means that the DNA samples from several individuals are pooled together and subsequently used for genotyping. However, this design only yields estimated allele frequencies across all case or control samples instead of individual genotypes. Therefore, analyses relying on individuals’ genotypes such as phenotype associations and imputation are not possible.
As opposed to a candidate gene study, a GWAS aims to, as the name implies, assay the whole genome. Therefore, the genetic coverage of the employed genotyping array is of crucial importance. At present, the dbSNP database (http://www.ncbi.nlm.nih.gov/sites/entrez) of known SNPs includes over 12 million entries[35], and of these almost 9 million are annotated as validated (for validation criteria see http://www.ncbi.nlm.nih.gov/projects/SNP/snp_legend.cgi?legend=validation). Due to LD, assaying all these SNPs in a GWAS is not necessary to achieve complete genome-wide coverage. The genetic information obtained at SNPs can depending on the rate of recombination within this genomic region - precisely predict the alleles of closely linked, un-genotyped markers. There are substantial differences in the coverage between the different commercially available SNP arrays, but simulation has demonstrated that this does not necessarily translate into an increased power for detecting disease-associated variants[36]. It is also important to keep in mind that a high overall coverage does not necessarily mean that an individual gene is well covered, and at the gene level, there are large differences in coverage between different genotyping platforms[37]. Coverage estimations also tend to be biased, as in most cases the HapMap is used as the reference which is, by itself, assumed to be partially biased in terms of selected SNPs and in terms of included populations. Therefore, SNP arrays that mostly include HapMap tagging SNPs will have a higher genetic coverage compared to arrays that include random SNPs when basing assumptions on the HapMap.
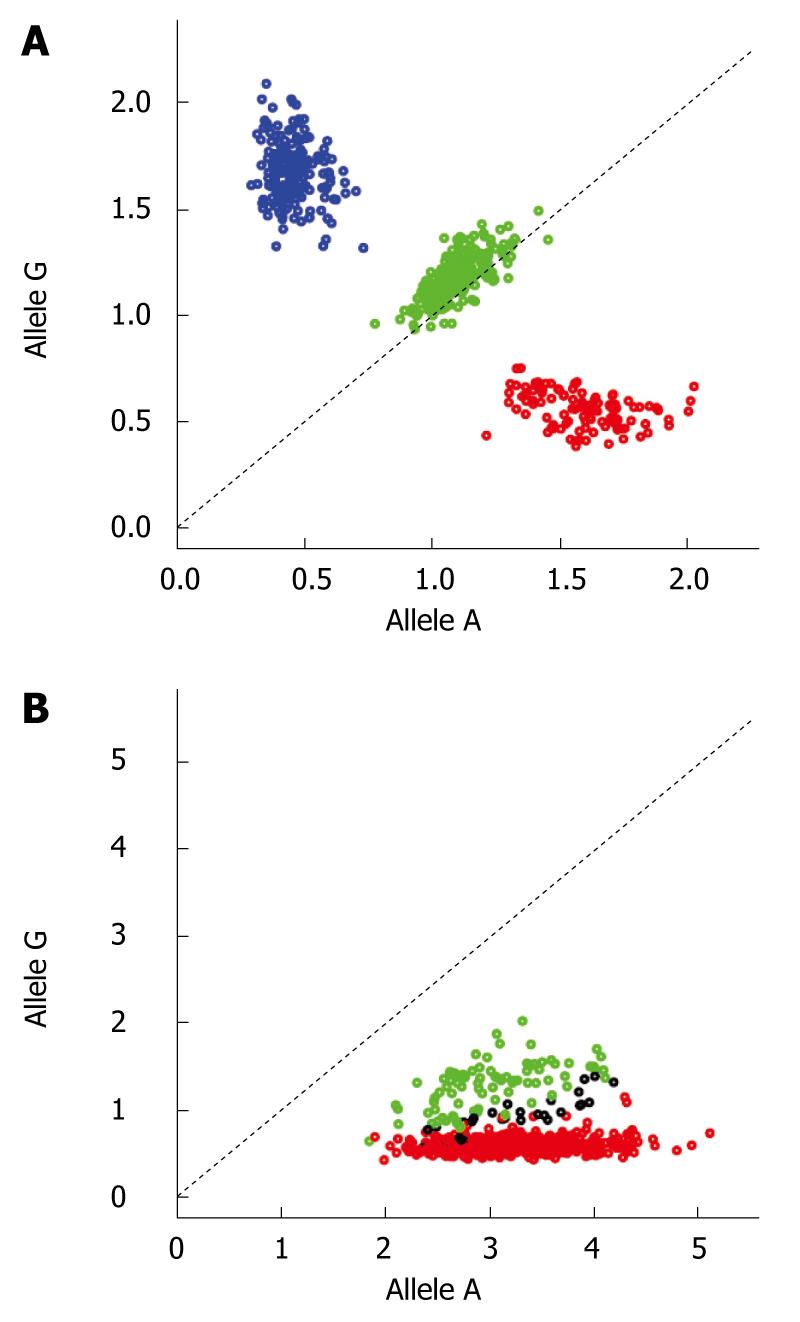
The sizes of modern genome-wide datasets are larger by many magnitudes than what was typical for a candidate gene study. For instance, in a GWAS with 1000 cases and 1000 controls using the Affymetrix SNP 6.0, the number of genotypes generated is 1.8 × 109. This huge amount of data renders automatic and semi-automatic procedures necessary, since manual processing is simply not feasible. In principle, this process can be divided into two steps (1) Exclusion of samples (low-performance samples, related individuals and population outliers) and (2) Exclusion of SNPs with evidence of bad performance. Partly this process has to be done iteratively as (a) influences (b) and vice-versa. Firstly, measures should be taken to ensure that the PCR and hybridization reaction are performed properly, and samples where this is not the case should be discarded at this stage or processed over again. Next, platform-specific quality measures should be applied (e.g. the QC-contrast for the Affymetrix SNP 6.0 chip[38]) to make sure the samples are within acceptable limits for the experiment as a whole. After this, genotype calling is performed, preferably in batches of similarly handled samples as batch effects can have an impact on the results[39]. Next, samples with a low call-rate (typically < 95%) and samples where there are mismatches between the gender recorded and the gender calculated based on the genotype data of the X chromosome are detected and should be excluded, since both of these measures can relate to poor performance on particular chips. The latter also can be due to sample mix-up. To avoid poor performing probes within otherwise acceptable arrays, SNPs with study-wise low genotyping rate (< 95%) should be removed along with SNPs with a low (< 1%) minor allele frequency. SNPs with a low minor allele frequency have a tendency to be mis-called by the clustering algorithm, since most likely only two instead of three clusters will be present in the signal intensity plots, with a significantly smaller heterozygote cloud. Even when one plans to use robust statistical tests, SNPs showing deviation (normal cut-off P-values 10-4-10-7) from Hardy-Weinberg Equilibrium in the healthy controls (not in the case/patient panel) should be removed, since this is also an indication of low genotyping quality[40]. After these measures have been applied, the resulting SNP set can be forwarded to the initial association analysis for removal of duplicates, related individuals software and ethnic outliers with, for instance, PLINK[41] and EIGENSTRAT[30]. After the removal of duplicates, related individuals and ethnic outliers, the SNP-specific quality measures should be performed over again on a fresh dataset without these samples. As the genotyping is an automatic process with little user input, one should go back to the raw data after the chosen statistical test has been performed to manually inspect the clustering plots of top hits. An example of two typical cluster plots, one good and one bad, can be found in Figure 1. The SNPs with evidence of bad clustering should be discarded and not followed up.
To increase the number of SNPs to test for association, the use of imputation to estimate genotypes at un-genotyped loci has become popular. Imputation will also increase call rates at all typed SNPs to 100%. Given the former feature, researchers may choose less dense and therefore cheaper arrays. Another advantage is that the chance to be closer to the possible functional variant, and hence statistical power - increases with a denser SNP data set.
In brief, imputation relies on the LD and haplotype information contained in a reference dataset. The HapMap datasets are most commonly used, which the algorithm aligns with the genotyped SNPs to use LD information to estimate the genotype at the target locus. Imputation has already been available in software packages such as PHASE and fastPHASE[42,43], however recent implementations such as MACH (http://www.sph.umich.edu/csg/abecasis/MaCH/), IMPUTE[44] and Beagle[45] specifically aimed at GWAS data are recommended[46] and have been shown to produce accurate and reliable results[47]. There are some differences between the software packages, with MACH and IMPUTE having the edge[47,48], but in general, they produce comparable results.
The goal of a GWAS is to identify the genetic variants that are statistically associated with the disease or trait in question. The first step to achieve this is normally to perform a single-locus association test, i.e. only a single SNP is considered at a time. As up to 1 million SNPs are assessed, it is impossible to have an a priori hypothesis about the genetic model expected. The statistical test used should therefore be robust and powerful to detect different genetic models (e.g. dominant, recessive and allele-dosage). As both the allele count and genotype count χ2 tests do not meet these requirements, a trend-based test is normally recommended, for instance the Cochrane-Armitage trend test. If it is sensible to include co-variables in the disease model (e.g. sex, age, BMI etc.), the best way is to use a logistic regression procedure and add these as covariates. For quantitative traits (e.g. enzyme levels), normal linear regression is applicable and could also include co-variables.
