Published online Sep 21, 2006. doi: 10.3748/wjg.v12.i35.5651
Revised: June 8, 2006
Accepted: June 14, 2006
Published online: September 21, 2006
AIM: To verify the expression and methylation status of the MAGE-A1 and MAGE-A3 genes in colorectal cancer tissues and cancer cell lines.
METHODS: We evaluated promoter demethylation status of the MAGE-A1 and MAGE-A3 genes by RT-PCR analysis and methylation-specific PCR (MS-PCR), as well as sequencing analysis, after sodium bisulfite modification in 32 colorectal cancer cell lines and 87 cancer tissues.
RESULTS: Of the 32 cell lines, MAGE-A1 and MAGE-A3 expressions were observed in 59% and 66%, respectively. Subsequent to sodium bisulfite modification and MS-PCR analysis, the promoter hypomethylation of MAGE-A1 and MAGE-A3 was confirmed in both at 81% each. Promoter hypomethylation of MAGE-A1 and MAGE-A3 in colorectal cancer tissues was observed in 43% and 77%, respectively. Hypomethylation of MAGE-A1 and MAGE-A3 genes in corresponding normal tissues were observed in 2% and 6%, respectively.
CONCLUSION: The promoter hypomethylation of MAGE genes up-regulates its expression in colorectal carcinomas as well as in gastric cancers and might play a significant role in the development and progression of human colorectal carcinomas.
-
Citation: Kim KH, Choi JS, Kim IJ, Ku JL, Park JG. Promoter hypomethylation and reactivation of
MAGE-A1 andMAGE-A3 genes in colorectal cancer cell lines and cancer tissues. World J Gastroenterol 2006; 12(35): 5651-5657 - URL: https://www.wjgnet.com/1007-9327/full/v12/i35/5651.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i35.5651
Human tumors often display changes in DNA methylation, which include both genome-wide hypomethylation and site-specific hypermethylation. Global hypomethylation and CpG island hypermethylation have been recognized as important contributors to the development of carcinogenesis in humans. Hypermethylation of promoter CpG islands is the signature of transcriptional silencing of their downstream genes, including RB, p16, VHL, BRCA1, E-cadherin, APC, hMLH1, FHIT, COX2, and CDX1 in various human cancers; and is as effective as inactivation by gene mutation or deletion[1-6]. Global DNA hypomethylation has been implicated in the activation of oncogenes such as c-myc, k-ras, and it may also contribute to tumor progression by the induction of genome instability[7,8].
The MAGE family of genes belongs to a group of germ line-specific genes that are activated in different types of tumors. This family of genes was reported to direct the expression of a tumor-specific antigen that was recognized in a melanoma cell by cytolytic T lymphocytes[9]. The MAGE-A1 gene has a CpG-rich promoter, which, unlike classical CpG-rich promoters, is methylated in all normal somatic tissues, except for the placenta and testis. In contrast, the promoter region of MAGE-A1 is completely unmethylated in testicular germ cells and in tumor cells that express the gene[10]. Demethylation, and therefore, activation of MAGE-A1 in tumors appears to be a consequence of the genome-wide demethylation process, since the expression of this gene in tumor cells correlates with a decreased level of overall DNA methylation[11]. A correlation between MAGE-A1 and MAGE-A3 expression and genome-wide hypomethylation has been observed in some types of carcinomas[12,13]. The human MAGE-A1 and MAGE-A3 genes, which are located on chromosome X, are expressed in 29% and 66%, respectively, of human gastric cancer cells due to the hypomethylation of the promoter region[12]. However, it is unknown if this relationship is present in colorectal carcinomas.
In this study, we investigated the promoter methylation status of MAGE-A1 and MAGE-A3 genes. A total of 32 colorectal cancer cell lines were tested for hypomethylation of the MAGE-A1 and MAGE-A3 genes promoter. In addition, we screened the methylation status of the MAGE-A1 and MAGE-A3 genes promoter in 87 paired colorectal cancers and normal mucosal tissue samples.
