Published online Jul 27, 2014. doi: 10.5411/wji.v4.i2.107
Revised: April 19, 2014
Accepted: June 14, 2014
Published online: July 27, 2014
Viral protein U (Vpu) is an accessory protein associated with two main functions important in human immunodeficiency virus type 1 (HIV-1) replication and dissemination; these are down-regulation of CD4 receptor through mediating its proteasomal degradation and enhancement of virion release by antagonizing tetherin/BST2. It is also well established that Vpu is one of the most highly variable proteins in the HIV-1 proteome. However it is still unclear what drives Vpu sequence variability, whether Vpu acquires polymorphisms as a means of immune escape, functional advantage, or otherwise. It is assumed that the host-pathogen interaction is a cause of polymorphic phenotype of Vpu and that the resulting functional heterogeneity of Vpu may have critical significance in vivo. In order to comprehensively understand Vpu variability, it is important to integrate at the population level the genetic association approaches to identify specific amino acid residues and the immune escape kinetics which may impose Vpu functional constraints in vivo. This review will focus on HIV-1 accessory protein Vpu in the context of its sequence variability at population level and also bring forward evidence on the role of the host immune responses in driving Vpu sequence variability; we will also highlight the recent findings that illustrate Vpu functional implication in HIV-1 pathogenesis.
Core tip: Viral protein U (Vpu) is a highly polymorphic human immunodeficiency virus type 1 (HIV-1) accessory protein; however factors that are attributable to Vpu sequence variability are not well defined. In this review we have focused on the immune responses both innate (natural killer cells) and adaptive (cellular and humoral) immunity that are directed towards HIV-1 Vpu and we also show the interaction between Vpu and host cellular factors. We also highlight evidence that suggests interaction between the host immune responses and Vpu may contribute to Vpu sequence variability. Finally we have summarized the current knowledge on HIV-1 Vpu functions including Vpu evasion activities from the host immune surveillance.
- Citation: Hasan Z, Kamori D, Ueno T. Role of host immune responses in sequence variability of HIV-1 Vpu. World J Immunol 2014; 4(2): 107-115
- URL: https://www.wjgnet.com/2219-2824/full/v4/i2/107.htm
- DOI: https://dx.doi.org/10.5411/wji.v4.i2.107
Human immunodeficiency virus type 1 (HIV-1) demonstrates a significant genetic diversity due to its high mutation rate; so far this extraordinary diversity has been a major setback in development of vaccine and antiretroviral drugs. Low fidelity of reverse transcriptase that give rise to error prone replication process, high progeny production, turnover rates and recombination of circulating HIV-1 strains are some of the viral factors that contributes to HIV-1 diversity[1-3]. The adaptive potential of HIV-1 is shaped by both virus and the host immune factors, in other words both the diversifying and purifying selection factors influence HIV-1 diversity. In fact, strong evidence has also indicated that the host immune responses influence HIV-1 diversity by selection of escape mutations[4-6]. Thus a comprehensive analysis of the dynamics of polymorphisms in HIV-1 proteins is a powerful tool to reveal actual interactions between HIV-1 and the host immune system[7-9].
HIV-1 viral protein U (Vpu) is a 16-kDa accessory protein[10] responsible for various functions such as CD4 down-regulation[11-13] and enhancement of virion release by antagonizing tetherin/BST2[14-17]. Interestingly, functionally competent Vpu (with respect to BST-2 antagonistic activity) were only found in the pandemic group M subtypes, suggesting that Vpu functional adaptation may confer pandemic spread of this HIV-1 subtype[18]. In general, the host genetic factor is one of the main driving force of sequence polymorphism in HIV-1[18], as evidenced in HIV-1 Nef[7,19-21] and Env[22,23] proteins whose highly polymorphic phenotype is mostly attributed by the host immune responses such as HLA class I-restricted CD8+ T lymphocytes and neutralizing antibodies, respectively. However, it is still unclear to what extent the host immune responses influence Vpu sequence variation. This review focuses on the role of host immune responses in Vpu sequence variability. Briefly, we also discuss the current understanding of Vpu functions including evasion of the immune system and their implication in viral pathogenesis.
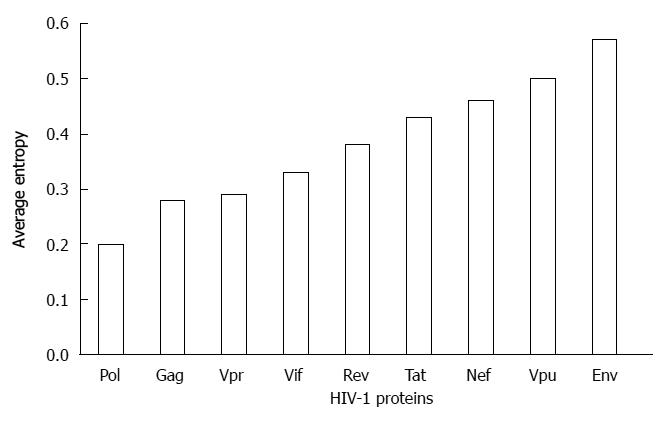
Vpu exhibit a stable reading frame in vivo despite being a highly variable protein, suggesting functional importance of Vpu in HIV-1 replication and persistence. Furthermore, it has evidently been shown that only HIV-1 strains of the pandemic M group evolved a fully functional Vpu that efficiently antagonizes human tetherin/BST-2; this suggests that Vpu evolutional adaptation may be associated with the pandemic spread of HIV-1[18]. Several studies have demonstrated the extent of Vpu sequence variability both at inter- and intra-patient level. By using the 101 aligned amino acid sequences of entire HIV-1 genome, one study showed that Vpu had the highest average entropy score in comparison to other proteins in HIV-1 genome[24]. Another study analyzing the intra-patient diversity and adaptation of non-structural genes in primary HIV-1 subtype C infection reported that vpu compared to vif, vpr, tat exon 1 and rev exon 1 genes has the highest mean of intra-patient diversity that increased gradually[25]. We retrieved full lengths clade B sequences (n = 544) of HIV-1 proteins (Gag, Pol, Env, Nef, Vif, Vpu, Vpr, Tat and Rev) from Los Alamos database and the average entropy score of each protein was determined. Vpu was observed to be one of the proteins with the highest average entropy score (Figure 1), confirming the highly variable nature of Vpu at population level. However, interestingly, a recent study has shown that despite extensive Vpu sequence variation in HIV-1 infected individuals, Vpu functions (CD4 cell surface downregulation and tetherin counteraction activity) were maintained[26].