After the initial test statistics have been calculated, the genomic inflation factor should be determined (i.e. the median χ2 test statistics observed divided by the expected test statistics) and a quantile-quantile plot (a plot of the observed vs the expected test statistics or the negative logarithm of the P-values) should be examined. The quantile-quantile plot should not, in the setting of a non-stratified dataset, show large deviations from the expected distribution in the lower tail. In the upper tail, deviation indicates possible disease association. A genomic inflation factor above 1 typically indicates the presence of population stratification (or a differential bias in genotyping)[49]. If the genomic inflation factor is above 1.1, methods for correcting for population heterogeneity should be considered. It is, however, important to note that a small increase in the inflation factor can be caused by disease-associated markers.
Most of the analytical methods described above are implemented in the software package, PLINK[41]. The analysis of a small number of cases and controls could in principle be analyzed on a standard desktop computer. However, for more computationally challenging analyses, e.g. imputation, and/or a large number of cases and controls, the use of a high-performance computing cluster with a comfortable batch job submission environment is desirable.
Methods looking at haplotypes[43,50] and gene-gene or SNP-SNP interactions (so called epistasis)[51] should also be applied. Recently, pathway-oriented analyses, i.e. analyzing SNPs that belong to genes in a common biological pathway[52-54], have been proposed. We would recommend that these methods are first applied after the main associations in the dataset have been explored and replicated.
Applying the traditional P-value cut-off of < 0.05 for statistical significance to a GWAS leads to the fact, even in the presence of no association, that 5% of the tested SNPs are reported as statistically significant. In a typical GWAS where around 650 000 SNPs are tested, this means that 30 000-35 000 SNPs are reported to associate significantly with disease. To avoid this very large number of false-positive results, a very conservative P-value cut-off that is robust to correction using Bonferroni’s method has been described, namely the term of a “genome-wide significant P-value”. In the Wellcome Trust case-control consortium landmark paper[5] this level was set to P < 5 × 10-7. However, with new chips assaying over 1 million markers and even imputed results with more than 2 million markers, this genome-wide significance threshold might not be conservative enough. On the other hand, simulation studies have suggested the effective number of tests to be around 106[55]; the reason for this number being lower than the actual number of tested SNPs being the correlation between SNPs due to LD. In Figure 2 the power estimates for different sample sizes for different ORs are shown. A few studies have already, in the first round of analyses, been able to identify novel disease loci at genome-wide significance[56-58]. As most studies do not achieve such robust associations in their initial phase, a two-staged design is applied. This means that the strongest associated SNPs are carried forward to another study panel and again tested for association. Corrections for multiple testing must be applied to the association results obtained in the second stage. Even when applying strict criteria for selection of SNPs for replication, normally only a few SNPs will replicate. While most often signifying that the original associations were due to type I errors, this can also be due to different effects of a disease variant in different populations, e.g. due to interaction with other genetic variants; so called epistasis[59].
Ulcerative colitis (UC) and Crohn’s disease are the two major phenotypes of IBD with a combined incidence rate of 2.2 to 28.9 per 100 000 person-years in Caucasian populations[60], giving rise to inflammation in the colon and the entire intestinal tract, respectively[61]. The sibling recurrence risks are estimated to be 15-35 for Crohn’s disase and 6-9 for UC[1]. Besides the associations seen within the human leukocyte antigen (HLA)-complex[62-64], there was one particularly notable and reproducible genetic discovery in gastroenterology before the advent of GWAS; the association of Crohn’s disease with the NOD2 (also known as CARD15) gene[65,66]. A thorough review of the relevant genetics, including the NOD2 gene in Crohn’s disease before the GWAS era, can be found elsewhere[67].
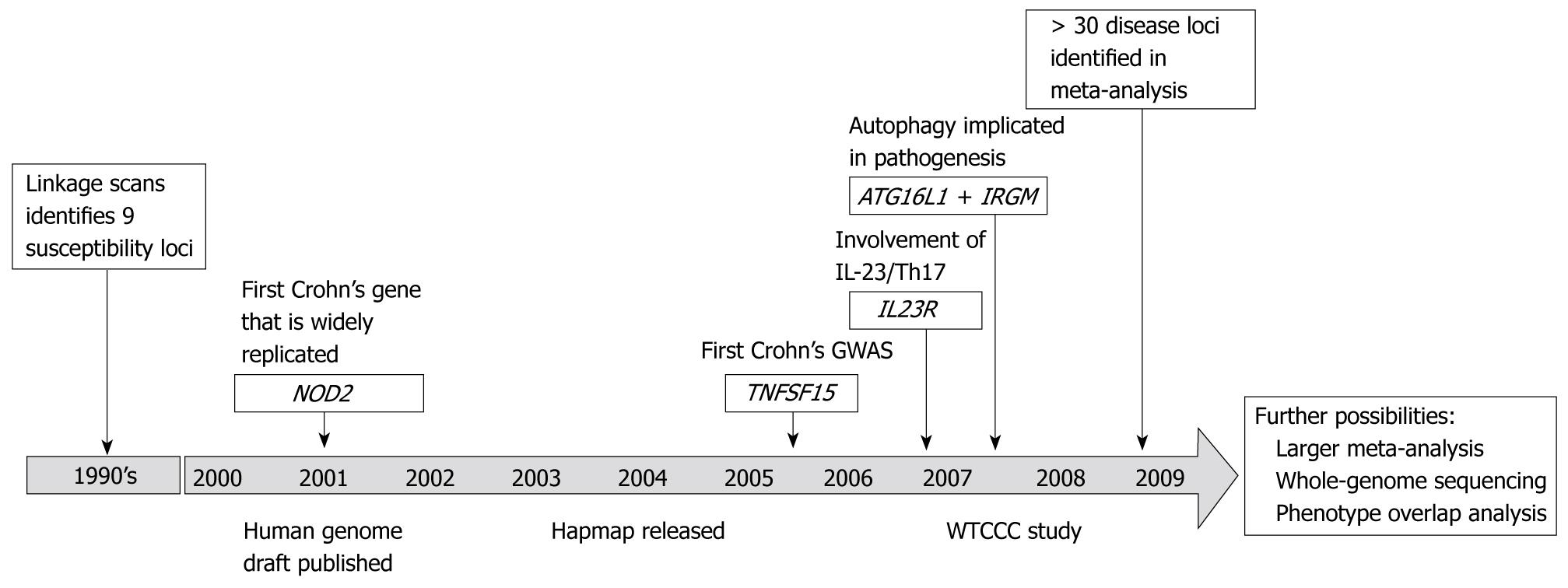
Figure 3 shows the development of Crohn’s disease genetics over the last 10 years. The eight GWAS performed in Crohn’s disease[3-9,15,17] have identified several loci influencing disease susceptibility and a recent meta-analysis implicated 20-30 additional loci[10]. It should be noted that the discovery panel and the replication panel used in the meta-analysis overlap with the previous studies and some of the confirmatory associations are biased, as the original study panel is also included. Many of the new findings in Crohn’s disease segregate into particular biological pathways and functions. Two of the key pathways are autophagy and the IL-23/Th17 pathway. Autophagy is responsible for recycling of cellular organelles and long-lived proteins, and plays an important role in tissue homeostasis and in the processing of intracellular bacteria, which is also known as xenophagy. ATG16L1 may participate in this process via the regulation of Paneth cells[68], whereas IRGM mediates autophagy of intracellular bacteria[69]. IL-23 stimulates the Th17 cell population to produce IL-17 and other pro-inflammatory cytokines involved in intestinal inflammation[70,71]. Interestingly, one of the first reported IBD genes by GWAS, IL-23R[4], participates in the IL-23/Th17 pathway. Through pathway analyses[53] of other components of this pathway the importance has been further demonstrated[54,72]. Importantly, a “pathway-based” analysis approach also takes into account variants that do not pass the formal threshold for being taking forward for replication in a traditional GWAS.
Several genes originally shown to associate with Crohn’s disease were recently shown to confer risk also for ulcerative colitis[73,74]. This indicates the presence of shared pathogenetic mechanisms in these closely-related conditions. However, disease genes that are specific for UC also exist, such as variants in the IL-10 gene[11]. This specific finding is supported by the fact that il-10 -/- mice develop colitis[75] closely resembling human UC. Other UC-specific regions are found at chromosome 1p36 and 12q15[12], however, at these loci, the exact disease genes remain to be identified. The association of classical HLA alleles and UC is well known[63], and also the GWAS SNPs near the HLA class II genes are among the most prominent findings[11,12]. Functional enquiries are needed to clarify how the general IBD genes and phenotype-specific UC and Crohn’s disease genes operate in defining the IBD phenotype and even in affecting extraintestinal manifestations such as primary sclerosing cholangitis[64].
Except for one study regarding Crohn’s disease[15], which identified the TNFSFI5 gene in a Japanese population, all GWAS in IBD have so far been performed in populations of Caucasian decent. Follow-up studies of TNFSFI5 have demonstrated differences in the effect across different populations[76,77]. This mirrors differences in IBD epidemiology in Asian populations compared to Caucasians[78], and further GWAS in Asia are likely to yield insight into differences in genetic susceptibility to IBD between these populations.
Notably, Crohn’s disease genes identified in adult patient populations show associations also in pediatric populations of the same disease[14,79-84].
Colorectal cancer is one of the most important malignancies world-wide, responsible for approximately 500 000 deaths annually[85]. The relative risk in siblings is estimated to be between 2-7, depending on the site in the colon, with the right colon showing the highest heritability[86].
