Published online Nov 21, 2007. doi: 10.3748/wjg.v13.i43.5692
Revised: August 8, 2007
Accepted: September 14, 2007
Published online: November 21, 2007
AIM: To screen out the differentially methylated DNA sequences between gastric primary tumor and metastatic lymph nodes, test the methylation difference of gene PTPRG between primary gastric tumor and metastatic lymph nodes, and test the regulatory function of 5-aza-2-deoxycytidine which is an agent with suppression on methylation and the level of methylation in gastric cancer cell line.
METHODS: Methylated DNA sequences in genome were enriched with methylated CpG islands amplification (MCA) to undergo representational difference analysis (RDA), with MCA production of metastatic lymph nodes as tester and that of primary tumor as driver. The obtained differentially methylated fragments were cloned and sequenced to acquire the base sequence, which was analyzed with bioinformatics. With methylation-specific PCR (MSP) and RT-PCR, methylation difference of gene PTPRG was detected between primary tumor and metastatic lymph nodes in 36 cases of gastric cancer. Methylation of gene PTPRG and its regulated expression were observed in gastric cancer cell line before and after being treated with methylation-suppressive agent.
RESULTS: Nineteen differentially methylated sequences were obtained and located at 5’ end, exons, introns and 3’ end, in which KL59 was observed to be located at 9p21 as the first exon of gene p16 and KL22 to be located at promoter region of PRPRG. KL22, as the probes, was hybridized with driver, tester and 3-round RDA products respectively with all positive signals except with the driver. Significant difference was observed in both methylation rate of gene PTPRG and PTPRG mRNA expression rate between primary tumor and metastatic lymph nodes. Demethylation of gene PTPRG, with recovered expression of PTPRG mRNA, was observed after gastric cancer cell line being treated with methylation-suppressive agent.
CONCLUSION: Difference exists in DNA methylation between primary tumor and metastatic lymph nodes of gastric cancer, with MCA-RDA as one of the good analytical methods. Significant difference exists in methylation of gene PTPRG between primary tumor and metastatic lymph nodes of gastric cancer. Methylation level in gastric cancer cell line can be decreased by 5-aza-2’-deoxycytidine, which is the methylation-suppressive agent, with PTPRG expression being recovered.
- Citation: Wang JF, Dai DQ. Metastatic suppressor genes inactivated by aberrant methylation in gastric cancer. World J Gastroenterol 2007; 13(43): 5692-5698
- URL: https://www.wjgnet.com/1007-9327/full/v13/i43/5692.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i43.5692
Metastasis of gastric cancer, at the genetic level, is caused by mutations or loss of corresponding cancer suppressor genes, while at epigenetic level it is caused by low expression of metastasis-suppression genes due to multiple reasons, in which gene hypermethylation is an important mechanism[1-3]. A genome with methylation at CpG is usually accompanied by inactivation of genes in that region. If not, on the contrary, it is usually accompanied by active expression of genes in the region. The aberrant hypermethylation of CpG islands in transcription regulatory region of metastasis-inhibition genes relating to gastric cancer causes these genes to be in silence and this fact will induce the metastasis of gastric cancer[4-6]. Most of previous analytic methods on methylation can only analyze the methylation condition of a known gene, but cannot analyze that of a whole genome. Methylated CpG islands amplification (MCA) methods, combined with representative difference analysis (RDA), can analyze effectively the condition of methylation in a whole genome, especially good at detecting unknown methylated fragments.
In this research, gene methylation difference was detected between primary tumor and metastatic lymph nodes of gastric cancer using MCA-RDA method, to screen out genes relating to metastasis of gastric cancer, which further underwent analysis on methylation difference with methods, including methylation-specific PCR, in order to define further the mechanism of metastasis of gastric cancer.
Pathological specimens, including tumor tissues and metastatic lymph nodes, taken from 36 gastric cancer patients hospitalized in Surgical Oncology Ward of the First Affiliated Hospital of China Medical University, were included in this study. The tumor tissues and suspected metastatic lymph nodes, after resecting from the patients, were promptly placed into a liquid nitrogen tank. Each tumor tissue and lymph node was cut into two pieces, one piece was kept in liquid nitrogen and the other underwent HE pathological staining to examine whether a true metastasis had occurred. In addition, gastric cancer line SGC7901 was also included in this study.
Hydroxybenzene-chloroform extraction method was adopted to extract DNA of the genome, and total RNA was extracted with TRIZOL reagent according to the manufacturer’s instruction.
MCA was adopted to obtain CpG islands enriched with methylation. The CpG island region of DNA mixture extracted from tumor tissues and metastatic lymph nodes of 5 cases of gastric cancer were enriched with MCA, respectively. Firstly, 5 μg of DNA of genome was digested with 100U SmaI endonuclease (provided by NEB, not functional on methylated sites) for 6 h to cut the unmethylated -CCCGGG- sites to form the blunt ends, and also digested with 20U XmaI enzyme (provided by NEB) for 16 h to cut the methylated -CCCGGG- sites to form the sticky -CCCGGG- ends. Then T4DNA (Promega) was used to connect corresponding adaptor RXMA 24/12, with RXMA24 fragment as the primer to amplify the DNA fragments with adaptor, which were incubated for 5 min at 72°C, followed by pre-denaturalization for 3 min at 95°C, 30 amplification cycles for 1 min at 95°C and 3 min at 72°C, and a final extension for 10 min at 72°C, to enrich methylated fragments of DNA from both tumor tissues and metastatic lymph nodes. The products were electrophoresed on 15 g/L agarose gel containing ethidium bromide[7].
