Published online Dec 7, 2005. doi: 10.3748/wjg.v11.i45.7142
Revised: February 18, 2005
Accepted: February 21, 2005
Published online: December 7, 2005
AIM: To investigate the use of PCR and DGGE to investigate the association between bacterial translocation and systemic inflammatory response syndrome in predicted severe AP.
METHODS: Patients with biochemical and clinical evidence of acute pancreatitis and an APACHE II score ≥8 were enrolled. PCR and DGGE were employed to detect bacterial translocation in blood samples collected on d 1, 3, and 8 after the admission. Standard microbial blood cultures were taken when there was clinical evidence of sepsis or when felt to be clinically indicated by the supervising team.
RESULTS: Six patients were included. Of all the patients investigated, only one developed septic complications; the others had uneventful illness. Bacteria were detected using PCR in 4 of the 17 collected blood samples. The patient with sepsis was PCR-positive in two samples (taken on d 1 and 3), despite three negative blood cultures. Using DGGE and specific primers, the bacteria in all blood specimens which tested positive for the presence of bacterial DNA were identified as E coli.
CONCLUSION: Our study confirmed that unlike traditional microbiological techniques, PCR can detect the presence of bacteria in the blood of patients with severe AP. Therefore, this latter method in conjunction with DGGE is potentially an extremely useful tool in predicting septic morbidity and evaluating patients with the disease. Further research using increased numbers of patients, in particular those patients with necrosis and sepsis, is required to assess the reliability of PCR and DGGE in the rapid diagnosis of infection in AP.
- Citation: Pearce CB, Zinkevich V, Beech I, Funjika V, Ruiz AG, Aladawi A, Duncan HD. Using the polymerase chain reaction coupled with denaturing gradient gel electrophoresis to investigate the association between bacterial translocation and systemic inflammatory response syndrome in predicted acute severe pancreatitis. World J Gastroenterol 2005; 11(45): 7142-7147
- URL: https://www.wjgnet.com/1007-9327/full/v11/i45/7142.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i45.7142
Infection and septic complications are the major factors contributing to the poor outcome in acute severe pancreatitis. They cause up to 80% of deaths and occur in 5-10% of patients[1-3]. It is thought that in the majority of cases infection is caused by bacterial translocation from the gut lumen[4-7], a hypothesis which animal experiments have generally supported[8-10]. Unfortunately attempts to confirm the link between bacterial translocation and morbidity and mortality in acute pancreatitis in human beings have been largely unsuccessful[11,12].
Conventional microbiological blood culture methods are currently used widely[13-15], but may fail to yield positive results, if the causative organism is fastidious in nature, cell dependent or has a fungal etiology. It is thought that 60-70% of the bacteria in the human intestinal tract cannot be cultured[16,17]. Molecular-based diagnostic approaches are therefore being increasingly employed, especially when a quick diagnosis is required[18-21].
The use of the polymerase chain reaction (PCR) to identify microbial DNA in clinical specimens has been described by many investigators[22-25]. PCR using 16S rRNA-specific primers has identified bacterial DNA in blood after the surgery[26]. 16S rRNA is a highly conserved region of bacterial DNA, found in all Gram-positive and Gram-negative bacteria[27]; if these primers are used, the majority of pathogenic bacteria can theoretically be detected and identified by subsequent cloning and sequencing. From a practical point of view blood cultures can take days to yield a result, whereas PCR can produce results within hours.
PCR without subsequent cloning can identify the bacterial genus leaving the species undefined, which may cause difficulties, for instance, if the therapeutic guidelines for the species are different. Polymicrobial infections can also be problematic due to the inability of PCR to identify several microorganisms in a single specimen[28]. These problems can be solved by denaturing gradient gel electrophoresis (DGGE)[27], which uses the presence of unique heterodiplexes in DNA affecting migration to separate different DNA fragments[29].
Our investigation was carried out to assess the potential of PCR and DGGE as rapid tools for the detection and identification of systemic bacterial translocation in blood samples of patients with acute severe pancreatitis. The study also aimed to elucidate the relationship between the presence or absence of clinical infection; manifested as either sepsis or infected necrosis, and the detection of translocated bacterial DNA.
