Published online May 15, 2020. doi: 10.4239/wjd.v11.i5.182
Peer-review started: December 12, 2019
First decision: January 6, 2020
Revised: March 16, 2020
Accepted: March 30, 2020
Article in press: March 30, 2020
Published online: May 15, 2020
Perinatal exposure to a poor nutritional environment predisposes the progeny to the development of metabolic disease at the adult age, both in experimental models and humans. Numerous adaptive responses to maternal protein restriction have been reported in metabolic tissues. However, the expression of glucose/fatty acid metabolism-related genes in adipose tissue and liver needs to be described.
To evaluate the metabolic impact of perinatal malnutrition, we determined malnutrition-associated gene expression alterations in liver and adipose tissue.
In the present study, we evaluated the alterations in gene expression of glycolytic/Krebs cycle genes (Pyruvate dehydrogenase kinase 4 and citrate synthase), adipogenic and lipolytic genes and leptin in the adipose tissue of offspring rats at 30 d and 90 d of age exposed to maternal isocaloric low protein (LP) diet throughout gestation and lactation. We also evaluated, in the livers of the same animals, the same set of genes as well as the gene expression of the transcription factors peroxisome proliferator-activated receptor gamma coactivator 1, forkhead box protein O1 and hepatocyte nuclear factor 4 and of gluconeogenic genes.
In the adipose tissue, we observed a transitory (i.e., at 30 d) downregulation of pyruvate dehydrogenase kinase 4, citrate synthase and carnitine palmitoyl transferase 1b gene expression. Such transcriptional changes did not persist in adult LP rats (90 d), but we observed a tendency towards a decreased gene expression of leptin (P = 0.052). The liver featured some gene expression alterations comparable to the adipose tissue, such as pyruvate dehydrogenase kinase 4 downregulation at 30 d and displayed other tissue-specific changes, including citrate synthase and fatty acid synthase upregulation, but pyruvate kinase downregulation at 30 d in the LP group and carnitine palmitoyl transferase 1b downregulation at 90 d. These gene alterations, together with previously described changes in gene expression in skeletal muscle, may account for the metabolic adaptations in response to maternal LP diet and highlight the occurrence of persistent transcriptional defects in key metabolic genes that may contribute to the development of metabolic alterations during the adult life as a consequence of perinatal malnutrition.
We conclude that perinatal malnutrition relays long-lasting transcriptional alterations in metabolically active organs, i.e., liver and adipose tissue.
Core tip: Perinatal exposure to a poor nutritional environment predisposes to metabolic disease. Here, the expression of metabolism-related genes in adipose tissue and liver was investigated. We evaluated the alterations in gene expression of glycolytic/Krebs cycle genes, adipogenic and lipolytic genes in adipose tissue of offspring rats at 30 d/90 d of age, exposed to maternal low protein diet throughout gestation/lactation. We also evaluated expression of liver transcription factors and gluconeogenic genes. Persistent gene alterations were observed that may account for the metabolic adaptations in response to maternal low protein diet, highlighting the occurrence of persistent transcriptional defects as a consequence of perinatal malnutrition.
- Citation: de Oliveira Lira A, de Brito Alves JL, Pinheiro Fernandes M, Vasconcelos D, Santana DF, da Costa-Silva JH, Morio B, Góis Leandro C, Pirola L. Maternal low protein diet induces persistent expression changes in metabolic genes in male rats. World J Diabetes 2020; 11(5): 182-192
- URL: https://www.wjgnet.com/1948-9358/full/v11/i5/182.htm
- DOI: https://dx.doi.org/10.4239/wjd.v11.i5.182
Perinatal malnutrition occurring during pregnancy and lactation not only has a negative impact on fetal development and neonatal growth but also relays long-lasting adverse effects resulting in increased susceptibility to cardiovascular and metabolic diseases in adulthood, as posited by the developmental origin of health and disease hypothesis[1,2]. Evidence from epidemiological cohorts, with the Dutch Famine Birth Cohort Study being the most relevant[3], and experimental animal models[4-6] support the idea that a poor nutritional environment during fetal and early postnatal life predisposes to cardiovascular and metabolic disease in adulthood. The occurrence of persistent epigenetic alterations has been proposed as one of the mechanisms linking in utero nutritional deprivation to increased risk of disease in adulthood. Individuals who were prenatally exposed to the Dutch famine during the 1944-1945 were shown six decades later to have lower DNA methylation of the imprinted IGF2 gene in comparison to their siblings not exposed to the famine period[7]. Observational studies have suggested a link between poor fetal growth and the development of impaired glucose tolerance at the adult age in both sexes[8], and final evidence that maternal nutrition during gestation affects glucose metabolism in adult life was provided by the observation that an oral glucose load in adults exposed prenatally to the Dutch famine led to higher glycemic concentrations as compared to individuals being born around the same years but not exposed in utero to the 1944-1945 famine[9].
Work in animal models suggested that the link between early-life malnutrition and the increased risk of developing metabolic disease in adulthood may be mediated by persistent biochemical alterations in the main insulin-responsive tissues, including glycolytic and oxidative skeletal muscle fibers[10]. Exposure of mice to a maternal protein-restricted diet during gestation and lactation was shown to impact the morphological features and body distribution of white adipose tissue and to reduce the protein expression levels of most of the key insulin signaling proteins, including IRS1, the PI3K subunits p110 and p85, Akt1 (v-akt murine thymoma viral oncogene homolog 1) and its phosphorylated form on serine 473 in male offspring[11], resulting in an altered distribution and morphology of white adipose tissue[12]. In a similar way, perinatal low protein (LP) diet consumption contributed to fatty liver phenotype at the adult age, which the magnitude was related to the period of exposure to the LP diet[13].
The occurrence of alterations of glycemic control in humans, associated to the observation that perinatal malnutrition induces defects in the insulin signaling pathways in rodent models, prompted us to evaluate whether the main metabolic pathways are affected by a LP diet administered to dams during pregnancy and lactation. In a previous study, we reported that gene and protein expression of enzymes participating in glucose and fatty acid metabolism in skeletal muscle were altered at short-term (30 d) and long-term (90 d) timepoints in male rat offspring exposed to a maternal LP diet during gestation and lactation[10]. Interestingly, we observed, both in soleus and extensor digitorum longus skeletal muscle, a LP-induced, long-lasting downregulation of pyruvate dehydrogenase kinase 4 (PDK4), an enzyme leading to allosteric deactivation of the pyruvate dehydrogenase complex via phosphorylation and hence a redirection of the metabolic flux from catabolic Krebs cycle to anabolic pathways[14].
To obtain a wider description of the effects of perinatal LP diet on insulin-responsive tissues, the main goal of the present study was to evaluate the short-term and long-term effects of a LP diet during gestation and lactation on the expression of key genes involved in the metabolism of glucose and fatty acid in the liver and adipose tissue of male rat offspring. We demonstrate the occurrence of long lasting alterations of gene expression in both tissues, up to 90 d of age, that reflect the persistence of altered metabolism in the offspring consequent to in utero and early-life exposure to deleterious nutritional conditions.
The experimental protocol was approved by the Ethical Committee of the Biological Sciences Centre (protocol 23076 062778/2014-38), Federal University of Pernambuco, Brazil. All efforts were made to minimize animal discomfort and the number of animals used; in addition, we followed the Guidelines for the Care and Use of Laboratory Animals.
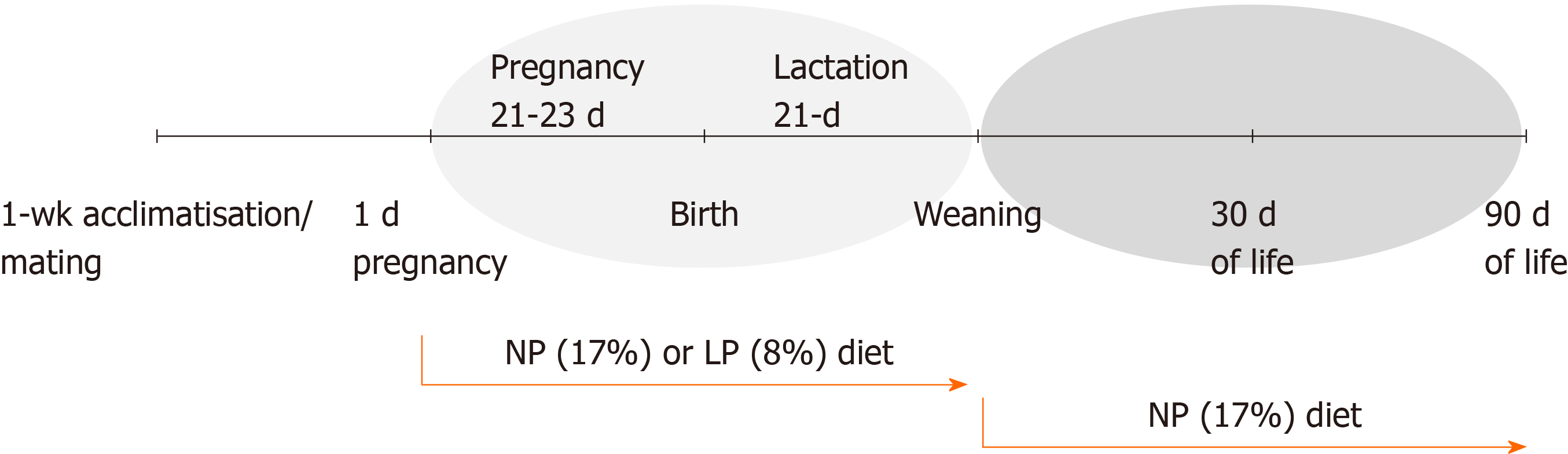
Male albino Wistar rats (Rattus novergicus) were obtained from the Academic Center of Vitoria de Santo Antão animal facility, Federal University of Pernambuco, Brazil. Animals were housed at 22 ± 1 °C with a controlled light–dark cycle (dark 18:00–06:00 h). Standard laboratory chow (52% carbohydrate, 21% protein and 4% lipids-Labina, Purina Agriband, São Paulo, Brazil) and water were administered ad libitum. Groups were divided according to their mother’s diet: Control pups from dams fed a 17% protein diet (n = 5, normal protein group, NP), and LP pups from dams fed an 8% casein diet (n = 5, low protein, LP) during gestation and lactation. Diets were prepared at the Laboratory of Experimental Nutrition-Center of Vitoria de Santo Antão, Federal University of Pernambuco, according to the American Institute of Nutrition–AIN-93 dietary guidelines[15].
During suckling, offspring was maintained as litters of eight pups of both sexes to ensure standardized nutrition until weaning. At weaning (21 d postpartum), three to four male offspring of each litter were housed in collective cages and received standard diet and water ad libitum. The experimental groups consisted of one or two male rats from each mother. Female offspring were not included in the present study. All experimental analyses were performed in adipose tissue and liver collected from male rats sacrificed either at 30 d old or 90 d old by decapitation. All rats were euthanized between 14:00-17:00 after a 4-5 h fasting period. The liver and visceral adipose tissue were carefully dissected, snap-frozen in liquid nitrogen and stored at –80 °C until RNA extraction.
Total RNA was extracted from liver and visceral adipose tissue with Tripure reagent [Sigma-Aldrich (Roche), St. Quentin Fallavier, France] according to the manufacturer’s instructions. Briefly, 10 µL of Tripure per milligram of tissue was added, and the resulting suspension was homogenized using a Precellys Lysing kit (Bertin, Montigny-le-Bretonneux, France) according to the manufacturer’s instructions. After grinding, 1/4 volume of chloroform was added. They were vortexed 3 × 15 s, incubated at room temperature for 5 min and centrifuged for 15 min at 15000 g at 4 °C.
RNA was precipitated by addition of 1/2 volume of isopropanol (Carlo Erba, Val-de-Reuil, France) and centrifugation (15 min at 15000 g at 4 °C). Supernatants were used for protein extraction and RNA-containing pellets were washed sequentially with 70% and 95% ethanol (Carlo Erba), dried and dissolved in 100 µL RNase-free water. RNA concentration and purity (260/280 nm absorbance ratio) was determined on a Nanodrop 2000 (Thermo-Fisher).
Reverse transcription was performed using an RT-Takara kit (Primescript TM, Takara) using 1 µg of RNA as template and following the manufacturer’s instructions. Briefly, samples were heated for 10 min at 65 °C. Samples were mixed with 4 μL PrimeScript Buffer 5 ×, 1 μL oligodT (50 µM), 4 μL random hexamers and 1 μL of PrimeScript RT Enzyme Mix followed by a 15 min incubation at 37 °C and 15 s at 85 °C. RNA was removed by incubation with 1 μL of RNase H for 20 min at 37 °C. Reverse transcription reactions were brought to 200 µL final volume by adding RNase free water and stored at -20 °C. Real-time quantitative PCR (qPCR) amplification was performed using a Rotor-Gene Real-Time PCR System (Labgene Scientific Instruments, Archamps, France). Sequences of primers used in this study are available upon request.
Reactions were incubated at 95 °C for 10 min, followed by 40 cycles of denaturation (95 °C, 10 s), annealing (58-62 °C depending on the primer sets, 30 s) and elongation (72 °C, 30 s). mRNA expression levels of PDK4, citrate synthase (CS), carnitine palmitoylacyltransferase 1b, acetyl-CoA carboxylase, fatty acid synthase, leptin, insulin receptor, phosphofructokinase, beta hydroxyacyl-coenzyme-A dehydrogenase, peroxisome proliferator-activated receptor-alpha coactivator 1 alpha, peroxisome proliferator-activated receptor-alpha, forkhead box protein O1, hepatocyte nuclear factor 4, glucose 6-phosphatase, phosphoenolpyruvate carboxykinase and pyruvate kinase L/R were measured on total RNA extracted from adipose tissue and liver. qPCR results from each gene (including the housekeeping gene) were expressed as arbitrary units derived from a standard calibration curve derived from a reference sample. Reference samples were generated by mixing 10 μL aliquots from ten cDNA samples, five from the NP group and five from the LP group. qPCR for each sample was carried out in duplicate. Gene expression data were normalized using ribosomal protein L19 as a housekeeping gene. As a further control, qPCR amplicons were analyzed by agarose gel to validate the amplicon size.
Statistical analysis was conducted with GraphPad Prism 5 program for Windows (GraphPad Software®. Inc., La Jolla, CA, United States). Exploratory data analysis was used to identify possible inaccurate information and the presence of outliers and to test the assumption of normality in all data distributions. Kolmogorov-Smirnov and Shapiro-Wilk normality tests were applied in total sample. Statistical significance was evaluated using analysis of variance ANOVA two-way test with maternal diet (low/normal protein) and age (30 d and 90 d) as factors. Bonferroni’s post hoc test was used. The values are presented as mean and standard error means, and P values < 0.05 were considered statistically significant. P values < 0.05 are denoted as “a” in figures; P values < 0.01 are denoted as “b” in figures.
We applied a model of perinatal protein restriction to rat dams throughout pregnancy and lactation followed by a switch to a NP diet after weaning as schematically represented in Figure 1.
In spite of the administration of a NP diet after weaning, the LP group displayed a lower bodyweight both at 30 d [NP: 106.3 ± 17.1 g, LP: 87.3 ± 6.0 g; P < 0.05, unpaired t-test, n = 12 (NP) and 6 (LP)] and 90 d of age (NP: 326.7 ± 22.6 g, LP: 306.2 g; P < 0.05, unpaired t-test, n = 8 for both groups).
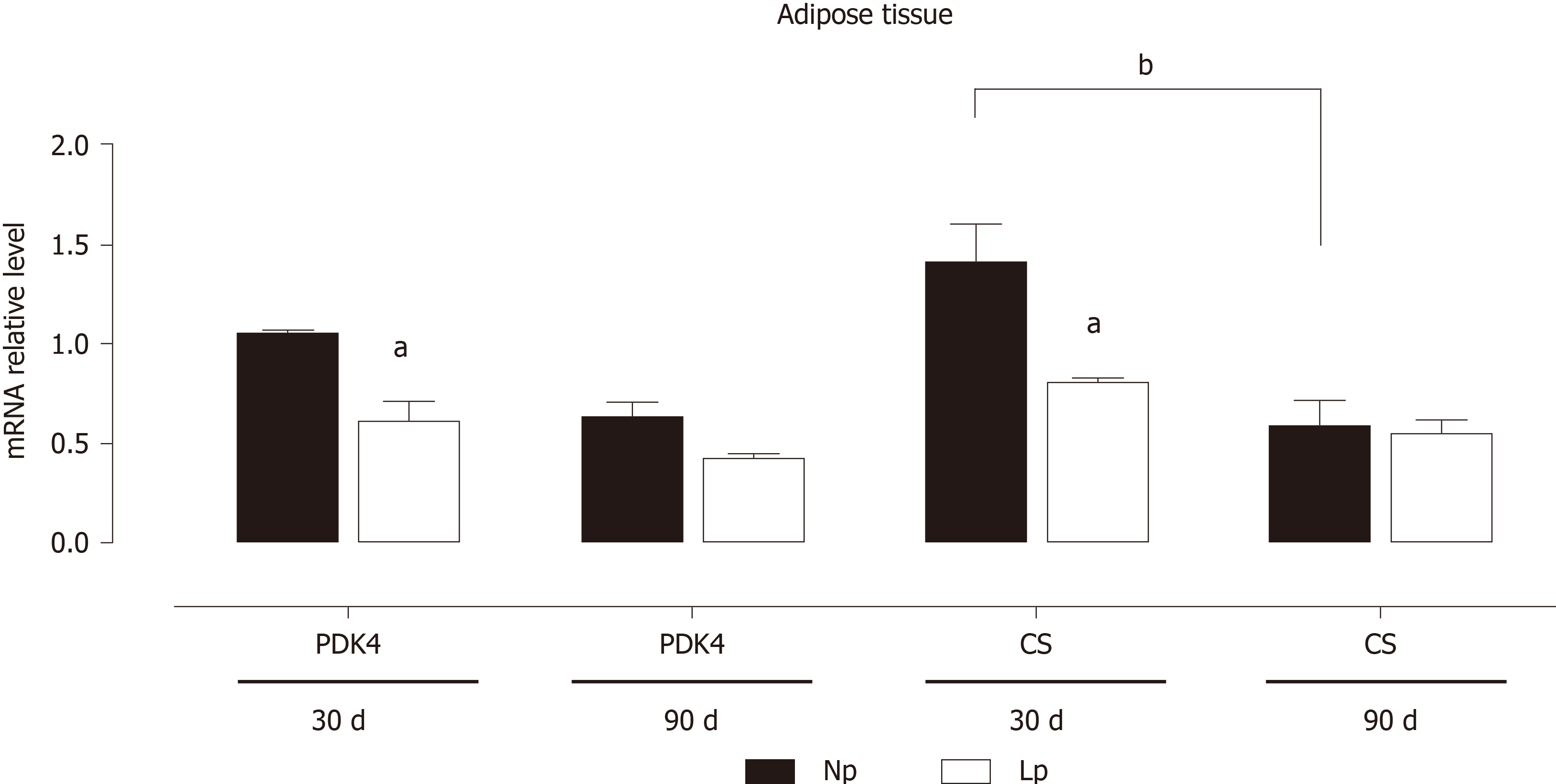
The gene expression of two key enzymes linking the glycolytic pathway to the Krebs cycle, PDK4 and CS, was evaluated in adipose tissue at different ages (Figure 2).
In NP rats, CS and PDK4 genes displayed a time-dependent downregulation, with mRNA expression at 90 d being lower than at 30 d (P = 0.057 for PDK4 and < 0.01 for CS; Figure 2). In comparison, the maternal LP diet, by inducing a significant downregulation of both genes at 30 d, abolished the time-dependent downregulation of both genes. These results indicate that the glycolytic flux may be altered in LP offspring.
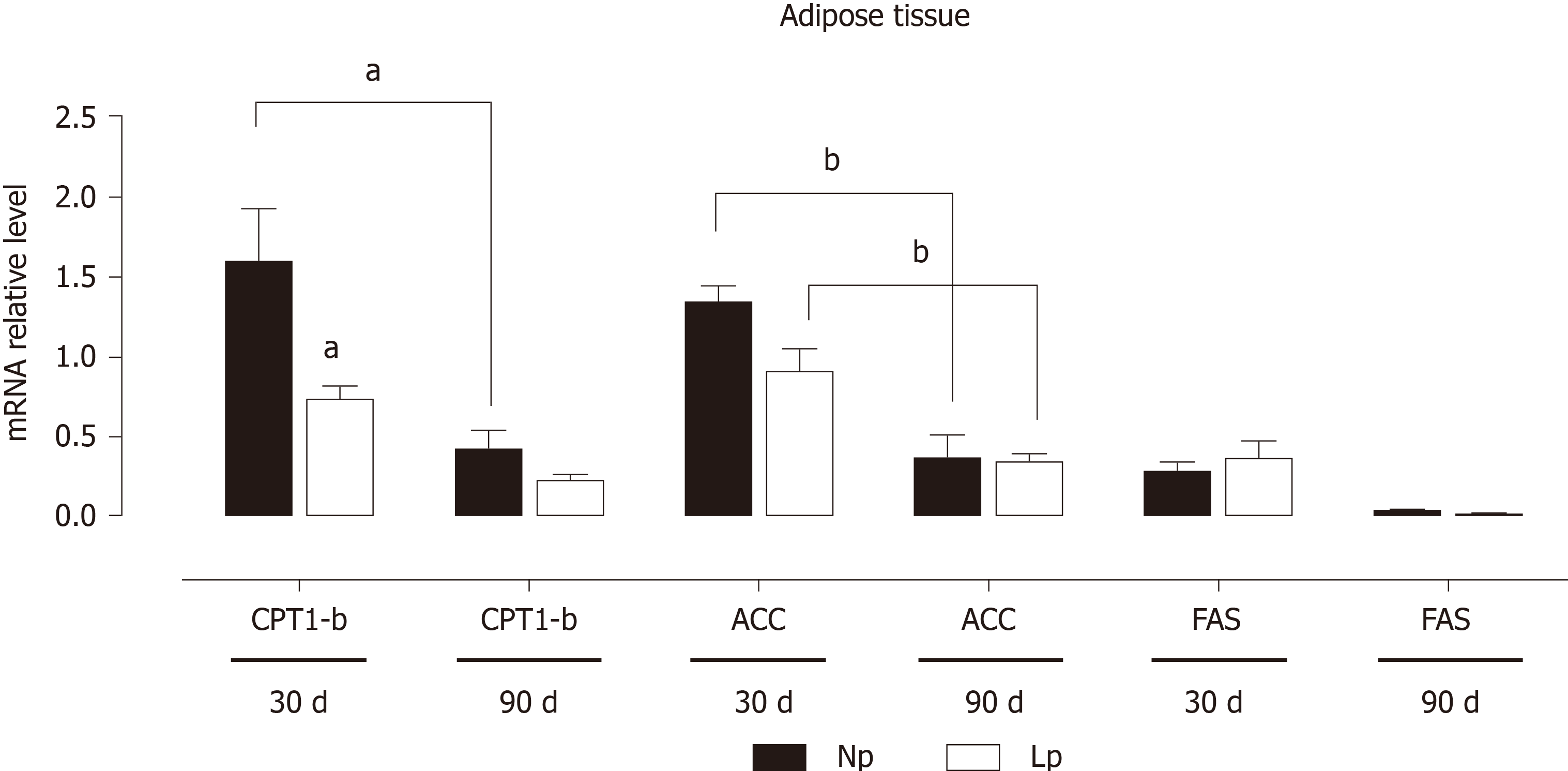
On the contrary, genes related to fatty acid metabolism, while showing a time-dependent downregulation with lower expression at 90 d, were not affected by the administration of a LP diet during pregnancy and lactation (Figure 3).
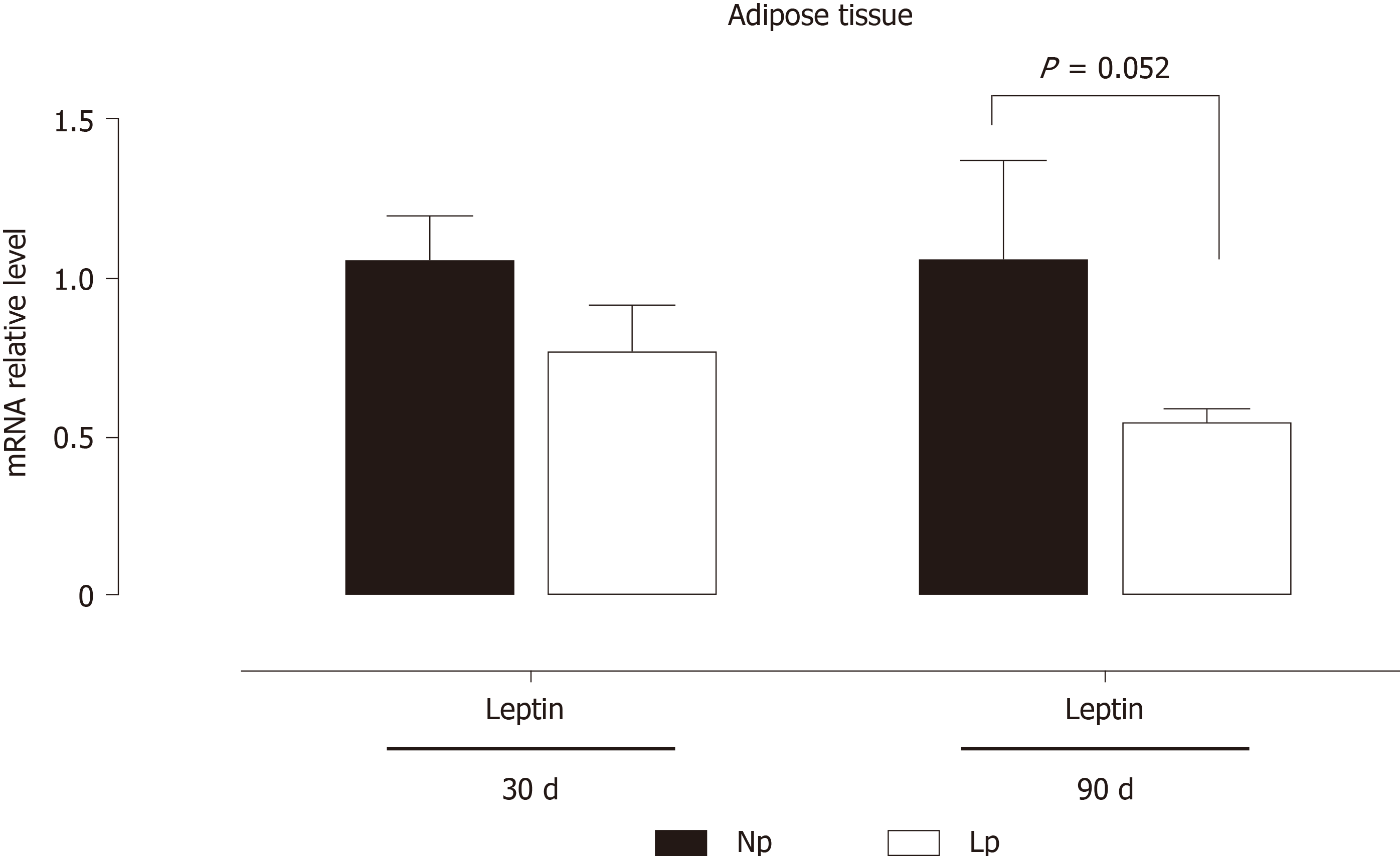
These results indicate that the glycolytic flux may be altered in the adipose tissue of LP offspring while fatty acid metabolism is not affected. As body weight of LP offspring remained lower at 30 d and 90 d as compared to NP offspring, we also evaluated the gene expression levels of leptin and observed a quasi-significant decrease of the hormone’s gene expression (P = 0.052; Figure 4).
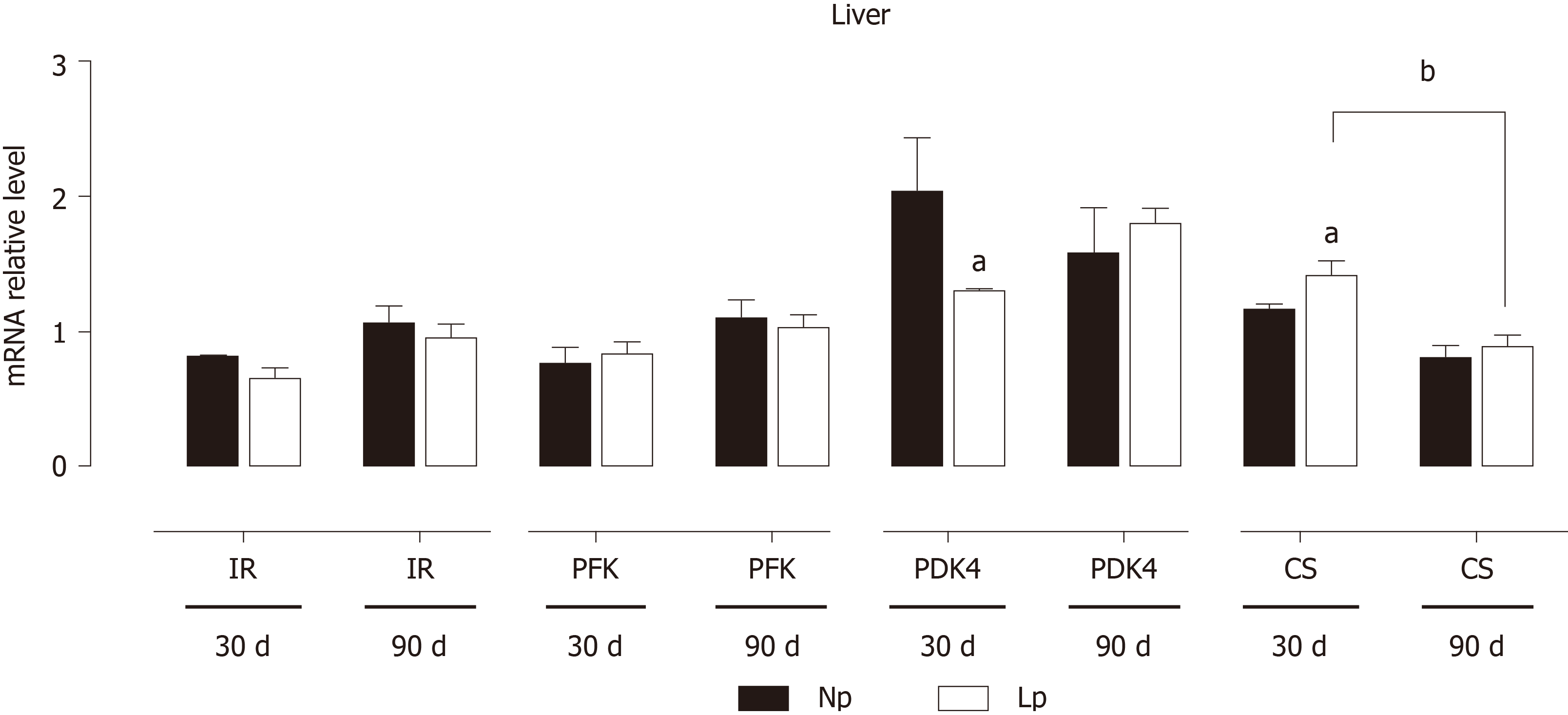
The expression of genes of the glycolytic pathway and Krebs cycle was evaluated in the liver at 30 d and 90 d (Figure 5). As observed in adipose tissue, PDK4 expression was reduced in the LP group at 30 d. Conversely, CS was significantly upregulated at 30 d in the LP group but then returned to levels comparable to the NP group at 90 d. The phosphofructokinase gene and insulin receptor gene did not show any difference between the two groups.
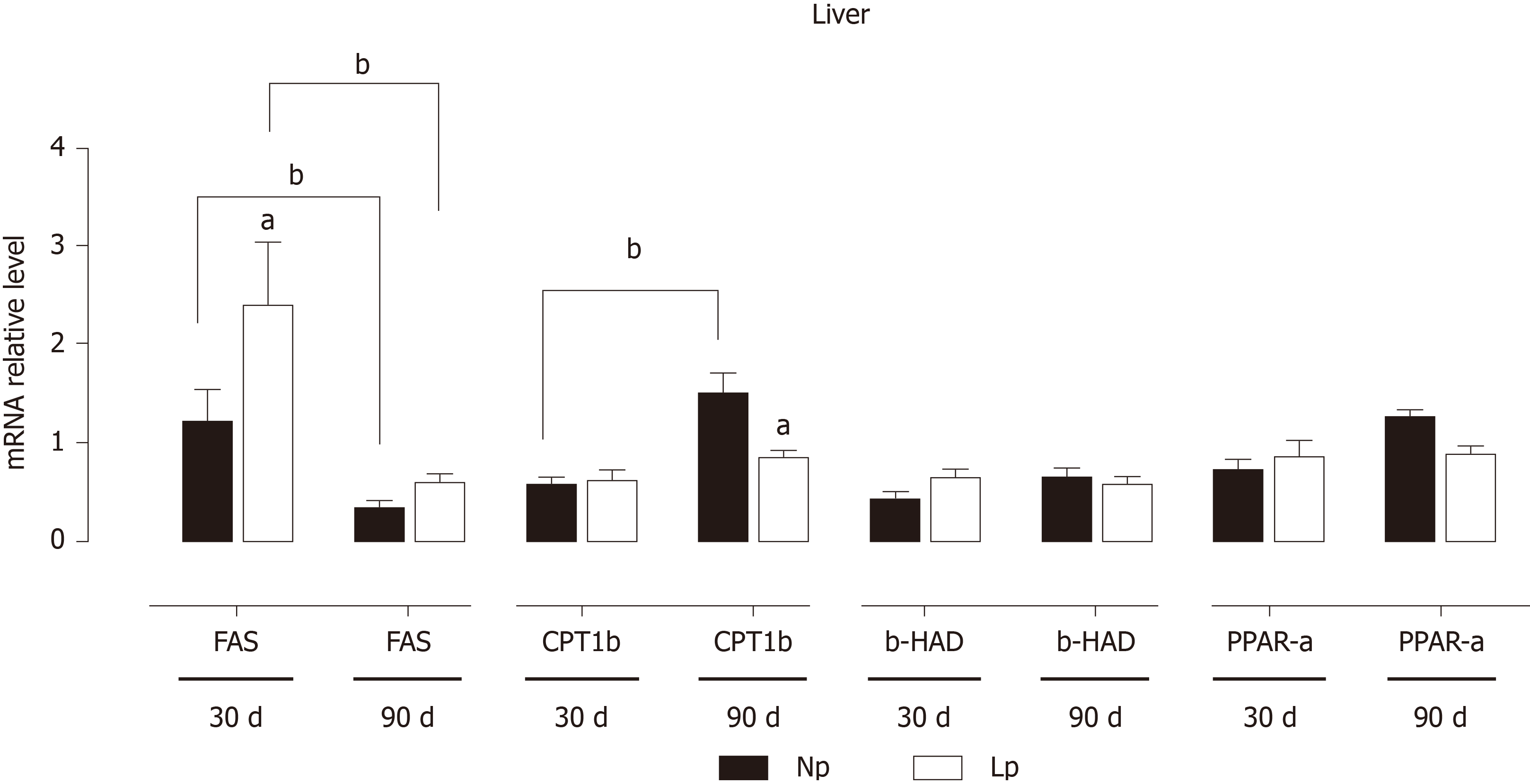
In the liver, genes related to fatty acid metabolism were also significantly modulated by the perinatal exposure to a LP diet (Figure 6). Fatty acid synthase, that displayed a strong age-dependent downregulation in both groups, is strongly upregulated in the LP group at 30 d, but then returned to levels comparable to the NP group at 90 d. On the contrary, carnitine palmitoylacyltransferase 1b (fatty acid transporter) showed a significant age-dependent increase in the NP group and was strongly downregulated at 90 d in the LP group.
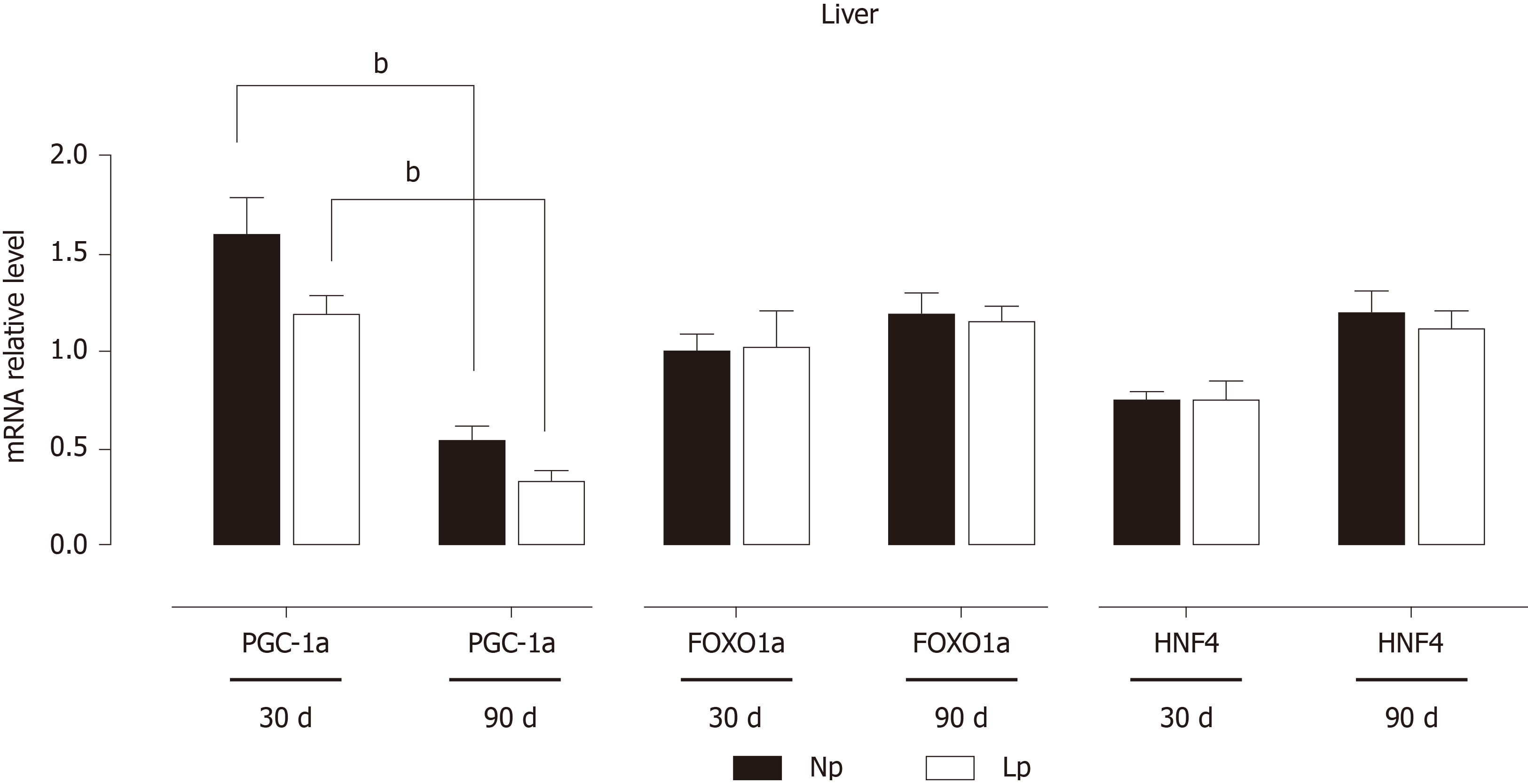
Transcriptional patterns in the liver are orchestrated by a group of key transcription factors that include peroxisome proliferator-activated receptor-alpha coactivator 1 alpha, forkhead box protein O1 and hepatocyte nuclear factor 4. None of these genes were significantly affected in the LP group, and the time-dependent decrease of the gene expression of peroxisome proliferator-activated receptor-alpha coactivator 1 alpha was maintained in both LP and NP groups (Figure 7).
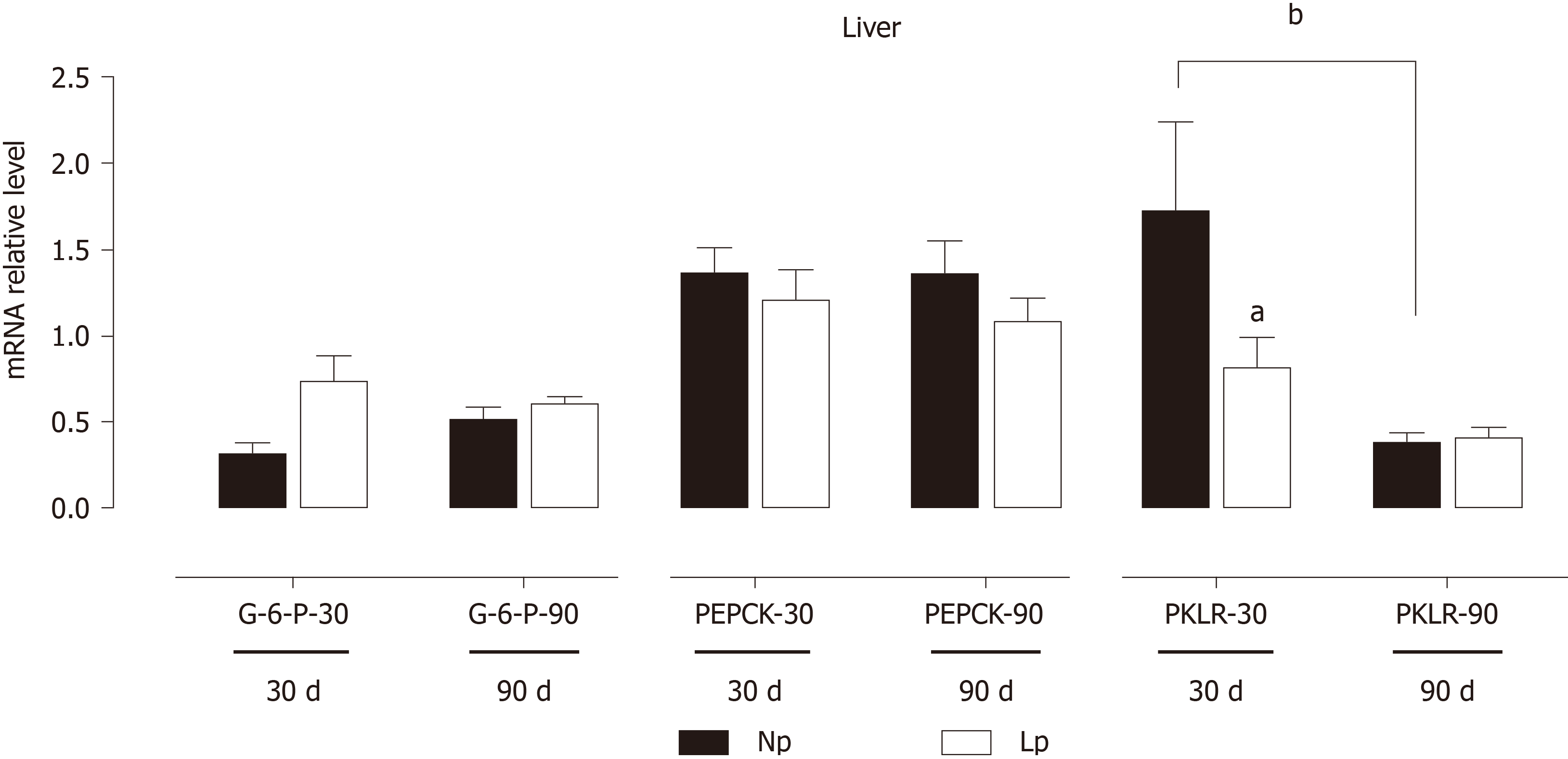
The liver is the quantitatively major organ responsible for gluconeogenesis, providing glucose supply during starvation. We evaluated the impact of the LP diet on gluconeogenic genes by measuring mRNA levels of glucose 6-phosphatase and PEPKC without detecting any LP-induced defect. However, pyruvate kinase L/R was significantly downregulated in the LP group at 30 d, suggesting an accumulation of phosphoenolpyruvate and potentially a higher gluconeogenic rate at 30 d due to higher abundance of the precursor (Figure 8).
The developmental origin of health and disease model supports the idea that exposure in the critical periods of development, represented by the prenatal and early postnatal life, to a poor nutritional status, toxic substances, drugs or other kind of stress can predispose to the development of disease states, including metabolic syndrome and diabetes during adult life[2]. In particular, early-life undernutrition, especially when associated to a nutritional transition leading to obesity, is associated to a higher incidence of diabetes in adult life[16]. One of the mechanisms proposed to explain such long lasting effects is the development of epigenetic alterations that sustain alterations of gene expression patterns from the young age into adulthood[17,18] by causing persistent alterations of DNA methylation patterns, histone post-translational modifications and microRNA patterns[19,20].
In a previous study, using a rat model of perinatal protein undernutrition throughout gestation and lactation, there were changes in the gene and protein expression levels of key enzymes of glycolysis and fatty acid oxidation pathways in the skeletal muscle of the progeny, observing both postnatal acute effects (at 30 d of age) and chronic effects (at 90 d of age), the latter being representative of adaptive processes[14]. Specifically, oxidative soleus muscle responded to a LP maternal diet by downregulating hexokinase 2 and PDK4 up to 90 d of age. For glycolytic extensor digitorum longus, the effects of a LP maternal diet were more pronounced at 30 d of age with a similar downregulation of genes coding for enzymes of the glycolytic pathway[10].
To obtain a more exhaustive description of the transcriptional disturbances induced by a perinatal LP diet in insulin-responsive tissues, we have now investigated the transcriptional changes resulting from a prenatal and postnatal exposure to LP in visceral adipose tissue and liver.
A key finding of our study is the short-term downregulation of PDK4, observed both in liver and adipose tissue and previously detected in soleus and extensor digitorum longus. PDK4 by phosphorylating the pyruvate dehydrogenase complex inhibits its activity and the resulting production of acetyl CoA. In the LP condition, PDK4 downregulation would therefore favor the activity of the pyruvate dehydrogenase complex and increase the glycolytic flux into the Krebs cycle[21,22].
Our observations pointing to a PDK4 modulation in the LP diet condition underscores the central role of this enzyme in regulating metabolic flexibility, which was also observed in the heart[23,24]. Interestingly, the first enzyme of the Krebs cycle, CS appears to be upregulated in the liver, while in the adipose tissue is downregulated, thus favoring the use of newly synthesized acetyl CoA as a lipogenic substrate[21]. To keep with this hypothesis, we observed a parallel decrease, at 30 d of age, of carnitine palmitoylacyltransferase 1b, the rate-controlling enzyme of long-chain fatty acid beta-oxidation pathway. At the more advanced age of 90 d, such transcriptional changes in the adipose tissue were lost, but a decrease in the expression of the gene coding for leptin was observed, which neared statistical significance (P = 0.052, Figure 4). We hypothesize that decreased leptin gene expression observed at 90 d of age in the LP group may be a compensatory mechanism to induce higher food uptake in the LP animals as this group has a significantly lower body weight both at 30 d and 90 d of age.
The liver also shows gene expression alterations that would favor anabolic pathways. At 30 d, fatty acid synthase is upregulated in the LP group suggesting increased hepatic lipogenesis. At the same time, gluconeogenesis may also be increased in the LP group. While phosphoenolpyruvate carboxykinase and glucose 6-phosphatase were not upregulated, we observed downregulation of the pyruvate kinase L/R at 30 d, which may favor the accumulation of phosphoenolpyruvate and thus funneling of this glycolytic intermediate into gluconeogenesis.
Taken together, the gene expression changes that we have observed in the liver and adipose tissue in male rats submitted to perinatal LP undernutrition suggest the occurrence of improved lipogenesis (in adipose tissue) and gluconeogenesis (in liver) that may provide a compensatory effect to counteract the early-life exposure to the perinatal LP diet.
Perinatal exposure to a poor nutritional environment predisposes the progeny to the development of metabolic disease at the adult age, both in experimental models and humans. Numerous adaptive responses to maternal protein restriction have been reported in metabolic tissues. However, the expression of glucose/fatty acid metabolism-related genes in adipose tissue and liver needs to be described.
To evaluate the metabolic impact of perinatal malnutrition, we determined malnutrition-associated gene expression alterations in liver and adipose tissue.
In the present study, we evaluated the alterations in gene expression of glycolytic/Krebs cycle genes (pyruvate dehydrogenase kinase 4 and citrate synthase), adipogenic and lipolytic genes and leptin in the adipose tissue of offspring rats at 30 d and 90 d of age exposed to maternal isocaloric low protein (LP) diet throughout gestation and lactation. We also evaluated these genes in the livers of the same animals as well as the gene expression of the transcription factors peroxisome proliferator-activated receptor gamma coactivator 1, forkhead box protein O1 and hepatocyte nuclear factor 4 and of gluconeogenic genes.
Research methods included animal husbandry, RNA extraction, reverse transcription and quantitative PCR and appropriate statistical analysis.
In the adipose tissue, we observed a transitory (i.e., at 30 d) downregulation of pyruvate dehydrogenase kinase 4, citrate synthase and carnitine palmitoylacyltransferase 1b gene expression. Such transcriptional changes did not persist in adult LP rats (90 d), but we observed a tendency towards a decreased gene expression of leptin (P = 0.052). The liver featured some gene expression alterations comparable to the adipose tissue, such as pyruvate dehydrogenase kinase 4 downregulation at 30 d, and displayed other tissue-specific changes, including citrate synthase and fatty acid synthase upregulation, but pyruvate kinase downregulation at 30 d in the LP group and carnitine palmitoylacyltransferase 1b downregulation at 90 d. These gene alterations, together with previously described changes in gene expression in skeletal muscle, may account for the metabolic adaptations in response to maternal LP diet and highlight the occurrence of persistent transcriptional defects in key metabolic genes that may contribute to the development of metabolic alterations during the adult life as a consequence of perinatal malnutrition.
We conclude that perinatal malnutrition relays long-lasting transcriptional alterations in metabolically active organs, i.e., the liver and adipose tissue.
Our observations lay the basis for possible future research directed to human studies.
Manuscript source: Invited manuscript
Specialty type: Endocrinology and metabolism
Country/Territory of origin: France
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Manenti A, Silva LD, Sun XT S-Editor: Zhang L L-Editor: Filipodia E-Editor: Liu MY
| 1. | Barker DJ. The fetal and infant origins of disease. Eur J Clin Invest. 1995;25:457-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 366] [Cited by in F6Publishing: 370] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 2. | Barker DJ. The origins of the developmental origins theory. J Intern Med. 2007;261:412-417. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1749] [Cited by in F6Publishing: 1700] [Article Influence: 100.0] [Reference Citation Analysis (0)] |
| 3. | Roseboom T, de Rooij S, Painter R. The Dutch famine and its long-term consequences for adult health. Early Hum Dev. 2006;82:485-491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 742] [Cited by in F6Publishing: 634] [Article Influence: 35.2] [Reference Citation Analysis (0)] |
| 4. | de Brito Alves JL, Nogueira VO, de Oliveira GB, da Silva GS, Wanderley AG, Leandro CG, Costa-Silva JH. Short- and long-term effects of a maternal low-protein diet on ventilation, O₂/CO₂ chemoreception and arterial blood pressure in male rat offspring. Br J Nutr. 2014;111:606-615. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 5. | Fragoso J, Carvalho Jurema Santos G, Thomaz da Silva H, Oliveira Nogueira V, Loizon E, Vidal H, Costa-Silva JH, da Silva Aragão R, Pirola L, Leandro CG. Maternal physical activity-induced adaptive transcriptional response in brain and placenta of mothers and rat offspring. J Dev Orig Health Dis. 2020;11:108-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Nogueira VO, Andrade LDS, Rocha-Júnior RL, Melo PED, Helvécio E, Fontes DAF, Romão TP, Leandro CG, Costa-Silva JH. Maternal physical activity prevents the overexpression of hypoxia-inducible factor 1-α and cardiorespiratory dysfunction in protein malnourished rats. Sci Rep. 2019;9:14406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046-17049. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2120] [Cited by in F6Publishing: 1806] [Article Influence: 112.9] [Reference Citation Analysis (0)] |
| 8. | Phipps K, Barker DJ, Hales CN, Fall CH, Osmond C, Clark PM. Fetal growth and impaired glucose tolerance in men and women. Diabetologia. 1993;36:225-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 378] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 9. | Ravelli AC, van der Meulen JH, Michels RP, Osmond C, Barker DJ, Hales CN, Bleker OP. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1102] [Cited by in F6Publishing: 966] [Article Influence: 37.2] [Reference Citation Analysis (0)] |
| 10. | de Brito Alves JL, Toscano AE, da Costa-Silva JH, Vidal H, Leandro CG, Pirola L. Transcriptional response of skeletal muscle to a low protein perinatal diet in rat offspring at different ages: The role of key enzymes of glucose-fatty acid oxidation. J Nutr Biochem. 2017;41:117-123. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Jones RH, Ozanne SE. Fetal programming of glucose-insulin metabolism. Mol Cell Endocrinol. 2009;297:4-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Martin-Gronert MS, Fernandez-Twinn DS, Bushell M, Siddle K, Ozanne SE. Cell-autonomous programming of rat adipose tissue insulin signalling proteins by maternal nutrition. Diabetologia. 2016;59:1266-1275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 13. | Campisano SE, Echarte SM, Podaza E, Chisari AN. Protein malnutrition during fetal programming induces fatty liver in adult male offspring rats. J Physiol Biochem. 2017;73:275-285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Pettersen IKN, Tusubira D, Ashrafi H, Dyrstad SE, Hansen L, Liu XZ, Nilsson LIH, Løvsletten NG, Berge K, Wergedahl H, Bjørndal B, Fluge Ø, Bruland O, Rustan AC, Halberg N, Røsland GV, Berge RK, Tronstad KJ. Upregulated PDK4 expression is a sensitive marker of increased fatty acid oxidation. Mitochondrion. 2019;49:97-110. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 63] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 15. | Reeves PG, Nielsen FH, Fahey GC. AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939-1951. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5738] [Cited by in F6Publishing: 5770] [Article Influence: 186.1] [Reference Citation Analysis (0)] |
| 16. | Stein AD, Obrutu OE, Behere RV, Yajnik CS. Developmental undernutrition, offspring obesity and type 2 diabetes. Diabetologia. 2019;62:1773-1778. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 17. | Portha B, Fournier A, Kioon MD, Mezger V, Movassat J. Early environmental factors, alteration of epigenetic marks and metabolic disease susceptibility. Biochimie. 2014;97:1-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Bianco-Miotto T, Craig JM, Gasser YP, van Dijk SJ, Ozanne SE. Epigenetics and DOHaD: from basics to birth and beyond. J Dev Orig Health Dis. 2017;8:513-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 176] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 19. | Ruchat SM, Hivert MF, Bouchard L. Epigenetic programming of obesity and diabetes by in utero exposure to gestational diabetes mellitus. Nutr Rev. 2013;71 Suppl 1:S88-S94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 90] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 20. | Fernandez-Twinn DS, Hjort L, Novakovic B, Ozanne SE, Saffery R. Intrauterine programming of obesity and type 2 diabetes. Diabetologia. 2019;62:1789-1801. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 144] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 21. | Zhang S, Hulver MW, McMillan RP, Cline MA, Gilbert ER. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab (Lond). 2014;11:10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 297] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 22. | Sugden MC, Holness MJ. Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab. 2003;284:E855-E862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 350] [Cited by in F6Publishing: 361] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 23. | Schafer C, Young ZT, Makarewich CA, Elnwasany A, Kinter C, Kinter M, Szweda LI. Coenzyme A-mediated degradation of pyruvate dehydrogenase kinase 4 promotes cardiac metabolic flexibility after high-fat feeding in mice. J Biol Chem. 2018;293:6915-6924. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Crewe C, Schafer C, Lee I, Kinter M, Szweda LI. Regulation of Pyruvate Dehydrogenase Kinase 4 in the Heart through Degradation by the Lon Protease in Response to Mitochondrial Substrate Availability. J Biol Chem. 2017;292:305-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |