Published online May 26, 2014. doi: 10.4330/wjc.v6.i5.327
Revised: February 10, 2014
Accepted: April 16, 2014
Published online: May 26, 2014
AIM: To provide an updated review on current genetic aspects possibly affecting essential hypertension (EH), and to further elucidate their role in EH.
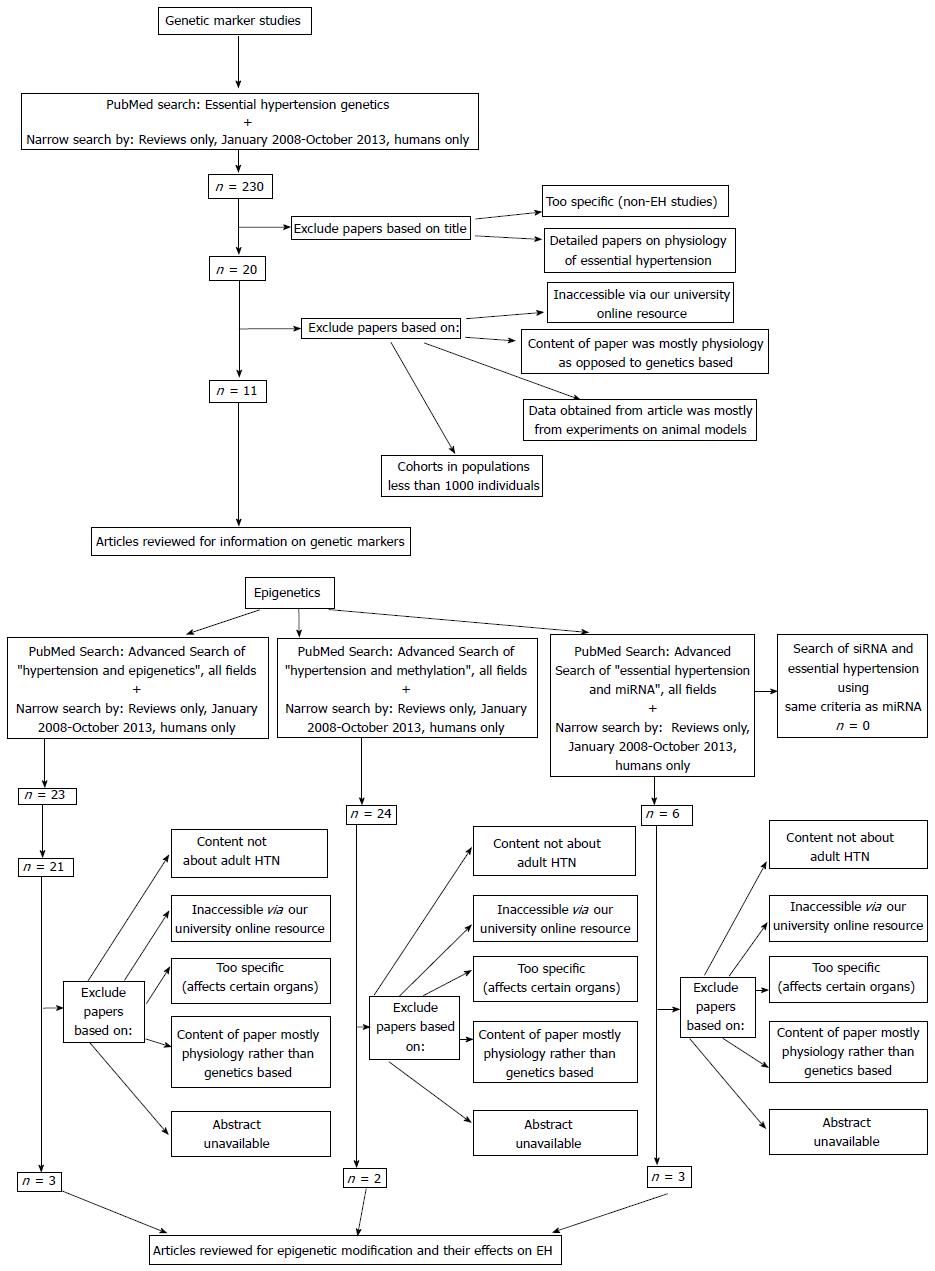
METHODS: We searched for genetic and epigenetic factors in major studies associated with EH between Jan 2008-Oct 2013 using PubMed. We limited our search to reviews that discussed mostly human studies, and were accessible through the university online resource. We found 11 genome wide association studies (GWAS), as well as five methylation and three miRNA studies that fit our search criteria. A distinction was not made between genes with protective effects or negative effects, as this article is only meant to be a summary of genes associated with any aspect of EH.
RESULTS: We found 130 genes from the studies that met our inclusion/exclusion criteria. Of note, genes with multiple study references include: STK39, CYP17A1, MTHFR-NPPA, MTHFR-NPPB, ATP2B1, CSK, ZNF652, UMOD, CACNB2, PLEKHA7, SH2B3, TBX3-TBX5, ULK4, CSK-ULK3, CYP1A2, NT5C2, CYP171A, PLCD3, SH2B3, ATXN2, CACNB2, PLEKHA7, SH2B3, TBX3-TBX5, ULK4, and HFE. The following genes overlapped between the genetic studies and epigenetic studies: WNK4 and BDKRB2. Several of the identified genes were found to have functions associated with EH. Many epigenetic factors were also correlated with EH. Of the epigenetic factors, there were no articles discussing siRNA and its effects on EH that met the search criteria, thus the topic was not included in this review. Among the miRNA targets found to be associated with EH, many of the genes involved were also identified in the GWAS studies.
CONCLUSION: Genetic hypertension risk algorithms could be developed in the future but may be of limited benefit due to the multi-factorial nature of EH. With emerging technologies, like next-generation sequencing, more direct causal relationships between genetic and epigenetic factors affecting EH will likely be discovered creating a tremendous potential for personalized medicine using pharmacogenomics.
Core tip: Essential hypertension (EH) is considered a multifactorial disease, indicating that many genetic, epigenetic, and environmental influences affect the initiation and continuance of the disease. Our goal is to provide an updated report on current genetic aspects possibly affecting EH by elucidating genetic factors’ role in EH. We found 130 genes meeting our inclusion/exclusion criteria. To our knowledge, this is the first review to discuss both genetic and epigenetic factors associated with EH in one article. With emerging technologies, more direct causal relationships between genetic and epigenetic factors with EH will likely be disdiscovered, creating tremendous potential for personalized medicine using pharmacogenomics.