Patients who were admitted with a diagnosis of acute pancreatitis predicted to have severe disease during the study period were enrolled. Patients were only considered if their duration of symptoms was 48 h or less on admission. Pancreatitis was defined as appropriate clinical signs and symptoms with hyperamylasemia of more than three times the upper limit of normal. Patients predicted to have severe disease were identified by an APACHE II score of eight or more on admission[30].
Blood samples were taken on d 1, 3, and 8 of admission for examination by PCR and DGGE techniques. Standard microbial blood cultures were taken only when there was a clinical evidence of sepsis or when felt to be clinically indicated by the supervising team to limit the number of venesections patients enrolled in the study would need to volunteer. The supervising teams were not aware of the PCR results.
Blood was transferred from Na2 EDTA tubes (Sigma, UK) to sterile 1.5 mL Eppendorf tubes (Sigma, UK), and purified using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. Whole blood samples were processed in aliquots of 400 µL for DNA extraction.
The DNA from human blood samples and from different bacterial strains was purified, and bacterial DNA was amplified using PCR with primers specific for (i) the bacterial 16S rRNA region, (ii) E coli, and (iii) Bacteroides spp., as outlined below. Enzymatic amplification for DGGE was performed on human blood samples tested PCR positive for the presence of bacterial DNA.
Prior to DNA extraction, bacterial cells were cultured in LB medium (10 mL) overnight at +37 °C inside an incubator shaker (New Brunswick Scientific). Chromosomal DNA was extracted using the QIAGEN Genomic DNA Purification Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instructions.
The oligonucleotide primers were synthesized by Sigma Aldrich Co. The primer pairs, sequences, gene targets and size of the product amplified after PCR are listed in Table 1. The primer pair designated BD-1 and BD-2 is specific for a highly conserved region of different bacterial DNA coding for 16S ribosomal RNA. The second primer pair BG-1 and BG-4 was derived from the β-galactosidase gene of E coli, which is found in most E coli strains. The third primer pair used BFR-1 and BFR-2 targets specifically for the ubiquitous glutamine synthase gene found in many Bacteroides spp[26]. The fourth primer pair F3 and Rev 2 was used for amplification of variable V3 region of 16S rDNA gene[29].
| Primer designate | Sequences of (+) and (-) primers (nucleotide) | Gene target | Size of amplicon (bp) |
| BG-1 (+ strand) | 5’CTT TGC CTG GTT TCC GGC ACC AGA A- 3’ (201-225) | b-Galactosidase gene of E coli | 762 |
| BG-4 (- strand) | 5’AAC CAC CGC ACG ATA GAG ATT CGG G- 3’ (983-939) | ||
| BD-1 (+ strand) | 5’AGT TTG ATC CTG GCT GAG- 3’ (8-27) | DNA coding for 16S rRNA | 798 |
| BD-2 (- strand) | 5’GGA CTA CCA GGG TAT CTA AT- 3’ (805-787) | ||
| BFR-1 (+ strand) | 5’ACT CTT TGT ATC CCG ACG ATT-3’ (484-504) | Glutamine synthase gene of Bacteroides spp. | 581 |
| BFR-2 (- strand) | 5’GAG GTT GAT GCC TGT ATC GGT-3’ (1 065-1 045) | ||
| F3 (+ strand) | 5’CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3’ | Variable V3 region of 16S rRNA | 233 |
| Rev-2 (- strand) | 5’ATTACCGCGGCTGCTGG-3’ |
PCR was performed with minor modification following the protocol of Kane et al[26]. Extracted DNA (20-30 µL) was placed in 0.5 mL sterile Eppendorf tube to which the PCR reaction mixture was added to make a final volume of 50 µL per tube. The mixture consisted of the following: 5 µL of 10× PCR reaction buffer, 1 µL each of (10 µmol/L) designated primers, 1 µL of a mixture of 10 mmol/L deoxynucleotide triphosphate (dNTP), 0.2 µL of 1 unit/µL Super Taq DNA polymerase (HT Biotechnology Ltd) and nuclease free dH2O (Sigma Chemical Ltd) up to the final volume.
PCR amplification was carried out using a Techne (Progene) machine through the cycles as follows: an initial cycle of 95 °C/3 min, 60 °C/45 s, and 72 °C/10 min was followed by 35 cycles of 95 °C/45 s, 60 °C/45 s, and 72 °C/1 min; an extension period of 72 °C/10 min completed the cycling sequence.
Enzymatic amplification for DGGE was performed on samples positive for bacterial DNA. PCR reaction quantifications were outlined as above. The amplification was performed as follows: an initial cycle of 95 °C for 15 min was followed by 35 cycles each of 1 min at 94, 60, and 70 °C, respectively; an extension period of 72 °C for 5 min completed the cycling sequence.
All PCR products were separated by electrophoresis on 1% agarose gel in 1× TAE buffer (0.04 mol/L Tris-acetate, 0.002 mol/L EDTA). The gels were stained with ethidium bromide (0.5 µg/mL) for 30 min, washed twice with distilled H2O and photographed under UV light using a UVP gel documentation system. A 1-kbp DNA ladder (Sigma) was used as a molecular weight marker. All reagents were purchased from Sigma, UK.
DGGE was performed using Ingeny Phos system (Leiden, The Netherlands). A 10 µL volume of each PCR product was applied directly onto 9% (wt/vol) polyacrylamide gels in 0.5× TAE (20 mmol/L Tris-acetate, 10 mmol/L sodium acetate, 0.5 mmol/L Na2-EDTA) with gradients which were formed with 9% (wt/vol) acrylamide, 37:1) and which contained 0 and 100% denaturant [7 mol/L urea (GIBCO BRL)] and 40% (wt/vol) formamide. The gel was subjected to 200 V for 10 min at 60 °C and 85 V for 16 h. After electrophoresis, the gel was stained in ethidium bromide solution (0.5 µg/mL) for 30 min, washed twice by distilled H2O and analyzed under UV light using UVP gel documentation system.
Conventional blood cultures were processed using the BacT/ALERT® system manufactured by BioMerieux©. BacT/ALERT® aerobic (SA) and anaerobic (SN) culture media bottles were taken peripherally from patients under aseptic conditions and a BacT/ALERT® 3D analyzer was used to process the samples. Positive samples were Gram stained, and subcultured onto appropriate solid culture media, which was examined at 24 and 48 h.
The study was performed as part of a larger study into enteral nutrition in acute pancreatitis. Written informed consent was a condition of entry into the study. The local ethics committee (Portsmouth, Hampshire, UK) approved the study.
Of the six patients tested one developed septic complications; pneumonia, respiratory failure and severe systemic inflammatory response syndrome, although this patient subsequently made a full recovery (patient no. 5, Table 2). This was the only patient in the group that retrospectively developed severe acute pancreatitis according to the Santorini Consensus definitions[31]. This patient did not, however, require ventilatory support, and did not develop necrosis, infected or otherwise. The other patients had uneventful illness.
| Patient no. | Day | 1 | 2 | 3 | 4 | 8 | Complications |
| 1 | CRP | 135 | 158 | 91 | 70 | 40 | None |
| SIRS | Y | Y | N | N | N | ||
| PCR | Negative | Negative | Positive | ||||
| 2 | CRP | 55 | 83 | 108 | 105 | 94 | None |
| SIRS | N | N | N | N | N | ||
| PCR | Negative | Negative | Negative | ||||
| 3 | CRP | 345 | 301 | 289 | 234 | 142 | None |
| SIRS | Y | Y | Y | N | N | ||
| PCR | Negative | Negative | Negative | ||||
| 4 | CRP | 301 | 284 | 248 | 169 | 122 | None |
| SIRS | Y | Y | Y | Y | N | ||
| PCR | Positive | Negative | Negative | ||||
| 5 | CRP | 59 | 201 | 193 | 155 | 114 | Pneumonia, sepsis |
| SIRS | Y | Y | Y | Y | N | (-ve blood culture) | |
| PCR | Positive | Positive | Negative | ||||
| 6 | CRP | 326 | 209 | 150 | 128 | None | |
| SIRS | Y | Y | Y | Y | |||
| PCR | Negative | Negative |
PCR detected the presence of bacteria in 4 of the 17 samples (23.6%). The blood samples from the patient with sepsis tested PCR-positive for bacterial DNA in specimens collected on d 1 and 3. This patient had three sets of standard blood cultures taken on d 1, 3, and 5, all of which were negative; he/she was the only patient in our study in whom standard blood cultures were taken.
When the incidence of the systemic inflammatory response syndrome (SIRS), C-reactive protein (CRP) levels and septic complications were compared with the PCR results there was no observed correlation between the presence of bacterial DNA in the blood, as confirmed by PCR, and SIRS or CRP data (Table 2).
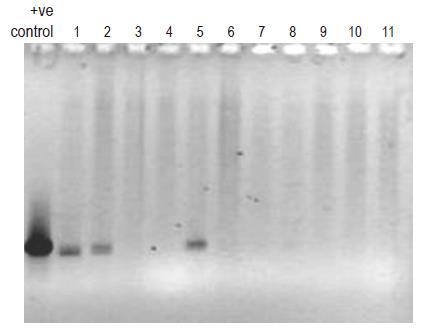
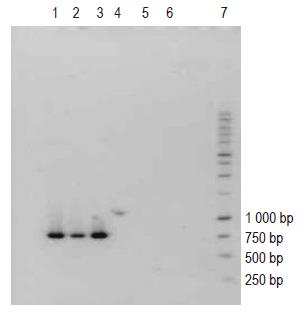
The PCR profiles of DNA recovered from human blood samples and of E coli DNA (positive control), treated with general bacterial primers (Table 1) are depicted in Figure 1 (lanes 1 and 2-13, respectively). The appearance of characteristic bands indicative of the presence of bacterial DNA (lane marked positive control) can be seen in human blood samples (lanes 1-11). Figure 2 (lanes 1-6) illustrates the profile of E coli DNA obtained using two different pairs of primers (Table 1); one pair specific only for E coli (lanes 1-3) and another pair for Bacteroides spp. (lanes 4-6, negative control). The treatment of E coli DNA with Bacteroides specific primers resulted in a blank PCR profile (i.e. no bands were observed in lanes 4-6), while the use of E coli specific primers resulted in the appearance of characteristic PCR product (lanes 1-3).
Human blood samples treated with Bacteroides specific primers did not reveal any bands when subjected to PCR (data not shown).
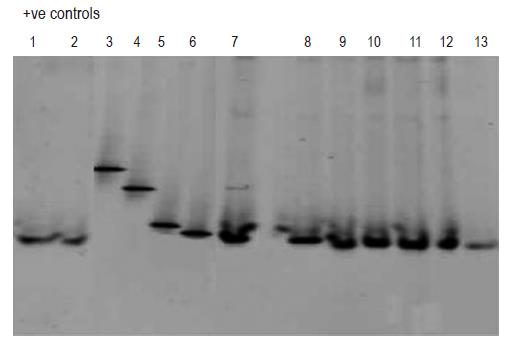
Subsequent analysis employing DGGE (Figure 3, lanes 1-13) revealed that the blood samples which tested PCR positive for bacterial DNA (lanes 1-2 and 7-13) contained DNA fragments of the identical size as the one characteristic for E coli DNA (lane 6). DGGE profiles of DNA from bacterial strains, such as Staphylococcus aureus (lane 3), Pseudomonas aeruginosa (lane 4) and Bacillus cereus (lane 5) served as negative controls.
These results from our preliminary investigation reveal that although conventional blood cultures techniques fail to demonstrate bacterial infection, using PCR and DGGE it is possible to detect bacterial DNA in the blood in patients with acute severe pancreatitis and also to identify the species present. The number of patients tested in the study is inadequate to make inferences regarding the connection between the presence of bacteria, systemic inflammatory response syndrome and morbidity.