A total of 32 colorectal cancer cell lines (Table 1) and 2 gastric cancer cell lines (SNU-1 and SNU-5) were obtained from either the Korean Cell Line Bank (KCLB; Seoul, Korea) or the American Type Culture Collection (ATCC; Manassas, VA, USA). Sixteen SNU-colorectal cancer cell lines were established and were reported upon previously by this laboratory[14,15]. SNU-1 and SNU-5 gastric carcinoma cell lines were used as methylation positive (SNU-1) and negative (SNU-5) controls for MAGE gene expression[12]. All the cell lines were maintained in RPMI1640, which was supplemented with 10% FBS, 100 kU/L penicillin, and 0.1 g/L streptomycin. The cultures were maintained in humidified incubators at 37°C in a 5% CO2 and 95% ambient air atmosphere.
| Cell line | MAGEs expression | MS-PCR | |||||||
| MAGE-A1 | MAGE-A3 | MAGE-A1 | MAGE-A3 | ||||||
| -5-aza/+5-aza | -5-aza/+5-aza | M/U | M/U | ||||||
| 1 | SNU-61 | - | ++ | ± | ++ | + | + | - | + |
| 2 | SNU-81 | - | ++ | - | +++ | + | - | + | - |
| 3 | SNU-175 | ++ | ++ | +++ | +++ | + | + | + | + |
| 4 | SNU-283 | +++ | NT | +++ | NT | + | + | + | - |
| 5 | SNU-407 | +++ | + | +++ | ++ | + | - | + | + |
| 6 | SNU-503 | ++ | NT | - | NT | + | + | + | + |
| 7 | SNU-769A | +++ | ++ | +++ | +++ | - | + | - | + |
| 8 | SNU-769B | +++ | +++ | +++ | +++ | - | + | - | + |
| 9 | SNU-1033 | ++ | + | ++ | +++ | + | + | + | + |
| 10 | SNU-1040 | - | - | ± | ++ | + | - | + | - |
| 11 | SNU-1047 | +++ | ++ | ++ | ++ | + | - | - | + |
| 12 | SNU-1197 | ++ | +++ | ++ | +++ | + | + | + | + |
| 13 | SNU-C1 | ± | NT | ++ | NT | + | + | - | + |
| 14 | SNU-C2A | ++ | NT | +++ | NT | + | + | + | + |
| 15 | SNU-C4 | - | NT | - | NT | + | - | + | + |
| 16 | SNU-C5 | ± | ++ | - | +++ | + | + | + | - |
| 17 | Caco-2 | - | ++ | ++ | +++ | + | + | + | + |
| 18 | COLO201 | - | ++ | - | +++ | - | + | - | + |
| 19 | COLO205 | - | - | - | ++ | + | + | + | + |
| 20 | COLO320 | +++ | NT | +++ | NT | + | + | - | + |
| 21 | DLD-1 | - | +++ | - | +++ | + | + | + | + |
| 22 | HCT-8 | - | ++ | - | +++ | + | - | + | - |
| 23 | HCT-15 | - | ++ | - | +++ | + | + | + | + |
| 24 | HCT-116 | +++ | NT | +++ | NT | + | + | + | + |
| 25 | HT-29 | +++ | ++ | +++ | +++ | + | + | + | + |
| 26 | Lovo | +++ | +++ | +++ | +++ | + | + | + | + |
| 27 | LS174T | +++ | NT | +++ | NT | + | + | - | + |
| 28 | NCI-H716 | +++ | +++ | +++ | +++ | + | + | - | + |
| 29 | SW403 | +++ | ++ | +++ | +++ | + | + | + | + |
| 30 | SW480 | +++ | NT | +++ | NT | + | + | + | - |
| 31 | SW1116 | - | NT | ++ | NT | + | + | - | + |
| 32 | WiDR | ++ | +++ | +++ | +++ | + | + | - | + |
Genomic DNA and total RNA were isolated from washed-cell pellets. Total genomic DNA was extracted in accordance with the standard SDS-proteinase K procedure; and total cellular RNA was extracted based on the manufacturer’s instructions (Intron Biotechnology; Seoul, Korea). For cDNA synthesis, 2 µg of total RNA was reverse transcribed with a random hexamer, dNTPs, and 1 µL (200 U) of SuperscriptTM II reverse transcriptase (Life Technologies; Gaithersburg, MD, USA) in a final volume of 20 µL for 1 h and 15 min at 42°C after a 10-min denaturation at 70°C. Eighty microliters of distilled water were added subsequent to the reverse-transcription reaction.