- Citation: Natekar A, Olds RL, Lau MW, Min K, Imoto K, Slavin TP. Elevated blood pressure: Our family’s fault? The genetics of essential hypertension. World J Cardiol 2014; 6(5): 327-337
- URL: https://www.wjgnet.com/1949-8462/full/v6/i5/327.htm
- DOI: https://dx.doi.org/10.4330/wjc.v6.i5.327
Approximately 1 in 3 American adults, or about 67 million people, have hypertension (HTN)[1]. According to the American Heart Association, the majority of Americans who have had first heart attacks, first strokes, or chronic heart failure had underlying HTN, a known risk factor for each condition[2]. HTN costs the United States approximately $47.5 billion annually in direct medical costs and roughly $3.5 billion annually in lost economic productivity[3].
Essential hypertension (EH), the most common form of HTN[4], is defined as an elevation in blood pressure of unknown cause and increases the risks for cerebral, cardiac, and renal complications[5]. EH is thought to be a multifactorial disease, indicating that many factors affect the initiation and continuance of the disease[6]. From a genetic perspective, many single nucleotide polymorphisms (SNPs), genes and epigenetic factors are associated with EH. This suggests that people with these hereditary factors might have a genetic predisposition to having high blood pressure. Additionally, since EH has idiopathic origins, environmental factors may also play an important role in the cause of the disease. Weight gain and dietary factors appear to have a major role in causing EH due to impaired renal function, though the mechanisms are not well understood[7].
There has been some discussion on the common disease, common variant (CDCV) and common disease, rare variant (CDRV) hypotheses and their relation to complex diseases, such as EH[8]. The CDCV hypothesis predicts that there are common disease-producing alleles/variants that are found in all human populations with a particular phenotype for a certain disease. However, insufficient data has led to scientists challenging the validity of this hypothesis and its compatibility with many diseases[9]. Meanwhile, the CDRV hypothesis predicts that diseases with genetic predispositions may not be found commonly in the diseased human population[10]. One study argued that with human lineage, diseases were more likely to favor multiple rare variations contributing to disease, rather than common variations contributing to disease[11]. This is because common variations might have external factors that would have eliminated these genes from the population, while rare variants are new, contributing to disease[12].
The purpose of this article is to provide an updated report on the current genetic aspects that could affect EH, and to further elucidate the role of genetic factors in EH. This includes summarizing genome-wide association studies (GWAS), as well as studies that identified genes with specific physiological functions. We also summarize current knowledge of the epigenetics in EH and/or HTN.
Since genetic factors that influence EH in the literature are broad, we looked at specific categories of genetic factors and their influence on EH. Genetic marker studies were chosen since these studies looked specifically at what genes were involved with EH, and if any had specific physiologic effects. As epigenetics has become an emerging field of interest in genetics, DNA modification related to EH is also included, specifically focusing on DNA methylation and RNA regulation studies. It is important to note that a distinction was not made between genes with protective effects or negative effects, as this article is only meant to be a summary of genes associated with any aspect of EH.
For the search criteria, specific keywords used for each category of genetic and epigenetic factors are listed below in Figure 1.
Reviews were selected if there was a primary focus on the genes and genetic factors associated with EH. Additionally, reviews between Jan 2008-Oct 2013 were chosen to obtain the most current information. Reviews were selected that discussed human studies, with little if any focus on animal studies. Reviews were also included if there was discussion of non-European populations since EH affects many ethnicities. Lastly, the results reported from the selected reviews were limited to reviews that discussed cohorts in populations greater than 1000 individuals. Cohorts with populations > 1000 people were chosen to reduce selection bias within the primary studies, and to ensure that the genes found could apply to large populations. From the articles that were selected to be in the study, the authors identified if the genes had known pathways related to EH.
For epigenetic factors associated with EH, we included articles that discussed various epigenetic modifications and their physiologic effects, as well as specific techniques such as methylation. If the studies had relevant animal data, this was included due to the fact that there is limited epigenetic information in human studies. Articles that discussed miRNA and the association with EH were also included to ensure a more thorough gathering of data. No articles for siRNA met our search criteria. Therefore, a discussion on siRNA as it relates to EH is not provided in this article.
Reviews were excluded if the reviews involved rare types of HTN and/or were too detailed on EH physiology. While EH physiology is important, it does not contribute to the purpose of this paper in understanding the genetic basis for EH. Additionally, reviews were eliminated if the articles were inaccessible or the reviews focused on animal models. Genome-wide linkage studies were also exxcluded, as there was no consistency in the results for genetic markers associated with EH. Also, articles were excluded if their abstracts were unavailable. Lastly, articles were excluded if there was no access available through the library at the University of Hawaii medical school.
A total of 11 genetic marker studies (genome-wide association studies) are found to contain relevant information with regards to gene associations with EH. Many of the studies identify genes within cohorts, and there are some genes identified in multiple cohorts. These can be found from references[12-21], identified in Table 1. Furthermore, some of the genes have specific phenotypic effects, or associate with other genes and/or proteins related to EH. Some of the genes found have no known function, or the authors do not list the function. These can be found in references[12-21], identified in Table 2. Genes listed with hyphens include all of the genes found inclusive of, and between, the genetic range listed.
