Published online Nov 21, 2005. doi: 10.3748/wjg.v11.i43.6780
Revised: February 15, 2005
Accepted: February 18, 2005
Published online: November 21, 2005
AIM: To investigate the effects of four different ingredients of zedoary (Curcuma aromatica oil, Curcumol, β-elemence, and Curcumin) on the gene expressions of hepatic stellate cells (HSCs), and to explore the molecular mechanism of zedoary against hepatic fibrosis at gene network level.
METHODS: We detected the mRNA sequences of 50 liver fibrosis-related genes in GenBank and designed oligonucleotide probes. We synthesized oligonucleotides with PE8909 DNA synthesizing instrument, and carried out oligonucleotide microarray with OGR-04 dropping instrument and aldehyded glass chip. Cultured HSC-T6 cells were treated with different concentrations of Colchicine, Curcuma aromatica oil, Curcumol, β-elemence, and Curcumin. According to the experiment of cell toxicity, we took the appropriate concentrations of medicines that resulted in over 50% of cell survival as experiment concentrations. We collected the cells at 1, 6, 12, and 24 h, and extracted total RNA with TRIzol reagent, then labeled cDNAs with Cy3-dUTP and Cy5-dUTP. These labeled cDNAs were hybridized to an oligonucleotide microarray which was washed several times and scanned by scanner GenePix 4000B. Different gene expressions of HSC-T6 cells were analyzed by ImaGene 4.2 software.
RESULTS: After HSC-T6 cells were cultured in a medium containing 6.25 μg/mL Colchicine for 12 h, expression of TIMP-1 decreased 2.2-folds. After HSC-T6 cells were cultured in a medium containing 78.125 μg/mL of Curcuma aromatica oil for 24 h, the expression of TIMP-2 and IL-6 decreased 2.3- and 2.2-folds, respectively. Moreover, after HSC-T6 cells were cultured in a medium containing 1.5625 μg/mL of Curcumol for 12 h, the expression of TGFβ1 and P450a decreased 2.3- and 2.1-folds, respectively.
CONCLUSION: Our results may show the possible molecular mechanism of Curcuma aromatica oil and Curcumol against hepatic fibrosis.
- Citation: Jiang Y, Li ZS, Jiang FS, Deng X, Yao CS, Nie G. Effects of different ingredients of zedoary on gene expression of HSC-T6 cells. World J Gastroenterol 2005; 11(43): 6780-6786
- URL: https://www.wjgnet.com/1007-9327/full/v11/i43/6780.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i43.6780
In the past 10 years or more, great progress has been made in treating liver fibrosis with Chinese herbal medicines such as compound 861[1] and Fuzheng Huayu Decoction[2] and Tidu Hugan Decoction[3] as well as single herbal medicines as red sage root, zedoary,Chinese caterpillar fungus,hanfangchin A,extracts from peach seeds. Although many medicines can be used to treat liver fibrosis,effective medicines are still hard to find. Herbal medicines have the characteristics of multiple targets and poly-functioning routes,but genes in the organisms alone form a strict and complicated network.Therefore,study of herbal medicines should focus on the molecular mechanism at the level of genetic network based on the integral bio-system. Genetic chip technology is characterized by high communication,low consumption and miniaturization,thus providing a technological platform to study the mechanism of herbal medicines against liver fibrosis[4].
We found that zedoary could inhibit the proliferation of hepatic stellate cells (HSCs). Curcumin can be used to treat inflammation and tumors and Curcuma aromatica oil functions as an anti-inflammation,anti-virus,anti-tumor and anti-thrombus agent. Now more than 20 chemical ingredients such as Curcumol,Epicurzerenone,β-elemence,Camphene,Isoborneol,Borneol,Cineole,and 4-methyl-pyrazine have been identified from Curcuma aromatica oil. It was reported that Curcumol and Elemence can function as anti-tumor and virus agents[5,6]. Xi et al[7] found that zedoary could protect hepatic cells against necrosis and degeneration as well as proliferation of fibrous tissues.
To study the molecular mechanism of zedoary against liver fibrosis, we used the genetic expression spectrum chips to represent 50 genes related to liver fibrosis and substituted HSC-T6 for original HSCs, and investigated the effects of four different ingredients of zedoary (Curcuma aromatica oil,Curcumol,β- elemence,Curcumin)on the gene expression of activated HSCs.