Based on the obtained results it seems plausible to propose that the bacteria detected on d 1 and 3 in patient no. 5 were related to the infectious complications and sepsis. It is unlikely that the pneumonia was caused by E coli (sputum cultures were negative). The detection of bacterial DNA was probably indicative of a higher degree of bacterial translocation, which led to other infections as well as the possible E coli sepsis. The positive PCR profile of blood samples taken on admission from patient no. 4 is also not surprising. This patient represented a case of relatively severe disease with a high CRP and probably also had bacterial translocation.
The DGGE profile of the samples from all the patients was exactly the same, i.e. positive exclusively for E coli. Interpretation of these results in view of the low numbers of subjects has to be treated with great caution, and it is not until more samples are taken from a larger cohort of patients that statistically valid interpretations can be offered.
To the best of our knowledge, prior to our study, reports on PCR detection of bacterial DNA in patients with acute pancreatitis are limited to two publications[11,12]; neither of these investigations used DGGE as a method of identifying bacteria.
Zhang et al[12] performed a PCR on blood specimens on patients only with acute necrotizing pancreatitis. They reported a PCR detection of bacterial DNA in 8 out of 22 tested samples (33.35%), but the samples were taken exclusively during periods of likely sepsis. The lower PCR positive rate of 23.6% in our study probably reflects the fact that blood specimens from patients were collected in a sequential fashion i.e. from the time of admission onwards, and from patients with predicted severe pancreatitis, rather than diagnosed necrotizing disease. Similarly to our study all of the positive blood samples tested were found to contain E coli.
Ammori et al[11] failed to find any bacterial DNA by performing PCR on blood samples from 26 patients with acute pancreatitis. Blood cultures that tested positive for bacterial infection revealed the presence of E coli and Enterococcus in blood of one patient, and coagulase negative Staphylococcus in the blood of another. The reasons for the negative PCR results reported by Ammori et al. are not clear, but may include contamination by substances which inhibit the polymerase reaction.
In acute pancreatitis it is perhaps what separates necrotizing from non-necrotizing disease and what differentiates infected necrosis from sterile necrosis which is of most interest. The controversy surrounding when to operate on necrotizing pancreatitis illustrates this[32-34]; a debate which centers on whether it is necessary to prove infection before operative therapy. The diagnosis of infection is not straightforward, and the currently recommended method of diagnosis, i.e. needle aspiration[34], carries its own risk due to the inherent interventional nature of the procedure. It is possible that more sensitive non-invasive methods of detecting infection such as PCR could improve diagnostic accuracy.
The presence of E coli DNA in blood specimens does not necessarily mean that the intestine is the source of infection. It is conceivable that these organisms arose from, for example, the biliary tract due to cholangitis. Moreover, the PCR detection method simply demonstrates the presence of bacterial DNA, and does not specify genera and species. DNA extraction methods provide total DNA that could originate from dead or living microorganisms in the blood or microbes that have been engulfed, or subsequently released by the phagocytes. This approach cannot differentiate between controlled and invasive infections.
The importance of PCR techniques with respect to clinical application will depend on establishing a relationship between the presence or absence of clinical infection and the presence of bacterial DNA in the blood. The PCR detection of bacterial DNA may provide information about the nature of the inflammatory response in acute pancreatitis when traditional methods fail to detect bacteria, even in the absence of culture-positive complications. This type of research may also reveal more about the nature of susceptibility towards infective complications in pancreatitis. In addition, the sensitivity and accuracy of PCR could help target antibiotic therapy in the future.
It is anticipated that in a cohort study including patients with sepsis or septic necrosis, and with the development of more sophisticated quantitative PCR techniques, the proposed approach could offer considerable diagnostic potential.
Science Editor Li WZ Language Editor Elsevier HK
| 1. | Howard TJ, Wiebke EA, Mogavero G, Kopecky K, Baer JC, Sherman S, Hawes RH, Lehman GA, Goulet RJ, Madura JA. Classification and treatment of local septic complications in acute pancreatitis. Am J Surg. 1995;170:44-50. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 2. | Runkel NS, Rodriguez LF, Moody FG. Mechanisms of sepsis in acute pancreatitis in opossums. Am J Surg. 1995;169:227-232. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Bassi C, Larvin M, Villatoro E. Antibiotic therapy for prophylaxis against infection of pancreatic necrosis in acute pancreatitis. Cochrane Database Syst Rev. 2003;4:CD002941. [PubMed] [Cited in This Article: ] |
| 4. | Danner RL, Elin RJ, Hosseini JM, Wesley RA, Reilly JM, Parillo JE. Endotoxemia in human septic shock. Chest. 1991;99:169-175. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 465] [Cited by in F6Publishing: 492] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 5. | Brisson-Noël A, Gicquel B, Lecossier D, Lévy-Frébault V, Nassif X, Hance AJ. Rapid diagnosis of tuberculosis by amplification of mycobacterial DNA in clinical samples. Lancet. 1989;2:1069-1071. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 274] [Cited by in F6Publishing: 287] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 6. | Rush BF, Sori AJ, Murphy TF, Smith S, Flanagan JJ, Machiedo GW. Endotoxemia and bacteremia during hemorrhagic shock. The link between trauma and sepsis? Ann Surg. 1988;207:549-554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 315] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 7. | Sibbald WJ, Vincent JL. Round table conference on clinical trials for the treatment of sepsis. Crit Care Med. 1995;23:394-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 44] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 8. | Cicalese L, Sahai A, Sileri P, Rastellini C, Subbotin V, Ford H, Lee K. Acute pancreatitis and bacterial translocation. Dig Dis Sci. 2001;46:1127-1132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Gianotti L, Munda R, Alexander JW, Tchervenkov JI, Babcock GF. Bacterial translocation: a potential source for infection in acute pancreatitis. Pancreas. 1993;8:551-558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 56] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 10. | Tarpila E, Nyström PO, Franzén L, Ihse I. Bacterial translocation during acute pancreatitis in rats. Eur J Surg. 1993;159:109-113. [PubMed] [Cited in This Article: ] |
| 11. | Ammori BJ, Fitzgerald P, Hawkey P, McMahon MJ. The early increase in intestinal permeability and systemic endotoxin exposure in patients with severe acute pancreatitis is not associated with systemic bacterial translocation: molecular investigation of microbial DNA in the blood. Pancreas. 2003;26:18-22 DOI : 10.1097/00006676-200301000-00004. [Cited in This Article: ] |
| 12. | Zhang WZ, Han TQ, Tang YQ, Zhang SD. Rapid detection of sepsis complicating acute necrotizing pancreatitis using polymerase chain reaction. World J Gastroenterol. 2001;7:289-292. [PubMed] [Cited in This Article: ] |
| 13. | Schwabe LD, Thomson RB, Flint KK, Koontz FP. Evaluation of BACTEC 9240 blood culture system by using high-volume aerobic resin media. J Clin Microbiol. 1995;33:2451-2453. [PubMed] [Cited in This Article: ] |
| 14. | Alfa MJ, Degagne P, Olson N, Harding GK. Improved detection of bacterial growth in continuous ambulatory peritoneal dialysis effluent by use of BacT/Alert FAN bottles. J Clin Microbiol. 1997;35:862-866. [PubMed] [Cited in This Article: ] |
| 15. | Viganò EF, Vasconi E, Agrappi C, Clerici P. Use of simulated blood cultures for time to detection comparison between BacT/ALERT and BACTEC 9240 blood culture systems. Diagn Microbiol Infect Dis. 2002;44:235-240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 16. | Badillo AT, Sarani B, Evans SR. Optimizing the use of blood cultures in the febrile postoperative patient. J Am Coll Surg. 2002;194:477-487; quiz 554-556. [PubMed] [Cited in This Article: ] |
| 17. | Mylotte JM, Tayara A. Blood cultures: clinical aspects and controversies. Eur J Clin Microbiol Infect Dis. 2000;19:157-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 95] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 18. | Murray PR, Traynor P, Hopson D. Critical assessment of blood culture techniques: analysis of recovery of obligate and facultative anaerobes, strict aerobic bacteria, and fungi in aerobic and anaerobic blood culture bottles. J Clin Microbiol. 1992;30:1462-1468. [PubMed] [Cited in This Article: ] |
| 19. | Weinstein MP, Mirrett S, Wilson ML, Reimer LG, Reller LB. Controlled evaluation of 5 versus 10 milliliters of blood cultured in aerobic BacT/Alert blood culture bottles. J Clin Microbiol. 1994;32:2103-2106. [PubMed] [Cited in This Article: ] |
| 20. | Wilson ML, Weinstein MP, Mirrett S, Reimer LG, Feldman RJ, Chuard CR, Reller LB. Controlled evaluation of BacT/alert standard anaerobic and FAN anaerobic blood culture bottles for the detection of bacteremia and fungemia. J Clin Microbiol. 1995;33:2265-2270. [PubMed] [Cited in This Article: ] |
| 21. | McDonald LC, Fune J, Gaido LB, Weinstein MP, Reimer LG, Flynn TM, Wilson ML, Mirrett S, Reller LB. Clinical importance of increased sensitivity of BacT/Alert FAN aerobic and anaerobic blood culture bottles. J Clin Microbiol. 1996;34:2180-2184. [PubMed] [Cited in This Article: ] |
| 22. | Bernet C, Garret M, de Barbeyrac B, Bebear C, Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol. 1989;27:2492-2496. [PubMed] [Cited in This Article: ] |
| 23. | Relman DA, Schmidt TM, MacDermott RP, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293-301. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 845] [Cited by in F6Publishing: 688] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 24. | Kane TD, Johnson SR, Alexander JW, Babcock GF, Ogle CK. Detection of intestinal bacterial translocation using PCR. J Surg Res. 1996;63:59-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Yamashita Y, Kohno S, Koga H, Tomono K, Kaku M. Detection of Bacteroides fragilis in clinical specimens by PCR. J Clin Microbiol. 1994;32:679-683. [PubMed] [Cited in This Article: ] |
| 26. | Kane TD, Alexander JW, Johannigman JA. The detection of microbial DNA in the blood: a sensitive method for diagnosing bacteremia and/or bacterial translocation in surgical patients. Ann Surg. 1998;227:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 96] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 27. | Mai V, Morris JG. Colonic bacterial flora: changing understandings in the molecular age. J Nutr. 2004;134:459-464. [PubMed] [Cited in This Article: ] |
| 28. | Rantakokko-Jalava K, Nikkari S, Jalava J, Eerola E, Skurnik M, Meurman O, Ruuskanen O, Alanen A, Kotilainen E, Toivanen P. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000;38:32-39. [PubMed] [Cited in This Article: ] |
| 29. | Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695-700. [PubMed] [Cited in This Article: ] |
| 30. | Larvin M, McMahon MJ. APACHE-II score for assessment and monitoring of acute pancreatitis. Lancet. 1989;2:201-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 446] [Cited by in F6Publishing: 405] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 31. | Dervenis C, Johnson CD, Bassi C, Bradley E, Imrie CW, McMahon MJ, Modlin I. Diagnosis, objective assessment of severity, and management of acute pancreatitis. Santorini consensus conference. Int J Pancreatol. 1999;25:195-210. [PubMed] [Cited in This Article: ] |
| 32. | Ramesh H, Prakash K, Lekha V, Jacob G, Venugopal A. Are some cases of infected pancreatic necrosis treatable without intervention? Dig Surg. 2003;20:296-299; discussion 300. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 29] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | Bradley EL. Necrotizing pancreatitis. Br J Surg. 1999;86:147-148. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 34. | Uhl W, Warshaw A, Imrie C, Bassi C, McKay CJ, Lankisch PG, Carter R, Di Magno E, Banks PA, Whitcomb DC. IAP Guidelines for the Surgical Management of Acute Pancreatitis. Pancreatology. 2002;2:565-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 397] [Cited by in F6Publishing: 322] [Article Influence: 14.6] [Reference Citation Analysis (0)] |