For mRNA expression analysis, the cDNA was amplified in 25 µL of a PCR reaction mix with 1 µL of the reverse-transcription reaction, the primers and 0.5 U of Taq DNA polymerase. The PCR conditions consisted of 10 min at 94°C for the initial denaturation, followed by 35 cycles of 94°C for 30 s, 54°C for 60 s, and 72°C for 60 s, and a final elongation of 7 min at 72°C. The primer sequences are as follows. MAGE-A1 cDNA was amplified by PCR with MG1 RT primers; MG1 RT sense, 5’-TGTGGGCAGGAGCTGGGCAA-3’, MG1 RT antisense, 5’- GCCGAAGGAACCTGACCCAG -3’. For the MAGE-A3 cDNA, the MG3 RT primers were used; MG3 RT sense, 5’-AAGCCGGCCCAGGCTCGGT-3’, MG3 RT antisense, 5’-GCTGGGCAATGGAGACCCAC-3’. PCR amplification was performed in a programmable thermal cycler (PCR System 9700, Applied Biosystems; Foster City, CA, USA). Primers for β-actin were used to confirm RNA integrity. Both MAGE-A1 and MAGE-A3 and β-actin RT-PCR reactions used the same cDNA synthesis. The amplified DNA fragments were fractionated in 2% agarose gel and stained with ethidium bromide.
A total of 87 paired tumor and normal mucosal tissue samples were obtained from 87 patients, who had primary colorectal adenocarcinoma. The normal mucosal tissue specimens were collected from each patient 10 cm or more away from the tumor areas. Approximately 2 g of the surgically removed tissues were frozen immediately and then stored in liquid nitrogen. The remaining sections of the samples were fixed with formalin and used for further histological examination in order to confirm the diagnosis postoperatively. Genomic DNA was isolated from the frozen-tissue biopsies with the standard SDS-proteinase K procedure.
With respect to the MS-PCR, the sodium bisulfite modification of genomic DNA was performed as reported previously[16]. A total of 2 µg of genomic DNA obtained from colorectal cancer cell lines, was denatured with NaOH and hydroquinone. Then, 3 mmol/L sodium bisulfite was added and the mixture was incubated at 55°C for 16 h. Following the bisulfite modification, the DNA was purified with a Wizard DNA purification system (Promega; Madison, WI, USA), ethanol precipitated, dried, and resuspended in 100 µL distilled water. The PCR was performed using the PCR primers that were described previously[12]. The amplified DNA fragments were fractionated in 2% agarose gel that was stained with ethidium bromide and visualized under UV light.
For 5-aza-2’-deoxycytidine treatment, the cells were seeded in two 2 × 105 cells/75 cm2 culture flasks on d 0. The cells were treated with and without 1-5 µmol/L of 5-aza-2’-deoxycytidine (Sigma Chemical Co.) for 24 h on d 2 and 5, and the medium was changed 24 h after addition of 5-aza-2’-deoxycytidine. The cells were harvested on d 8 for the analysis of the MAGEs expression. Subsequently, the RNA was prepared, and RT-PCR was performed to detect MAGE-A1 and MAGE-A3 expression with the MAGE-A1 and MAGE-A3 RT-PCR primers as described above.