Colchicine was purchased from American ALEXIS Co, Elemence injection and Curcumin were obtained from Jingang Pharmaceutic Corporation Ltd,Dalian,China. Curcumol and Curcuma aromatica oil were from Pharmaceutical University,Shenyang,China. HSC-T6 was provided by the Insititute of Hepatology,Shanghai University of Traditional Chinese Medicine and pharmacology. AXSys Probe Punctum-controlling software was purchased from Cartesian Technologies Co., and ImaGene 4.2 figure-analyzing softwares was from American Biodiscovery Co.
Design of oligonucleotide probe Oligonucleotide probes were designed by the design software of oligonucleotide probe . The coding region near the 3 end was selected for BLAST analysis. One or two probes related to liver fibrosis whose homology was less than 70% were used as spare probes.
Synthesis of oligonucleotide Oligonucleotides were synthesized by the chemical method of standard subphosphorus imide using PE8909 DNA synthesizer. N-MMTr-6- ammonia-2-cyanogen-N and N-diisopropyl-subimide ammonia were modified by 5- or 3-amino-group. Dense ammonia was deprotected at 55 °C and incised for 15 h, and purified by ordinary portland cement column.
Preparation of probes In brief, 0.5 µg/µL oligonucleotide probe was resolved into 3×SSC solution, glass chip was aldehyded and stayed overnight, processed with 2 g/L SDS de-ion water for 10 min, and then dried for later use.
Preparation of medical culture medium Colchicine was dissolved in double-vaporing water to get the original solution (3.2 mg/mL). Curcumin was mixed with 950 mL/L alcohol to get the original solution (320 mg/L). Curcumol was mixed with 950 mL/L alcohol plus Tween-80 to get the original solution (3.2 mg/mL). Curcuma aromatica oil was mixed with Tween-80 (ratio, 1:10) plus 950 mL/L alcohol to get the original solution (2.5 mg/mL). Elemence injection (5 mg/mL) was used. All the medicines were filtrated through 0.45 micropores and stored at 4 °C.
The ampoule was taken out of the liquid nitrogen jar(wearing protective glasses and gloves)and put into a porcelain enamel vessel containing 36-37 °C water with shaking. The pocket was cut and the ampoule was taken out,sterilized with 700 mL/L alcohol. The cell suspension was aspirated and put into centrifuge tube,then 10 mL culture medium was added,centrifuged for 5 min at 500-1 000 r/min and rinsed . The culture medium was changed on the next day.
The 96-well plates were incubated at 37 °C until HSC-T6 cells were grown in a single layer. The culture medium was incubated for 48 h, and then 5 mg/mL MTT was added and incubated for 48 h. The A value of the solution was tested in enzymatic marking instrument (wave length on the light-filtrating slice is 492 nm). According to the experiment of cell toxicity, we took the appropriate concentrations of medicines that resulted in over 50% of cell survival. The formula of cell survival rate: cell survival rate (%) = (medicine group/control group)×100%.
HSC-T6 cells were incubated with different concentrations of Colchicine, Curcuma aromatica oil, Curcumol, β-elemence, and Curcumin, followed by collecting them at 1, 6, 12, and 24 h, respectively.
Extraction of cellular tRNA HSC-T6 cells were washed softly with germ-free PBS. One-milliliter of TRIzol reagent was used to blow the cells and to make them dissolve completely. Then 0.2 mL of methylene trichloride was added, followed by centrifugation at 12 000 g for 15 min at 4 °C. The supernatant was aspirated with 200 µL tip (dealt with DEPC) and moved to another EP tube. Then 0.5 mL of isopropylalcohol was added and put aside for 15 min, and centrifuged at 12 000 g for 10 min at 4 °C. The RNA was washed with 750 mL/L alcohol (dealt with DEPC), and then 1 mL of 750 mL/L alcohol (dealt with DEPC) was added, centrifuged at 7 500 r/min for 5 min at 4 °C, and stored at -20 °C.
Evaluation of purity DEPC water was added to 2 µL of RNA to make a total volume of 100 µL. Ultraviolet spectrophotometer was used to measure the value of A260 and A280 as well as A260/A280. The formula of concentration of RNA: RNA (µg/µL) = A260×40×dilution multiple/1 000.
Evaluation of integrity Five-microliter of RNA samples were put into 10 mL/L formaldehyde degeneration sepharose for cataphoresis, followed by painting with EB and examination under ultraviolet light.