Several studies have reported Vpu-specific humoral immune responses during HIV-1 infection[27-31]. However there has been some controversy on correlation between the presence of anti-Vpu Ab responses in HIV-infected patients’ sera and clinical outcome. Some studies have indicated that anti-Vpu Ab responses may influence the clinical outcomes in HIV-1 infected individuals[27,28,30,31]; while on the other hand other studies have showed no correlation[29]. These findings indicate that Vpu is indeed a target of antibodies although no evidence yet support that such antibody responses influence the Vpu variability. The epitopic regions for such antibodies reported include 37-50[30] and 68-81[28] of Vpu; nonetheless there is no specific Vpu activity mapped to these regions so far. However, considering that Vpu is a small protein (81 amino acids); it is intriguing to test whether such Vpu-specific antibodies can inhibit Vpu functions and subvert viral replication.
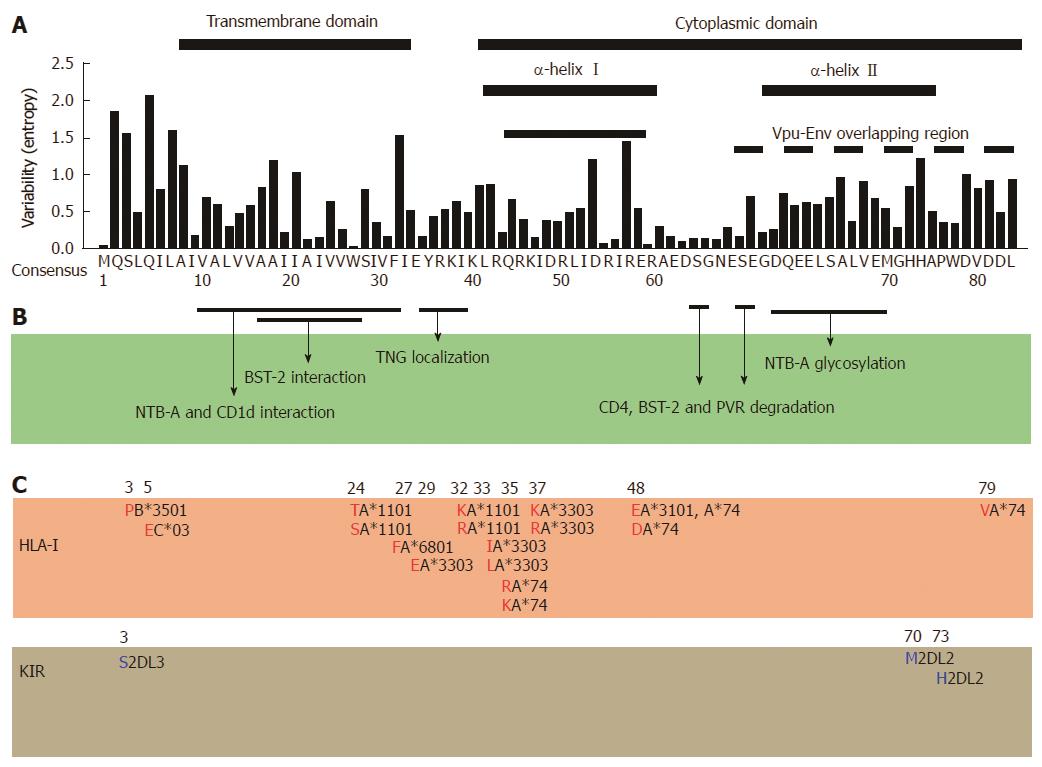
A growing number of clinical evidence has suggested that HLA-restricted, HIV-specific CD8+ cytotoxic T lymphocytes (CTL) is mainly involved in controlling HIV-1 replication[32-34]. CTL responses have been well appreciated in SIV-infected macaque’s model[32,33] and in HIV-1 infected patients of both acute[35,36] and chronic[37] phases as well as in elite controllers who spontaneously suppress viral replication below detection limit[38,39]. HLA-restricted CTL responses are thought to be the main driving force of HIV-1 control and viral evolution[40-43]. The viral polymorphism in response to immune selective pressures follows predictable patterns and kinetics at the population and these immune “footprints/landscape” could be predictable based on the autologous viral sequences and the host immune genetics[9,42,44]. However, Vpu has been reported to be a poor target for CD8+ T cells as revealed by interferon (IFN)-γ Elispot assay[45], because only some few epitopes were identified and less than 3% of patients showed detectable Vpu-specific CD8+ T cell responses. Although several HLA-restricted CTL epitopes of Vpu are reported[45-49], this protein is less targeted by CTLs at least compared to the Nef protein. Consistently, our previous study showed only three HLA-associated polymorphisms in Vpu at Glu-5 with HLA-C*03 and Arg-37, Lsy-37 with HLA-A*3303 in a chronic HIV-infected patient cohort in Japan (n = 216), indicating that the HLA class I has minor contribution (2% of the total codons) towards Vpu variability[50]. The increased numbers of subjects to 516 showed similar results (DK, ZH, and TU: unpublished observation). Furthermore, an international large IHAC cohort (International HIV Adaptation Collaborative, n = 1888) identified that only 26.3% of the highly variable Vpu codons exhibited statistically significant HLA class I associations[20]. Although the HLA class I-associated viral polymorphisms observed in the two cohorts suggested to be influenced by several factors such as the host genetic profiles, mixture of multiethnic populations, studied sample size, geographical location and circulating HIV-strains, these results suggest that HLA-associated polymorphisms are only partly attributable to the Vpu variability (Figure 2). However, it is of note that the low CTL responses observed in the previous studies[45,51] and subtle numbers of HLA-associated polymorphisms[20,50] may be an underestimation due to the current technical limitation toward a highly variable protein, even though a number of studies reported a plenty of CTL targeting[52,53] and HLA-associated polymorphisms in Nef[19,20,42], which showed comparable variability with Vpu at a population level (Figure 1).
A number of evidence suggests that natural killer (NK) cells have an important role in control of HIV-1 infection[54-56]. Assuming that NK cells may act as a selective force, as similar to CTLs, HIV-1 may leave footprints as viral polymorphisms in association with polymorphic NK cell ligand such as killer-cell immunoglobulin-like receptors (KIR). In fact, one study identified 22 amino-acid polymorphisms within the HIV-1 clade B sequence that are significantly associated with the expression of specific KIR genes in chronically HIV-1 infected, treatment naïve patients (n =91)[44]. Three (13.6%) of these KIR associated polymorphisms were located in Vpu at positions Ser-3 and Vpu-Env overlapping region (Met-71 and His-74) (Figure 2)[44]. In addition, the HIV-1-specific antibody-dependent NK cell cytotoxicity is identified towards a 13-mer Vpu peptide (69EMGHHAPWDVDDL81)[57]. Such responses are also observed toward Env[58] and Nef[59] in HIV-1 infected patients as well. However, there is no evidence at the moment that show antibody-dependent NK cell cytotoxicity associates with viral polymorphisms.
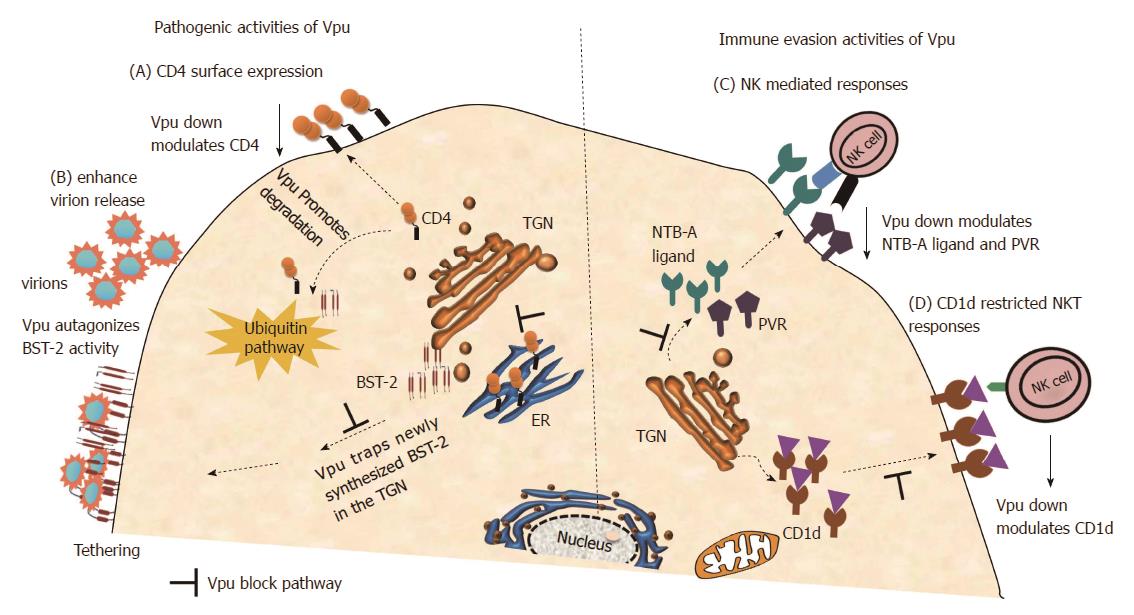
In order to conquer the hostile host environment, viruses need to evolve and develop critical interactions with the host cellular factors. Vpu does not only play important role in HIV-1 pathogenesis through CD4 receptor degradation[11] and enhancement of virion release from infected cells by antagonizing tetherin/BST-2[60-62]; but Vpu has also evolved to interact with and modulate other host surface receptors and factors (Figure 3).
Vpu induces the rapid degradation of newly synthesized CD4 receptor molecules that are retained together with Env precursor protein (gp160) in the endoplasmic reticulum[13]. The cytoplasmic domain of Vpu and the DSGxxS motif are critical in interaction with and degradation of CD4, respectively[12,63] (Figure 2). The degradation process is achieved by Vpu recruiting β-TrCP and then interacts with CD4 cytoplasmic domain and subsequently subject CD4 to degradation by the ubiquitin-proteasome pathway[11,64]. In doing so Vpu contributes to the suppression of HIV-1 primary receptor at the surface of the infected cell.