| Cohort | Genes |
| Framingham offspring cohort | CCL20-WDR69, CDH13, TGFBR2, STK39 |
| Amish cohort | STK39 |
| AGEN | NPR3, CYP17A1, FGF5, MTHFR, NPPA, NPPB, ATP2B1, CSK, ZNF652 |
| BP-extremes | UMOD |
| BRIGHT | BCAT1 |
| CARe | c21orf91, GPR98 and ARRDC3 |
| CBPgen | CYP17A1, CACNB2, PLEKHA7, SH2B3, TBX3, TBX 4, TBX5, ULK4 |
| CHARGE | CPLX3, PLEKHA7, TBX3, UMOD, CYP17A1, CSK-ULK3, CYP1A2, NT5C2, CYP171A, PLCD3, SH2B3-ATXN2, CACNB2, SH2B3, TBX3, TBX4, TBX5, ULK4, c10orf107, BLK-GATA4, CASZ1, FGF5, MTHFR, NPPA, NPPB, ATP2B1, CSK |
| FHS | ANKMY, FOXD3 |
| GBPgen | UMOD, CSK-ULK3, CYP1A2, NT5C2, CYP171A, PLCD3, SH2B3-ATXN2,ATXN2, c10orf107, GNAS-EDN3, MECOM (MDS1 locus), FGF5, MTHFR, NPPA, NPPB, ATP2B1, CSK, ZNF652 |
| GENE-centric | SOX6, AGT, LSP1-TNNT3, MTHFR, NPPA, NPPB, ATP2B1, HFE |
| Health2 | ATP2B1 |
| HUFS | IPO7, MYLIP, PMS1, SLC24A4, YWHAZ, CACANA1H |
| Hypergenes | NOS3 |
| ICBP | ADAMTS-8, ADM, BAT2-BAT5, CHIC2-PDGFRA1, EBF1, FES, FIGN, FLJ32810-TMEM133, GOSR2, GUCY1A3-GUCY1B3, JAG1, MOV10, NOV, NPR3-c5orf23, PIK3CG, PLCE1, SLC39A8, SLC4A7, NPR3, CYP17A1, CACNB2, PLEKHA7, SH2B3, TBX3-TBX5, ULK4, GNAS-EDN3, MECOM (MDS1 locus), FGF5, MTHFR, NPPA, NPPB, ATP2B1, CSK, ZNF652, HFE |
| KARE | ATP2B1 |
| KORA S3 | CCNG1 |
| Suita study | CCBE1 |
| WGHS | BLK-GATA4, CASZ1 |
| Study reference not mentioned in article | ADD1, ADD2, ADRB1, ADRB2, APOB, CACNA1A, CACNA1C, CLCNKB, CYBA, CYP11B2, CYP2C8, EDN1, EDNRA, GNB3, SCNN1A, SCNN1B, SCNN1G, SGK1, KCNJ1, ACE, ADRB2, AGT, APLNR, BDKRB2, CAPN13, CYP11B2, CYP19A, GNB3, MMP3 |
| Genes | Pathway related to EH |
| NOS3 | RAAS pathway[22] |
| SH2B3 | Endothelial cell function[17] |
| AGT | Renal electrolyte balance[17] |
| NPPA | Control of extracellular fluid volume and electrolyte homeostasis[23] |
| NPPB | Involved in vasorelaxation and inhibition of renin and aldosterone[24] |
| NPR3 | Involved with regulating blood volume and pressure, pulmonary hypertension, and cardiac function[25] |
| UMOD | Constitutive inhibitor of calcium crystallization in renal fluids[26] |
| CYP17A1 | Involved with steroid/aldosterone synthesis. Enzyme dysfunction leads to increased levels of mineralocorticoid activating hormones[17] |
| ATP2B1 | Codes for enzymes that have a critical role in intracellular calcium homeostasis[27] |
| CACNB2 | Encodes for a subunit of a voltage-dependent calcium channel protein that is a member of the voltage-gated calcium channel superfamily[28] |
| SLC24A4 | Encodes for a member of the potassium-dependent sodium/calcium exchanger protein family[29] |
| YWHAZ | Protein interacts with insulin receptor substrate 1 protein, suggesting a role in regulating insulin sensitivity[30] |
| ADAMTS-8 | Enzyme encoded by the gene disrupts angiogenesis in vivo[31] |
| ADM | Protein encoded by gene may function as a hormone in circulation control[32] |
| c5 site between SUB1 and NPR3 | SNP associated with SBP |
| NPR3 encodes natriuretic peptide receptor C/guanylate cyclase C for natriuretic peptide clearance[33-35] | |
| Also found relationship with DBP | |
| CACANA1H | Codes for α1 subunit of voltage-dependent calcium channel for heart contractions and associated with SBP in African Americans[36] |
| ENPEP | Facilitates production of angiotensinII in RAAS pathway and associated with SBP and DBP[33] |
| ADD1 and ACE | ADD1 codes for α-adducin protein that interacts with sodium channel of Na/K co-transporter and Na/K ATPase[37] |
| Angiotensin converting enzyme produces angiotensin-converting enzyme which converts angiotensin I to angiotensin II in RAAS pathway[38] | |
| ADD2 | β-adducin is a cytoskeletal actin-binding protein implicated in glomerular lesions[39] |
| CYP11B2 | Contributes to aldosterone synthesis in RAAS pathway[40] |
| AGT | Encodes angiotensinogen in RAAS pathway[41] |
| LOC344371 and RASGRP3 | Activation decreases vascular responsiveness to endothelin-1 and angiotensin II in rats[41] |
| EDN3 | Endothelin-3 involved in vasoconstriction[42] |
| BCAT1 | Associated with salt sensitivity[43] |
| CASZ1 | Zinc-finger transcription factor that is associated with DBP[33] |
| ADRB2 | Ion channel involved with regulation of vasoconstriction[12] |
| CYP11B2 | Enzymatic defects results in decreased aldosterone and increased salt-wasting[12,17] |
| MMP3 | Gene variants affect arterial stiffness and endothelial function[44] |
| NR3C2 | Involved with aldosterone signaling[12] |
| SCNN1B | C terminus deletion leads to reduced ENaC clearance and increased ENaC activity[12] |
| APLNR | Mediator of cardiovascular disease[45] |
| BDKRB2 | Involved in catecholamine synthesis[46] |
| MTHFS | Involved with catecholamine binding[47] |
| SOX6 | Required in transcription for maintenance of cardiac and skeletal muscle cells[17] |
| CACNA1A | Involved with regulating SBP[48] |
| CCNG1 | Involved with regulation of SBP and DBP and is component of regulating hypertension[15] |
| CPLX3 | Involved with regulating DBP[15] |
| CSK | Cytoplasmic tyrosine kinase involved with angiotensin II-dependent vascular smooth muscle cell contraction[17] |
| CACNA1C | Regulates calcium influx after depolarization[49] |
| CLCNKB | Involved in renal salt absorption[50] |
| EDN1 | Endothelin-1 involved in vasoconstriction[51] |
| EDNRA | Endothelin receptor type A involved in vasoconstriction[52] |
| KCNJ1 | Potassium channel involved with potassium homeostasis[53] |
| SCNN1A | Involved with renal sodium regulation[54] |
| SCNN1B | Involved with renal sodium regulation[55] |
| SCNN1G | Involved with renal sodium regulation[56] |
| SGK1 | Activation of certain potassium, sodium and chloride channels, playing a role in cellular stress response[57] |
| SLC12A1 | Cotransporter involved in sodium and chloride reabsorption in the distal convoluted tubule[58] |
| SLC12A3 | Cotransporter involved in sodium and chloride reabsorption in the loop of Henle[59] |
| TNNT3 | Involved in calcium-induced muscle contraction[60] |
| WNK1 | Kinase involved with sodium and chloride transport[61] |