Reverse transcription system and conditions Ten-microliter of TRNA (10 µg/µL), 0.3 µL of positive control, 4.0 µL of oligo (dT)16 (0.5 µg/µL) and 0.5 µL of RNase inhibitor (40 U/µL) were mixed together, incubated for 10 min and then put into ice bath. Then 5.0 µL of 5× first chain buffer, 2.0 µL of DTT (0.1 mol/L), 1.0 µL of Cy3-dUTP or Cy5-dUTP (1 mmol/L), 0.5 µL of dTTP (10 mmol/L, and 0.1 µL of each dATP, dCTP and dGTP (100 mmol/L) were mixed together and incubated for 2 min at 42 °C. Then 1.0 µL of SuperScript II RNase H-transcriptase (10 U/µL) was added and incubated for 2 h. Then 1.0 µL of RNaseH was added and incubated for 0.5 h at 30 °C, followed by inactivation of anti-transcriptase at 70 °C for 15 min. Then 5.0 µL of NaOH (1 mol/L) was added and incubated at 65 °C for 1 h, followed by addition of 5.0 µL of 1 mol/L HCl (pH 6.8) and 6.0 µL of 5 mol/L NaCl. The sediment was stayed overnight at –20 °C, centrifuged at 12 000 g for 10 min at 4 °C. The supernatant was abandoned, the sedimentation was washed once with 750 mL/L alcohol and then dried, followed by resolving in 2 µL of aseptic water and stored at -20 °C.
Evaluation of cDNA One-microliter of the above product of reverse transcription was put into 10 mL/L formaldehyde degeneration sepharose for cataphoresis, and then the quality of probes was evaluated.
Two microliters of probes labeled by fluorescence were diluted to 8 µL for hybridization, and the hybridization liquor was moved onto the cover glass chip with the density of 2 µL/cm2. The solution was put onto the carry sheet glass chip equally by the capillarity between cover glass chip and carry sheet glass chip. Then the glass chips were put into a hybridization box and 5 µL of 3× SSC was added to keep the humidity. The probe washing temperatures varied according to the different probes, usually under room temperature. The order of washing liquor was lotion A–C.
After hybridization,the genetic chips were scanned by Scanner Genepix 4 000B,ImaGene4.2 was used to analyze the ratio of Cy3,Cy5 and the intensity of two kinds of fluorescence signals.Housekeeping gene and positive control were taken to balance the data of fluorescence of Cy3 and Cy5. Ratio of Cy3,Cy5>2 or <0.5 was used to evaluate the differences of genetic expression.
After treatment of HSC-T6 cells with different concentrations of Colchicine, Curcuma aromatica oil, Curcumol, β-elemence, and Curcumin for 48 h, along with deduction of the concentrations, the survival ratio of HSC-T6 increased.
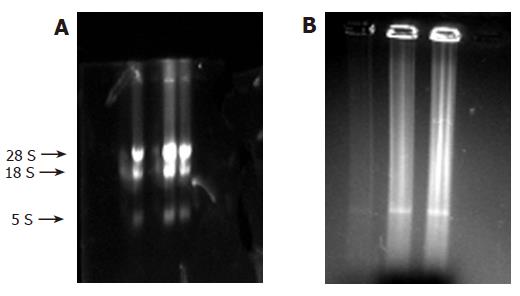
RNA was put on 10 mL/L formaldehyde agarose gels for cataphoresis, and the results were recorded with gel photography. Three strips (28S, 18S, and 5S) can be seen in Figure 1A. After the analysis by software, the ratio of 28S and 18S was found to be between 1.5 and 2.0, showing that the RNA was integral and without degradation. Using ultraviolet spectrophotometer, the value of A260/A280 was found to be between 1.7 and 2.0, showing that the RNA was pure and without protein pollution or phenol.
In addition, most of the cDNAs were observed between 0.5 and 2 kb (Figure 1B). Distribution of probes is shown in Table 1.