Enhancement of virion release by Vpu has been shown to be achieved through antagonizing tetherin/BST-2, an IFN regulated host restriction factor. BST-2 directly binds to virions and hence retains them on the surface of infected cells[61,62]. Vpu through AxxxAxxxA motif in transmembrane domain directly interacts with BST-2 transmembrane domain, the Vpu DSGxxS and [D/E]XXXL[L/I/V] motifs in the cytoplasmic domain also play crucial role in ensuring BST-2 downmodulation[15,65,66] (Figure 2). Previous studies indicated BST-2 downmodulation is through β-TrCP-dependent proteasomal degradation pathway[67] while others suggested the β-TrCP-dependent endo-lysosomal pathway[65,68]. In contrast, recent studies showed that BST-2 antagonistic activity by Vpu takes place in the trans-Golgi networks (TGN)[14]. Vpu interferes with anterograde transport of BST-2 to the cell surface subsequently leading to BST-2 trapping in the TGN[15-17,69].
Recent studies have indicated that Vpu is emerging as a viral factor with a range of activities devoted to counteracting host innate and adaptive immunity including the modulation of NK cell co-activation ligand NK-T and B cell antigen (NTB-A)[70], PVR activating ligand of NK cells[71], and CD1d[72,73] (Figure 3).
NTB-A triggering is necessary for induction of efficient lysis of target cells upon engagement of the activating receptor NKG2D[74]. The Ser-52 and Ser-56 residues important for CD4 and BST-2 degradation did not affect NTB-A expression, indicating that the down modulation of NTB-A by Vpu is mediated by different domains[70]. A recent study has shown that downmodulation of NTB-A is achieved by Vpu interfering with the anterograde transport of NTB-A by retaining it within the Golgi compartment and hence affects its glycosylation pattern that subsequently reduces surface expression of NTB-A[75].
PVR (CD155, Necl-5) is a ligand for the activating receptor DNAM-1 (CD226) expressed by NK cells[76,77]. PVR downmodulation by Nef and Vpu is another strategy evolved by HIV-1 to avoid NK cell-mediated lysis of infected cells[71]. PVR downregulation alters multiple important PVR-mediated innate cellular immune processes such as adhesion and migration, and therefore may influence HIV-1 pathogenesis.
CD1d molecules are important in dendritic cells for lipid antigen presentation to CD1d-restricted NKT cells[78,79]. CD1d and CD1d-restricted NKT cells are present at pathogen entry sites thus play a crucial role in early immune responses[80]. Vpu has been shown to be the major viral factor that inhibit recycling of CD1d from the endosomal compartment back to cell surface through retaining CD1d in early endosomes[72].
Vpu has also been implicated in inhibition of ubiquitination and degradation of p53 (a substrate of SCFβ-TrCP ligase complex). The successful interaction of SCFβ-TrCP complex with β-TrCP binding motif (DS52GNES56) present in Vpu has been shown to be essential[81]. It was observed that Vpu mutants with alanine substitutions (DA52GNEA56) failed to stabilize p53 and did not prevent its ubiquitination. This suggested that Vpu is able to achieve modulation of p53 through competing efficiently with p53 protein for the β-TrCP subunit of the SCF complex and hence inhibits subsequent ubiquitination of p53 protein. The modulation of p53 positively correlated with apoptosis during the late stages of HIV-1 infection[81].
Finally, although Vpu showed multiple functions in vitro and ex vivo, it is yet clear how and what functions of Vpu are important in viral pathogenesis in vivo.
The current knowledge on factors that are attributed to Vpu polymorphism has not been quite sufficient; therefore this prompt for further analysis to reveal the unresolved questions of why Vpu is so variable and what factors drive Vpu polymorphism. In order to define the complex dynamics of HIV-1 Vpu evolution, immune escape patterns, and functional adaptation during the course of infection, further insight is needed on the role of host genetics and other immune selection pressures towards shaping HIV-1 Vpu diversity. The emergence of advanced DNA sequencing technologies such as ultra-deep sequence which is superior and more sensitive than Sanger sequence methods has made it possible to accurately detect and analyze minor variants of HIV-1 within a host[82-85]. Furthermore, the establishment of different contemporaneous cohorts of HIV-1-infected individuals worldwide enables us to examine to what extent the host immune components play a role on viral adaptation and/or evolution at both intra- and inter-patients’ level.
So far the current studies have indicated that the host immune responses directed towards Vpu is not entirely attributable to HIV-1 Vpu variation (Figure 2), it is therefore crucial to apprehend other factors that may explain Vpu variation. Of note previous studies have identified immune responses directed towards Vpu, using peptides of HIV-1 consensus sequences[45,57]. However, ironically due to Vpu polymorphic nature itself, these results may mask the exact extent to which immune responses contribute to Vpu sequence variation. Alternatively, HIV-1 like other RNA viruses has evolved to shorten its genome length through overlapping its genes[86]. The overlapping region of Vpu and Env is one of promising aspect to consider when we focus on Vpu variation. Because host immune responses (neutralizing antibodies) contribute to Env polymorphic nature[87,88], it is enticing to assume that immune responses directed towards Env may influence Vpu polymorphisms through Vpu-Env overlapping region. KIR associated polymorphisms within Vpu-Env overlapping region have been reported previously[44]. Although it is still unknown whether NK cells recognize Vpu or Env protein, nonetheless these findings indicate the importance of this region for Vpu variability. Furthermore, it is reported that X4- and R5-tropic HIV-1 showed differential amino acid polymorphisms in Vpu[89], suggesting that cellular compartment influences Vpu variability.
The current increase in number of new findings of Vpu from pandemic HIV-1 group M strain and other HIV-1 strains, enlighten us the precise role or mechanisms of how Vpu degrade the viral receptor CD4, antagonize tetherin/BST-2, enhance p53 stability and modulate NK-cell activities through modulation of PVR, NTB-A and CD1d receptors (Figure 3). Understanding the mode of action of Vpu and association of the immune factors certainly open plenty of new windows to deciphering the intricate mechanisms associated with HIV-1 immune pathogenesis in vivo. Also, understanding pathways of Vpu intra- and inter-patients sequence variability and adaptation may provide us with an alternative approach for prospects of viral persistence and Vpu contributions in vivo.