The chromosome 8q24 locus is the most widely-replicated region for colorectal cancer discovered by GWAS[87-99]. This region has proven to harbor variants that predispose to several cancer types (e.g. prostate cancer and breast cancer[100-103]). As suggested by a recent publication[98], there is most likely more than one cancer-associated mutation at this locus. It was recently shown that one of the lead SNPs (rs6983267) is related to MYC expression and the activity of key Wnt signaling pathways[104,105]. In prostate cancer, it has been noted that the effect sizes of the risk variants differ among populations, necessitating the need for further characterization, e.g. systematic re-sequencing[106] of this locus, to be performed in multiple ethnicities in parallel[107]. The association between colorectal cancer and genetic variants at chromosome 18q21, most probably in the SMAD7 gene, has been subjected to extensive characterization. This characterization has led to the identification of a novel SNP which influences expression levels of SMAD7, highlighting the importance of SMAD7 expression in colorectal carcinogenesis[108]. Interestingly, a variant at chromosome 5p15 (rs401681), originally shown to confer risk for basal cell carcinoma, was recently reported to protect against colorectal carcinoma[109]. Most likely the causative variant(s) at this latter locus remain(s) to be defined. This finding is an example of the complexity of allelic associations at a disease locus.
Most of the disease loci discovered in colorectal cancer exhibit low ORs (~1.1-1.2). Identification of further disease loci would therefore necessitate a large number of cases and controls to achieve a sufficient power. In a recent meta-analysis of two colorectal cancer GWAS studies[88,110], totaling 13 315 individuals, and subsequent replication in 27 418 individuals[18], as little as four novel loci (at chromosomes 14q22.2, 16q22.1 19q13.1 and 20p12.3, respectively) were identified. This picture contrasts with the situation in Crohn’s disease where smaller study panels have detected more than 30 disease loci, and highlights the challenges of detecting disease loci in conditions with a low degree of heritability.
When exposed to gluten, a protein found in wheat, rye and barley, celiac disease patients develop inflammatory lesions with villi destruction in the small intestine[111]. The main genetic factor predisposing to celiac disease, the HLA-DQB1*0201 variant, has been known for 20 years[62]. However, since this HLA allele is present at high prevalence also in individuals who do not develop celiac disease[62], other genetic risk factors are likely to exist. One GWAS in celiac disease has been performed and published in three stages[112-114]. The first of these publications demonstrated that, besides markers in the HLA-complex, variants in the IL2-IL21 region are associated[112]. Evidence concerning the involvement of a specific variant in this region was not possible due to strong LD in the region. Interestingly, this region has also been shown to associate with type I diabetes, rheumatoid arthritis and recently UC, hinting at the presence of a common factor for immune-mediated diseases[115-117]. To increase the likelihood of disease-gene identification in this GWAS data, additional non-HLA SNPs were subsequently subjected to genotyping in an even larger replication panel[113,114]. Solid evidence for association at several novel regions was obtained, several of which harbor genes of relevance to immunological components of celiac disease pathogenesis (such as the chemokine-receptor cluster at chromosome 3p21, the IL12A locus at 3q25-3q26 and TNFAIP3). For other associations, e.g. the LPP gene at chromosome 3q28, further studies are required to define how defective gene function could contribute to the pathogenesis. As for the IL2-IL21 region, several of the loci also show associations with type I diabetes[118].
For Hirschsprung’s disease, a disease characterized by lack of ganglia in the colon, there are large ethnic differences in incidence and clear cases of family aggregation, both of which are good indicators of a genetic contribution to pathogenesis. For a large proportion of familial cases and a considerable amount of sporadic cases, it has long been known that mutations in the RET gene are important[119]. In a small GWAS, Garcia-Barcelo et al[120] recently made several important discoveries. Firstly, they confirmed the associations previously detected at the RET locus. Secondly, they identified strong associations at the NRG1 gene. Thirdly, they reported a significant and strong interaction effect between NRG1 and RET polymorphisms in the combined discovery and replication panel. The NRG1 gene is probably important in the development of the enteric nervous system and is thus a plausible biological candidate for a Hirschsprung’s disease gene. Another interesting aspect of this study is that robust gene findings are possible even in small patient panels. This is probably more likely when the phenotype is clearly defined and shows early life debut, i.e environmental factors are less likely to be influential.
One of the most important clinical tools for detecting, diagnosing and monitoring liver diseases is the use of different biochemical parameters (often called ‘liver enzymes’). It has long been known that there are genetic factors influencing the level of these enzymes, described in genetic terms as a genuine quantitative trait[121] that could either be due to (a) variants influencing the levels without any pathological condition present and/or (b) variants that are truly associated with liver disease and which indicates that undiagnosed cases of disease are the cause of the increased blood levels. Both of these entities are important to discover as: (a) may have practical implications for the handling of apparently increased levels in healthy individuals, while (b) may serve as early markers for liver disease in apparently healthy individuals. Yuan et al[122] were able to identify 6 different loci associated with the levels of these enzymes (ALT: CPN1-ERLIN1-CHUK and PNPLA3-SAMM50, ALP: ALPL, GPLD and JMJD1C-REEP, GGT: HNF1A). A large meta-analysis of several GWAS datasets recently suggested highly significant associations with bilirubin levels and variants at the UGT1A1 and SLCO1B1 loci[123].
In a very small genome-wide analysis in terms of sample size, a highly significant association of variants at the HLA-B locus was detected in 51 cases of flucloxacillin-induced liver injury[124]. The identified SNP was in LD with the HLA-B*5701 allele and subsequent direct genotyping confirmed this with an effect size equaling an OR of 80.6 (95% CI: 22.8-284.9). Although no formal replication panel was investigated, as many as 20 out of 23 additional cases of flucloxacillin-induced liver injury were HLA-B*5701 positive. This shows that development of drug-induced liver injury is clearly dependent upon host factors of which HLA variants are probably key determinants[125].
There is an increased number of subjects with NAFLD in siblings of overweight children with NAFLD, indicating the presence of genetic risk factors in this condition[126]. In a study evaluating the genetics of NAFLD using a quantitative measure (hepatic fat content measured by magnetic resonance imaging) an association with markers in the PNPLA3 gene was recently reported[127]. Importantly, the association was independent of key confounders such as body mass index, alcohol consumption and diabetes. Noteworthy is that the low number of SNPs assayed in the GWAS (approximately 10 000) was sufficient to generate this highly interesting finding[128]. Whereas the function of the PNPLA3 gene is not known, the finding is substantiated by differential regulation of the gene in different metabolic states[129]. Since NAFLD is an increasingly common condition, it is interesting to note that this locus also shows associations with alanine transaminase levels in the liver enzyme population-based study mentioned above[122].
Hepatitis B is one of the major causes of world-wide liver morbidity and mortality[130]. Part of the variability in clinical course[131] is related to viral properties, but there is strong reason to believe that host genetics are of importance. In a three-staged GWAS design, particular variants of the class II HLA-DPA and HLA-DPB genes were recently shown to be associated with chronic hepatitis B infection in Asian populations[132]. The HLA-complex is characterized by strong LD[133] and, as hinted at by other studies, distinct HLA genes may prove to be important[134]. To clarify this, verification of the Asian findings in Caucasian populations and supplementary mechanistic studies are needed. Chronic hepatitis C is a progressive liver disease, complicated by development of cirrhosis and hepatocellular carcinoma[135]. Treatment with pegylated interferon and ribavirin offers the potential of viral eradication in up to 80% of the patients[136]. In a GWAS of treatment response, variants in the IL28B gene were found to be associated in the three (European-Americans, African-Americans and Hispanics) ethnicities tested[137], shedding light on the biological mechanisms operating to define treatment success.
Defects of bile formation are likely to be important in the development of biliary calculi[138], inspiring candidate gene studies of important transport proteins involved in this process[139]. Given this a priori knowledge, it was not surprising that the main disease locus in a German gallstone GWAS was ABCG8, encoding a component of the cholesterol transporter heterodimer ABCG5/G8[140]. Interestingly, the findings in previous linkage studies support the GWAS results[141]. The ABCG8 effect is not specific to Caucasian populations, as is evident from a Taiwanese report[142].
Primary biliary cirrhosis (PBC) is a chronic cholestatic liver disease characterized by autoimmune destruction of the biliary canaliculi. There is evidence for genetic factors being involved in pathogenesis with a sibling recurrence risk reported at approximately 10.5[2]. PBC has long been known to exhibit strong HLA associations[143,144], so it was not surprising that SNPs in the HLA-complex were among the top hits of a recent GWAS[27]. Of particular interest were associations detected at the IL12A and IL12RB loci, strongly supporting the presence of autoimmune mechanisms in PBC pathogenesis. Genetic defects of the IL-12 pathway have been proposed in other autoimmune conditions and the findings in PBC are thus in line with a paradigm where the majority of GWAS findings in these conditions seem to be common denominators rather than disease specific-findings[145].
The advent of imputation has facilitated integration of data from different genotyping platforms[10,146,147]. A meta-analysis of GWAS data sets in type 2 diabetes has highlighted the importance of risk variants with low effect sizes[148,149]. In this condition, one of the loci with a low effect size (OR < 1.2) is the PPARG locus, where the biological implications in terms of glitazone therapeutics in type II diabetes has long been proven. In gastroenterology, a large GWAS meta-analysis has so far only been performed in Crohn’s disease, where more than 30 loci were detected[10]. There is also a potential for combining different but related phenotypes to increase power in discovering factors common to both entities. A common risk factor for Crohn’s disease and sarcoidosis, both of which are characterized by granulomas, was discovered with such an approach[16], and similar approaches can be defined for other clinical features of otherwise unrelated conditions[109,115].