| Genetic probe matrix | ||||||||
| TIMP1 | TIMP1 | TIMP2 | TIMP2 | TIMP3 | TIMP3 | MMP2 | MMP2 | |
| PDGFA | PDGFA | PDGFC | PDGFC | MMP3 | MMP3 | IL-6 | IL-6 | IL-10 |
| HGF2 | HGF2 | HGF1 | HGF1 | VEGFA | VEGFA | VEGFB | VEGFB | VEGFC |
| IGF2 | IGF2 | IGF1 | IGF1 | TGFβRI | TGFβRI | TGFβRII | TGFβRII | PDGFRα |
| N | N | P | P | N | N | P | P | N |
| C-myc | C-myc | P-450d | P-450d | P-450a | P-450a | P-450e | P-450e | P450-4A3 |
| TNFα | TNFα | TNFR1 | TNFR1 | CJUNB | CJUNB | CJUND | CJUND | |
| CYP2D4 | CYP2D4 | CYP1B1 | CYP1B1 | ESTSUL | ESTSUL | FGF1 | FGF1 | STAA |
| ICAM-1 | ICAM-1 | PAFR | PAFR | VCAM-1 | VCAM-1 | MIP-2 | MIP-2 | MCP-1 |
| β-actin | β-actin | GAPDH | GAPDH | |||||
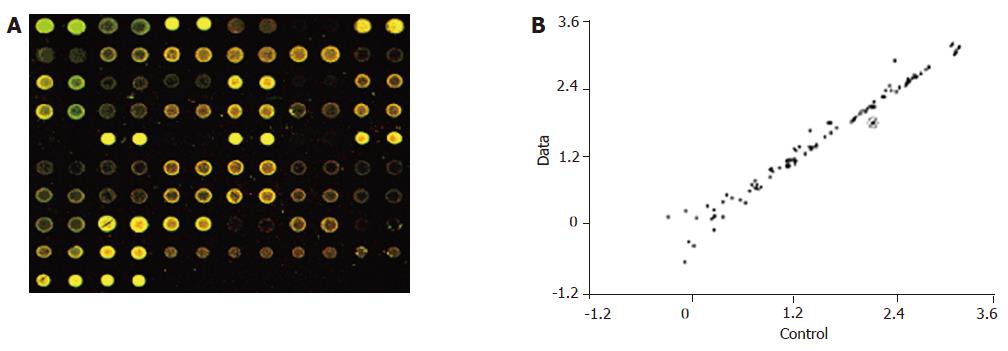
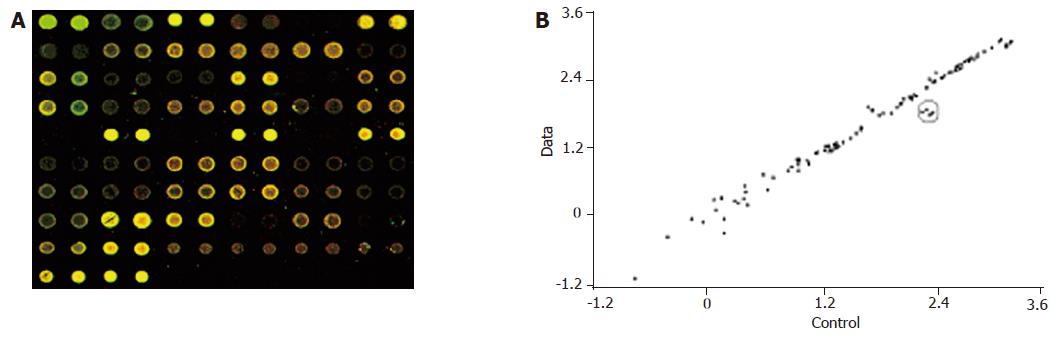
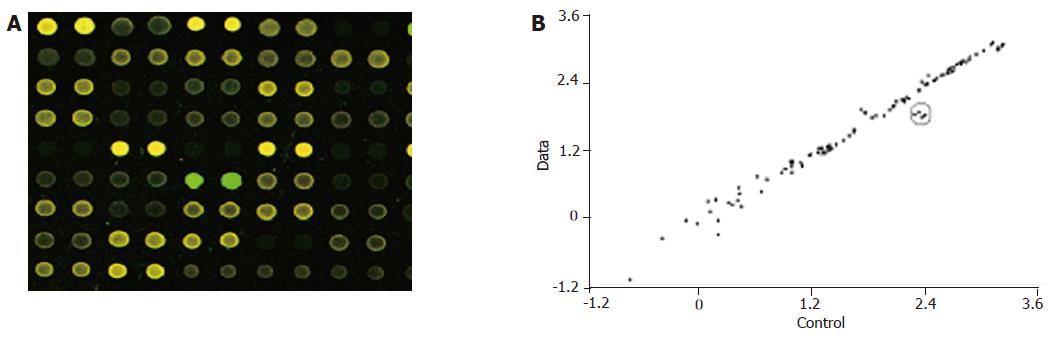
In the hybridization scanning figures of probes (Figures 2A, 3A, and 4A), the green spots represent the locations of downregulated genetic expression. ImaGene 4.2 software was used to analyze the intensity and ratio of Cy3 and Cy5. After correction of housekeeping gene and positive control, the two conditions were taken to evaluate the difference of genetic expression as follows: (1) the ratio of Cy3 and Cy5 >2 or <0.5; and (2) one of Cy3 and Cy5 >1 000. In the scattering diagram (Figures 2B, 3B, and 4B), the spots on the opposite angles show that the intensity of two groups (treatment group and control group) was the same. The farther they depart from the opposite angles, the bigger the difference of genetic expression would be. The different effects of medicines on genetic expression are shown in Tables 2-4. After culture of HSC-T6 cells in a medium containing 6.25 μg/mL of Colchicine for 12 h, the expression of TIMP-1 decreased 2.2-folds, which was in agreement with a previous report[9], suggesting that the genetic probe testing system in the experiment was dependable. Furthermore, Figures 3 and 4 stand for the scattering diagram and the scattering diagram of Curcuma aromatica oil group and Curcumol group, respectively, and the data of analysis are shown in Tables 3 and 4.