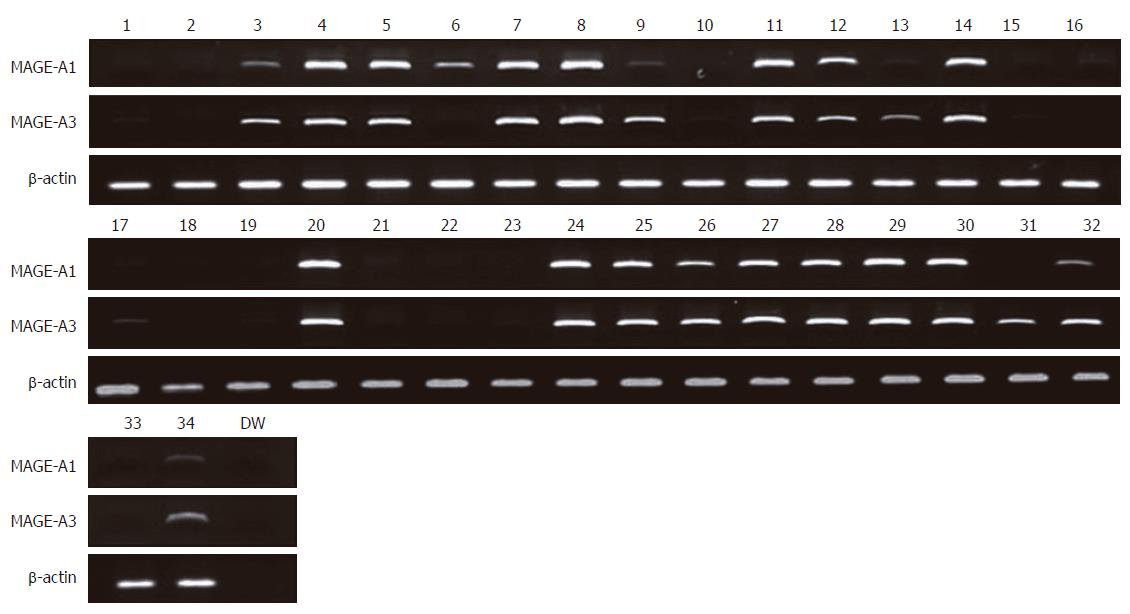
Expression of MAGE-A1 and MAGE-A3 mRNA in 32 colorectal cancer cell lines was analyzed by RT-PCR, and MAGE-A1 and MAGE-A3 expressions were observed in 19 (59%) and 21 (66%) of the cell lines, respectively (Figure 1 and Table 1). PCR for β-actin confirmed the integrity of the RNA.
Of the 87 colorectal carcinomas, 57 (66%) were obtained from the proximal colon (cecum to splenic flexure), and 30 (34%) from the distal colorectum (splenic flexure to rectum). Randomly selected patients aged 16-81 years, including 55 males and 32 females. Of the 32 colorectal cancer cell lines, 7 originated from the proximal colon and 8 from the distal colorectum. The origin of the remaining 17 colorectal cancer cell lines was unknown.
By using primers for unmethylated MAGE-A1 DNA amplification on bisulfite modified DNA, amplified DNA fragments were observed in 26 (81%) cell lines (SNU-61, SNU-175, SNU-283, SNU-503, SNU-769A, SNU-769B, SNU-1033, SNU-1197A, SNU-C1, SNU-C2A, SNU-C5, Caco2, COLO201, COLO205, COLO320, DLD-1, HCT-15, HCT-116, HT-29, Lovo, LS174T, NCI-H716, SW403, SW480, SW1116, and WiDR) (Figure 2 and Table 1). And by using primers for unmethylated MAGE-A3 DNA amplification on bisulfite modified DNA, amplified DNA fragments were found also in 26 (81%) cell lines (SNU-61, SNU-175, SNU-407, SNU-503, SNU-769A, SNU-769B, SNU-1033, SNU-1047, SNU-1197A, SNU-C1, SNU-C2A, SNU-C4, Caco2, COLO201, COLO205, COLO320, DLD-1, HCT-15, HCT-116, HT-29, Lovo, LS174T, NCI-H716, SW403, SW1116, and WiDR) (Figure 2 and Table 1). By using primers for amplification of unmethylated or methylated DNA, amplified DNA fragments were found in all 32 cell lines. MAGE-A1 unmethylated DNA products were observed in 37 out of 87 tumor tissue samples (43%; Figure 3). In the normal tissue samples, the methylated DNA was amplified in all 87 samples. However, the unmethylated DNA was amplified in 2 normal tissues (2%). MAGE-A3 unmethylated DNA products were observed in 67 out of 87 tumor tissue samples (77%; Figure 3). In the normal tissue samples, the methylated DNA was amplified in all 87 samples. However, the unmethylated DNA was amplified in 5 normal tissues (6%).
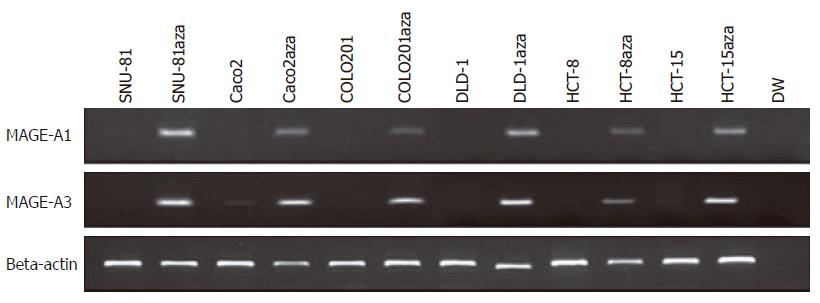
We investigated whether MAGE-A1 mRNA was re-expressed after 5-aza-2’-deoxycytidine treatment in 22 cell lines, including 10 cell lines (SNU-61, SNU-81, SNU-1040, SNU-C5, Caco-2, COLO201, COLO205, DLD1, HCT-8, and HCT-15) that did not express MAGE-A1 mRNA. After an RT-PCR analysis, we observed that all of the MAGE-A1 mRNAs were re-expressed, except for the SNU-1040 and COLO205 cell lines (Table 1). This suggested that the inactivation of MAGE-A1 expression was caused by another mechanism. Further, we investigated whether MAGE-A3 mRNA was re-expressed after 5-aza-2’-deoxycytidine treatment in 22 cell lines, including 9 cell lines (SNU-61, SNU-81, SNU-1040, SNU-C5, COLO201, COLO205, DLD1, HCT-8, and HCT-15) that did not express MAGE-A3 mRNA. After an RT-PCR analysis, we observed that all of the MAGE-A3 mRNAs were re-expressed (Table 1 and Figure 4).
Genome-wide hypomethylation and site-specific hypermethylation are common features of cancer cells. DNA hypomethylation in cancer cells is accompanied by the activation of germ line-specific genes, such as the MAGE-A1 gene, the repression of which, in normal somatic tissues, is dependent upon DNA methylation[17]. Recent studies have reported the presence of very high MAGE-A1 and MAGE-A3 expressions in colorectal carcinomas[18,19]. Although previous reports have shown the expression of MAGE genes, the mechanism of MAGE genes expression in colorectal carcinomas was unclear. This led us to question whether it could be associated with decreased genomic methylation. It has been reported that MAGE-A1 and MAGE-A3 expression was related to gene hypomethylation in gastric carcinoma, hepatocarcinoma, and melanoma[12,20,21]. However, such a relationship has still not been confirmed in colorectal carcinoma. Accordingly, we analyzed the methylation status of the promoter region on the MAGE-A1 and MAGE-A3 genes in 32 colorectal cancer cell lines and estimated its association with MAGE-A1 and MAGE-A3 mRNA expression. We first examined the expression pattern of the MAGE-A1 and MAGE-A3 genes in these cell lines by an RT-PCR and observed that MAGE-A1 and MAGE-A3 were over-expressed significantly in 19 (59%) and 21 (66%) cell lines, respectively. This expression ratio of MAGE-A1 and MAGE-A3, obtained by an RT-PCR in colorectal cancer cell lines, is similar to that observed in gastric cancer cell lines[12]. On the other hand, previous reports have revealed a much lower expression of MAGE in colorectal carcinomas ranging between 5%-39%[18,19,22]. In the literature, the expression of the MAGE genes was studied in colorectal carcinoma tissues; however, we have tested that in cancer cell lines. We assume that the major discrepancy of expression rates of MAGE genes between other reports and our findings might result from this. Since RNA or DNA was extracted from surgically removed frozen tissue biopsies, the tumor tissues may have been contaminated with normal stromal cells, therefore, masking the true levels of hypomethylation or expression of the MAGE genes in cancer tissues[6]. For further analysis of the methylation status of the MAGE genes in colorectal cancer tissues, laser capture microdissection techniques would allow more precise isolation of cancer cells and normal cells. It has already been reported that cancer cell lines have much higher levels of CpG island hypermethylation than corresponding malignant tissues, which may explain our lower incidence of hypomethylation in tissues versus cell lines. Moreover, cancer cells might be clonally selected with growth advantages over cancer cell lines. However, cancer cell lines often preserve hypermethylation or hypomethylation from the tumors they originate, thus they are indeed useful tools to study methylation status.