In this way, GWAS studies also herald collaboration between research groups in different countries and across scientific traditions, a trend which can possibly generate scientific initiatives and discoveries, even beyond the meta-analyses. In the Genetic Association Information Network (GAIN) consortium (http://www.genome.gov/19518664), the collaboration has been formalized and has led to a series of successful publications[150]. Interestingly, researchers have also released their datasets into the public domain, partly due to requirements from their funding sources (e.g. NIH). The release of data has been facilitated through the dbGAP interface[151]; however caution needs to be exercised in terms of data protection and privacy issues[152].
In the present review we have focused on the typical findings of a GWAS; the associations between particular SNPs and a disease trait. The importance of further genetic and functional characterization of SNP findings is highlighted by the association between a deletion polymorphism at the IRGM locus and Crohn’s disease[153], a mutation which was later shown to alter the expression level of IRGM[154]. Interestingly, this deletion was perfectly correlated with a SNP in the first study reporting an association in this region[6]. With dense SNP arrays and the aid of imputation of un-genotyped markers, there is a good chance of detecting CNV associations through LD in this manner. In addition, the genotyping chips have separate probes for CNVs and specific computer algorithms can use these probes (and even the intensities from the SNP probes) to generate genotypes for CNVs at a given position. Most likely we will see a number of studies, even in gastroenterology and hepatology, where the application of these algorithms will lead to the identification of disease-associated CNVs[155,156].
Due to the inherent design of the SNP arrays and the available SNPs in the HapMap, rare variants (frequency < 1%-10%) are not easily detected in a GWAS analysis[157]. Also for statistical reasons, the power for detecting such variants in the unbiased GWAS design is low. In addition to common variants influencing gene expression or protein function, a disease locus can be constituted by a multitude of rare variants with a high penetrance. This is a well-known phenomenon in monogenic diseases such as cystic fibrosis, where hundreds of rare disease-causing mutations have been defined[158]. Probably the typical situation at a disease locus is a combination of common and rare disease variants, as highlighted for the NOD2 locus (20% rare, 80% common)[159]. Only the common variants are “visible” in the GWAS design, but this does not thus exclude the presence of additional disease-causing variants at a locus only detectable by careful investigation of the identified disease genes. The identification of a disease-related variant with a functional effect, even if only present in singular patients[160], can yield important insight into the pathogenesis of a condition. Over the last few years second generation sequencing technology (also called next-generation) has become available. This technology is able to resequence large regions of DNA or even complete human individual genomes[161]. There are now three commercial platforms available (SOLiD from Applied Biosystems, 454/FLX from Roche and Genome Analyzer from Illumina), all of which offer unprecedented throughput when compared to traditional Sanger sequencing (which has been the gold standard since 1977[162]). The application of a combined re-sequencing/association study approach was recently demonstrated in type 1 diabetes with the identification of IFIH1 variants with likely functional effects[163]. No such extensive re-sequencing studies have so far been performed in gastroenterology or hepatology.
GWAS have proven to be an important tool for dissecting the genetic architecture of common diseases. The genotyping arrays and statistical tools are now mature and their application has been a huge success in gastroenterology. In crude numbers, in only 2-3 years, the approach has delivered many times the number of genes that were discovered during the first 15-20 years of gastroenterological disease genetics. As illustrated by several of the findings we have summarized in this review, the effect sizes are often low and can only be discovered with large case-control collections. Clearly, for some diseases (such as primary sclerosing cholangitis) this situation may not be achievable and complementary approaches (e.g. pathway analyses[52-54]) may be needed to identify genetically defined disease mechanisms.
As we have argued in the present review, and which is also a view shared by the editors of prominent journals, the size of the study panels is important. Interestingly, however, several of the genes so far confirmed were initially detected in what was an apparently under-powered discovery panel (e.g. TNFSF15 discovered in 94 cases[15]). In addition, clinically highly important findings have been made in small case-control panels[57]. Therefore, GWAS in smaller disease populations should also be welcomed, since identifying as little as a single disease gene can open broad avenues for future mechanistic studies[127].
In addition to the results from recent GWAS, it is worth mentioning a gene identified through a classical candidate gene approach. After identification of the role of XBP1 in endoplasmatic reticulum stress in a mouse xbp1 knock-out model, polymorphisms in this gene were tested in a large panel of IBD patients[164]. In terms of P-values, several of the IBD associations detected at this locus are below the detection limit of the “GWAS radar” and highlight why hypothesis-based candidate gene approaches still have a role in disease genetics.
Even in the case of large effect sizes, the new GWAS findings may not have immediate implications for clinical practice[124], however, and what seems to be the bottom line of the present role of these studies in biology serves as a glimpse of the intricate pathology involved. Whereas few, if any, of the GWAS have provided a comprehensive mechanistic explanation as to how the detected polymorphisms affect disease risk, they have defined the priorities for basic research for decades to come. Ultimately, the challenge and goal for this research will be to define relevant diagnostic and prognostic markers as well as novel therapeutic options.
Peer reviewer: Shu Zheng, Professor, Scientific Director of Cancer Institute, Zhejiang University, Secondary Affiliated Hospital, Zhejiang University, 88# Jiefang Road, Hangzhou 310009, Zhejiang Province, China
S- Editor Tian L L- Editor Logan S E- Editor Yin DH
| 1. | Mathew CG, Lewis CM. Genetics of inflammatory bowel disease: progress and prospects. Hum Mol Genet. 2004;13 Spec No 1:R161-R168. [Cited in This Article: ] |
| 2. | Jones DE, Watt FE, Metcalf JV, Bassendine MF, James OF. Familial primary biliary cirrhosis reassessed: a geographically-based population study. J Hepatol. 1999;30:402-407. [Cited in This Article: ] |
| 3. | Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207-211. [Cited in This Article: ] |
| 4. | Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461-1463. [Cited in This Article: ] |
| 5. | Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661-678. [Cited in This Article: ] |
| 6. | Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830-832. [Cited in This Article: ] |
| 7. | Franke A, Hampe J, Rosenstiel P, Becker C, Wagner F, Häsler R, Little RD, Huse K, Ruether A, Balschun T. Systematic association mapping identifies NELL1 as a novel IBD disease gene. PLoS One. 2007;2:e691. [Cited in This Article: ] |
| 8. | Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596-604. [Cited in This Article: ] |
| 9. | Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. [Cited in This Article: ] |
| 10. | Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955-962. [Cited in This Article: ] |
| 11. | Franke A, Balschun T, Karlsen TH, Sventoraityte J, Nikolaus S, Mayr G, Domingues FS, Albrecht M, Nothnagel M, Ellinghaus D. Sequence variants in IL10, ARPC2 and multiple other loci contribute to ulcerative colitis susceptibility. Nat Genet. 2008;40:1319-1323. [Cited in This Article: ] |
| 12. | Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216-220. [Cited in This Article: ] |
| 13. | Fisher SA, Tremelling M, Anderson CA, Gwilliam R, Bumpstead S, Prescott NJ, Nimmo ER, Massey D, Berzuini C, Johnson C. Genetic determinants of ulcerative colitis include the ECM1 locus and five loci implicated in Crohn's disease. Nat Genet. 2008;40:710-712. [Cited in This Article: ] |
| 14. | Kugathasan S, Baldassano RN, Bradfield JP, Sleiman PM, Imielinski M, Guthery SL, Cucchiara S, Kim CE, Frackelton EC, Annaiah K. Loci on 20q13 and 21q22 are associated with pediatric-onset inflammatory bowel disease. Nat Genet. 2008;40:1211-1215. [Cited in This Article: ] |
| 15. | Yamazaki K, McGovern D, Ragoussis J, Paolucci M, Butler H, Jewell D, Cardon L, Takazoe M, Tanaka T, Ichimori T. Single nucleotide polymorphisms in TNFSF15 confer susceptibility to Crohn's disease. Hum Mol Genet. 2005;14:3499-3506. [Cited in This Article: ] |