| WNK4 | Kinase regulates balance between sodium chloride and potassium reabsorption in kidneys[62] |
| GOSR2 | Interacts with target-localized SNAREs, allowing angiotensinogen to move between Golgi compartments, possibly leading to vasoconstriction[63] |
| GUCY1B3 | Receptor for nitric oxide involved with vasodilation[64] |
| ATXN2 | Possible association with regulation of GFR[65] |
| SLC4A7 | Possible transporter of sodium and bicarbonate ions[66] |
| CDH13 | Regulates endothelial cell growth[67] |
| Identifier information | Gene |
| Non- European genes | NPR3, IPO7, MYLIP, PMS1, SLC24A4, TBX3, YWHAZ, FIGN-GRB14, ALDH2, c5 site between SUB1 and NPR3, CACANA1H, SNP upstream of CCBE1, ENPEP, ST7L-CAPZA1 |
| Gene-gene interaction | ADD1 and ACE, ADD1 and ADD2, ADD1 and CYP11B2, AGT and ACE, c20q12, IMPG1, LOC344371 and RASGRP3, PCDH15, NPR3-c5orf23, CSK-ULK3, BAT2-BAT5, BLK-GATA4, GNAS-EDN3 |
| Gene- environment interaction | Body Mass Index: ADD1, ADRB2, CAPN13, CYP11B2, CYP19A1, MMP3 Black, Male: AGT Level of physical activity: GNB3, NR3C2, SCNN1B, APLNR, BDKRB2 Oral contraceptive use: COL25A1 Preterm birth: MTHFS |
| Unknown function/ function could not be determined | GNAS-EDN3, NPR3-c5orf23, BLK-GATA4, ST7L-CAPZA1, CSK-ULK3, FIGN-GRB14, c10orf107, c21orf91, LSP1-TNNT3, GNAS-EDN3, BAT2, IPO7, MYLIP, PMS1, TBX3, TBX4, TBX5, ANKMY, BAT2, BAT3, BAT4, BAT5, ALDH2, SNP upstream of CCBE1, BCAT1, PCDH15, c20q12, IMPG1, CAPN13, CYP19A1, GNB3, COL25A1, PCDH15, IMPG1, c5 site between SUB1 and NPR3, CHIC2-PDGRA1, APOB, HFE, CYPBA, CYP1A2, CYP2C8, EBF1, FES, FGF5, FIGN, FLJ32810, GNB3, LSP1, NOS3, TMEM133, FOXD3, GPR98, ARRDC3, GUCY1A3, JAG1, MECOM (MD1 locus), MOV10, NOV, NPR3-c5orf23, NT5C2-CYP171A, PIK3CG, PLCD3, PLCE1, PLEKHA7, RPL6-PTPN11-ALDH2, SLC39A8, ULK4, ZNF652, CCL20, WDR69, TGFBR2, STK39 |
Table 1 demonstrates the numerous amount of genes found to affect populations greater than 1000 individuals. There are several cohorts identified, each with multiple genes that are associated with EH. Also, there are some genes that are repeated in different cohorts, indicating that different populations have some genes in common with respect to EH.
Tables 1 and 2 contain the meta-analysis of two large studies with European subjects, Cohorts for Heart and Aging Research in Genetic Epidemiology Consortium and GlobalBPGen[12], which reveal fourteen loci that reached genome-wide significance. These are thought to account for 1.5% of the observed variance in blood pressure[12]. Many of the related genes have now been matched to physiologic functions (see “Known Pathway”, rows 1-6) that play a role in blood pressure (BP) regulation. Further studies were done on subjects of non-European descent, including African American, Japanese, Korean, and Han Chinese populations, which are listed as “Non-European Genes”. Table 2 specifically identifies the genes with known pathways related to EH regulation. Table 2 lists genes without a current known pathway to explain their influence on EH regulation.
Tables 3 and 4 identify many correlations between DNA and histone modifications, as well as miRNA-gene interactions and their effect on EH. Many of the genes identified were also identified through GWAS, indicating a possible mechanism for how the identified genes affect EH. It is important to note that the authors found no articles that discussed siRNA and its association with EH after conducting the literature search, thus the epigenetic section does not include siRNA.
| Ref. | Study | Subjects | Results | Site of modification and type |
| Smolarek et al[68] | Humans | 5-mC significantly higher in healthy subjects than entire group of patients with EH | N/A | |
| Wang et al[69] | Humans | Increased methylation levels observed at 2-CpG sites in comparison with normotensive controls | SULF1: Methylation | |
| Liang et al[70] | Humans | Regulation of renal sodium reabsorption β-2 adrenergic stimulation → inhibition of histone deacetylase-8 in kidney → increased histone acetylation and decreased genetic transcription of WNK4 caused increased blood pressure 11β-hydroxysteroid dehydrogenase type 2–converts active glucocorticoids to inactive glucocorticoids Promoter methylation of HSD11B2 gene decreased expression of renal 11β-hydroxysteroid dehydrogenase type 2 affects regulation of volume and BP homeostasis ENaCα-epithelial sodium channel–affects Na+ reabsorption in the distal nephron Proposed mechanism: Methylation of Lys79 of histone H3 suppresses ENaCα transcription ACE1-Angiotensin-converting enzyme ACE1-up-regulated in association with increased binding of histone 3 acetylation (H3Ac) and 4th lysine trimethylation (H3K4me3) and in association with decreased binding of histone ninth lysine residue demethylation (H3K9me2) | WNK4: Decreased transcription and increased histone acetylation HSD11B2: Promoter methylation ENaCα: Methylation of Lys79 of histone H3 H3K4me3: Histone 3 acetylation and 4th lysine trimethylation. H3K9me2: Decreased binding of histone 9th lysine residue demethylation | |
| Udali et al[71] | Friso et al[72] | Humans | 11β-hydroxysteroid dehydrogenase 2 methylation at HSD11B2 promoter in DNA of PBMCs of hypertensive patients inversely related to enzyme function Promoter methylation of HSD11B2 gene plays a role in HTN | HSD11B2: Methylation in promoter region |
| Lee et al[73] | Rats | Na+-K+-2 Cl- cotransporter 1 (NKCC1) Methylation status of NKCC1 promoter–elevated in hearts of spontaneously hypertensive rats SHRs–significant hypomethylation of NKCC1 associated with increase in gene expression contributing to HTN | NKCC1: Methylation in promoter region NKCC1: Hypomethylation in promoter region | |
| Riviere et al[74] | Human endothelial cell lines and rats in vivo | Somatic angiotensin-converting enzyme (= ACE1) Promoter methylation levels: Higher levels of methylation associated with transcriptional repression Therefore hypomethylation of promoter region of sACE could contribute to HTN | sACE: Methylation in promoter region | |