| Gene | GenBank | Ratio |
| TIMP-1 | U06179 | 2.2 |
| Gene | GenBank | Ratio |
| TIMP-2 | NM-021989 | 2.3 |
| IL-6 | M26744 | 2.2 |
| Gene | GenBank | Ratio |
| TGFβ1 | NM-021578 | 2.3 |
| P-450a | J02669 | 2.1 |
These results showed that after HSC-T6 cells were cultured in a medium containing 6.25 μg/mL Colchicine for 12 h, the expression of TIMP-1 decreased 2.2-folds. The expression of TIMP-2 and IL-6 decreased 2.3- and 2.2-folds, respectively, after HSC-T6 cells were cultured in a medium containing 78.125 μg/mL of Curcuma aromatica oil for 24 h. Moreover, the expression of TGFβ1 and p450a decreased 2.3- and 2.1-folds, respectively, after HSC-T6 cells were cultured in a medium containing 1.5625 μg/mL Curcumol for 12 h. Other concentrations of neither Curcuma aromatica oil nor Curcumol could bring forth gene expression differences, nor did β-elemence and Curcumin.
Genetic chip technology is characterized by high commun-ication, low consumption and miniaturization[10-13], providing a technological platform to study the mechanism of herbal medicines against liver fibrosis. Genetic expression spectrum chips can be categorized into two kinds, namely cDNA chips and oligonucleotide chips, the probe of the former is cDNA, the probe of the latter is fragment of oligonucleotide. To study medicines with genetic expression spectrum chips can help us acknowledge the target genes of herbal medicines, western medicine and some foods with medical functions. It is also an effective way to study the toxicity of medicines, and the mechanism of causing deformity and genetic mutation of medicines.
According to the key role of HSCs in hepatic fibrosis, 50 genes related to liver fibrosis are chosen. They can be categorized into five groups as follows: (1) expressing cytokines and receptors of cytokines, such as transforming growth factor β1 (TGF β1), platelet-derived growth factor A (PDGFA), platelet-derived growth factor C (PDGFC), interleukin-1, interleukin-6, interleukin-10, hepatocyte growth factor 1 (HGF1), hepatocyte growth factor 2 (HGF2), vascular endothelial growth factor A (VEGFA), vascular endothelial growth factor B (VEGFB), vascular endothelial growth factor C (VEGFC), vascular endothelial growth factor D (VEGFD), fibroblast growth factor 1 (FGF1), fibroblast growth factor 2 (FGF2), platelet- activating factor (PAF), tumor necrosis factor α (TNFα), insulin-like growth factor 1 (IGF1), insulin-like growth factor 2 (IGF2), endothelin 1 (ET-1), intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), murine macrophage inflammatory protein 2 (MIP-2), monocyte chemotactic protein 1 (MCP-1), transforming growth factor β receptor I (TGFβRI), transforming growth factor β receptor II (TGFβRII), platelet-derived growth factor receptor α (PDGFRα), platelet-derived growth factor receptor β (PDGFRβ), tumor necrosis factor α receptor 1 (TNFαR1), platelet- activating factor receptor (PAFR), and endothelin receptor A (ETRA). TGFβ1 is the strongest factor which promotes the synthesis of extracellular matrix (ECM). PDGF can accelerate the proliferation of HSCs; (2) expressing MMPs (matrix metalloproteinases) and TIMPs (tissue inhibitor of metalloproteinases), such as MMP2, MMP3, MMP8, TIMP-1, TIMP-2, and TIMP-3, which take part in the degradation of ECM; (3) gene expression related to the preliminary activation of HSCs, such as c-myc, Ets-1, STAT1, c-jun B, c-jun D; (4) involving in gene expression related to the biological oxidation[14], such as cytochrome P450d, cytochrome P450a, cytochrome P450e, cytochrome P450-LA, and cytochrome P450 (4A3), CYP1B1, CYP2D4; and (5) house keeping gene, such as β-actin and GAPDH.
How to choose the genes related to liver fibrosis is the key of the design of oligonucleotide probes. The mRNA sequences which contain 40 amino acids are selected and then oligonucleotide probes are designed using the design software of oligonucleotide probe. By and large, the length of oligonucleotide probe is 15-80 nt, the content of GC is 45-55%. Probes of the coding region approaching end 3 are chosen for BLAST analysis. One to two probes whose homology of sequences was less than 70% are chosen as gene distinctive oligonucleotide probes.
Because some sequences of mRNA related to collagen I, III, IV are so short, the probes designed through software are easy to cause cross reactions which may result in false positive results, and such genes, therefore, cannot be chosen. However, the aforementioned 50 genes, in general, can represent approximately the changes which take place in the process of preliminary and persistent activation of HSCs in liver fibrosis.
HSC-T6 cells are SV40 transfected HSCs of Sprague-Dawley rats. The cells can be steadily cultured and their phenotype is activated HSCs which can express high-level collagen I and TIMP-1 mRNA, etc. In our study, HSC-T6 was substituted for the original HSC. Yin et al[15,16] took HSC-T6 as model cells to investigate the influence of compound 861 on the gene expression of MMP-3 and TIMP-1 and they found that 0.25, 0.5, and 1.0 mg/mL of compound 861 could increase the expression of MMP3 and inhibit the expression of TIMP1.
TIMPs can prevent MMPs from degrading ECM[17-21]. Using genetic chip technology, we found that after HSC-T6 cells were cultured in a medium containing 6.25 μg/mL Colchicine for 12 h, the expression of TIMP-1 decreased 2.2-folds. Expression of TIMP-2 decreased 2.3-folds after HSC-T6 cells were cultured in a medium containing
78.125 μg/mL Curcuma aromatica oil for 24 h. These results showed that Curcuma aromatica oil, like Colchicines, can also inhibit the expression of TIMPs and reduce the inhibition of MMPs which, in turn, help MMPs to degrade ECM. This may be one of the mechanisms for zedoary against liver fibrosis.
IL-6 is also called hepatic cell stimulating factor which can directly stimulate hepatic cells proliferation, induce the expression of IL-6 receptors in liver, and stimulate fibroblastic cells to synthesize collagens[22-26]. In our study, 78.125 μg/mL Curcuma aromatica oil could decrease the expressions of IL-6, TIMP1 and other genes related to hepatic fibrosis, thereby enhancing MMPs to degrade ECM. It might be another mechanism of zedoary against hepatic fibrosis.
TGFβ1 plays an important role in hepatic fibrosis which can activate HSCs and, as a transcription factor of collagen, accelerate its expression[27]. In our study, the expression of TGFβ1 decreased 2.3-folds after culture of HSC-T6 cells in a medium containing 1.5625 μg/L Curcumol for 12 h, suggesting that Curcumol can inhibit the synthesis of ECM through the inhibition of TGFβ1 which might be an another important mechanism of zedoary against hepatic fibrosis.
Cytochrome P450 (or Cyt P450) involves in the synthesis of steroid hormone, bile acid and bile pigments as well as the process of the bio-transformation of medicine and poison[14]. Yang et al[28] have shown that lipid peroxidation takes part in the activation of HSCs. Svegliati Baroni et al[29] have indicated that HCM/Fe might induce a significant increase in collagen type I accumulation in HSC culture media, and HSCs proliferation may be associated with changes in the Na+/H+ exchanger activity. Nieto et al[30] transfected CYP2E1 to HSC-T6 cells, and found that there was an increase in the level of reactive oxygen species and type I collagen mRNA. It has been reported that when HSCs were cultured with HepG2 cells which overexpress CYP2E1 together, the level of collagen markedly increased, suggesting that the solved oxidants can activate HSCs[31]. In our study, we found that after culture of HSC-T6 cells in a medium containing 1.5625 μg/mL Curcumol for 12 h, the expression of P450a decreased 2.1-folds, suggesting that the metabolism of Curcumol in HSC-T6 cells might bring forth oxidation-conjugation reaction through the P450 enzyme system, induce a decline of oxidative stress and lipid peroxidation, and thus inhibit the activation of HSCs. This may be one of the mechanisms of zedoary against hepatic fibrosis.