The authors wish to thank the Ministry of Education, Science, Sports, and Culture (MEXT) of Japan and the Ministry of Health, Labor, and Welfare of Japan for their grant-in aid for the AIDS research. We also wish to thank Dr. J Carlson (Microsoft Research, Redmond, Washington, United States) and M. Mahiti (Kumamoto University, Japan) for their helpful discussion.
P- Reviewer: Brett TJ, Fackler OT S- Editor: Wen LL L- Editor: A E- Editor: Wang CH
| 1. | Ji JP, Loeb LA. Fidelity of HIV-1 reverse transcriptase copying RNA in vitro. Biochemistry. 1992;31:954-958. [PubMed] [Cited in This Article: ] |
| 2. | Shriner D, Rodrigo AG, Nickle DC, Mullins JI. Pervasive genomic recombination of HIV-1 in vivo. Genetics. 2004;167:1573-1583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 3. | Ramratnam B, Bonhoeffer S, Binley J, Hurley A, Zhang L, Mittler JE, Markowitz M, Moore JP, Perelson AS, Ho DD. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet. 1999;354:1782-1785. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 391] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 4. | Snoeck J, Fellay J, Bartha I, Douek DC, Telenti A. Mapping of positive selection sites in the HIV-1 genome in the context of RNA and protein structural constraints. Retrovirology. 2011;8:87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 5. | O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565-574. [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 245] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 6. | An P, Winkler CA. Host genes associated with HIV/AIDS: advances in gene discovery. Trends Genet. 2010;26:119-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | Brumme ZL, Walker BD. Tracking the culprit: HIV-1 evolution and immune selection revealed by single-genome amplification. J Exp Med. 2009;206:1215-1218. [PubMed] [Cited in This Article: ] |
| 8. | Brumme ZL, Art FYP, Jonathan MC, Walkerd BD. Identifying HLA-Associated Polymorphisms in HIV-1. HIV Molecular Immunology 2010. Los Alamos, New Mexico: Los Alamos National Laboratory, Theoretical Biology and Biophysics 2010; 3-16. [Cited in This Article: ] |
| 9. | Moore CB, John M, James IR, Christiansen FT, Witt CS, Mallal SA. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 2002;296:1439-1443. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 597] [Cited by in F6Publishing: 584] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 10. | Maldarelli F, Chen MY, Willey RL, Strebel K. Human immunodeficiency virus type 1 Vpu protein is an oligomeric type I integral membrane protein. J Virol. 1993;67:5056-5061. [PubMed] [Cited in This Article: ] |
| 11. | Schubert U, Antón LC, Bacík I, Cox JH, Bour S, Bennink JR, Orlowski M, Strebel K, Yewdell JW. CD4 glycoprotein degradation induced by human immunodeficiency virus type 1 Vpu protein requires the function of proteasomes and the ubiquitin-conjugating pathway. J Virol. 1998;72:2280-2288. [PubMed] [Cited in This Article: ] |
| 12. | Tiganos E, Yao XJ, Friborg J, Daniel N, Cohen EA. Putative alpha-helical structures in the human immunodeficiency virus type 1 Vpu protein and CD4 are involved in binding and degradation of the CD4 molecule. J Virol. 1997;71:4452-4460. [PubMed] [Cited in This Article: ] |
| 13. | Willey RL, Maldarelli F, Martin MA, Strebel K. Human immunodeficiency virus type 1 Vpu protein regulates the formation of intracellular gp160-CD4 complexes. J Virol. 1992;66:226-234. [PubMed] [Cited in This Article: ] |
| 14. | Dubé M, Roy BB, Guiot-Guillain P, Mercier J, Binette J, Leung G, Cohen EA. Suppression of Tetherin-restricting activity upon human immunodeficiency virus type 1 particle release correlates with localization of Vpu in the trans-Golgi network. J Virol. 2009;83:4574-4590. [PubMed] [Cited in This Article: ] |
| 15. | Dubé M, Roy BB, Guiot-Guillain P, Binette J, Mercier J, Chiasson A, Cohen EA. Antagonism of tetherin restriction of HIV-1 release by Vpu involves binding and sequestration of the restriction factor in a perinuclear compartment. PLoS Pathog. 2010;6:e1000856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 176] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 16. | Dube M, Paquay C, Roy BB, Bego MG, Mercier J, Cohen EA. HIV-1 Vpu Antagonizes BST-2 by Interfering Mainly with the Trafficking of Newly Synthesized BST-2 to the Cell Surface. Traffic. 2011;12:1714-1729. [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 17. | Schmidt S, Fritz JV, Bitzegeio J, Fackler OT, Keppler OT. HIV-1 Vpu blocks recycling and biosynthetic transport of the intrinsic immunity factor CD317/tetherin to overcome the virion release restriction. MBio. 2011;2:e00036-e00011. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 18. | Sauter D, Unterweger D, Vogl M, Usmani SM, Heigele A, Kluge SF, Hermkes E, Moll M, Barker E, Peeters M. Human tetherin exerts strong selection pressure on the HIV-1 group N Vpu protein. PLoS Pathog. 2012;8:e1003093. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 19. | Brumme ZL, John M, Carlson JM, Brumme CJ, Chan D, Brockman MA, Swenson LC, Tao I, Szeto S, Rosato P. HLA-associated immune escape pathways in HIV-1 subtype B Gag, Pol and Nef proteins. PLoS One. 2009;4:e6687. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 138] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 20. | Carlson JM, Brumme CJ, Martin E, Listgarten J, Brockman MA, Le AQ, Chui CK, Cotton LA, Knapp DJ, Riddler SA. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol. 2012;86:13202-13216. [PubMed] [Cited in This Article: ] |