| Millis[75] | Human | Methyl CpG binding protein-2 (MECP-2) Methylates and thereby silences the expression of the norepinephrine transporter gene Phenyl-ethanolamine N-methyltransferase (PMNT)–converts Norepinephrine into Epinephrine Also mimics gene-silencing actions of MECP-2 Leads to increased synaptic levels of catecholamines (increased Epinephrine release and decreased Norepinephrine reuptake) CTGF Lysine methyltransferase that methylates the histone H3K79 site of nucleosomes that inhibits the expression of CTGF (in the cells of the collecting ducts) | MECP-2: Methylation PMNT: Methylation H3K79: Methylation of histone site of nucleosomes |
| Ref. | Subjects | Results | miRNA targets |
| Xu et al[76] | Human plasma | hcmv-miR-UL112; miR-605; miR-623; let-7e;miR-516b; miR-600; kshv-miR-K12-6-3p; miR-602; miR-1252 miR-296-5p; miR-133b; miR-30 d; miR-625*; miR- 1236; miR-518b; miR-1227; miR-664; miR-615-5p; miR-18b*; miR- 1249; miR-324-3p; ebv-miRBART17-3p; miR-634; ebvmiR-miRBART19-5p; miR-486-5p; kshvmiR-K12-10a; kshv-miR-K12-10b | INF-1 is direct target of hmcv-miR-UL112 Indicates link between CMV infection and EH |
| Batkai et al[77] | Human | Endothelial miRNA miR-126 miR-217 miR-122 miR-21 miR-24 miR-27b, -130a, -210, -378, -17–92, let-7f miR-15, -16, -20a, -20b, -24, -221, -222 Renal miRNA miR-29b miR-200a, miR-200b, miR-141, miR-429, miR-205, miR-192 miRNA targeting RAAS miR-155 miR-526b and -578 miR-34a, and -34c miR-765 miR-383 miR-9 miR-124 and miR-135a miRNA targeting smooth muscle cells miR-143 and miR-145 miR-21 miR-21, -26b, -98, and -1826 miR-221 and -222 miRNA in other etiologic factors miR-296-5p, let-7e, hcmv-miR-UL112 hcmv-miR-UL1 miR-637 | SPRED-1; PIK3 regulatory subunit-2; VCAM-1; CXCL12; RhoB SirT1 SLC7A1 Nitric oxide pathway Hypoxia-induced mechanism Pro-angiogenic Anti-angiogenic Fibrotic pathway; collagen genes; Mmp2; Itgb1 Biomarkers of nephrosclerosis AGTR1AVPR1ABDKRB2TBXA2RNR3C2NFATc3NR3C2 Actin stress fibers; ACE; KLF5; myocardin; MRTF-B; calmodulin kinase II-δ PTEN; Bcl-2; cGMP signaling Nitric oxide and ANP pathway p27(Kip1), p57(Kip2) and/or c-kit Association with hypertension IRF-1 ATP6V0A1, chromaffin granule function |
| Fung et al[78] | Human | miR-155 | Suppress expression of AGTR1 |
To our knowledge, this is the first review to discuss both genetic and epigenetic factors associated with EH in one article. As one can see, many genetic factors are involved with EH. There are many genes from genetic marker studies that are found to have some association with EH, as seen in Table 2. Some genes do have known physiologic pathway associated with EH, however, many do not. Our literature review herein denotes 129 genes. Of note, genes/gene regions with multiple study references include: STK39, CYP17A1, MTHFR-NPPA, MTHFR-NPPB, ATP2B1, CSK, ZNF652, UMOD, CACNB2, PLEKHA7, SH2B3, TBX3-TBX5, ULK4, CSK-ULK3, CYP1A2, NT5C2-CYP171A, PLCD3, SH2B3-ATXN2, CACNB2, PLEKHA7, SH2B3, TBX3-TBX5, ULK4 and HFE. The following genes overlap between the genetic studies and epigenetic studies: WNK4 and BDKRB2. While WNK4 and BDKRB2 are found in both genetic and epigenetic studies, it appears that WNK4 (kinase regulates balance between sodium chloride and potassium reabsorption in kidneys), and BDKRB2 (involved in catecholamine synthesis) may be associated with EH through interactions with miRNA.
Prior to GWAS, studies were somewhat successful in isolating genes associated with rare monogenic forms of hypertension that are inherited in a classic Mendelian fashion. The introduction of GWAS has made it possible to identify novel loci that could not be predicted physiologically, using non-family cohorts.
This review shows that no Mendelian variants or epigenetic factors are consistently associated with EH in the large cohort studies examined. Furthermore, it was not possible for the authors to correlate the epigenetic factors associated with the pathways identified, as there were no clear relationships between EH and the individual genes. Therefore, it can be inferred that EH follows multifactorial inheritance and insinuates that it follows the CDRV genetic hypothesis. In regards to identifying rare variants, GWAS is used for polymorphism detection, and is not set up to identify SNPs with low mean allele frequencies (MAFs) (low MAFs are usually under 1%, and sometimes even as high as 5%). Therefore, other techniques will need to be used to identify rare variants. Next-generation sequencing has revolutionized our ability to sequence thousands of genes at one time in a cost-effective manner. Using full exome or full genome sequencing of EH cohorts, next-generation sequencing will help to identify rare, as well as low-MAF, variants associated with regulating blood pressure[12]. This will likely show the exact genetic factors responsible for EH instead of mere associations which have been the mainstay of our genetic search using GWAS. Similar high throughput techniques will likely also improve our identification of epigenetic regulators.
Insufficient evidence was found in this study to pursue single site genetic marker or epigenetic testing to provide a simple genetic risk assessment for EH. Genetic algorithms comprised of information from multiple genes and epigenetic factors, along with family history and environmental variables, could potentially be developed to provide a genetic risk assessment for EH. However, it will be difficult to know what to do with this data, since preventative factors such as exercise and a healthy diet would be recommended to anyone at any level of personal and/or family history risk for EH. A similar concept was examined in a recent publication evaluating genetic testing and type 2 diabetes[79]. The evaluation of genomic applications in practice and prevention (EGAPP) consortium recommend against using genetic diabetic markers for risk assessments since it would be of limited benefit[79]. Additionally, for cardiovascular morbidity, current non-genetic algorithms already exist[80,81] that assess the risk of heart disease using a patient’s medical profile.
Although risk assessments may be difficult, pharmacogenomic utility may be found by studying risk alleles in individuals and treating their HTN in a personalized manner based on the pathway affected to obtain optimal blood pressure control[13].
To our knowledge, this is the first review to discuss both genetic and epigenetic factors associated with EH in one article. Insufficient evidence was found in this review to pursue any one single genetic test to provide a genetic risk assessment for EH.
In conclusion, while there exist genetic and epigenetic associations that play a role in EH, there are still no well-established cause-and-effect relationships for the development of EH. With emerging technologies, such as next-generation sequencing, a more direct relationship may be established between genetic and epigenetic factors and EH. Extensive algorithms for EH will likely need to be developed to incorporate these genetic risk factors, in concert with a patient’s personal risk factors. However, the utility of this approach will need to be proven. There is a large potential for personalized medicine through pharmacogenomics that will come from our better understanding of the genetic factors and pathways involved in EH.
The authors would like to thank all of the patients who participated in the studies that were cited throughout this article. Without them, we would not be able to further our collective knowledge.
Essential hypertension (EH) is thought to be a multifactorial disease, meaning that environmental and genetic factors affect the initiation and continuation of the disease. While there have been several publications discussing the genetic factors involved with EH, to date there has been no single publication that has discussed both genetic and epigenetic factors in one article.
While EH is thought to be a multifactorial disease, several genetic factors have been associated with EH. In the area of genetic and epigenetic factors associated with EH, their remains a need to review the most updated information regarding genetic and epigenetic factors and discuss both in one article.
Previously, scientists would have to refer to Genome-wide association studies and epigenetic studies to understand how genetic and epigenetic factors are associated with EH. This is the first review article to discuss both genetic and epigenetic factors in one article. Also, this article discusses the most current up-to-date literature, providing a more recent understanding of genetic factors associated with EH.
Next-generation sequencing will allow scientists to analyze thousands of genes in a cost-effective manner. Using full exome or full genome sequencing of EH cohorts, next-generation sequencing will help to identify rare, as well as, low- mean allele frequency variants associated with regulating blood pressure. This will be useful in the growing field of pharmacogenomics, where medical regimens are being tailored to individuals based on specific genetic polymorphisms. This will help to personalize treatment regimens and improve the care given to patients with EH.
Essential hypertension is a form of hypertension that has no known cause, but is responsible for most cases of hypertension. Genome-wide association studies look at the whole genome of populations of individuals who suffer from a specific condition to see if these individuals have any genes that differ from the general population without the condition in question. Pharmacogenomics is an emerging field where scientists and doctors use someone’s genetic code to determine appropriate doses for medications to ensure fewer side effects and the best possible therapy.
The present study appears well conducted for design and contents. Inclusion criteria and exclusion criteria are reasonable.
P- Reviewers: Okumura K, Wang M S- Editor: Gou SX L- Editor: A E- Editor: Wu HL
| 1. | Centers for Disease Control and Prevention (CDC). Vital signs: prevalence, treatment, and control of hypertension--United States, 1999-2002 and 2005-2008. MMWR Morb Mortal Wkly Rep. 2011;60:103-108. [PubMed] [Cited in This Article: ] |
| 2. | Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS. Executive summary: heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation. 2012;125:188-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 912] [Cited by in F6Publishing: 1004] [Article Influence: 83.7] [Reference Citation Analysis (0)] |
| 3. | Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123:933-944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2162] [Cited by in F6Publishing: 2169] [Article Influence: 166.8] [Reference Citation Analysis (0)] |
| 4. | Oparil S, Zaman MA, Calhoun DA. Pathogenesis of hypertension. Ann Intern Med. 2003;139:761-776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 397] [Cited by in F6Publishing: 377] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 5. | Messerli FH, Williams B, Ritz E. Essential hypertension. Lancet. 2007;370:591-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 441] [Cited by in F6Publishing: 433] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 6. | Nakayama T. Genetic factors of hypertension. Rinsho Byori. 2013;61:144-149. [PubMed] [Cited in This Article: ] |
| 7. | Hall JE, Granger JP, do Carmo JM, da Silva AA, Dubinion J, George E, Hamza S, Speed J, Hall ME. Hypertension: physiology and pathophysiology. Compr Physiol. 2012;2:2393-2442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 132] [Cited by in F6Publishing: 136] [Article Influence: 12.4] [Reference Citation Analysis (1)] |
| 8. | Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212-219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 487] [Cited by in F6Publishing: 403] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 9. | Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502-510. [PubMed] [Cited in This Article: ] |
| 10. | Iyengar SK, Elston RC. The genetic basis of complex traits: rare variants or “common gene, common disease”? Methods Mol Biol. 2007;376:71-84. [PubMed] [Cited in This Article: ] |
| 11. | Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69:124-137. [PubMed] [Cited in This Article: ] |
| 12. | Basson J, Simino J, Rao DC. Between candidate genes and whole genomes: time for alternative approaches in blood pressure genetics. Curr Hypertens Rep. 2012;14:46-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Delles C, Padmanabhan S. Genetics and hypertension: is it time to change my practice? Can J Cardiol. 2012;28:296-304. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Delles C, McBride MW, Graham D, Padmanabhan S, Dominiczak AF. Genetics of hypertension: from experimental animals to humans. Biochim Biophys Acta. 2010;1802:1299-1308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 15. | Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17-25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 16. | El Shamieh S, Visvikis-Siest S. Genetic biomarkers of hypertension and future challenges integrating epigenomics. Clin Chim Acta. 2012;414:259-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Padmanabhan S, Newton-Cheh C, Dominiczak AF. Genetic basis of blood pressure and hypertension. Trends Genet. 2012;28:397-408. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 85] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 18. | Rafiq S, Anand S, Roberts R. Genome-wide association studies of hypertension: have they been fruitful? J Cardiovasc Transl Res. 2010;3:189-196. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 19. | Simino J, Rao DC, Freedman BI. Novel findings and future directions on the genetics of hypertension. Curr Opin Nephrol Hypertens. 2012;21:500-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 20. | Wang X, Prins BP, Sõber S, Laan M, Snieder H. Beyond genome-wide association studies: new strategies for identifying genetic determinants of hypertension. Curr Hypertens Rep. 2011;13:442-451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Xi B, Chen M, Chandak GR, Shen Y, Yan L, He J, Mou SH. STK39 polymorphism is associated with essential hypertension: a systematic review and meta-analysis. PLoS One. 2013;8:e59584. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 22. | Kimura L, Angeli CB, Auricchio MT, Fernandes GR, Pereira AC, Vicente JP, Pereira TV, Mingroni-Netto RC. Multilocus family-based association analysis of seven candidate polymorphisms with essential hypertension in an african-derived semi-isolated brazilian population. Int J Hypertens. 2012;2012:859219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 23. | Cannone V, Huntley BK, Olson TM, Heublein DM, Scott CG, Bailey KR, Redfield MM, Rodeheffer RJ, Burnett JC. Atrial natriuretic peptide genetic variant rs5065 and risk for cardiovascular disease in the general community: a 9-year follow-up study. Hypertension. 2013;62:860-865. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in F6Publishing: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 24. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/4879. [Cited in This Article: ] |
| 25. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/4883. [Cited in This Article: ] |
| 26. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/7369. [Cited in This Article: ] |
| 27. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/490. [Cited in This Article: ] |
| 28. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/783. [Cited in This Article: ] |
| 29. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/123041. [Cited in This Article: ] |
| 30. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/7534. [Cited in This Article: ] |
| 31. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/11095. [Cited in This Article: ] |
| 32. | Available from: http://www.ncbi.nlm.nih.gov.eres.library.manoa.hawaii.edu/gene/133. [Cited in This Article: ] |
| 33. | Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, Tay WT, Chen CH, Zhang Y, Yamamoto K. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531-538. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 435] [Cited by in F6Publishing: 452] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 34. | Anand-Srivastava MB. Natriuretic peptide receptor-C signaling and regulation. Peptides. 2005;26:1044-1059. [PubMed] [Cited in This Article: ] |
| 35. | Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, Tayo B, Adeyemo A, Sun YV, Li Y. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet. 2011;20:2285-2295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 36. | Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 294] [Cited by in F6Publishing: 350] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 37. | Kalita J, Somarajan BI, Kumar B, Mittal B, Misra UK. A study of ACE and ADD1 polymorphism in ischemic and hemorrhagic stroke. Clin Chim Acta. 2011;412:642-646. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 38. | Ferrandi M, Cusi D, Molinari I, Del Vecchio L, Barlassina C, Rastaldi MP, Schena FP, Macciardi F, Marcantoni C, Roccatello D. alpha- and beta-Adducin polymorphisms affect podocyte proteins and proteinuria in rodents and decline of renal function in human IgA nephropathy. J Mol Med (Berl). 2010;88:203-217. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 39. | Hu Q, Yin L, Hartmann RW. Aldosterone Synthase Inhibitors as Promising Treatments for Mineralocorticoid Dependent Cardiovascular and Renal Diseases. J Med Chem. 2014;Epub ahead of print. [PubMed] [Cited in This Article: ] |
| 40. | Ji L, Cai X, Zhang L, Fei L, Wang L, Su J, Lazar L, Xu J, Zhang Y. Association between polymorphisms in the renin-angiotensin-aldosterone system genes and essential hypertension in the Han Chinese population. PLoS One. 2013;8:e72701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 41. | Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103-109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1554] [Cited by in F6Publishing: 1500] [Article Influence: 115.4] [Reference Citation Analysis (0)] |
| 42. | Carey RM, Schoeffel CD, Gildea JJ, Jones JE, McGrath HE, Gordon LN, Park MJ, Sobota RS, Underwood PC, Williams J. Salt sensitivity of blood pressure is associated with polymorphisms in the sodium-bicarbonate cotransporter. Hypertension. 2012;60:1359-1366. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 43. | Huang R, Deng L, Shen A, Liu J, Ren H, Xu DL. Associations of MMP1, 3, 9 and TIMP3 genes polymorphism with isolated systolic hypertension in Chinese Han population. Int J Med Sci. 2013;10:840-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 44. | Li YY. α-Adducin Gly460Trp gene mutation and essential hypertension in a Chinese population: a meta-analysis including 10,960 subjects. PLoS One. 2012;7:e30214. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 45. | Jin W, Su X, Xu M, Liu Y, Shi J, Lu L, Niu W. Interactive association of five candidate polymorphisms in Apelin/APJ pathway with coronary artery disease among Chinese hypertensive patients. PLoS One. 2012;7:e51123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 46. | Nostramo R, Tillinger A, Serova L, Kvetnansky R, Sabban EL. Bradykinin B2 receptor in the adrenal medulla of male rats and mice: glucocorticoid-dependent increase with immobilization stress. Endocrinology. 2013;154:3729-3738. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 47. | Anguera MC, Stover PJ. Methenyltetrahydrofolate synthetase is a high-affinity catecholamine-binding protein. Arch Biochem Biophys. 2006;455:175-187. [PubMed] [Cited in This Article: ] |
| 48. | Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, Rice K, Verwoert GC, Launer LJ, Gudnason V. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57:903-910. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 157] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 49. | Sun Q, Li QX, Song XF, Zheng SG, Yan F, Chen P, Tang JF, Niu YX, Bao QY, Zhang GQ. [Impact of CACNA1C polymorphisms on antihypertensive efficacy of calcium channel blocker]. Zhonghua Xinxue Guanbing Zazhi. 2012;40:3-7. [PubMed] [Cited in This Article: ] |
| 50. | Su X, Chang P, Liu Z, Yan M, Liu G, Cui H. Association of CLCNKB haplotypes and hypertension in Mongolian and Han populations. Clin Exp Hypertens. 2012;34:482-487. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 51. | Tobe SW, Baker B, Hunter K, Kiss A, Perkins N, Gomez L, Feng Y, Wigg K, Barr CL. The impact of endothelin-1 genetic analysis and job strain on ambulatory blood pressure. J Psychosom Res. 2011;71:97-101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 52. | Calabrò P, Limongelli G, Maddaloni V, Vizza CD, D’Alto M, D’Alessandro R, Poscia R, Argiento P, Ziello B, Badagliacca R. Analysis of endothelin-1 and endothelin-1 receptor A gene polymorphisms in patients with pulmonary arterial hypertension. Intern Emerg Med. 2012;7:425-430. [PubMed] [Cited in This Article: ] |
| 53. | Nüsing RM, Pantalone F, Gröne HJ, Seyberth HW, Wegmann M. Expression of the potassium channel ROMK in adult and fetal human kidney. Histochem Cell Biol. 2005;123:553-559. [PubMed] [Cited in This Article: ] |
| 54. | Yu Z, Kong Q, Kone BC. Sp1 trans-activates and is required for maximal aldosterone induction of the αENaC gene in collecting duct cells. Am J Physiol Renal Physiol. 2013;305:F653-F662. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 55. | Nguyen KD, Pihur V, Ganesh SK, Rakha A, Cooper RS, Hunt SC, Freedman BI, Coresh J, Kao WH, Morrison AC. Effects of rare and common blood pressure gene variants on essential hypertension: results from the Family Blood Pressure Program, CLUE, and Atherosclerosis Risk in Communities studies. Circ Res. 2013;112:318-326. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 56. | Büsst CJ, Bloomer LD, Scurrah KJ, Ellis JA, Barnes TA, Charchar FJ, Braund P, Hopkins PN, Samani NJ, Hunt SC. The epithelial sodium channel γ-subunit gene and blood pressure: family based association, renal gene expression, and physiological analyses. Hypertension. 2011;58:1073-1078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 57. | Lang F, Stournaras C, Alesutan I. Regulation of transport across cell membranes by the serum- and glucocorticoid-inducible kinase SGK1. Mol Membr Biol. 2014;31:29-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 58. | Monette MY, Rinehart J, Lifton RP, Forbush B. Rare mutations in the human Na-K-Cl cotransporter (NKCC2) associated with lower blood pressure exhibit impaired processing and transport function. Am J Physiol Renal Physiol. 2011;300:F840-F847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 59. | Zhang F, Yang Y, Hu D, Lei H, Wang Y. Lack of an association between TSC gene Arg904Gln polymorphisms and essential hypertension risk based on a meta-analysis. Genet Mol Res. 2012;11:3511-3517. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 60. | Johnson T, Gaunt TR, Newhouse SJ, Padmanabhan S, Tomaszewski M, Kumari M, Morris RW, Tzoulaki I, O’Brien ET, Poulter NR. Blood pressure loci identified with a gene-centric array. Am J Hum Genet. 2011;89:688-700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 142] [Cited by in F6Publishing: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 61. | Liu F, Zheng S, Mu J, Chu C, Wang L, Wang Y, Xiao H, Wang D, Cao Y, Ren K. Common variation in with no-lysine kinase 1 (WNK1) and blood pressure responses to dietary sodium or potassium interventions- family-based association study. Circ J. 2013;77:169-174. [PubMed] [Cited in This Article: ] |
| 62. | Cao FF, Han H, Wang F, Chen XD, Lu M, Wang XF, Lin RY, Wen H, Jin L. [Study on the association between genetic polymorphism on WNK4 genes and essential hypertension among Kazakhs ethnic population, in Xinjiang]. Zhonghua Liu Xing Bing Xue Za Zhi. 2010;31:375-378. [PubMed] [Cited in This Article: ] |
| 63. | Pan S, Nakayama T, Sato N, Izumi Y, Soma M, Aoi N, Ma Y. A haplotype of the GOSR2 gene is associated with essential hypertension in Japanese men. Clin Biochem. 2013;46:760-765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 64. | Zabel U, Weeger M, La M, Schmidt HH. Human soluble guanylate cyclase: functional expression and revised isoenzyme family. Biochem J. 1998;335:51-57. [PubMed] [Cited in This Article: ] |
| 65. | Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 590] [Cited by in F6Publishing: 623] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 66. | Boedtkjer E, Moreira JM, Mele M, Vahl P, Wielenga VT, Christiansen PM, Jensen VE, Pedersen SF, Aalkjaer C. Contribution of Na+,HCO3(-)-cotransport to cellular pH control in human breast cancer: a role for the breast cancer susceptibility locus NBCn1 (SLC4A7). Int J Cancer. 2013;132:1288-1299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 67. | Philippova M, Joshi MB, Pfaff D, Kyriakakis E, Maslova K, Erne P, Resink TJ. T-cadherin attenuates insulin-dependent signalling, eNOS activation, and angiogenesis in vascular endothelial cells. Cardiovasc Res. 2012;93:498-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 68. | Smolarek I, Wyszko E, Barciszewska AM, Nowak S, Gawronska I, Jablecka A, Barciszewska MZ. Global DNA methylation changes in blood of patients with essential hypertension. Med Sci Monit. 2010;16:CR149-CR155. [PubMed] [Cited in This Article: ] |
| 69. | Wang X, Falkner B, Zhu H, Shi H, Su S, Xu X, Sharma AK, Dong Y, Treiber F, Gutin B. A genome-wide methylation study on essential hypertension in young African American males. PLoS One. 2013;8:e53938. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 70. | Liang M, Cowley AW, Mattson DL, Kotchen TA, Liu Y. Epigenomics of hypertension. Semin Nephrol. 2013;33:392-399. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 71. | Udali S, Guarini P, Moruzzi S, Choi SW, Friso S. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol Aspects Med. 2013;34:883-901. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 72. | Friso S, Pizzolo F, Choi SW, Guarini P, Castagna A, Ravagnani V, Carletto A, Pattini P, Corrocher R, Olivieri O. Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene promoter is related to human hypertension. Atherosclerosis. 2008;199:323-327. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 73. | Lee HA, Baek I, Seok YM, Yang E, Cho HM, Lee DY, Hong SH, Kim IK. Promoter hypomethylation upregulates Na+-K+-2Cl- cotransporter 1 in spontaneously hypertensive rats. Biochem Biophys Res Commun. 2010;396:252-257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 74. | Rivière G, Lienhard D, Andrieu T, Vieau D, Frey BM, Frey FJ. Epigenetic regulation of somatic angiotensin-converting enzyme by DNA methylation and histone acetylation. Epigenetics. 2011;6:478-489. [PubMed] [Cited in This Article: ] |
| 75. | Millis RM. Epigenetics and hypertension. Curr Hypertens Rep. 2011;13:21-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 69] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 76. | Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl). 2012;90:865-875. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 77. | Bátkai S, Thum T. MicroRNAs in hypertension: mechanisms and therapeutic targets. Curr Hypertens Rep. 2012;14:79-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 78. | Fung MM, Zhang K, Zhang L, Rao F, O’Connor DT. Contemporary approaches to genetic influences on hypertension. Curr Opin Nephrol Hypertens. 2011;20:23-30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 79. | Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group. The EGAPP initiative: lessons learned. Genet Med. 2014;16:217-224. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 80. | Chen Q, Li G, Leong TY, Heng CK. Predicting coronary artery disease with medical profile and gene polymorphisms data. Stud Health Technol Inform. 2007;129:1219-1224. [PubMed] [Cited in This Article: ] |