The methylated DNA fragments obtained from tumor and metastatic lymph nodes were underwent representative difference analysis. The adaptor of methylated CpG fragments taken from tumor and metastatic lymph nodes was cut-off with SmaI being used for tumor tissue to form blunt ends as the driver, and with XmaI being used for metastatic lymph nodes to form sticky ends to be connected with new ends as the tester. Tester and driver were underwent 3 cycles of hybridization RDA analysis in a ratio of 1:80, 1:400 and 1:800, respectively. After each analysis, the adaptor was cut off with XmaI, with new adaptor being added. The adaptors used in the 3 cycles of analysis were NMCA24/12, JXMA24/12 and NMCA24/12, with different extension temperature for different connectors. The sequence of each connector is listed in Table 1. The products were analyzed on 15 g/L agarose gel containing ethidium bromide[8].
| Adaptor | Sequence |
| RXMA24 | 5'-AGCACTCTCCAGCCTCTCACCGAC-3' |
| RXMA12 | 5'-CCGGGTCGGTGA-3' |
| JXMA24 | 5'-ACCGACGTCGACTATCCATGAACC-3' |
| JXMA12 | 5'-CCGGGGTTCATG -3' |
| NMCA24 | 5'-GTTAGCGGACACAGGGCGGGTCAC-3' |
| NMCA12 | 5'-CCGGGTGACCCG-3' |
Products of the 3rd cycle of RDA analysis as well as pCAT®3-Control carrier (Promega) were treated with XmaI to cut their ends into sticky ones, which were connected with T4 ligase and transformed into competent bacteria JM109 for incubation with matrix containing Ampicillin. Positive clones were selected and cultivated in matrix containing antibiotics at 37°C. Then plasmid DNA was extracted, and was underwent to cleavage with XmaI, and to electrophoresis; then the more than 100 bp and the clones of more than 100 bp cleavage products were selected and delivered to bio-company (Combined Gene Company) for sequencing. The obtained sequence were underwent repeated sequence analysis with Repeatmasker. BLAST system was used to carry out similarity analysis, with relationship between cloned sequence and corresponding genes being analyzed via GenBank.
The differentially methylated fragments of KL22 obtained from MCA-RDA analysis were labeled with digoxin, using random primer method to form the probe. With this latter hybridization analysis was carried out on the 1st, 2nd, 3rd round RDA. MCA products of tumor or metastatic lymph nodes, respectively, in a volume of 5 μL for each sample, were dotted onto nylon membrane with positive electricity.
Gastric cancer cell line was subcultured according to standard methods and then randomized into two groups, one of them was treated with 5 μmol/L 5-Aza-2’-deoxycytidine and cultured for 5 d.
Sodium hydrogen sulfite was used for DNA modification, and then sodium hydrogen sulfite was eliminated from DNA with Wizard DNA Clean-up kit (Promega). The samples were amplified through 30 cycles, each amplification cycle consisting of denaturation at 95°C for 40 s, primers annealing at 65°C (unmethylation) or at 60°C (methylation) for 40 s and extension at 72°C for 60 s.
Cycles were preceded by incubation at 95°C for 3 min to ensure full denaturation of the target gene, and finally by an extra incubation at 72°C for 10 min to ensure full extension of the products. PCR was carried out with methylated primer and unmethylated primer, respectively. The primers adopted are listed in Table 2. The PCR products were analyzed on 20 g/L agarose gel[9].
| Methylated primers | 5'-GTTCGTTCGTTTTTTCGTTC-3' |
| 5'-CATACTCCTAAAAATTATAACTCCGAC-3' | |
| Unmethylated primers | 5'-TTTTGTTTGTTTGTTTTTTTGTTTG-3' |
| 5'-AATCCATACTCCTAAAAATTATAACTCCA-3' |
RNA was reverse transcribed into cDNA as the template, which was amplified through 30 cycles, each amplification cycle consisting of denaturation at 94°C for 40 s, primers annealing at 57°C for 30 s and extension at 72°C for 60 s. Cycles were preceded by incubation at 94°C for 2 min to ensure full denaturation of the target gene, and finally by an extra incubation at 72°C for 10 min to ensure full extension of the products.
Meanwhile, β-actin was adopted as internal control. The sequences of primers are listed in Table 3. The products were electrophoresed on 15 g/L agarose gel.
| PTPRG primer | 5'-CTAATAAGGGATGTTCACATGAAGC-3' |
| 5'-CTGTATTTTAATGGAGTGGATAGCA-3' | |
| β-actin primer | 5'-AAATCGTGCGTGACATTAA-3' |
| 5'-CTCGTCATACTCCTGCTTG-3' |
Chi-square test was adopted to verify the difference of PTPRG methylation rate and PTPRG mRNA expression between gastric tumor and metastatic lymph nodes, as well as the difference on absent expression of PTPRG mRNA between negative and positive group of methylated nodular PTPRG. Rectilinear regression was used to test the correlation between PTPRG methylation rate and metastatic lymph nodes number. SPSS11.0 software was used to process the data.
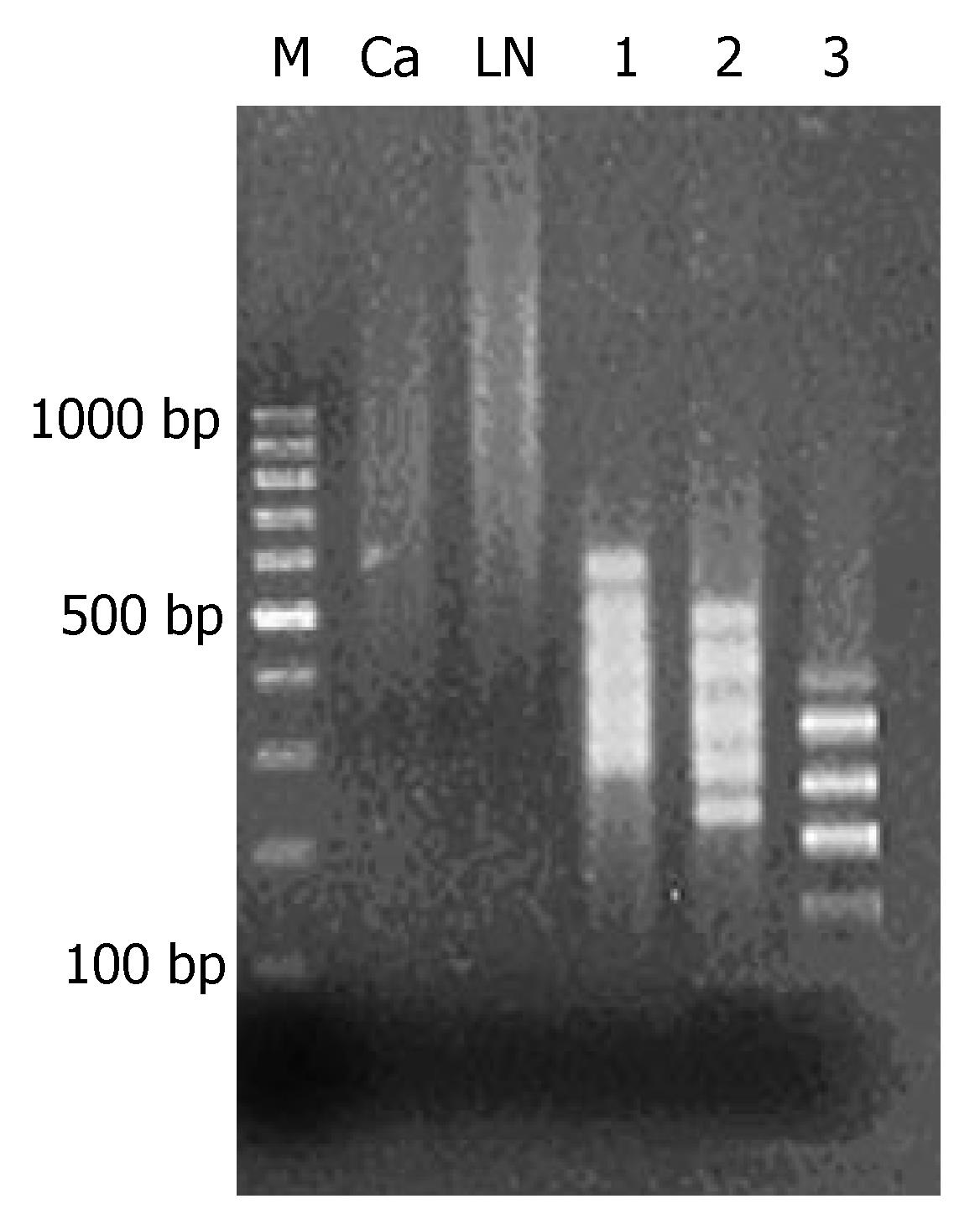
After methylated CpG islands amplification (MCA) of genome DNA of primary tumor and metastatic lymph nodes, bright smear was observed between 300 and 2000 bp, which were the concentrated methylated CpG islands (Figure 1).
MCA products of metastatic lymph nodes were adopted as the tester and MCA products of primary tumor as the driver to carry out 3 cycles of RDA analysis, which resulted in 100-500-bp fragments with methylation difference. From the 1st to the 3rd cycle of analysis, fragments with methylation difference decreased gradually and the straps gradually became clear. In the 3rd RDA analysis, 5 straps of different methylation were observed (Figure 1).
Ninety-six positive clones were selected to undergo sequencing analysis, 19 of them demonstrated the sequence longer than 100 bp. KL8 appeared for 21 times, while KL22 11 times, KL59 4 times, and both KL40 and KL71 for twice. No repeated sequence, such as ALU, was found after repeated sequencing analysis. Table 4 shows the 19 sequence characteristics. All sequences were of length between 100 bp and 400 bp, with GC content beyond 50%. Analysis showed these fragments to be distributed into various regions in the genome, including 5’ ends, exons, introns and 3’ ends, in which KL59 was situated at 9p21 as the first extron of gene p16, with 100% similarity rate, and KL22 was situated at 3p21, in the promoter region of gene PTPRG.
| Fragment | Length (bp) | GC % | CpG/GpC | Chromosome Positioning | Similarity rate % | S | E |
| KL2 | 198 | 55.0 | 0.5625 | 1q23.1 | 100 | 404 | e-110 |
| KL5 | 106 | 71.2 | 0.7142 | 15 | 100 | 222 | e-56 |
| KL6 | 159 | 59.4 | 1.0714 | 1p36.31-36.23 | 98 | 141 | 2e-31 |
| KL8 | 194 | 61.1 | 0.8461 | 2q33.3 | 99 | 396 | e-108 |
| KL14 | 347 | 72.3 | 0.9811 | 4p15 | 100 | 585 | e-165 |
| KL19 | 258 | 58.3 | 0.7692 | 5p15.1 | 97 | 480 | e-133 |
| KL22 | 332 | 64.1 | 0.7027 | 3p21 | 99 | 527 | e-147 |
| KL23 | 287 | 70.7 | 1.0606 | 1q42.1-43 | 98 | 458 | e-126 |
| KL33 | 136 | 53.2 | 0.9629 | 2p24.3-24.1 | 100 | 129 | 6e-28 |
| KL40 | 255 | 66.2 | 1.1428 | 4p16.1 | 99 | 404 | e-110 |
| KL55 | 268 | 65.6 | 0.7894 | 18 | 92 | 231 | e-152 |
| KL59 | 282 | 64.5 | 0.6471 | 9p21 | 100 | 571 | e-160 |
| KL68 | 251 | 62.5 | 0.6000 | 4 | 87 | 173 | 4e-39 |
| KL71 | 213 | 69.9 | 0.8260 | 10p12.1 | 100 | 434 | e-119 |
| KL74 | 341 | 69.5 | 0.7317 | 9p21 | 99 | 668 | 0 |
| KL79 | 225 | 61.3 | 0.4782 | 13q13 | 98 | 458 | e-127 |
| KL82 | 403 | 67.4 | 0.8043 | 8q21.2 | 99 | 383 | e-104 |
| KL87 | 275 | 51.9 | 1.0000 | 11 | 97 | 515 | e-144 |
| KL95 | 360 | 70.8 | 0.7608 | Xp22.3 | 100 | 726 | 0 |
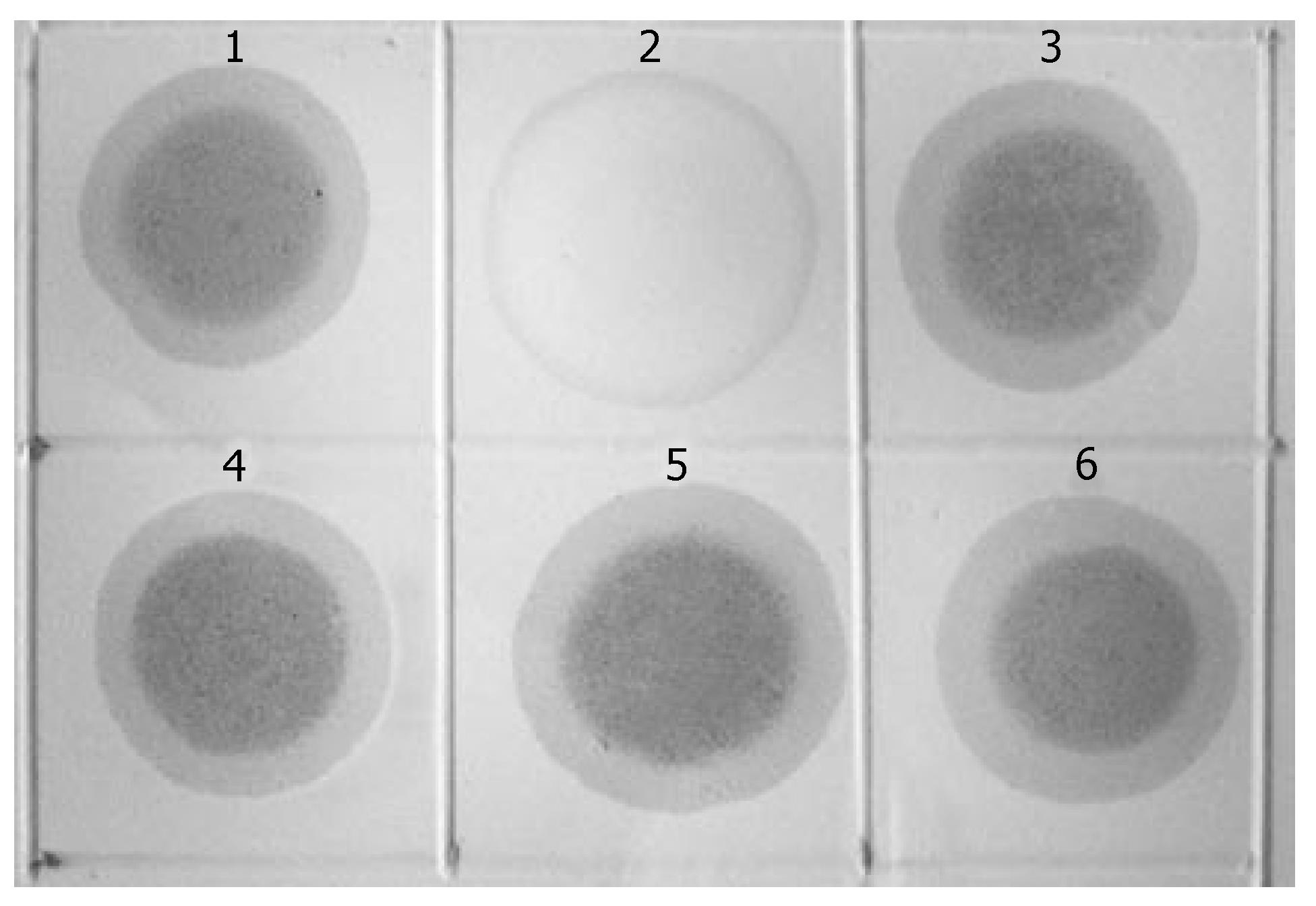
KL122 sequence was labeled with digoxin to form the probe, with which the three rounds MCA-RDA products underwent hybridization analysis with testers and drivers. Positive results were observed in all products of the 3-round RDA as well as in testers, while negative one in drivers (Figure 2).
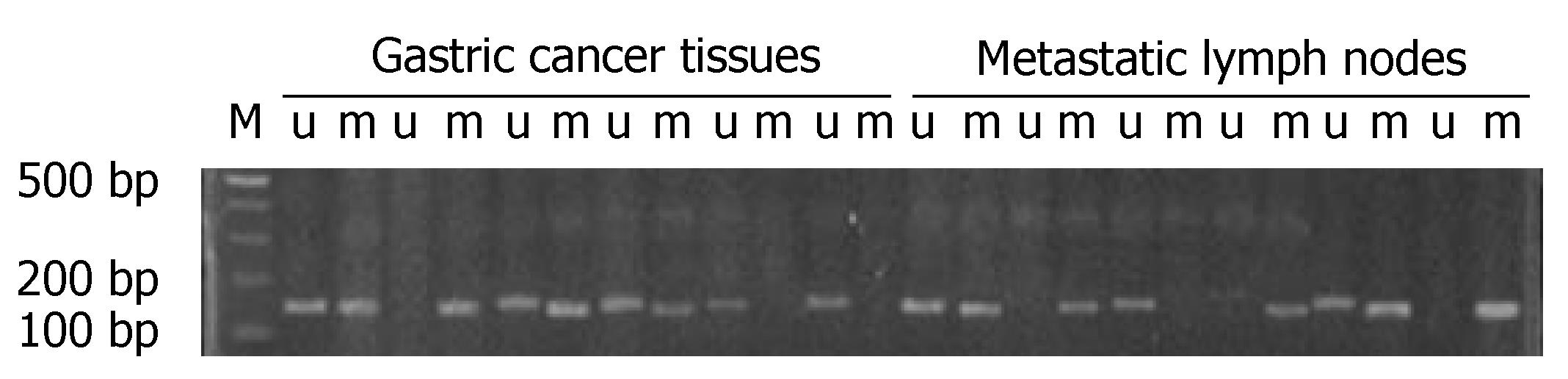
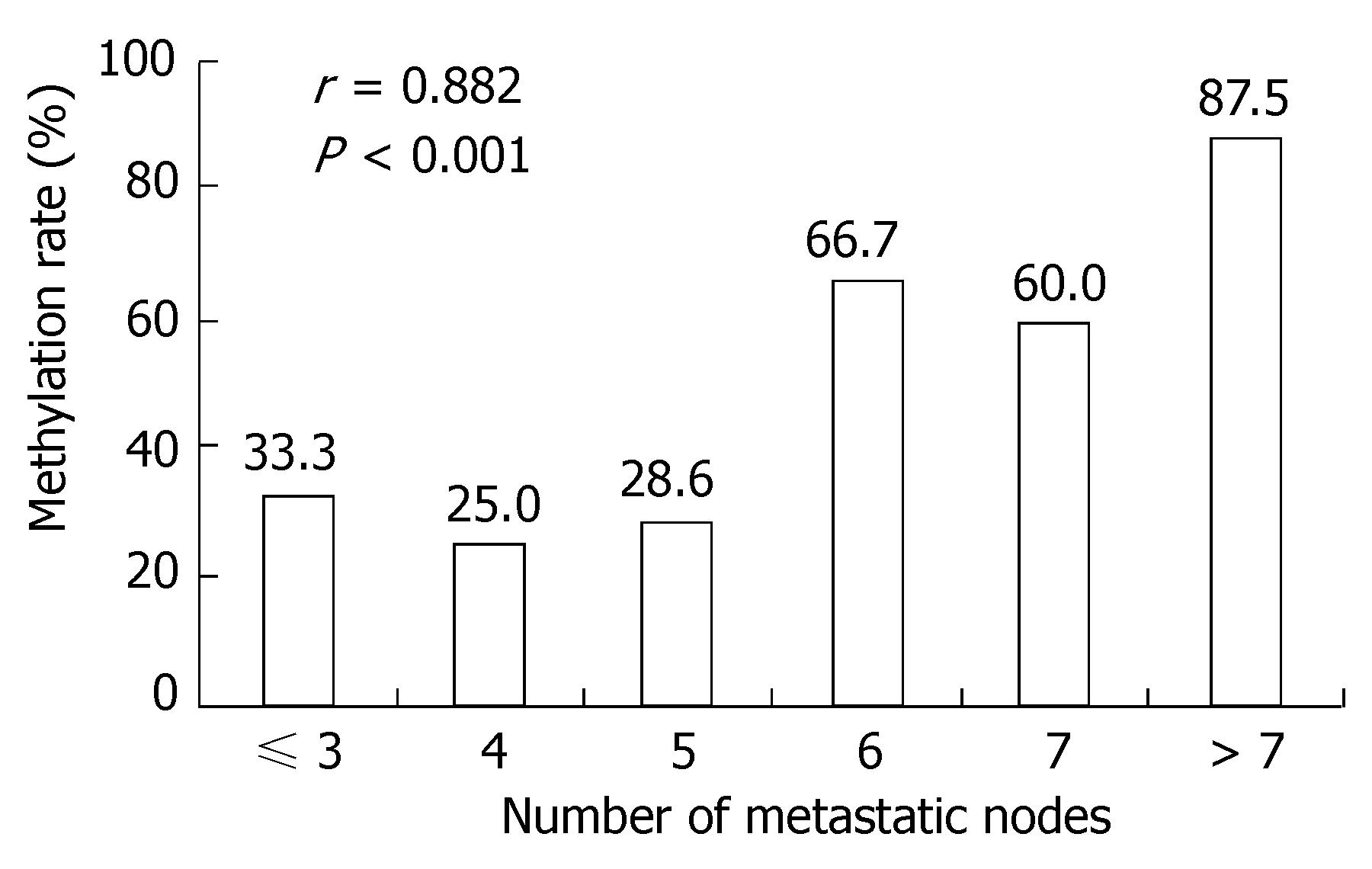
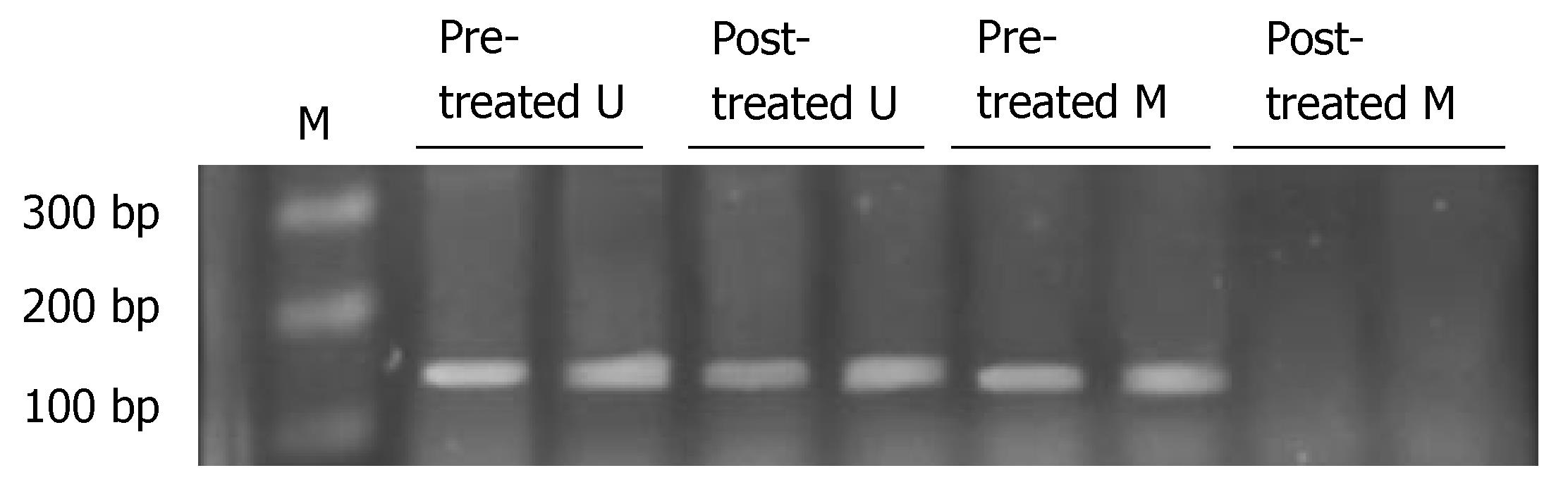
A positive band was observed at 158 bp of non-methylation PCR of primary tumor, with a positive rate of 77.78% (28/36), while that of metastatic lymph nodes was 63.89% (23/36) (P > 0.05). A positive strap was observed at 150 bp of methylation PCR of metastatic lymph nodes, with a positive rate of 52.78% (19/36), while that of primary tumor was 25.0% (9/36) (P < 0.05) (Figure 3 and Table 5). Linear correlation was observed between MSP positive rate of metastatic lymph nodes and the number of metastatic nodes (r = 0.882, P < 0.001, Figure 4).