We analyzed promoter unmethylation of the MAGE-A1 and MAGE-A3 genes with a methylation-specific PCR after sodium-bisulfite modification and by direct sequencing analysis. Of the 32 cell lines analyzed, the promoter hypomethylation of MAGE-A1 and MAGE-A3 was observed in both at 26 cell lines each. Further, 23 cell lines (SNU-61, SNU-175, SNU-503, SNU-769A, SNU-769B, SNU-1033, SNU-1197A, SNU-C1, SNU-C2A, Caco2, COLO201, COLO205, COLO320, DLD-1, HCT-15, HCT-116, HT-29, Lovo, LS174T, NCI-H716, SW403, SW1116, and WiDR) were simultaneously unmethylated in both the MAGE-A1 and MAGE-A3 genes. With exception, there were two cell lines (SNU-61, COLO201) with negative gene expression for either MAGE-A1 or MAGE-A3, but unmethylated MAGE-A1 or MAGE-A3 promoter was detected. On the contrary, there were four cell lines (SNU-283, SNU-407, SNU-1047 and SW480) in which MAGE genes were strongly expressed, although no unmethylated MAGE promoter could be detected, suggesting the activation or inactivation of MAGE expression by another mechanism.
In our study, the rates of hypomethylation of the MAGE-A1 and MAGE-A3 genes in colorectal cancer cell lines were 81% in both and those in colorectal cancer tissues were 43% and 77%, respectively. The DNA was extracted from the surgically removed frozen-tissues; however, the tumor tissues might have been contaminated with some normal stromal cells. Therefore, the levels of hypomethylation of MAGE-A1 and MAGE- A3 genes in cancer tissues might be affected by the DNA from normal cells. To obtain a better understanding of the promoter hypomethylation status of the MAGE-A1 and MAGE-A3 genes in colorectal cancer tissues, expression analysis of MAGE-A1 and MAGE-A3 (such as, in situ hybridization or immunostaining) and the more precise methylation analysis method (such as, laser capture microdissection techniques) to isolate cancer cells from normal cells need to be performed. The hypomethylation of the MAGE-A1 and MAGE-A3 genes in corresponding normal tissues was detected in only 2 and 5 samples (2% and 6%), respectively (Table 2).