| 16. | Franke A, Fischer A, Nothnagel M, Becker C, Grabe N, Till A, Lu T, Müller-Quernheim J, Wittig M, Hermann A. Genome-wide association analysis in sarcoidosis and Crohn's disease unravels a common susceptibility locus on 10p12.2. Gastroenterology. 2008;135:1207-1215. [Cited in This Article: ] |
| 17. | Raelson JV, Little RD, Ruether A, Fournier H, Paquin B, Van Eerdewegh P, Bradley WE, Croteau P, Nguyen-Huu Q, Segal J. Genome-wide association study for Crohn's disease in the Quebec Founder Population identifies multiple validated disease loci. Proc Natl Acad Sci USA. 2007;104:14747-14752. [Cited in This Article: ] |
| 18. | Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, Chandler I, Vijayakrishnan J, Sullivan K, Penegar S. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426-1435. [Cited in This Article: ] |
| 20. | Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851-861. [Cited in This Article: ] |
| 21. | Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590-1605. [Cited in This Article: ] |
| 22. | Sauer U, Heinemann M, Zamboni N. Genetics. Getting closer to the whole picture. Science. 2007;316:550-551. [Cited in This Article: ] |
| 23. | Moum B, Ekbom A, Vatn MH, Aadland E, Sauar J, Lygren I, Schulz T, Stray N, Fausa O. Inflammatory bowel disease: re-evaluation of the diagnosis in a prospective population based study in south eastern Norway. Gut. 1997;40:328-332. [Cited in This Article: ] |
| 24. | Garner C. The use of random controls in genetic association studies. Hum Hered. 2006;61:22-26. [Cited in This Article: ] |
| 25. | Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623-630. [Cited in This Article: ] |
| 26. | Xiao R, Boehnke M. Quantifying and correcting for the winner's curse in genetic association studies. Genet Epidemiol. 2009;33:453-462. [Cited in This Article: ] |
| 27. | Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Lu Y, Gu X, Walker EJ, Jing K, Juran BD. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544-2555. [Cited in This Article: ] |
| 28. | Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657-665. [Cited in This Article: ] |
| 29. | Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357:1199-1209. [Cited in This Article: ] |
| 30. | Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904-909. [Cited in This Article: ] |
| 31. | Li Q, Yu K. Improved correction for population stratification in genome-wide association studies by identifying hidden population structures. Genet Epidemiol. 2008;32:215-226. [Cited in This Article: ] |
| 32. | Gudmundsson J, Sulem P, Gudbjartsson DF, Jonasson JG, Sigurdsson A, Bergthorsson JT, He H, Blondal T, Geller F, Jakobsdottir M, Magnusdottir DN, Matthiasdottir S, Stacey SN, Skarphedinsson OB, Helgadottir H, Li W, Nagy R, Aguillo E, Faure E, Prats E, Saez B, Martinez M, Eyjolfsson GI, Bjornsdottir US, Holm H, Kristjansson K, Frigge ML, Kristvinsson H, Gulcher JR, Jonsson T, Rafnar T, Hjartarsson H, Mayordomo JI, de la Chapelle A, Hrafnkelsson J, Thorsteinsdottir U, Kong A, Stefansson K. Common variants on 9q22.33 and 14q13.3 predispose to thyroid cancer in European populations. Nat Genet. 2009;41:460-464. [Cited in This Article: ] |
| 33. | Anderson CA, Pettersson FH, Barrett JC, Zhuang JJ, Ragoussis J, Cardon LR, Morris AP. Evaluating the effects of imputation on the power, coverage, and cost efficiency of genome-wide SNP platforms. Am J Hum Genet. 2008;83:112-119. [Cited in This Article: ] |
| 34. | Bossé Y, Bacot F, Montpetit A, Rung J, Qu HQ, Engert JC, Polychronakos C, Hudson TJ, Froguel P, Sladek R. Identification of susceptibility genes for complex diseases using pooling-based genome-wide association scans. Hum Genet. 2009;125:305-318. [Cited in This Article: ] |
| 35. | Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308-311. [Cited in This Article: ] |
| 36. | Spencer CC, Su Z, Donnelly P, Marchini J. Designing genome-wide association studies: sample size, power, imputation, and the choice of genotyping chip. PLoS Genet. 2009;5:e1000477. [Cited in This Article: ] |
| 37. | Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635-643. [Cited in This Article: ] |
| 38. | Appendix E in Affymetrix® Genotyping Console 3. 0.1 User Manual. 2008;. [Cited in This Article: ] |
| 39. | Hong H, Su Z, Ge W, Shi L, Perkins R, Fang H, Xu J, Chen JJ, Han T, Kaput J. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500 K array set using 270 HapMap samples. BMC Bioinformatics. 2008;9 Suppl 9:S17. [Cited in This Article: ] |
| 40. | Teo YY, Fry AE, Clark TG, Tai ES, Seielstad M. On the usage of HWE for identifying genotyping errors. Ann Hum Genet. 2007;71:701-703; author reply 704. [Cited in This Article: ] |
| 41. | Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559-575. [Cited in This Article: ] |
| 42. | Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629-644. [Cited in This Article: ] |
| 43. | Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978-989. [Cited in This Article: ] |
| 44. | Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906-913. [Cited in This Article: ] |
| 45. | Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084-1097. [Cited in This Article: ] |
| 46. | Ellinghaus D, Schreiber S, Franke A, Nothnagel M. Current software for genotype imputation. Hum Genomics. 2009;3:371-380. [Cited in This Article: ] |
| 47. | Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. A comprehensive evaluation of SNP genotype imputation. Hum Genet. 2009;125:163-171. [Cited in This Article: ] |
| 48. | Pei YF, Li J, Zhang L, Papasian CJ, Deng HW. Analyses and comparison of accuracy of different genotype imputation methods. PLoS One. 2008;3:e3551. [Cited in This Article: ] |
| 49. | Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243-1246. [Cited in This Article: ] |
| 50. | Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263-265. [Cited in This Article: ] |
| 51. | Cordell HJ. Genome-wide association studies: Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392-404. [Cited in This Article: ] |
| 52. | Hong MG, Pawitan Y, Magnusson PK, Prince JA. Strategies and issues in the detection of pathway enrichment in genome-wide association studies. Hum Genet. 2009;126:289-301. [Cited in This Article: ] |
| 53. | Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007;81:1278-1283. [Cited in This Article: ] |
| 54. | Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399-405. [Cited in This Article: ] |
| 55. | Hoggart CJ, Clark TG, De Iorio M, Whittaker JC, Balding DJ. Genome-wide significance for dense SNP and resequencing data. Genet Epidemiol. 2008;32:179-185. [Cited in This Article: ] |
| 56. | Edwards AO, Ritter R 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421-424. [Cited in This Article: ] |
| 57. | Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789-799. [Cited in This Article: ] |
| 58. | Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385-389. [Cited in This Article: ] |
| 59. | Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4:e5639. [Cited in This Article: ] |
| 60. | Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504-1517. [Cited in This Article: ] |
| 61. | Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet. 2007;369:1641-1657. [Cited in This Article: ] |
| 62. | Sollid LM, Markussen G, Ek J, Gjerde H, Vartdal F, Thorsby E. Evidence for a primary association of celiac disease to a particular HLA-DQ alpha/beta heterodimer. J Exp Med. 1989;169:345-350. [Cited in This Article: ] |
| 63. | Stokkers PC, Reitsma PH, Tytgat GN, van Deventer SJ. HLA-DR and -DQ phenotypes in inflammatory bowel disease: a meta-analysis. Gut. 1999;45:395-401. [Cited in This Article: ] |
| 64. | Karlsen TH, Schrumpf E, Boberg KM. Genetic epidemiology of primary sclerosing cholangitis. World J Gastroenterol. 2007;13:5421-5431. [Cited in This Article: ] |
| 65. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606. [Cited in This Article: ] |
| 66. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [Cited in This Article: ] |
| 67. | Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376-388. [Cited in This Article: ] |
| 68. | Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259-263. [Cited in This Article: ] |
| 69. | Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438-1441. [Cited in This Article: ] |
| 70. | Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473-2483. [Cited in This Article: ] |
| 71. | Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310-1316. [Cited in This Article: ] |
| 72. | McGovern DP, Rotter JI, Mei L, Haritunians T, Landers C, Derkowski C, Dutridge D, Dubinsky M, Ippoliti A, Vasiliauskas E. Genetic epistasis of IL23/IL17 pathway genes in Crohn's disease. Inflamm Bowel Dis. 2009;15:883-889. [Cited in This Article: ] |
| 73. | Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713-715. [Cited in This Article: ] |
| 74. | Anderson CA, Massey DC, Barrett JC, Prescott NJ, Tremelling M, Fisher SA, Gwilliam R, Jacob J, Nimmo ER, Drummond H. Investigation of Crohn's disease risk loci in ulcerative colitis further defines their molecular relationship. Gastroenterology. 2009;136:523-529. [Cited in This Article: ] |
| 75. | Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263-274. [Cited in This Article: ] |
| 76. | Picornell Y, Mei L, Taylor K, Yang H, Targan SR, Rotter JI. TNFSF15 is an ethnic-specific IBD gene. Inflamm Bowel Dis. 2007;13:1333-1338. [Cited in This Article: ] |
| 77. | Thiébaut R, Kotti S, Jung C, Merlin F, Colombel JF, Lemann M, Almer S, Tysk C, O'Morain M, Gassull M. TNFSF15 polymorphisms are associated with susceptibility to inflammatory bowel disease in a new European cohort. Am J Gastroenterol. 2009;104:384-391. [Cited in This Article: ] |
| 78. | Thia KT, Loftus EV Jr, Sandborn WJ, Yang SK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol. 2008;103:3167-3182. [Cited in This Article: ] |
| 79. | Amre DK, Mack D, Israel D, Morgan K, Lambrette P, Law L, Grimard G, Deslandres C, Krupoves A, Bucionis V. Association between genetic variants in the IL-23R gene and early-onset Crohn's disease: results from a case-control and family-based study among Canadian children. Am J Gastroenterol. 2008;103:615-620. [Cited in This Article: ] |
| 80. | Amre DK, Mack DR, Morgan K, Krupoves A, Costea I, Lambrette P, Grimard G, Dong J, Feguery H, Bucionis V. Autophagy gene ATG16L1 but not IRGM is associated with Crohn's disease in Canadian children. Inflamm Bowel Dis. 2009;15:501-507. [Cited in This Article: ] |
| 81. | Baldassano RN, Bradfield JP, Monos DS, Kim CE, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Kanterakis S, Shaner JL. Association of variants of the interleukin-23 receptor gene with susceptibility to pediatric Crohn's disease. Clin Gastroenterol Hepatol. 2007;5:972-976. [Cited in This Article: ] |
| 82. | Leshinsky-Silver E, Karban A, Dalal I, Eliakim R, Shirin H, Tzofi T, Boaz M, Levine A. Evaluation of the interleukin-23 receptor gene coding variant R381Q in pediatric and adult Crohn disease. J Pediatr Gastroenterol Nutr. 2007;45:405-408. [Cited in This Article: ] |
| 83. | Dubinsky MC, Wang D, Picornell Y, Wrobel I, Katzir L, Quiros A, Dutridge D, Wahbeh G, Silber G, Bahar R. IL-23 receptor (IL-23R) gene protects against pediatric Crohn's disease. Inflamm Bowel Dis. 2007;13:511-515. [Cited in This Article: ] |
| 84. | Essers JB, Lee JJ, Kugathasan S, Stevens CR, Grand RJ, Daly MJ. Established genetic risk factors do not distinguish early and later onset Crohn's disease. Inflamm Bowel Dis. 2009;15:1508-1514. [Cited in This Article: ] |
| 85. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [Cited in This Article: ] |
| 86. | Hemminki K, Chen B. Familial risk for colorectal cancers are mainly due to heritable causes. Cancer Epidemiol Biomarkers Prev. 2004;13:1253-1256. [Cited in This Article: ] |
| 87. | Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984-988. [Cited in This Article: ] |
| 88. | Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631-637. [Cited in This Article: ] |
| 89. | Schafmayer C, Buch S, Völzke H, von Schönfels W, Egberts JH, Schniewind B, Brosch M, Ruether A, Franke A, Mathiak M. Investigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sample. Int J Cancer. 2009;124:75-80. [Cited in This Article: ] |
| 90. | Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, Bishop DT, Cox A, Camp NJ. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev. 2009;18:616-621. [Cited in This Article: ] |
| 91. | Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989-994. [Cited in This Article: ] |
| 92. | Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, Wu AH, Reich D, Henderson BE. A common genetic risk factor for colorectal and prostate cancer. Nat Genet. 2007;39:954-956. [Cited in This Article: ] |
| 93. | Li L, Plummer SJ, Thompson CL, Merkulova A, Acheson LS, Tucker TC, Casey G. A common 8q24 variant and the risk of colon cancer: a population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:339-342. [Cited in This Article: ] |
| 94. | Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, Fink SR, Foster NR, Kabat BF, Halling KC, Cunningham JM. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:2492-2500. [Cited in This Article: ] |
| 95. | Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, Siegmund KD, Casey G, Thibodeau SN, Jenkins MA, Hopper JL. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128-11132. [Cited in This Article: ] |
| 96. | Berndt SI, Potter JD, Hazra A, Yeager M, Thomas G, Makar KW, Welch R, Cross AJ, Huang WY, Schoen RE. Pooled analysis of genetic variation at chromosome 8q24 and colorectal neoplasia risk. Hum Mol Genet. 2008;17:2665-2672. [Cited in This Article: ] |
| 97. | Wokołorczyk D, Lubiński J, Narod SA, Cybulski C. Genetic heterogeneity of 8q24 region in susceptibility to cancer. J Natl Cancer Inst. 2009;101:278-279. [Cited in This Article: ] |
| 98. | Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, Pooley KA, Ramus SJ, Kjaer SK, Hogdall E. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962-966. [Cited in This Article: ] |
| 99. | Gruber SB, Moreno V, Rozek LS, Rennerts HS, Lejbkowicz F, Bonner JD, Greenson JK, Giordano TJ, Fearson ER, Rennert G. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol Ther. 2007;6:1143-1147. [Cited in This Article: ] |
| 100. | Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631-637. [Cited in This Article: ] |
| 101. | Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638-644. [Cited in This Article: ] |
| 102. | Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645-649. [Cited in This Article: ] |
| 103. | Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087-1093. [Cited in This Article: ] |
| 104. | Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet. 2009;41:882-884. [Cited in This Article: ] |
| 105. | Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Björklund M, Wei G, Yan J, Niittymäki I. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885-890. [Cited in This Article: ] |
| 106. | Yeager M, Xiao N, Hayes RB, Bouffard P, Desany B, Burdett L, Orr N, Matthews C, Qi L, Crenshaw A. Comprehensive resequence analysis of a 136 kb region of human chromosome 8q24 associated with prostate and colon cancers. Hum Genet. 2008;124:161-170. [Cited in This Article: ] |
| 107. | Waters KM, Le Marchand L, Kolonel LN, Monroe KR, Stram DO, Henderson BE, Haiman CA. Generalizability of associations from prostate cancer genome-wide association studies in multiple populations. Cancer Epidemiol Biomarkers Prev. 2009;18:1285-1289. [Cited in This Article: ] |
| 108. | Pittman AM, Naranjo S, Webb E, Broderick P, Lips EH, van Wezel T, Morreau H, Sullivan K, Fielding S, Twiss P. The colorectal cancer risk at 18q21 is caused by a novel variant altering SMAD7 expression. Genome Res. 2009;19:987-993. [Cited in This Article: ] |
| 109. | Rafnar T, Sulem P, Stacey SN, Geller F, Gudmundsson J, Sigurdsson A, Jakobsdottir M, Helgadottir H, Thorlacius S, Aben KK. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41:221-227. [Cited in This Article: ] |
| 110. | Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315-1317. [Cited in This Article: ] |
| 111. | Di Sabatino A, Corazza GR. Coeliac disease. Lancet. 2009;373:1480-1493. [Cited in This Article: ] |
| 112. | van Heel DA, Franke L, Hunt KA, Gwilliam R, Zhernakova A, Inouye M, Wapenaar MC, Barnardo MC, Bethel G, Holmes GK. A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet. 2007;39:827-829. [Cited in This Article: ] |
| 113. | Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395-402. [Cited in This Article: ] |
| 114. | Trynka G, Zhernakova A, Romanos J, Franke L, Hunt KA, Turner G, Bruinenberg M, Heap GA, Platteel M, Ryan AW. Coeliac disease-associated risk variants in TNFAIP3 and REL implicate altered NF-kappaB signalling. Gut. 2009;58:1078-1083. [Cited in This Article: ] |
| 115. | Zhernakova A, Alizadeh BZ, Bevova M, van Leeuwen MA, Coenen MJ, Franke B, Franke L, Posthumus MD, van Heel DA, van der Steege G. Novel association in chromosome 4q27 region with rheumatoid arthritis and confirmation of type 1 diabetes point to a general risk locus for autoimmune diseases. Am J Hum Genet. 2007;81:1284-1288. [Cited in This Article: ] |
| 116. | Festen EA, Goyette P, Scott R, Annese V, Zhernakova A, Lian J, Lefèbvre C, Brant SR, Cho JH, Silverberg MS. Genetic variants in the region harbouring IL2/IL21 associated with ulcerative colitis. Gut. 2009;58:799-804. [Cited in This Article: ] |
| 117. | Glas J, Stallhofer J, Ripke S, Wetzke M, Pfennig S, Klein W, Epplen JT, Griga T, Schiemann U, Lacher M. Novel genetic risk markers for ulcerative colitis in the IL2/IL21 region are in epistasis with IL23R and suggest a common genetic background for ulcerative colitis and celiac disease. Am J Gastroenterol. 2009;104:1737-1744. [Cited in This Article: ] |
| 118. | Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N Engl J Med. 2008;359:2767-2777. [Cited in This Article: ] |
| 119. | Amiel J, Sproat-Emison E, Garcia-Barcelo M, Lantieri F, Burzynski G, Borrego S, Pelet A, Arnold S, Miao X, Griseri P. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1-14. [Cited in This Article: ] |
| 120. | Garcia-Barcelo MM, Tang CS, Ngan ES, Lui VC, Chen Y, So MT, Leon TY, Miao XP, Shum CK, Liu FQ. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung's disease. Proc Natl Acad Sci USA. 2009;106:2694-2699. [Cited in This Article: ] |