| 21. | Ueno T, Motozono C, Dohki S, Mwimanzi P, Rauch S, Fackler OT, Oka S, Takiguchi M. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J Immunol. 2008;180:1107-1116. [PubMed] [Cited in This Article: ] |
| 22. | Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1944] [Cited by in F6Publishing: 1879] [Article Influence: 89.5] [Reference Citation Analysis (0)] |
| 23. | Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144-4149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 938] [Cited by in F6Publishing: 910] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 24. | Yusim K, Kesmir C, Gaschen B, Addo MM, Altfeld M, Brunak S, Chigaev A, Detours V, Korber BT. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J Virol. 2002;76:8757-8768. [PubMed] [Cited in This Article: ] |
| 25. | Rossenkhan R, Novitsky V, Sebunya TK, Musonda R, Gashe BA, Essex M. Viral diversity and diversification of major non-structural genes vif, vpr, vpu, tat exon 1 and rev exon 1 during primary HIV-1 subtype C infection. PLoS One. 2012;7:e35491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 26. | Pickering S, Hué S, Kim EY, Reddy S, Wolinsky SM, Neil SJ. Preservation of tetherin and CD4 counter-activities in circulating Vpu alleles despite extensive sequence variation within HIV-1 infected individuals. PLoS Pathog. 2014;10:e1003895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 48] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Matsuda Z, Chou MJ, Matsuda M, Huang JH, Chen YM, Redfield R, Mayer K, Essex M, Lee TH. Human Immunodeficiency Virus Type-1 Has an Additional Coding Sequence in the Central Region of the Genome. P Natl Acad Sci USA. 1988;85:6968-6972. [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 63] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Schneider T, Hildebrandt P, Rönspeck W, Weigelt W, Pauli G. The antibody response to the HIV-1 specific “out” (vpu) protein: identification of an immunodominant epitope and correlation of antibody detectability to clinical stages. AIDS Res Hum Retroviruses. 1990;6:943-950. [PubMed] [Cited in This Article: ] |
| 29. | Reiss P, Lange JMA, Deronde A, Dewolf F, Dekker J, Danner SA, Debouck C, Goudsmit J. Antibody-Response to Viral Protein-U (Vpu) and Protein-R (Vpr) in Hiv-1-Infected Individuals. J Acq Immun Def Synd. 1990;3:115-122. [Cited in This Article: ] |
| 30. | Kusk P, Lindhardt BO, Bugge TH, Holmbäck K, Hulgaard EF. Mapping of a new immunodominant human linear B-cell epitope on the vpu protein of the human immunodeficiency virus type 1. J Acquir Immune Defic Syndr. 1993;6:334-338. [PubMed] [Cited in This Article: ] |
| 31. | Chen YM, Rey WY, Lan YC, Lai SF, Huang YC, Wu SI, Liu TT, Hsiao KJ. Antibody reactivity to HIV-1 Vpu in HIV-1/AIDS patients on highly active antiretroviral therapy. J Biomed Sci. 2003;10:266-275. [PubMed] [Cited in This Article: ] |
| 32. | Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991-998. [PubMed] [Cited in This Article: ] |
| 33. | Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857-860. [PubMed] [Cited in This Article: ] |
| 34. | Altfeld M, Rosenberg ES. The role of CD4(+) T helper cells in the cytotoxic T lymphocyte response to HIV-1. Curr Opin Immunol. 2000;12:375-380. [PubMed] [Cited in This Article: ] |
| 35. | Streeck H, Nixon DF. T cell immunity in acute HIV-1 infection. J Infect Dis. 2010;202 Suppl 2:S302-S308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 36. | Liu Y, McNevin JP, Holte S, McElrath MJ, Mullins JI. Dynamics of viral evolution and CTL responses in HIV-1 infection. PLoS One. 2011;6:e15639. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 37. | Klein MR, van Baalen CA, Holwerda AM, Kerkhof Garde SR, Bende RJ, Keet IP, Eeftinck-Schattenkerk JK, Osterhaus AD, Schuitemaker H, Miedema F. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J Exp Med. 1995;181:1365-1372. [PubMed] [Cited in This Article: ] |
| 38. | Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406-416. [PubMed] [Cited in This Article: ] |
| 39. | Miura T, Brockman MA, Schneidewind A, Lobritz M, Pereyra F, Rathod A, Block BL, Brumme ZL, Brumme CJ, Baker B. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphocyte [corrected] recognition. J Virol. 2009;83:2743-2755. [PubMed] [Cited in This Article: ] |
| 40. | Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797-1800. [PubMed] [Cited in This Article: ] |
| 41. | Brumme ZL, Brumme CJ, Chui C, Mo T, Wynhoven B, Woods CK, Henrick BM, Hogg RS, Montaner JS, Harrigan PR. Effects of human leukocyte antigen class I genetic parameters on clinical outcomes and survival after initiation of highly active antiretroviral therapy. J Infect Dis. 2007;195:1694-1704. [PubMed] [Cited in This Article: ] |
| 42. | Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. [PubMed] [Cited in This Article: ] |
| 43. | Sewell AK, Price DA, Oxenius A, Kelleher AD, Phillips RE. Cytotoxic T lymphocyte responses to human immunodeficiency virus: control and escape. Stem Cells. 2000;18:230-244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 44. | Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96-100. [PubMed] [Cited in This Article: ] |
| 45. | Addo MM, Altfeld M, Rathod A, Yu M, Yu XG, Goulder PJ, Rosenberg ES, Walker BD. HIV-1 Vpu represents a minor target for cytotoxic T lymphocytes in HIV-1-infection. AIDS. 2002;16:1071-1073. [PubMed] [Cited in This Article: ] |