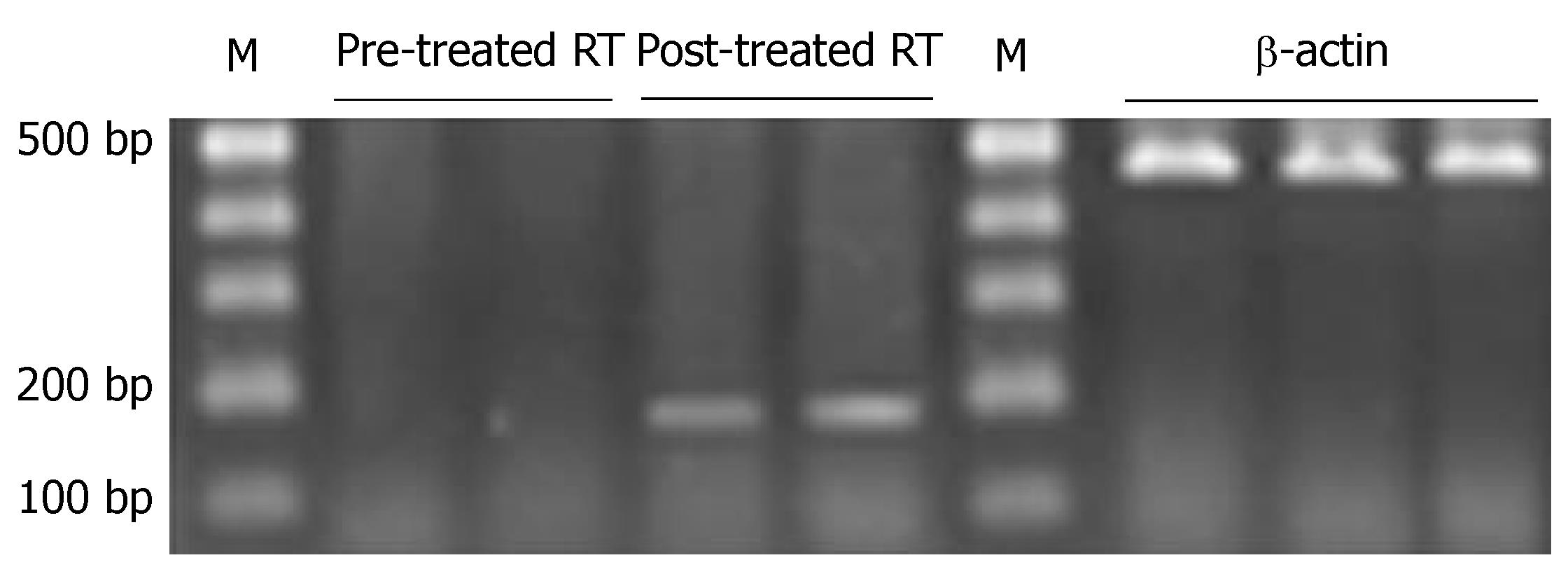
A positive strap was observed at 177 bp in RT-PCR of gene PTPRG of primary tumor, with a positive rate of 50.0% (18/36) and a 177-bp positive band was observed in metastatic lymph nodes, with a positive rate of 25.0% (9/36) (P < 0.05), (Figure 5, Table 5).
Among 19 cases of positive PTPRG methylation in metastatic lymph nodes, there was only one case of positive expression of PTPRG mRNA, with the positive rate of 5.26%, while 9 cases of positive expression existed among 17 cases of negative PTPRG methylation, with the positive rate of 52.9% (P < 0.01), (Table 6).
| PTPRG methylation | PTPRG mRNA expression (%) | |
| + | - | |
| + | 1 (5.26) | 18 (94.73) |
| - | 9 (52.94) | 8 (47.05) |
Before the treatment, a positive band was observed at 158 bp in unmethylated PCR, while a positive band was seen at 150 bp in methylated PCR of gastric cancer cell line. After the treatment, a positive band was also observed at 158 bp in unmethylated PCR, but no positive band was seen in methylated PCR (Figure 6, Table 5). PTPRG mRNA expression of cell line was negative before treatment, while a weak positive band was seen at 177 bp after treatment (Figure 7, Table 5).