| 121. | Bathum L, Petersen HC, Rosholm JU, Hyltoft Petersen P, Vaupel J, Christensen K. Evidence for a substantial genetic influence on biochemical liver function tests: results from a population-based Danish twin study. Clin Chem. 2001;47:81-87. [Cited in This Article: ] |
| 122. | Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83:520-528. [Cited in This Article: ] |
| 123. | Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, Lin JP, van Duijn CM, Harris TB, Cupples LA. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 2009;18:2700-2710. [Cited in This Article: ] |
| 124. | Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Pe'er I, Floratos A, Daly MJ, Goldstein DB, John S, Nelson MR. HLA-B*5701 genotype is a major determinant of drug-induced liver injury due to flucloxacillin. Nat Genet. 2009;41:816-819. [Cited in This Article: ] |
| 125. | Andrade RJ, Lucena MI, Alonso A, García-Cortes M, García-Ruiz E, Benitez R, Fernández MC, Pelaez G, Romero M, Corpas R. HLA class II genotype influences the type of liver injury in drug-induced idiosyncratic liver disease. Hepatology. 2004;39:1603-1612. [Cited in This Article: ] |
| 126. | Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, Yokoo T, Chavez A, Middleton MS. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585-1592. [Cited in This Article: ] |
| 127. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [Cited in This Article: ] |
| 128. | Karlsen TH. Genome-wide association studies reach hepatology. J Hepatol. 2009;50:1278-1280. [Cited in This Article: ] |
| 129. | Lake AC, Sun Y, Li JL, Kim JE, Johnson JW, Li D, Revett T, Shih HH, Liu W, Paulsen JE. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005;46:2477-2487. [Cited in This Article: ] |
| 130. | Williams R. Global challenges in liver disease. Hepatology. 2006;44:521-526. [Cited in This Article: ] |
| 131. | Fattovich G, Bortolotti F, Donato F. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. J Hepatol. 2008;48:335-352. [Cited in This Article: ] |
| 132. | Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591-595. [Cited in This Article: ] |
| 133. | Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889-899. [Cited in This Article: ] |
| 135. | Seeff LB. The history of the "natural history" of hepatitis C (1968-2009). Liver Int. 2009;29 Suppl 1:89-99. [Cited in This Article: ] |
| 136. | Nash KL, Bentley I, Hirschfield GM. Managing hepatitis C virus infection. BMJ. 2009;338:b2366. [Cited in This Article: ] |
| 137. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [Cited in This Article: ] |
| 138. | Lambou-Gianoukos S, Heller SJ. Lithogenesis and bile metabolism. Surg Clin North Am. 2008;88:1175-1194, vii. [Cited in This Article: ] |
| 139. | Schafmayer C, Tepel J, Franke A, Buch S, Lieb S, Seeger M, Lammert F, Kremer B, Fölsch UR, Fändrich F. Investigation of the Lith1 candidate genes ABCB11 and LXRA in human gallstone disease. Hepatology. 2006;44:650-657. [Cited in This Article: ] |
| 140. | Buch S, Schafmayer C, Völzke H, Becker C, Franke A, von Eller-Eberstein H, Kluck C, Bässmann I, Brosch M, Lammert F. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995-999. [Cited in This Article: ] |
| 141. | Grünhage F, Acalovschi M, Tirziu S, Walier M, Wienker TF, Ciocan A, Mosteanu O, Sauerbruch T, Lammert F. Increased gallstone risk in humans conferred by common variant of hepatic ATP-binding cassette transporter for cholesterol. Hepatology. 2007;46:793-801. [Cited in This Article: ] |
| 142. | Kuo KK, Shin SJ, Chen ZC, Yang YH, Yang JF, Hsiao PJ. Significant association of ABCG5 604Q and ABCG8 D19H polymorphisms with gallstone disease. Br J Surg. 2008;95:1005-1011. [Cited in This Article: ] |
| 143. | Donaldson PT, Baragiotta A, Heneghan MA, Floreani A, Venturi C, Underhill JA, Jones DE, James OF, Bassendine MF. HLA class II alleles, genotypes, haplotypes, and amino acids in primary biliary cirrhosis: a large-scale study. Hepatology. 2006;44:667-674. [Cited in This Article: ] |
| 144. | Invernizzi P, Selmi C, Poli F, Frison S, Floreani A, Alvaro D, Almasio P, Rosina F, Marzioni M, Fabris L. Human leukocyte antigen polymorphisms in Italian primary biliary cirrhosis: a multicenter study of 664 patients and 1992 healthy controls. Hepatology. 2008;48:1906-1912. [Cited in This Article: ] |
| 145. | Lettre G, Rioux JD. Autoimmune diseases: insights from genome-wide association studies. Hum Mol Genet. 2008;17:R116-R121. [Cited in This Article: ] |
| 146. | de Bakker PI, Ferreira MA, Jia X, Neale BM, Raychaudhuri S, Voight BF. Practical aspects of imputation-driven meta-analysis of genome-wide association studies. Hum Mol Genet. 2008;17:R122-R128. [Cited in This Article: ] |
| 147. | Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703-707. [Cited in This Article: ] |
| 148. | Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336-1341. [Cited in This Article: ] |
| 149. | Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638-645. [Cited in This Article: ] |
| 150. | Manolio TA, Rodriguez LL, Brooks L, Abecasis G, Ballinger D, Daly M, Donnelly P, Faraone SV, Frazer K, Gabriel S. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045-1051. [Cited in This Article: ] |
| 151. | Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181-1186. [Cited in This Article: ] |
| 152. | Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. [Cited in This Article: ] |
| 153. | McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, Zody MC, Hall JL, Brant SR, Cho JH. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107-1112. [Cited in This Article: ] |
| 154. | Huett A, McCarroll SA, Daly MJ, Xavier RJ. On the level: IRGM gene function is all about expression. Autophagy. 2009;5:96-99. [Cited in This Article: ] |
| 155. | Helbig I, Mefford HC, Sharp AJ, Guipponi M, Fichera M, Franke A, Muhle H, de Kovel C, Baker C, von Spiczak S. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat Genet. 2009;41:160-162. [Cited in This Article: ] |
| 156. | Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232-236. [Cited in This Article: ] |
| 157. | Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695-701. [Cited in This Article: ] |
| 158. | Terwilliger JD, Weiss KM. Linkage disequilibrium mapping of complex disease: fantasy or reality? Curr Opin Biotechnol. 1998;9:578-594. [Cited in This Article: ] |
| 159. | Lesage S, Zouali H, Cézard JP, Colombel JF, Belaiche J, Almer S, Tysk C, O'Morain C, Gassull M, Binder V. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J Hum Genet. 2002;70:845-857. [Cited in This Article: ] |
| 160. | Reddy S, Jia S, Geoffrey R, Lorier R, Suchi M, Broeckel U, Hessner MJ, Verbsky J. An autoinflammatory disease due to homozygous deletion of the IL1RN locus. N Engl J Med. 2009;360:2438-2444. [Cited in This Article: ] |
| 161. | Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872-876. [Cited in This Article: ] |
| 162. | Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463-5467. [Cited in This Article: ] |
| 163. | Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387-389. [Cited in This Article: ] |
| 164. | Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, Nieuwenhuis EE, Higgins DE, Schreiber S, Glimcher LH. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743-756. [Cited in This Article: ] |
| 165. | Adamovic S, Amundsen SS, Lie BA, Gudjónsdóttir AH, Ascher H, Ek J, van Heel DA, Nilsson S, Sollid LM, Torinsson Naluai A. Association study of IL2/IL21 and FcgRIIa: significant association with the IL2/IL21 region in Scandinavian coeliac disease families. Genes Immun. 2008;9:364-367. [Cited in This Article: ] |
| 166. | Romanos J, Barisani D, Trynka G, Zhernakova A, Bardella MT, Wijmenga C. Six new coeliac disease loci replicated in an Italian population confirm association with coeliac disease. J Med Genet. 2009;46:60-63. [Cited in This Article: ] |
| 167. | Amundsen SS, Rundberg J, Adamovic S, Gudjónsdóttir AH, Ascher H, Ek J, Nilsson S, Lie BA, Naluai AT, Sollid LM. Four novel coeliac disease regions replicated in an association study of a Swedish-Norwegian family cohort. Genes Immun. 2009;Epub ahead of print. [Cited in This Article: ] |
| 168. | Koskinen LL, Einarsdottir E, Dukes E, Heap GA, Dubois P, Korponay-Szabo IR, Kaukinen K, Kurppa K, Ziberna F, Vatta S. Association study of the IL18RAP locus in three European populations with coeliac disease. Hum Mol Genet. 2009;18:1148-1155. [Cited in This Article: ] |
| 169. | Yang SK, Lim J, Chang HS, Lee I, Li Y, Liu J, Song K. Association of TNFSF15 with Crohn's disease in Koreans. Am J Gastroenterol. 2008;103:1437-1442. [Cited in This Article: ] |
| 170. | Tremelling M, Berzuini C, Massey D, Bredin F, Price C, Dawson C, Bingham SA, Parkes M. Contribution of TNFSF15 gene variants to Crohn's disease susceptibility confirmed in UK population. Inflamm Bowel Dis. 2008;14:733-737. [Cited in This Article: ] |
| 171. | Kakuta Y, Kinouchi Y, Negoro K, Takahashi S, Shimosegawa T. Association study of TNFSF15 polymorphisms in Japanese patients with inflammatory bowel disease. Gut. 2006;55:1527-1528. [Cited in This Article: ] |
| 172. | Cummings JR, Cooney R, Pathan S, Anderson CA, Barrett JC, Beckly J, Geremia A, Hancock L, Guo C, Ahmad T. Confirmation of the role of ATG16L1 as a Crohn's disease susceptibility gene. Inflamm Bowel Dis. 2007;13:941-946. [Cited in This Article: ] |
| 173. | Latiano A, Palmieri O, Valvano MR, D'Incà R, Cucchiara S, Riegler G, Staiano AM, Ardizzone S, Accomando S, de Angelis GL. Replication of interleukin 23 receptor and autophagy-related 16-like 1 association in adult- and pediatric-onset inflammatory bowel disease in Italy. World J Gastroenterol. 2008;14:4643-4651. [Cited in This Article: ] |
| 174. | Dusatkova P, Hradsky O, Lenicek M, Bronsky J, Nevoral J, Kotalova R, Bajerova K, Vitek L, Lukas M, Cinek O. Association of IL23R p.381Gln and ATG16L1 p.197Ala With Crohn Disease in the Czech Population. J Pediatr Gastroenterol Nutr. 2009;Epub ahead of print. [Cited in This Article: ] |
| 175. | Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, Bagnall R, Mirza MM, Sanderson J, Forbes A. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665-1671. [Cited in This Article: ] |
| 176. | Baldassano RN, Bradfield JP, Monos DS, Kim CE, Glessner JT, Casalunovo T, Frackelton EC, Otieno FG, Kanterakis S, Shaner JL. Association of the T300A non-synonymous variant of the ATG16L1 gene with susceptibility to paediatric Crohn's disease. Gut. 2007;56:1171-1173. [Cited in This Article: ] |
| 177. | Márquez A, Núñez C, Martínez A, Mendoza JL, Taxonera C, Fernández-Arquero M, Díaz-Rubio M, de la Concha EG, Urcelay E. Role of ATG16L1 Thr300Ala polymorphism in inflammatory bowel disease: a Study in the Spanish population and a meta-analysis. Inflamm Bowel Dis. 2009;15:1697-1704. [Cited in This Article: ] |
| 178. | Weersma RK, Zhernakova A, Nolte IM, Lefebvre C, Rioux JD, Mulder F, van Dullemen HM, Kleibeuker JH, Wijmenga C, Dijkstra G. ATG16L1 and IL23R are associated with inflammatory bowel diseases but not with celiac disease in the Netherlands. Am J Gastroenterol. 2008;103:621-627. [Cited in This Article: ] |
| 179. | Glas J, Konrad A, Schmechel S, Dambacher J, Seiderer J, Schroff F, Wetzke M, Roeske D, Török HP, Tonenchi L. The ATG16L1 gene variants rs2241879 and rs2241880 (T300A) are strongly associated with susceptibility to Crohn's disease in the German population. Am J Gastroenterol. 2008;103:682-691. [Cited in This Article: ] |
| 180. | Okazaki T, Wang MH, Rawsthorne P, Sargent M, Datta LW, Shugart YY, Bernstein CN, Brant SR. Contributions of IBD5, IL23R, ATG16L1, and NOD2 to Crohn's disease risk in a population-based case-control study: evidence of gene-gene interactions. Inflamm Bowel Dis. 2008;14:1528-1541. [Cited in This Article: ] |
| 181. | Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Anderson NH, Davies G, Gillett PM, McGrogan P, Weaver LT. Autophagy gene ATG16L1 influences susceptibility and disease location but not childhood-onset in Crohn's disease in Northern Europe. Inflamm Bowel Dis. 2008;14:338-346. [Cited in This Article: ] |
| 182. | Lakatos PL, Szamosi T, Szilvasi A, Molnar E, Lakatos L, Kovacs A, Molnar T, Altorjay I, Papp M, Tulassay Z. ATG16L1 and IL23 receptor (IL23R) genes are associated with disease susceptibility in Hungarian CD patients. Dig Liver Dis. 2008;40:867-873. [Cited in This Article: ] |
| 183. | Fowler EV, Doecke J, Simms LA, Zhao ZZ, Webb PM, Hayward NK, Whiteman DC, Florin TH, Montgomery GW, Cavanaugh JA. ATG16L1 T300A shows strong associations with disease subgroups in a large Australian IBD population: further support for significant disease heterogeneity. Am J Gastroenterol. 2008;103:2519-2526. [Cited in This Article: ] |
| 184. | Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, Dijkstra G, Franke A, Nolte IM, Rutgeerts P. Confirmation of multiple Crohn's disease susceptibility loci in a large Dutch-Belgian cohort. Am J Gastroenterol. 2009;104:630-638. [Cited in This Article: ] |
| 185. | Newman WG, Zhang Q, Liu X, Amos CI, Siminovitch KA. Genetic variants in IL-23R and ATG16L1 independently predispose to increased susceptibility to Crohn's disease in a Canadian population. J Clin Gastroenterol. 2009;43:444-447. [Cited in This Article: ] |
| 186. | Peterson N, Guthery S, Denson L, Lee J, Saeed S, Prahalad S, Biank V, Ehlert R, Tomer G, Grand R. Genetic variants in the autophagy pathway contribute to paediatric Crohn's disease. Gut. 2008;57:1336-1337. [Cited in This Article: ] |
| 187. | Palomino-Morales RJ, Oliver J, Gómez-García M, López-Nevot MA, Rodrigo L, Nieto A, Alizadeh BZ, Martín J. Association of ATG16L1 and IRGM genes polymorphisms with inflammatory bowel disease: a meta-analysis approach. Genes Immun. 2009;10:356-364. [Cited in This Article: ] |
| 188. | Roberts RL, Gearry RB, Hollis-Moffatt JE, Miller AL, Reid J, Abkevich V, Timms KM, Gutin A, Lanchbury JS, Merriman TR. IL23R R381Q and ATG16L1 T300A are strongly associated with Crohn's disease in a study of New Zealand Caucasians with inflammatory bowel disease. Am J Gastroenterol. 2007;102:2754-2761. [Cited in This Article: ] |
| 189. | Lin Z, Poritz L, Franke A, Li TY, Ruether A, Byrnes KA, Wang Y, Gebhard AW, Macneill C, Thomas NJ. Genetic Association of Nonsynonymous Variants of the IL23R with Familial and Sporadic Inflammatory Bowel Disease in Women. Dig Dis Sci. 2009;Epub ahead of print. [Cited in This Article: ] |
| 190. | Glas J, Seiderer J, Wetzke M, Konrad A, Török HP, Schmechel S, Tonenchi L, Grassl C, Dambacher J, Pfennig S. rs1004819 is the main disease-associated IL23R variant in German Crohn's disease patients: combined analysis of IL23R, CARD15, and OCTN1/2 variants. PLoS One. 2007;2:e819. [Cited in This Article: ] |
| 191. | Oliver J, Rueda B, López-Nevot MA, Gómez-García M, Martín J. Replication of an association between IL23R gene polymorphism with inflammatory bowel disease. Clin Gastroenterol Hepatol. 2007;5:977-981, 981.e1-e2. [Cited in This Article: ] |
| 192. | Van Limbergen J, Russell RK, Nimmo ER, Drummond HE, Smith L, Davies G, Anderson NH, Gillett PM, McGrogan P, Hassan K. IL23R Arg381Gln is associated with childhood onset inflammatory bowel disease in Scotland. Gut. 2007;56:1173-1174. [Cited in This Article: ] |
| 193. | Büning C, Schmidt HH, Molnar T, De Jong DJ, Fiedler T, Bühner S, Sturm A, Baumgart DC, Nagy F, Lonovics J. Heterozygosity for IL23R p.Arg381Gln confers a protective effect not only against Crohn's disease but also ulcerative colitis. Aliment Pharmacol Ther. 2007;26:1025-1033. [Cited in This Article: ] |
| 194. | Lappalainen M, Halme L, Turunen U, Saavalainen P, Einarsdottir E, Färkkilä M, Kontula K, Paavola-Sakki P. Association of IL23R, TNFRSF1A, and HLA-DRB1*0103 allele variants with inflammatory bowel disease phenotypes in the Finnish population. Inflamm Bowel Dis. 2008;14:1118-1124. [Cited in This Article: ] |
| 195. | Tremelling M, Cummings F, Fisher SA, Mansfield J, Gwilliam R, Keniry A, Nimmo ER, Drummond H, Onnie CM, Prescott NJ. IL23R variation determines susceptibility but not disease phenotype in inflammatory bowel disease. Gastroenterology. 2007;132:1657-1664. [Cited in This Article: ] |
| 196. | Borgiani P, Perricone C, Ciccacci C, Romano S, Novelli G, Biancone L, Petruzziello C, Pallone F. Interleukin-23R Arg381Gln is associated with susceptibility to Crohn's disease but not with phenotype in an Italian population. Gastroenterology. 2007;133:1049-1051; author reply 1051-1052. [Cited in This Article: ] |
| 197. | Márquez A, Mendoza JL, Taxonera C, Díaz-Rubio M, De La Concha EG, Urcelay E, Martínez A. IL23R and IL12B polymorphisms in Spanish IBD patients: no evidence of interaction. Inflamm Bowel Dis. 2008;14:1192-1196. [Cited in This Article: ] |
| 198. | Latiano A, Palmieri O, Cucchiara S, Castro M, D'Incà R, Guariso G, Dallapiccola B, Valvano MR, Latiano T, Andriulli A. Polymorphism of the IRGM gene might predispose to fistulizing behavior in Crohn's disease. Am J Gastroenterol. 2009;104:110-116. [Cited in This Article: ] |
| 199. | Yamazaki K, Takahashi A, Takazoe M, Kubo M, Onouchi Y, Fujino A, Kamatani N, Nakamura Y, Hata A. Positive association of genetic variants in the upstream region of NKX2-3 with Crohn's disease in Japanese patients. Gut. 2009;58:228-232. [Cited in This Article: ] |
| 200. | Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561-565. [Cited in This Article: ] |
| 201. | Goyette P, Lefebvre C, Ng A, Brant SR, Cho JH, Duerr RH, Silverberg MS, Taylor KD, Latiano A, Aumais G. Gene-centric association mapping of chromosome 3p implicates MST1 in IBD pathogenesis. Mucosal Immunol. 2008;1:131-138. [Cited in This Article: ] |
| 202. | Amre DK, Mack DR, Morgan K, Fujiwara M, Israel D, Deslandres C, Seidman EG, Lambrette P, Costea I, Krupoves A. Investigation of Reported Associations Between the 20q13 and 21q22 Loci and Pediatric-Onset Crohn's Disease in Canadian Children. Am J Gastroenterol. 2009;Epub ahead of print. [Cited in This Article: ] |
| 203. | Pittman AM, Webb E, Carvajal-Carmona L, Howarth K, Di Bernardo MC, Broderick P, Spain S, Walther A, Price A, Sullivan K. Refinement of the basis and impact of common 11q23.1 variation to the risk of developing colorectal cancer. Hum Mol Genet. 2008;17:3720-3727. [Cited in This Article: ] |