| % of expression | % of hypomethylation | |||||
| MAGE-A1 | MAGE-A3 | MAGE-A1 | MAGE-A3 | |||
| Cancer cell lines | 59 | 66 | 81 | 81 | ||
| Cancer tissues | NT1 | NT | 43 | 77 | ||
| Normal tissues2 | NT | NT | 2 | 6 | ||
| MAGE-A1 hypomethylation | MAGE-A3 hypomethylation | |||||
| + | - | P | + | - | P | |
| Location | ||||||
| Proximal | 14 (31.1%) | 31(68.9%) | 34 (75.6%) | 11 (24.4%) | ||
| Distal | 17 (56.7%) | 13(43.3%) | 0.028 | 26 (86.7%) | 4 (13.3%) | 0.239 |
| Sex | ||||||
| Male | 17 (30.9%) | 38(69.1%) | 38 (69.1%) | 17 (30.9%) | ||
| Female | 20 (62.5%) | 12(18.4%) | 0.004 | 29 (90.6%) | 3 (9.4%) | 0.021 |
To evaluate the association between the clinical parameters and MAGE expression, the Pearson χ2 test was used to evaluate differences in tumor location (proximal or distal) or gender, and significance was determined using 95% confidence intervals. In our study, unmethylated MAGE-A1 DNA expression was significantly different in respect of tumor location and gender. Unmethylated MAGE-A1 DNA expression was significantly higher in distal location (P = 0.028) and in females (P = 0.004). However, unmethylated MAGE-A3 DNA expression was not significantly associated with tumor location (P = 0.239), while it was only related to female gender (P = 0.021).
Our results supported the role of the promoter methylation in maintaining a silent phenotype of the MAGE-A1 and MAGE-A3 genes, as the MAGE gene was re-expressed after treatment with 5-aza-2'-deoxycytidine. This agent reactivates gene expression when methylation of CpG islands is the cause of reduced gene expression. We demonstrated that the MAGE-A1 and MAGE-A3 mRNAs were re-expressed after 5-aza-2'-deoxycytidine treatment in all 8 and 9 cell lines that did not express MAGE-A1 and MAGE-A3 mRNAs, respectively. However, the SNU-1040 and COLO 205 cell lines did not show re-expression, suggesting the inactivation of MAGE-A1 expression by another mechanism.
In conclusion, we observed hypomethylation in the promoter region of both the MAGE-A1 and MAGE-A3 genes in 23 of 32 colorectal cancer cell lines. This methylation was confirmed by MS-PCR, treatment with 5-aza-2’-deoxycytidine, and bisulfite direct sequencing analysis. Hypomethylation of the promoter region appears to be a frequent phenomenon in human colorectal cancers and upregulates transcription of the MAGE-A1 and MAGE-A3 genes in colorectal cancer cells. In addition, out of 87 colorectal cancer tissues, we observed hypomethylation in the promoter regions of the MAGE-A1 and MAGE-A3 genes in 37 (43%) and 67 (77%) tissues, respectively. This suggests that promoter hypomethylation of MAGE-A1 and MAGE-A3 genes up-regulates its expression in colorectal carcinomas as well as in gastric cancers, and might play a significant role in the development and progression of human colorectal carcinomas.
S- Editor Pan BR L- Editor Zhu LH E- Editor Liu WF
| 1. | Zöchbauer-Müller S, Fong KM, Maitra A, Lam S, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD. 5' CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res. 2001;61:3581-3585. [PubMed] [Cited in This Article: ] |
| 2. | Song SH, Jong HS, Choi HH, Inoue H, Tanabe T, Kim NK, Bang YJ. Transcriptional silencing of Cyclooxygenase-2 by hyper-methylation of the 5' CpG island in human gastric carcinoma cells. Cancer Res. 2001;61:4628-4635. [PubMed] [Cited in This Article: ] |
| 3. | Suh ER, Ha CS, Rankin EB, Toyota M, Traber PG. DNA methylation down-regulates CDX1 gene expression in colorectal cancer cell lines. J Biol Chem. 2002;277:35795-35800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 4. | Melki JR, Vincent PC, Brown RD, Clark SJ. Hypermethylation of E-cadherin in leukemia. Blood. 2000;95:3208-3213. [PubMed] [Cited in This Article: ] |
| 5. | Yang Q, Nakamura M, Nakamura Y, Yoshimura G, Suzuma T, Umemura T, Shimizu Y, Mori I, Sakurai T, Kakudo K. Two-hit inactivation of FHIT by loss of heterozygosity and hypermethylation in breast cancer. Clin Cancer Res. 2002;8:2890-2893. [PubMed] [Cited in This Article: ] |