In conclusion, two different ingredients of zedoary (Curcuma aromatica oil, Curcumol), when treated with HSC-T6 cells for 24 and 12 h, can decrease the expression of TIMP-2, IL-6, TGFβ1 and P450a by different degrees, indicating the molecular mechanisms of zedoary against hepatic fibrosis at gene network level. But the changes of genes and the expression of proteins might not be the same events. Along with the development of protein histology, further research needs to testify the proteins related to liver fibrosis, for example, to examine the content of type I collagen using ELISA or investigate the influence of Curcuma aromatica oil and Curcumol on protein expression of HSC-T6 cells by protein chip technology.
Co-first-author: Yuan Jiang
Co-Correspondent: Yuan Jiang
Science Editor Kumar M, Wang XL and Guo SY Language Editor Elsevier HK
| 1. | Wang L, Wang J, Wang BE, Xiao PG, Qiao YJ, Tan XH. Effects of herbal compound 861 on human hepatic stellate cell proliferation and activation. World J Gastroenterol. 2004;10:2831-2835. [PubMed] [Cited in This Article: ] |
| 2. | Liu C, Wang X, Liu P. Serapharmacological effect of fuzheng huayu 319 Decoction on expression of type I collagen and transforming growth factor beta 1 in hepatic stellate cells. Zhongguo Zhongxiyi Jiehe Zazhi. 1999;19:412-414. [Cited in This Article: ] |
| 3. | Zhu QJ, Nie G, Li HW, Xiao L, Hu CQ, Yang L, Ming AP, Wang BX. Inhibitory effect of Tidu Hugan Decoction on experimental hepatic fibrosis in ducks. Zhongxiyi Jiehe Ganbing Zazhi. 1998;8:84-87. [Cited in This Article: ] |
| 4. | Abelson PH. A third technological revolution. Science. 1998;279:2019. [PubMed] [Cited in This Article: ] |
| 5. | Wang Y, Wang MZ. Reseach of commonly used Chinese traditional medicine: Curcuma categories. Zhongguo Yaoxue Zazhi. 2001;36:80. [Cited in This Article: ] |
| 6. | Li GD, Xiu F, Shen AJ. Reseach of Curcuma aramatica oil. Zhongguo Yaoxue Zazhi. 2002;37:806-809. [Cited in This Article: ] |
| 7. | Xi ZT, Shan CM, Jiang XL, Luan XY, Li KK. Experimental study on Rhizoma sparganii and Radices zedoariae treating hepatic fibrosis. Zhongguo ZhongYao ZaZhi. 2002;27:929-932. [PubMed] [Cited in This Article: ] |
| 8. | Bian XY, Yin XN. The method and application of MTT assay. Gouwai Yixue Linchuang Shengwu Huaxue Yu Jianyanxue Fence. 1998;19:83-85. [Cited in This Article: ] |
| 9. | Yang CQ, Hu GL, Zhou WH. Effect of Colchicine on Matrix metalloproteinase-1 and tissue inhibitor of metalloproteinase-1 expression in rat liver fibrosis. Zhonghua Chuanran Bing Zazhi. 2000;18:176-179. [Cited in This Article: ] |
| 10. | Schena M, Heller RA, Theriault TP, Konrad K, Lachenmeier E, Davis RW. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301-306. [PubMed] [Cited in This Article: ] |
| 11. | Persidis A. Biochips. Nat Biotechnol. 1998;16:981-983. [PubMed] [Cited in This Article: ] |
| 12. | Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614-10619. [PubMed] [Cited in This Article: ] |
| 13. | Debouck C, Goodfellow PN. DNA microarrays in drug discovery and development. Nat Genet. 1999;21:48-50. [PubMed] [Cited in This Article: ] |
| 14. | Zhou AR. Biochemistry. Fifth edition. Beijing: Renming Weisheng Chuban She 2001; 160. [Cited in This Article: ] |
| 15. | Yin C, Ma H, Wang A, Ma X, Jia J, Wang B. Effect of compound 861 on tissue inhibitor of metalloprotenase 1 gene expression of HSC-T6 cells. Zhonghua Gan Zang Bing Za Zhi. 2002;10:197-199. [PubMed] [Cited in This Article: ] |
| 16. | Yin C, Ma H, Wang A, Ma X, Jia J, Wang B. Effect of compound 861 on stromelysin gene expression of HSC-T6 cells. Linchuang Gandanbing Zazhi. 2002;18:168-170. [PubMed] [Cited in This Article: ] |
| 17. | Iredale JP. Tissue inhibitors of metalloproteinases in liver fibrosis. Int J Biochem Cell Biol. 1997;29:43-54. [PubMed] [Cited in This Article: ] |
| 18. | McCrudden R, Iredale JP. Liver fibrosis, the hepatic stellate cell and tissue inhibitors of metalloproteinases. Histol Histopathol. 2000;15:1159-1168. [PubMed] [Cited in This Article: ] |
| 19. | Murawaki Y, Ikuta Y, Koda M, Okamoto K, Mimura K. The proMMP-2 activation rate in patients with chronic viral liver disease. Clin Chim Acta. 2002;324:99-103. [PubMed] [Cited in This Article: ] |
| 20. | Nie QH, Zhou YX, Xie YM, Chen YQ. The localization and expression of TIMP-2 in experi mental hepatic fibrosis in rats. Jiefangjun Yixue Zazhi. 2002;27:208-209. [Cited in This Article: ] |
| 21. | Luo XD, Nie QH. The value of tissue inhibitors of metalloproteinases in the study of hepatic fibrosis. Ganzang. 2003;8:40-42. [Cited in This Article: ] |
| 22. | Li D, Zhang LJ, Chen ZX, Huang YH, Wang XZ. Effects of TNFα,IL-6 and IL-10 on the development of experimental rat liver fibrosis. Shijie Huaren Xiaohua Zazhi. 2001;9:1242-1245. [Cited in This Article: ] |
| 23. | Llorent L, Richaud-Patin Y, Alcocer-Castillejos N, Ruiz-Soto R, Mercado MA, Orozco H, Gamboa-Domínguez A, Alcocer-Varela J. Cytokine gene expression in cirrhotic and non-cirrhotic human liver. J Hepatol. 1996;24:555-563. [PubMed] [Cited in This Article: ] |
| 24. | Liu HL, Li XH, Wang DY, Yang SP. Matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-1 expression in fibrotic rat liver. World J Gastroenterol. 2000;6:881-884. [PubMed] [Cited in This Article: ] |
| 25. | Ramadori G, Christ B. Cytokines and the hepatic acute-phase response. Semin Liver Dis. 1999;19:141-155. [PubMed] [Cited in This Article: ] |
| 26. | Moshage H. Cytokines and the hepatic acute phase response. J Pathol. 1997;181:257-266. [PubMed] [Cited in This Article: ] |
| 27. | Wang LS, Chen YW, Li DG. Activation of TGF-β1 and liver fibrosis. Gouwai Yixue Xiaohua Xitong Fence. 2003;23:222-225. [Cited in This Article: ] |
| 28. | Yang WF, Chen HC, Jiang YP. Some factors on hepatic stellate cells activation. Zhonghu GanZangBing ZaZhi. 2004;12:121-123. [PubMed] [Cited in This Article: ] |
| 29. | Svegliati Baroni G, D'Ambrosio L, Ferretti G, Casini A, Di Sario A, Salzano R, Ridolfi F, Saccomanno S, Jezequel AM, Benedetti A. Fibrogenic effect of oxidative stress on rat hepatic stellate cells. Hepatology. 1998;27:720-726. [PubMed] [Cited in This Article: ] |
| 30. | Nieto N, Friedman SL, Greenwel P, Cederbaum AI. CYP2E1-mediated oxidative stress induces collagen type I expression in rat hepatic stellate cells. Hepatology. 1999;30:987-996. [PubMed] [Cited in This Article: ] |
| 31. | Marí M, Cederbaum AI. Induction of catalase, alpha, and microsomal glutathione S-transferase in CYP2E1 overexpressing HepG2 cells and protection against short-term oxidative stress. Hepatology. 2001;33:652-661. [PubMed] [Cited in This Article: ] |