| 46. | Addo MM, Yu XG, Rosenberg ES, Walker BD, Altfeld M. Cytotoxic T-lymphocyte (CTL) responses directed against regulatory and accessory proteins in HIV-1 infection. DNA Cell Biol. 2002;21:671-678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 43] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 47. | Kloverpris H, Karlsson I, Bonde J, Thorn M, Vinner L, Pedersen AE, Hentze JL, Andresen BS, Svane IM, Gerstoft J. Induction of novel CD8+ T-cell responses during chronic untreated HIV-1 infection by immunization with subdominant cytotoxic T-lymphocyte epitopes. AIDS. 2009;23:1329-1340. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Corbet S, Nielsen HV, Vinner L, Lauemoller S, Therrien D, Tang S, Kronborg G, Mathiesen L, Chaplin P, Brunak S. Optimization and immune recognition of multiple novel conserved HLA-A2, human immunodeficiency virus type 1-specific CTL epitopes. J Gen Virol. 2003;84:2409-2421. [PubMed] [Cited in This Article: ] |
| 49. | Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, Reddy S, de Pierres C, Mncube Z, Mkhwanazi N. CD8( ) T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46-53. [DOI] [Cited in This Article: ] [Cited by in Crossref: 811] [Cited by in F6Publishing: 830] [Article Influence: 46.1] [Reference Citation Analysis (0)] |
| 50. | Hasan Z, Carlson JM, Gatanaga H, Le AQ, Brumme CJ, Oka S, Brumme ZL, Ueno T. Minor contribution of HLA class I-associated selective pressure to the variability of HIV-1 accessory protein Vpu. Biochem Biophys Res Commun. 2012;421:291-295. [PubMed] [Cited in This Article: ] |
| 51. | Altfeld M, Addo MM, Eldridge RL, Yu XG, Thomas S, Khatri A, Strick D, Phillips MN, Cohen GB, Islam SA. Vpr is preferentially targeted by CTL during HIV-1 infection. J Immunol. 2001;167:2743-2752. [Cited in This Article: ] |
| 52. | Lucchiari M, Niedermann G, Leipner C, Meyerhans A, Eichmann K, Maier B. Human immune response to HIV-1-Nef. I. CD45RO- T lymphocytes of non-infected donors contain cytotoxic T lymphocyte precursors at high frequency. Int Immunol. 1994;6:1739-1749. [PubMed] [Cited in This Article: ] |
| 53. | Novitsky V, Cao H, Rybak N, Gilbert P, McLane MF, Gaolekwe S, Peter T, Thior I, Ndung’u T, Marlink R. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J Virol. 2002;76:10155-10168. [PubMed] [Cited in This Article: ] |
| 54. | Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, Streeck H, Waring M, Meier A, Brander C. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027-3036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 333] [Cited by in F6Publishing: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 55. | Funke J, Dürr R, Dietrich U, Koch J. Natural killer cells in HIV-1 infection: a double-edged sword. AIDS Rev. 2011;13:67-76. [PubMed] [Cited in This Article: ] |
| 56. | Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med. 2009;265:29-42. [DOI] [Cited in This Article: ] [Cited by in Crossref: 138] [Cited by in F6Publishing: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 57. | Stratov I, Chung A, Kent SJ. Robust NK cell-mediated human immunodeficiency virus (HIV)-specific antibody-dependent responses in HIV-infected subjects. J Virol. 2008;82:5450-5459. [PubMed] [Cited in This Article: ] |
| 58. | Alsmadi O, Herz R, Murphy E, Pinter A, Tilley SA. A novel antibody-dependent cellular cytotoxicity epitope in gp120 is identified by two monoclonal antibodies isolated from a long-term survivor of human immunodeficiency virus type 1 infection. J Virol. 1997;71:925-933. [PubMed] [Cited in This Article: ] |
| 59. | Yamada T, Watanabe N, Nakamura T, Iwamoto A. Antibody-dependent cellular cytotoxicity via humoral immune epitope of Nef protein expressed on cell surface. J Immunol. 2004;172:2401-2406. [PubMed] [Cited in This Article: ] |
| 60. | Nomaguchi M, Fujita M, Adachi A. Role of HIV-1 Vpu protein for virus spread and pathogenesis. Microbes Infect. 2008;10:960-967. [PubMed] [Cited in This Article: ] |
| 61. | Dubé M, Bego MG, Paquay C, Cohen ÉA. Modulation of HIV-1-host interaction: role of the Vpu accessory protein. Retrovirology. 2010;7:114. [PubMed] [Cited in This Article: ] |
| 62. | Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425-430. [PubMed] [Cited in This Article: ] |
| 63. | Vincent MJ, Raja NU, Jabbar MA. Human immunodeficiency virus type 1 Vpu protein induces degradation of chimeric envelope glycoproteins bearing the cytoplasmic and anchor domains of CD4: role of the cytoplasmic domain in Vpu-induced degradation in the endoplasmic reticulum. J Virol. 1993;67:5538-5549. [PubMed] [Cited in This Article: ] |
| 64. | Bour S, Schubert U, Strebel K. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J Virol. 1995;69:1510-1520. [PubMed] [Cited in This Article: ] |