| 6. | Ku JL, Kang SB, Shin YK, Kang HC, Hong SH, Kim IJ, Shin JH, Han IO, Park JG. Promoter hypermethylation downregulates RUNX3 gene expression in colorectal cancer cell lines. Oncogene. 2004;23:6736-6742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 89] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 7. | Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47-54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 355] [Cited by in F6Publishing: 314] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 8. | Fang JY, Zhu SS, Xiao SD, Jiang SJ, Shi Y, Chen XY, Zhou XM, Qian LF. Studies on the hypomethylation of c-myc, c-Ha-ras oncogenes and histopathological changes in human gastric carcinoma. J Gastroenterol Hepatol. 1996;11:1079-1082. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 33] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Van Der Bruggen P, Zhang Y, Chaux P, Stroobant V, Panichelli C, Schultz ES, Chapiro J, Van Den Eynde BJ, Brasseur F, Boon T. Tumor-specific shared antigenic peptides recognized by human T cells. Immunol Rev. 2002;188:51-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 291] [Cited by in F6Publishing: 278] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 10. | De Smet C, Lurquin C, Lethé B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327-7335. [PubMed] [Cited in This Article: ] |
| 11. | De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci USA. 1996;93:7149-7153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 397] [Cited by in F6Publishing: 384] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 12. | Honda T, Tamura G, Waki T, Kawata S, Terashima M, Nishizuka S, Motoyama T. Demethylation of MAGE promoters during gastric cancer progression. Br J Cancer. 2004;90:838-843. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 13. | Liu J, Wang G, Okutomi T, Chen Z. Expression of MAGE-A1 and MAGE-A3 genes in human salivary gland carcinomas. Chin Med J (Engl). 2003;116:897-900. [PubMed] [Cited in This Article: ] |
| 14. | Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, Gazdar A. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710-6718. [PubMed] [Cited in This Article: ] |
| 15. | Oh JH, Ku JL, Yoon KA, Kwon HJ, Kim WH, Park HS, Yeo KS, Song SY, Chung JK, Park JG. Establishment and characterization of 12 human colorectal-carcinoma cell lines. Int J Cancer. 1999;81:902-910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 16. | Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821-9826. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4183] [Cited by in F6Publishing: 4203] [Article Influence: 150.1] [Reference Citation Analysis (0)] |
| 17. | De Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5' region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781-4790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 154] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 18. | Park MS, Park JW, Jeon CH, Lee KD, Chang HK. Expression of melanoma antigen-encoding genes (MAGE) by common primers for MAGE-A1 to -A6 in colorectal carcinomas among Koreans. J Korean Med Sci. 2002;17:497-501. [PubMed] [Cited in This Article: ] |
| 19. | Koketsu S, Watanabe T, Kazama S, Ishihara S, Komuro Y, Nagawa H. What types of colorectal cancer overexpress the MAGE protein. Hepatogastroenterology. 2004;51:1648-1652. [PubMed] [Cited in This Article: ] |
| 20. | Xiao J, Chen HS, Fei R, Cong X, Wang LP, Wang Y, Jiang D, Wei L, Wang Y. Expression of MAGE-A1 mRNA is associated with gene hypomethylation in hepatocarcinoma cell lines. J Gastroenterol. 2005;40:716-721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 21. | Zendman AJ, de Wit NJ, van Kraats AA, Weidle UH, Ruiter DJ, van Muijen GN. Expression profile of genes coding for melanoma differentiation antigens and cancer/testis antigens in metastatic lesions of human cutaneous melanoma. Melanoma Res. 2001;11:451-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 22. | Li M, Yuan YH, Han Y, Liu YX, Yan L, Wang Y, Gu J. Expression profile of cancer-testis genes in 121 human colorectal cancer tissue and adjacent normal tissue. Clin Cancer Res. 2005;11:1809-1814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 71] [Article Influence: 3.7] [Reference Citation Analysis (0)] |