Based on previous researches, a group containing methyl exists in every 100 nucleotide acids in human DNA, which is usually combined onto 5’-C position. Almost all methylated cytosine residues appear on the 5’-GC-3’ nucleotide acid in the symmetrical sequence. This kind of sequence is not randomly distributed, but concentrated to GC-enriched islands (CpG islands), which usually situates at the position in or near transcription regulatory region. Sensitivity of methylation on all CpG is not the same, and the methylation level at the site of CpG can be changed[10-12]. Prevalent hypomethylation and local hypermethylation exist in genome of cancer tissue, for example, hypermethylation on promoters of p16, E-cadherin, and genes encoding hormone receptors and genes of DNA repair, and genes inhibiting the genesis of blood vessels may induce the absent or low expression of these gene and improve the oncogenesis and metastasis. Therefore, research on methylation of genome provides a new route to study the oncogenesis and metastasis of cancer[13-17].
MCA technique is a new approach which has recently been adopted for research on gene methylation, which can be applied promptly and efficiently to the research on the whole genome methylation, with specific advantage especially for research on methylation condition of various unknown genes. Through the optimization of PCR condition, it is almost fit for every gene containing two adjacent SmaI restriction sites. The content of CpG is different in CpG islands, so different PCR primers and reaction conditions are needed, such as RXMA and RMCA. Thus RXMA seems to be more fit for this research. Through the optimization on the reaction condition, RXMA may yield MCA product steadily. It needs to point out that high quality sample of DNA is needed for MCA experiment, so generally wax-embedded sample is not considered in the case, in which only some sites can be detected, but not in total, among all sites in the CpG islands, which is only sensitive to partially digested products by SmaI, with distance shorter than 1000 bp between every two SmaI constriction sites. Generally speaking, GC content of MCA’s products is high; in this case if only an optimized PCR reaction system is adopted with high GC content, a satisfactory result can be made. RDA is a relatively mature technique adopted to screen out accurately the different fragments between two groups of DNA. In this research, we combined MCA and RDA to screen out the differentially methylated fragments between primary tumor and metastatic lymph nodes of gastric cancer, to explore the gene with alteration in methylation involved within process of metastatic lymph nodes. It provides a relatively accurate method with high efficiency, and it is fit for the primary screening out large quantities of metastasis-suppressive genes. It may be considered a high flux analytical method.
Phosphorylation of tyrosine residue is the important characteristic of many cellular signals transmission, influencing a number of vital biological processes including growth and differentiation of cells, adjustment of cell cycle, cell apoptosis and transference. Phosphorylation and dephosphorylation of tyrosine are adjusted by tyrosine kinase (TK) and tyrosine phosphatase (TP), respectively. Despite several tyrosine kinases have been recognized to correlate to oncogenesis directly through activating the mutant of cells in vivo, only a small amount of tyrosine phosphatase is known to correlate oncogenesis[18,19]. Gene PTPRG is a member belonging to the classic tyrosine phosphatase gene family, which includes receptor genes and non-receptor genes. PTPRG is a member of receptor gene, and is situated at 3p14 chromosome[20-22]. Previous researches observed methylation extinguishments of other genes that belong to the same family of PTPRG, such as PTPRN2, PTPRO, etc, in hepatic cancer[23]. PTPRG mutation is often found in colon cancer, lung cancer and kidney cancer, with conclusive identification on the role of PTPRG as one of tumor-inhibition genes[18,24]. However, there is a little information in literature regarding the extinguishments of PTPRG on epigenetic level. Based on recent studies, its deactivation on epigenetic level occurs in skin T-cell lymphoma. A study has demonstrated that a significant difference exists on PTPRG methylation in metastatic lymph nodes compared to primary tumor[9]. The silencing of genomic induced by methylation mainly consists of the methylation in promoter and in the first exon[9].
Methylation-specific PCR (MSP) revealed a significant difference in the positive methylation rate of PRPGR in primary tumor of gastric cancer (25.0%) compared to in metastatic lymph nodes (52.78%) (P < 0.05)[25,26]; this fact which further proved that the differentially methylated fragments we screened out were accurate. The significant linear correlation existing between positive rates of methylated PTPRG and numbers of metastatic nodes suggests that there is certain relationship between PTPRG methylation and metastatic lymph nodes of gastric cancer. In addition, a significant difference in PTPRG mRNA expression was observed between primary gastric tumor and metastatic lymph nodes, suggesting that product of PTPRG gene exerts suppressive effect against metastatic lymph nodes of gastric cancer. It is because of the down-regulated PTPRG, that the metastatic lymph nodes of gastric cancer was promoted.
PTPRG methylation exists in gastric cancer cell line. RT-PCR analysis demonstrated that PTPRG is not expressed in the cell line. However, MSP result of the cell line was negative and RT-PCR result was weakly positive after treatment with 5-aza-2'-deoxycytidine as the methylation suppressor; this fact, suggested that methylation of this gene was suppressed after the treatment with 5-aza-2'-deoxycytidine, so the MSP result was negative and the expression of PTPRG was partially recovered[27,28].
Among all 19 cases of positive methylation of PTPRG in metastatic lymph nodes, there was only one case of positive expression of PTPRG mRNA (positive rate of 5.26%), while among 17 cases of negative methylation of PTPRG, there were 9 cases of positive expression of PTPRG mRNA (positive rate of 52.9%) (P < 0.05); this result further implies that methylation in promoter region of PTPRG might be the mechanism of its being distinguished. However, the concrete mechanism of this gene involved in metastatic lymph nodes of gastric cancer is not clear yet, and it is waiting for the further research.
Metastasis of gastric cancer, at the genetic level, is caused by mutation or loss of corresponding cancer suppressor genes, while at epigenetic level it is caused by low expression of metastasis-suppression genes due to multiple reasons, among which gene hypermethylation is an important mechanism.
Methylated CpG islands amplification (MCA) methods, combined with representative difference analysis (RDA), can analyze wholly and effectively the condition of methylation in a whole genome, especially good at detecting unknown methylated fragments.
In this research, gene methylation difference was detected between primary tumor and metastatic lymph nodes of gastric cancer using MCA-RDA method, to screen out genes relating to metastasis of gastric cancer, which further underwent analysis on methylation difference by methylation specific PCR, in order to define further the mechanism of metastasis of gastric cancer.
This observation might be of potential value in gene therapy of gastric cancer.
The manuscript looked at the potential role of the PTPRG gene as metastatic suppressor gene, by comparing methylation status and LOI between the primary tumor and lymph nodes metastasis in 36 cases. Additionally, gastric cancer cell line was treated with 5-aza-2’-deoxycytidine (inhibitor of methylation) and gene expression was investigated. This study proved the hypothesis by demonstrating a significant difference in the level of methylation between the primary tumor and metastasis.
S- Editor Liu Y L- Editor Kumar M E- Editor Li HY
| 1. | Toyota M, Itoh F, Imai K. DNA methylation and gastrointestinal malignancies: functional consequences and clinical implications. J Gastroenterol. 2000;35:727-734. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Rashid A, Shen L, Morris JS, Issa JP, Hamilton SR. CpG island methylation in colorectal adenomas. Am J Pathol. 2001;159:1129-1135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 165] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 3. | Cowled P, Kanter I, Leonardos L, Jackson P. Uroplakin Ib gene transcription in urothelial tumor cells is regulated by CpG methylation. Neoplasia. 2005;7:1091-1103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Teodoridis JM, Hall J, Marsh S, Kannall HD, Smyth C, Curto J, Siddiqui N, Gabra H, McLeod HL, Strathdee G. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65:8961-8967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 190] [Cited by in F6Publishing: 189] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 5. | Costello JF, Frühwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomäki P, Lang JC. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 980] [Cited by in F6Publishing: 932] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 6. | Toyota M, Ahuja N, Suzuki H, Itoh F, Ohe-Toyota M, Imai K, Baylin SB, Issa JP. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 1999;59:5438-5442. [PubMed] [Cited in This Article: ] |
| 7. | Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307-2312. [PubMed] [Cited in This Article: ] |
| 8. | Lisitsyn N, Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946-951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 883] [Cited by in F6Publishing: 865] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 9. | Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793-797. [PubMed] [Cited in This Article: ] |
| 10. | Siegfried Z, Cedar H. DNA methylation: a molecular lock. Curr Biol. 1997;7:R305-R307. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 131] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 11. | Tycko B. Epigenetic gene silencing in cancer. J Clin Invest. 2000;105:401-407. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 125] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 12. | Catteau A, Harris WH, Xu CF, Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene. 1999;18:1957-1965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 229] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 13. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4367] [Cited by in F6Publishing: 4062] [Article Influence: 193.4] [Reference Citation Analysis (0)] |
| 14. | Dong C, Yoon W, Goldschmidt-Clermont PJ. DNA methylation and atherosclerosis. J Nutr. 2002;132:2406S-2409S. [PubMed] [Cited in This Article: ] |
| 15. | Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis. 2000;21:461-467. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 418] [Cited by in F6Publishing: 393] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 16. | Choi IS, Wu TT. Epigenetic alterations in gastric carcinogenesis. Cell Res. 2005;15:247-254. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Issa JP. Epigenetic variation and human disease. J Nutr. 2002;132:2388S-2392S. [PubMed] [Cited in This Article: ] |
| 18. | Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, Ptak J, Silliman N, Peters BA, van der Heijden MS. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304:1164-1166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 395] [Cited by in F6Publishing: 385] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 19. | van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, van der Velden PA, Vermeer MH, Willemze R, Yan PS. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. J Clin Oncol. 2005;23:3886-3896. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 181] [Cited by in F6Publishing: 192] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 20. | Van Poucke M, Yerle M, Chardon P, Jacobs K, Genêt C, Mattheeuws M, Van Zeveren A, Peelman LJ. A refined comparative map between porcine chromosome 13 and human chromosome 3. Cytogenet Genome Res. 2003;102:133-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 21. | Kholodnyuk ID, Szeles A, Yang Y, Klein G, Imreh S. Inactivation of the human fragile histidine triad gene at 3p14.2 in monochromosomal human/mouse microcell hybrid-derived severe combined immunodeficient mouse tumors. Cancer Res. 2000;60:7119-7125. [PubMed] [Cited in This Article: ] |
| 22. | Matsuyama A, Shiraishi T, Trapasso F, Kuroki T, Alder H, Mori M, Huebner K, Croce CM. Fragile site orthologs FHIT/FRA3B and Fhit/Fra14A2: evolutionarily conserved but highly recombinogenic. Proc Natl Acad Sci USA. 2003;100:14988-14993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 46] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Motiwala T, Ghoshal K, Das A, Majumder S, Weichenhan D, Wu YZ, Holman K, James SJ, Jacob ST, Plass C. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003;22:6319-6331. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 90] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 24. | LaForgia S, Morse B, Levy J, Barnea G, Cannizzaro LA, Li F, Nowell PC, Boghosian-Sell L, Glick J, Weston A. Receptor protein-tyrosine phosphatase gamma is a candidate tumor suppressor gene at human chromosome region 3p21. Proc Natl Acad Sci USA. 1991;88:5036-5040. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 147] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 25. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4183] [Cited by in F6Publishing: 4203] [Article Influence: 150.1] [Reference Citation Analysis (0)] |
| 26. | Deng G, Chen A, Hong J, Chae HS, Kim YS. Methylation of CpG in a small region of the hMLH1 promoter invariably correlates with the absence of gene expression. Cancer Res. 1999;59:2029-2033. [PubMed] [Cited in This Article: ] |
| 27. | Fang JY, Lu J, Chen YX, Yang L. Effects of DNA methylation on expression of tumor suppressor genes and proto-oncogene in human colon cancer cell lines. World J Gastroenterol. 2003;9:1976-1980. [PubMed] [Cited in This Article: ] |
| 28. | Liu LH, Xiao WH, Liu WW. Effect of 5-Aza-2'-deoxycytidine on the P16 tumor suppressor gene in hepatocellular carcinoma cell line HepG2. World J Gastroenterol. 2001;7:131-135. [PubMed] [Cited in This Article: ] |