| 65. | Mitchell RS, Katsura C, Skasko MA, Fitzpatrick K, Lau D, Ruiz A, Stephens EB, Margottin-Goguet F, Benarous R, Guatelli JC. Vpu antagonizes BST-2-mediated restriction of HIV-1 release via beta-TrCP and endo-lysosomal trafficking. PLoS Pathog. 2009;5:e1000450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 263] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 66. | Rong L, Zhang J, Lu J, Pan Q, Lorgeoux RP, Aloysius C, Guo F, Liu SL, Wainberg MA, Liang C. The transmembrane domain of BST-2 determines its sensitivity to down-modulation by human immunodeficiency virus type 1 Vpu. J Virol. 2009;83:7536-7546. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 67. | Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 68. | Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. HIV-1 Vpu neutralizes the antiviral factor Tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 224] [Cited by in F6Publishing: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 69. | Hauser H, Lopez LA, Yang SJ, Oldenburg JE, Exline CM, Guatelli JC, Cannon PM. HIV-1 Vpu and HIV-2 Env counteract BST-2/tetherin by sequestration in a perinuclear compartment. Retrovirology. 2010;7:51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 143] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 70. | Shah AH, Sowrirajan B, Davis ZB, Ward JP, Campbell EM, Planelles V, Barker E. Degranulation of natural killer cells following interaction with HIV-1-infected cells is hindered by downmodulation of NTB-A by Vpu. Cell Host Microbe. 2010;8:397-409. [PubMed] [Cited in This Article: ] |
| 71. | Matusali G, Potestà M, Santoni A, Cerboni C, Doria M. The human immunodeficiency virus type 1 Nef and Vpu proteins downregulate the natural killer cell-activating ligand PVR. J Virol. 2012;86:4496-4504. [PubMed] [Cited in This Article: ] |
| 72. | Moll M, Andersson SK, Smed-Sörensen A, Sandberg JK. Inhibition of lipid antigen presentation in dendritic cells by HIV-1 Vpu interference with CD1d recycling from endosomal compartments. Blood. 2010;116:1876-1884. [PubMed] [Cited in This Article: ] |
| 73. | Kelly H, Mandraju R, Coelho-dos-Reis JG, Tsuji M. Effects of HIV-1-induced CD1c and CD1d modulation and endogenous lipid presentation on CD1c-restricted T-cell activation. BMC Immunol. 2013;14:4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 74. | Flaig RM, Stark S, Watzl C. Cutting edge: NTB-A activates NK cells via homophilic interaction. J Immunol. 2004;172:6524-6527. [PubMed] [Cited in This Article: ] |
| 75. | Bolduan S, Hubel P, Reif T, Lodermeyer V, Höhne K, Fritz JV, Sauter D, Kirchhoff F, Fackler OT, Schindler M. HIV-1 Vpu affects the anterograde transport and the glycosylation pattern of NTB-A. Virology. 2013;440:190-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 76. | Shibuya A, Campbell D, Hannum C, Yssel H, Franz-Bacon K, McClanahan T, Kitamura T, Nicholl J, Sutherland GR, Lanier LL. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573-581. [PubMed] [Cited in This Article: ] |
| 77. | Takai Y, Miyoshi J, Ikeda W, Ogita H. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat Rev Mol Cell Biol. 2008;9:603-615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 394] [Cited by in F6Publishing: 417] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 78. | Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525-529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 873] [Cited by in F6Publishing: 863] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 79. | Tupin E, Kinjo Y, Kronenberg M. The unique role of natural killer T cells in the response to microorganisms. Nat Rev Microbiol. 2007;5:405-417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 346] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 80. | Kawana K, Matsumoto J, Miura S, Shen L, Kawana Y, Nagamatsu T, Yasugi T, Fujii T, Yang H, Quayle AJ. Expression of CD1d and ligand-induced cytokine production are tissue specific in mucosal epithelia of the human lower reproductive tract. Infect Immun. 2008;76:3011-3018. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 81. | Verma S, Ali A, Arora S, Banerjea AC. Inhibition of β-TrcP-dependent ubiquitination of p53 by HIV-1 Vpu promotes p53-mediated apoptosis in human T cells. Blood. 2011;117:6600-6607. [PubMed] [Cited in This Article: ] |
| 82. | Bushman FD, Hoffmann C, Ronen K, Malani N, Minkah N, Rose HM, Tebas P, Wang GP. Massively parallel pyrosequencing in HIV research. AIDS. 2008;22:1411-1415. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 57] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 83. | Bimber BN, Burwitz BJ, O’Connor S, Detmer A, Gostick E, Lank SM, Price DA, Hughes A, O’Connor D. Ultradeep pyrosequencing detects complex patterns of CD8+ T-lymphocyte escape in simian immunodeficiency virus-infected macaques. J Virol. 2009;83:8247-8253. [PubMed] [Cited in This Article: ] |
| 84. | Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 287] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 85. | Henn MR, Boutwell CL, Charlebois P, Lennon NJ, Power KA, Macalalad AR, Berlin AM, Malboeuf CM, Ryan EM, Gnerre S. Whole genome deep sequencing of HIV-1 reveals the impact of early minor variants upon immune recognition during acute infection. PLoS Pathog. 2012;8:e1002529. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 271] [Cited by in F6Publishing: 287] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 86. | Chirico N, Vianelli A, Belshaw R. Why genes overlap in viruses. Proc Biol Sci. 2010;277:3809-3817. [PubMed] [Cited in This Article: ] |
| 87. | Fenyö EM, Albert J, McKeating J. The role of the humoral immune response in HIV infection. AIDS. 1996;10 Suppl A:S97-106. [PubMed] [Cited in This Article: ] |
| 88. | McKeating JA. Biological consequences of human immunodeficiency virus type 1 envelope polymorphism: does variation matter? 1995 Fleming Lecture. J Gen Virol. 1996;77:2905-2919. [PubMed] [Cited in This Article: ] |
| 89. | Dimonte S, Babakir-Mina M, Aquaro S, Perno CF. Specific VpU codon changes were significantly associated with gp120 V3 tropic signatures in HIV-1 B-subtype. Virol Sin. 2012;27:360-368. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |