Published online Nov 15, 2016. doi: 10.4251/wjgo.v8.i11.772
Peer-review started: April 1, 2016
First decision: June 11, 2016
Revised: August 3, 2016
Accepted: August 30, 2016
Article in press: August 31, 2016
Published online: November 15, 2016
Gastrointestinal malignancies are among the leading causes of cancer-related deaths worldwide. Like all human malignancies they are characterized by accumulation of mutations which lead to inactivation of tumor suppressor genes or activation of oncogenes. Advances in Molecular Biology techniques have allowed for more accurate analysis of tumors’ genetic profiling using new breakthrough technologies such as next generation sequencing (NGS), leading to the development of targeted therapeutical approaches based upon biomarker-selection. During the last 10 years tremendous advances in the development of targeted therapies for patients with advanced cancer have been made, thus various targeted agents, associated with predictive biomarkers, have been developed or are in development for the treatment of patients with gastrointestinal cancer patients. This review summarizes the advances in the field of molecular biomarkers in tumors of the gastrointestinal tract, with focus on the available NGS platforms that enable comprehensive tumor molecular profile analysis.
Core tip: Gastrointestinal cancers are among the leading causes of cancer morbidity and mortality worldwide. So far, various targeted agents associated with predictive biomarkers are available or are under development for the selection of treatment in patients with gastrointestinal cancer. Advances in high-throughput technologies such as next generation sequencing and the use of noninvasive materials for tumor characterization, such as liquid biopsies, will facilitate tumor molecular profiling and lead to the establishment of further targeted treatment therapies.
- Citation: Papadopoulou E, Metaxa-Mariatou V, Tsaousis G, Tsoulos N, Tsirigoti A, Efstathiadou C, Apessos A, Agiannitopoulos K, Pepe G, Bourkoula E, Nasioulas G. Molecular predictive markers in tumors of the gastrointestinal tract. World J Gastrointest Oncol 2016; 8(11): 772-785
- URL: https://www.wjgnet.com/1948-5204/full/v8/i11/772.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v8.i11.772
The comprehension of the importance of tumor biology has led to the development of new drugs that target specific molecules involved in carcinogenesis. The efficacy of such targeted therapies often depends on the presence or absence of gene alterations that encode for the protein-target or for proteins involved in the molecular pathway targeted by the specific medication. This targeted therapeutical approach is based on the tumor’s molecular analysis in order to select patients with increased probability to respond to the treatment given. Advances in Molecular Biology techniques have permitted comprehensive tumor genomic profiling using new breakthrough technologies such as next generation sequencing (NGS)[1-3].
Nowadays, biomarkers are used in the management of patients with cancer and can be divided into predictive and prognostic. Prognostic biomarkers are defined as those that provide information on the possible outcome of cancer in a particular patient regardless of treatment. Predictive biomarkers provide information on the potential benefit of the administrated treatment (whether this relates to the tumor’s volume shrinkage or survival). Predictive biomarkers can be used to identify subpopulations of patients that are likely to respond to a particular treatment[1]. They can be subdivided in positive and negative predictive biomarkers. The first are used for positive selection of patients who are likely to benefit from targeted therapy, whereas the latter for resistance prediction[1].
The number of genes involved in targeted therapy (predictive biomarkers), is increasing continuously. The simultaneous analysis of these biomarkers is feasible using molecular biology technologies that allow accurate, fast and cost effective genomic analysis with limited requirements concerning the quantity of the biological material used[1,4]. ΝGS has all the features required to carry out such analysis and provides simultaneous information on a large number of actionable alterations in tumor tissues and thus a more precise molecular characterization of the tumor. The massive amount of genetic information produced is the main advantage of this technology. However, it also constitutes its main challenge, requiring usage of appropriate software and bioinformatics tools, along with web-based tools for data analysis, management and interpretation[5].
The human gastrointestinal (GI) tract is an organ system which includes all structures between mouth and anus and is divided into upper (buccal cavity, pharynx, esophagus, stomach and duodenum) and lower (small and large intestine) GI tracts. GI cancers are complex diseases and refer to malignant conditions that affect the digestive system. The current review will focus on the advances in the field of molecular biomarkers and the application of high throughput technologies, in the most common tumors of the gastrointestinal tract.
Esophageal cancer is one of the most aggressive malignancies with a rapidly increasing incidence rate in the recent decades. There are two predominant histological types: Esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) of the distal esophagus and the gastroesophageal junction. Smoking and heavy alcohol consumption are associated with increased risk of ESCC, while gastroesophageal reflux disease and Barrett esophagus may of increase the risk of EAC[6].
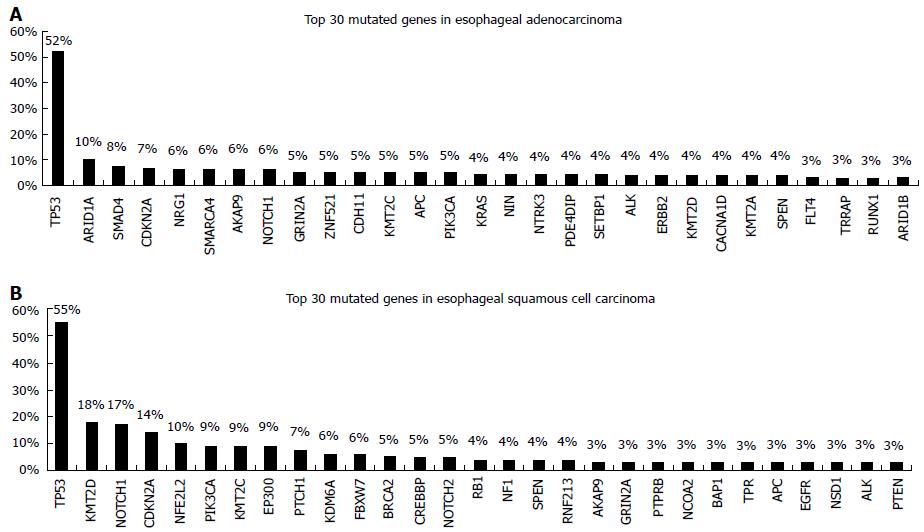
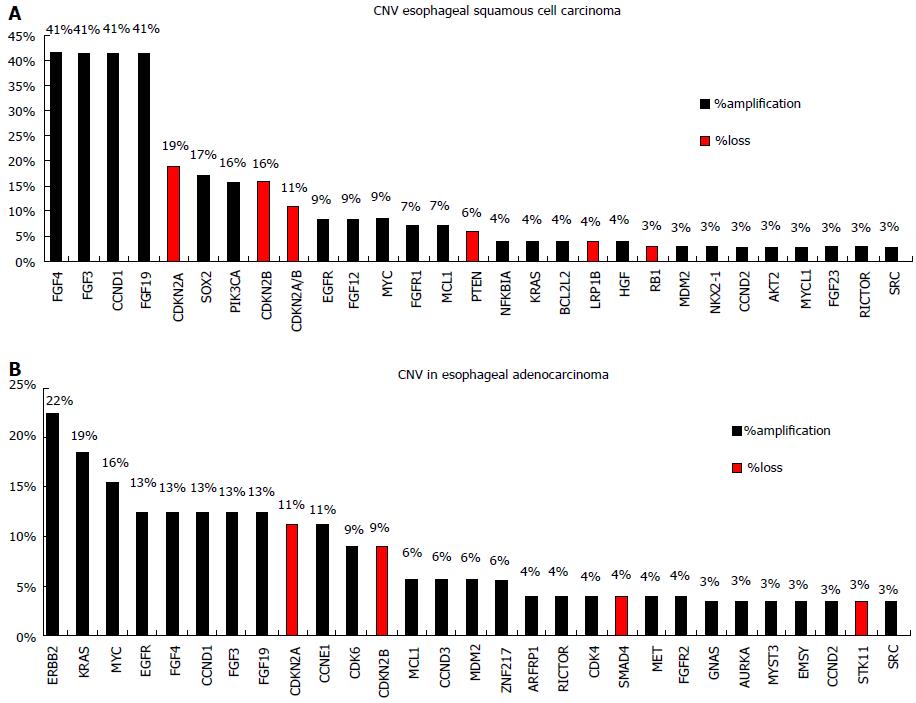
TP53 mutations are identified in about 50% of esophageal cancers and are associated with poorer survival[7]. Apart from mutations in TP53 ESCC and EAC seem to differ significantly in the genetic alterations pattern. Agrawal et al[8] using NGS reported a substantial disparity in the spectrum of mutations, with more insertions/deletions in ESCCs, A:T>C:G transversions in EACs, and C:G>G:C transversions in ESCCs. Inactivating mutations of NOTCH1 are identified in about 20% of ESCCs but not in EACs. Somatic aberrations in EACs are mainly identified in the Wnt, cell cycle and Notch pathways[9,10]. A number of genes that can be used as predictive markers for targeted therapy have been explored for somatic mutations in esophageal adenocarcinoma, including genes of the RAF/MEK/ERK (MAPK) kinase pathway such as EGFR, BRAF, KRAS, PIK3CA[11]. However the reported frequency of somatic mutations identified appears to be low and this is obvious when accessing data from the Catalogue of Somatic Mutations in Cancer database (COSMIC, cancer.sanger.ac.uk) (Figure 1), which is currently the most comprehensive global resource accessing the world literature on somatic mutations in human cancer[12]. In a recent study, NGS-based comprehensive genomic profiling was used to analyze ESCC and EAC tumors[13]. The analysis showed that the esophageal histotypes differ significantly in genomic alterations profile, with KRAS and ERBB2 far more frequently altered in EAC compared to ESCC. In contrast, genes of the mechanistic target of rapamycin (MTOR) pathway (PIK3CA and PTEN) and NOTCH1 are more frequently altered in ESCC compared to EAC. They also have different amplification patterns (Figure 2).
ESCC and EAC also differ in the gene amplification and/or protein (over)expression of the receptor tyrosin kinases (RTKs) EGFR and HER2 making them possible prognostic markers and as therapeutic targets[7,14]. EGFR is frequently overexpressed in ESCCs, while HER2 overexpression occurs mainly in EACs. Thus the trastuzumab-platinum regimen is currently used for the 15% of the EACs patients that test positive for HER2 (ERBB2) amplification or overexpression[13,14].
Numerous preclinical studies addressed EGFR and HER2 inhibition in esophageal cancer cell lines and there are various phase II/III clinical trials testing EGFR, HER2, and VEGF targeting therapies for esophageal cancer[7,15]. However, the results obtained to date do not allow the use of these agents in clinical practice. Upon trial completion several clinical studies have concluded, that in order to select patients who will respond to RTK-targeted therapy, there is a need for molecular patient stratification before treatment.
In a disease with historically poor outcomes and limited options, comprehensive genomic profiling of relapsed and refractory cancers, including distinct evaluation for EAC and ESCC has led to promising information suggesting targeted therapies for future consideration.
Gastric cancer (GC) develops from the inner lining of the stomach and is a very aggressive malignancy, with poor prognosis and very high cancer related mortality. The high mortality rate is largely due to the late stages of cancer diagnosis and to the lack of effective medical treatment for advanced stages of this disease[16,17]. The majority of these cancers are adenocarcinomas and can be further classified as diffuse (poorly differentiated) or intestinal (well-differentiated) types that have distinct molecular profiles[16,17].
Concerning the causes of GC, it can be viewed as a multifactorial disease since many inherited and environmental factors play a role in its development. Infectious agents such as Helicobacter pylori and EBV, dietary habits and the genetic background are considered as causative agents[17].
Given the variety of causes of the disease, it is not surprising that these tumors present a high level of biological heterogeneity, with distinct molecular profile for each patient. Genetic and epigenetic alterations play important role in GCs therefore, targeted therapy based on the biology of the individual patient could improve treatment outcome[17-19].
ERBB2 amplifications occur frequently in gastric tumors (2%-27%)[12,20]. Trastuzumab, a monoclonal antibody against HER2/neu receptor, was the first targeted agent to be used in the treatment of ERBB2-positive advanced gastric and gastroesophageal junction (GEJ) adenocarcinoma[20]. Several molecular targeted agents associated with a survival benefit in other cancer types are now under clinical investigation for the treatment of gastric cancer, including inhibitors of EGFR, MET, FGFR, VEGF, and PI3K[18,20]. Additionally, CDH1 gene mutations at the somatic level are considered of prognostic significance[19].
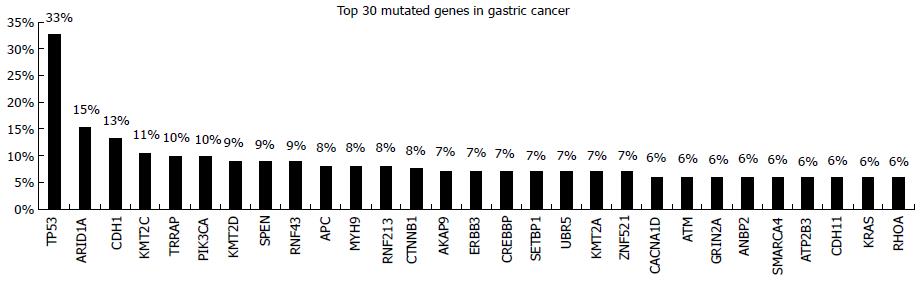
Several studies have investigated gastric cancer’s molecular profile using whole genome as well as targeted NGS approaches[19,21-23]. The presence of somatic mutations and copy number variations (CNV) in many cancer driver genes has been revealed. Among the cancer genes frequently mutated in gastric cancer P53, ARD1A, CDH1, PIK3CA, APC, CTNNB1, ERBB3, ATM, KRAS are the most important prognostic and/or predictive markers (Figure 3)[12]. Consequently, molecular profile-directed therapy seems to be a promising strategy for the improvement of standard chemotherapy effectiveness.
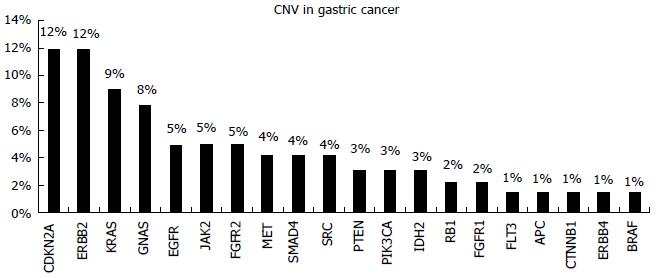
CNVs have been observed for HER2, FGFR2, and MET that represent viable treatment targets for which therapeutics are already approved or are currently under investigation[24] (Figure 4).
The high heterogeneity of these tumors triggered scientists to attempt their molecular characterization. In a study conducted by The Cancer Genome Atlas, molecular classification four major genomic subtypes of gastric cancer were defined: EBV-infected tumors; MSI tumors; genomically stable tumors; and chromosomally unstable tumors[21].
In a recent study, Li et al[19], using whole genome NGS data were able to classify gastric cancers into regular (86.8%) and hyper-mutated (13.2%) subtypes based on mutation burden. Additionally, in the “regular” mutated cohort a further classification, using 40 significantly mutated genes, could be obtained, separating the patients to S1 and S2 subtypes with distinct prognostic outcomes.
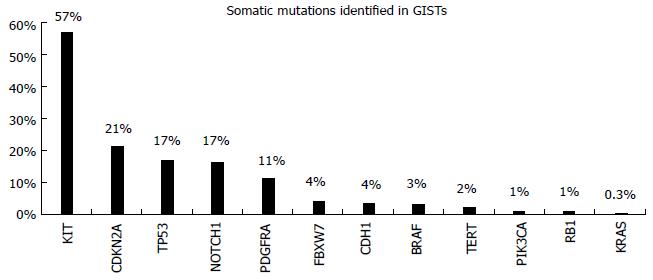
Gastrointestinal stromal tumors (GISTs) are rare tumors of the gastrointestinal tract. They are mesenchymal in origin and are characterized by overexpression of the KIT protein[25]. Morphological diagnosis based on microscopic examination is the standard for GIST diagnosis. They occur anywhere within the GI tract, but they are most common in the stomach (60%) or small intestine (30%)[26]. Their diagnosis is based on the expression of the transmembrane tyrosine kinase (TK) receptor, KIT, since 95% of GISTs express CD 117 antibody. In 80% of the cases, somatic mutations in the cKIT gene are observed, resulting in constitute receptor activation. Additionally, in 5%-10% of the cases without cKIT mutations, the TK receptor PDGFRA is mutated[27]. The mutation spectrum of these tumors is very limited as we observe in COSMIC database (Figure 5)[12]. Tyrosine kinase inhibitors (TKIs) like imatinib, sunitinib, and more recently regorafenib, have proven effectiveness in suppressing the growth of metastatic GISTs, allowing patients to live far longer than during the previous era of ineffective chemotherapy[28-30]. The response to targeted therapy with TKIs is mainly dependent on the presence and type of mutation. Patients with mutations in exon 11 of the cKIT are highly responsive to imatinib, while the presence of a mutation in exon 9 of this gene implies intermediate response rates and necessitates a double dose of drug administration. Furthermore, resistance mutations to imatinib are also observed in the cΚΙΤ/PDGFRA genes. These mutations can be present in the primary tumor or arise as a result of the drug administration (secondary mutations)[29].
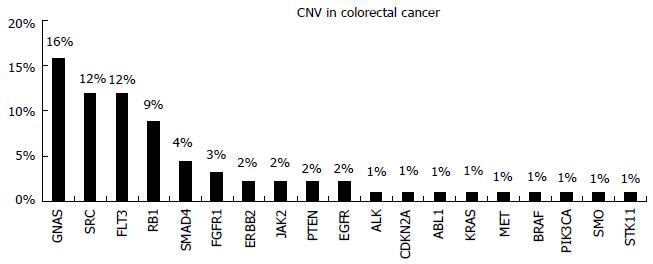
The cancer of colon and rectum (colorectal, CRC) is the third most common cancer worldwide with 95% of these tumors being classified as adenocarcinomas. It’s a leading cause of cancer related deaths; however, colorectal cancer mortality is declining in the last decades, mainly due to early diagnosis and the presence of new therapy strategies[31,32]. This malignancy is one of the first paradigms of the benefits that can be derived from the application of personalized treatment in cancer therapy[33-35].
Genetic alterations in colorectal cancer include mainly single-base substitutions (SBS). Nevertheless, small insertions and deletions (indels), amplifications, homozygous deletions and translocations can also be observed[36].
Five hundred and seventy-two cancer relevant genes are included in the Catalogue of Somatic Mutations in Cancer (COSMIC, cancer.sanger.ac.uk)[12]. Somatic mutations in CRC cancer are observed in the majority of these genes. The pattern of genomic alterations was identified through Massively Parallel Sequencing studies, revealing the inter- and intra-tumor genetic heterogeneity of these tumors[37-39]. Apart from single base substitutions, gene amplifications are also observed (Figure 6). Mutations in many important biologic pathways occur. In Table 1 the frequency of mutations in important molecular signaling pathways and the related therapies are represented. The gene mutation frequency is calculated using data from samples analysed by whole genome screening in the COSMIC database. The information concerning the therapies targeting each pathway was retrieved from MyCancer Genome knowledge database (http://www.mycancergenome.org/), that provides reliable information concerning important cancer related genes and their correlation with treatment options (Table 1)[40].
| Biologic pathway | Frequency of mutation in genes involved in each pathway | Therapies that target the pathway |
| Beta-catenin/WNT signaling | 75% | FZD, GSK inhibitors |
| Cell cycle control | 68% | CDK, CDK1, CDK2, CDK4/6 inhibitors |
| Receptor tyrosine kinase/growth factor signaling | 67% | Therapeutic antibodies/tyrosine kinase inhibitors |
| MAP kinase signaling | 61% | BRAF, ERK, MEK AND SRC inhibitors |
| PI3K/AKT1/MTOR | 52% | Allosteric mTORC1 inhibitors/mTORC1/2 catalytic inhibitors |
| DNA damage/repair | 48% | PARP INHIBITORS |
| TGFbeta signaling | 37% | TGFBR1 inhibitors |
| Chromatin remodeling/DNA methylation | 32% | DNMT inhibitors, Histone deactylase |
| Immune checkpoints | 26% | Anti-CTLA4 antibodies, anti-PD-1 antibodies, Anti-PD-L1 antibodies, Immunotherapies |
| JAK/STAT signaling | 23% | JAK inhibitors |
| Hedgehog signaling | 12% | SMO inhibitors |
The RAS proto-oncogenes (HRAS, KRAS and NRAS) encode a family of highly homologous proteins. They participate in a signal transduction cascade, namely the RAS/RAF/MEK/ERK pathway, which regulates the growth and survival properties of the cells. They are controlled by extracellular signals transmitted by the transmembrane receptor tyrosine kinase (TK), EGFR[34]. This TK and the RAS/RAF/MEK/ERK pathways it controls, play an important role in colorectal carcinogenesis, making it a good target for biological therapy of this disease.
Two monoclonal antibodies were designed as effective inhibitors of EGFR. Cetuximab (Erbitux, Merck KgaA, Darmstadt, Germany) is a chimeric mouse/human antibody, and Panitumumab (Vectibix, Amgen Thousand Oaks, CA, United States), is a fully human antibody[33-35]. They both target the extracellular domain of the EGFR protein and compete with ligands, blocking ligand induced intracellular signal transmission. However, anti-EGFR treatment is not effective in patients harboring activating mutations in genes that participate in the intracellular transduction RAS/RAF/MEK/ERK pathway. This is due to the constitutive, independent of ligand, activation of the mutated proteins[33].
In total, activating mutations in the RAS genes, mainly in codons 12, 13 or 61, occur in approximately 20% of all human cancers. Mutations in KRAS account for about 85% of all RAS mutations in human tumors, NRAS for about 15%, and HRAS for less than 1%[41,42]. Which particular RAS gene is mutated seems to be tumor specific. Colonic, pancreatic and lung cancers have high frequencies of KRAS mutations[41,42].
Acquired mutations in KRAS and NRAS are commonly used to identify colorectal cancer patients who are unlikely to benefit from anti-EGFR therapy. Approximately 40% of colorectal cancer tumors harbor mutations in the KRAS gene, with the majority of the mutations occurring in codons 12, 13 and 61. In 5% of the colorectal cancer cases a mutation occurs in the NRAS gene[35,41].
Another important gene of the RAS/RAF/MEK/ERK pathway is BRAF. Mutations in the BRAF gene (exons 11 and 15) have been detected in about 12% of colorectal cancers and are mutually exclusive with RAS mutations[12,43,44]. The BRAF activating aberrations, result in constitutive BRAF kinase activity, ERK signaling, proliferation and transformation[44]. The majority of BRAF mutations are observed in exon 15 (codon 600) and a minority of mutations are observed in exon 11[44,45].
Several studies have reported that patients with metastatic CRC (mCRC) that harbor BRAF mutations do not respond to the anti-EGFR antibody agents cetuximab or panitumumab[43,46,47]. However it is unclear if the presence of BRAF mutations in CRC cancer can be used as a predictive marker or if it has only a prognostic value, independent of treatment, since different studies arrive at controversial conclusions concerning its clinical significance[45,48].
The PIK3CA gene encodes the catalytic subunit of phosphatidylinositol 3-kinase while belongs to a family of lipid kinases. These kinases regulate a diverse range of cellular processes including cell proliferation, adhesion, survival, and migration[49]. Mutations in PIK3CA stimulate downstream AKT-mTOR signaling pathways, thereby promoting growth-factor independent growth, cell invasion and metastasis. PIK3CA mutations have been reported in multiple malignancies, including approximately 25% of gastric, 4% of lung, 25% of breast, and 20% of colorectal cancers[50]. The majority (80%) of PIK3CA mutations cluster in 2 “hotspot” regions, the helical domain (exon 9) and the kinase domain (exon 20). Concomitant PIK3CA mutations in exons 9 and 20 seem to be linked to significantly worse cancer-specific survival[51]. PIK3CA mutations may also be associated with clinical resistance to EGFR-targeted monoclonal antibodies, but there have been conflicting results[52-55]. A meta-analysis comprising 864 patients, from 11 studies, with colorectal cancer treated with cetuximab or panitumumab-based therapy showed that PIK3CA mutations, particularly in exon 20, are significantly associated with worse response and shorter progression-free and overall survival[51]. Somatic PIK3CA mutations have also been associated with superior colorectal cancer-specific survival in patients who regularly intake aspirin after diagnosis[56]. PIK3CA activating mutations may also predict sensitivity to inhibitors of the PI3K-AKT-mTOR pathway[57]. Inhibitors of mTOR, PI3K, and AKT, alone or in combination with other therapies are in clinical trials in solid tumors[58,59].
A number of rare gene mutations occurring in the PI3K/AKT/mTOR pathway are potentially actionable in colorectal cancer. PTEN is a key negative regulator of the PI3K pathway. PTEN gene mutations occur in about 5% of colorectal cancers[12,60,61]. PTEN inactivating mutations and PTEN loss have as a consequence the upregulation of the PI3K/ AKT pathway[54,61].
Currently, the prognostic and predictive significance of PTEN mutations or PTEN loss of expression is under investigation. In retrospective studies, PTEN loss was associated with decreased sensitivity of colorectal cancer tumors to anti-EGFR antibodies[60-62]. Preclinical data and in vitro studies suggest that it may be associated with sensitivity to PI3K and mTOR inhibitors. Based on these data, several PI3K and mTOR inhibitors are currently in clinical trials for the treatment of patients with PTEN-deficient cancers[61,63].
AKT (Protein kinase B, PKB) is a serine/threonine kinase that is encoded by three genes AKT1, 2 and 3. Somatic mutations in the AKT1 gene occur in colorectal cancer in about 1% of the cases according to the COSMIC database[12]. The only mutation observed is the activating mutation E17K, which is also observed in other types of cancer[64]. AKT1 is a critical component of the PI3K/AKT/mTOR pathway, thus it has become an attractive target for therapeutic intervention[49,65]. AKT1 E17K mutations have also been associated with primary resistance to cetuximab[66].
In colorectal cancer, DNA mismatch repair (MMR) system deficiency occurs frequently and leading to microsatellite instability (MSI). These are small changes in the DNA sequence that occur during DNA replication and are usually additions or deletions of one or two nucleotide bases[67,68]. This phenomenon is most common in areas of the genome that contain repetitive DNA sequences with a repeat unit, from one to four bases, and are known as Microsatellite regions[67,69]. The presence of microsatellite instability (MSI High) is a good prognostic marker[67,70]. It is found in 90% of cases of tumors arising patients with hereditary Lynch syndrome and in 10%-15% of the sporadic cancers[71]. Sporadic MSI-H tumors can be distinguished from the hereditary ones through somatic mutation analysis of the BRAF gene or loss of MLH1 expression[72,73]. Somatic mutations in the BRAF gene occurs only in sporadic MSI-H tumors but not in Lynch-associated CRC cancers. Similarly, hMLH1 promoter methylation rarely occurs in Lynch syndrome-associated cancers, while is common in sporadic MSI-high cancers[73].
Recent studies have indicated that MSI high tumors, both sporadic and hereditary, are less aggressive and are related with low probability of lymph node and distant recurrences[70]. In addition they respond differently to chemotherapy, since they are less sensitive to Topoisomerase inhibitors and to the treatment with 5-fluorouracil[74-76]. Additionally, it has been proposed that MMR-deficient tumors are more responsive to PD-1 blockade than the mismatch repair-proficient tumors[77].
Gene expression profiling (GEP) is an emerging tool which aims to identify differentially expressed subsets of genes (gene signatures) in groups of patients with distinct clinical outcomes. Several commercial GEP tests are currently available for stage II/IIIa colorectal cancer patients. Oncotype DX® Colon Cancer Assay (Genomic Health, Inc., Redwood City, CA) and ColoPrint (Agendia, BV, Amsterdam, Holland) are the most promising gene signature tests[78]. Both tests provide a risk of recurrence, but OncotypeDx has the advantage of being applicable to Formalin Fixed Paraffin Embedded (FFPE) tissue for analysis, while ColoPrint requires fresh tissue which is not easily available. Oncotype DX® Colon Cancer Assay is a quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay on RNA extracted from FFPE tumor tissue, used to assess risk of recurrence in stage II colon cancer patients at three years after surgery[79-81]. The test uses gene expression profiling of 12 genes that include seven prognostic genes and five reference genes, in order to provide a Recurrence Score (RS). The RS allows patients and physicians to determine the risk of developing a distant metastasis. In a retrospectively performed study by Yothers et al[82], RS was the strongest predictor of disease recurrence independent of other factors, such as T-stage, mismatch repair status, number of nodes examined, tumor grade, and lymphovascular invasion. The greatest utility of this test seems to be in the prediction of recurrence risk in T3, mismatch repair-proficient (MMR-P) stage II colon cancer patients[82]. However, it has also been validated in stage III patients with very promising results[83]. The continuous RS predicted recurrence as well as disease free survival (DFS) and overall survival (OS) in all three patient subgroups (stage II, IIIA/B, and IIIC). The use of this assay could lead to overall reduction in adjuvant chemotherapy use in this subgroup of stage II/III colon cancer patients[83].
Until recently, the best material for somatic mutation analysis was considered formalin fixed paraffin embedded (FFPE) tumor tissue. FFPE tissue is a widely available material, easy to use and maintain. In addition, the cancer tissue can be selected and mutation analysis can be performed without contamination by normal tissues[42]. This increases the sensitivity of mutation detection assays which is very important because, due to tumor heterogeneity, somatic mutations can sometimes be present at a very low percentage. However, FFPE tissue material also has several disadvantages[84,85]. First of all in some cases it is not available. This is the case of non-operable tumors. Furthermore, the examination of a limited tumor area present in a paraffin block doesn’t take into account tumor molecular heterogeneity and does not necessarily reflect the molecular profile of other tumors or metastasis that are eventually present in the patient’s body[85]. Additionally, the genetic material obtained, due to the paraffinization process, is sometimes of very bad quality and not suitable for molecular analysis[86]. Most importantly, tumor molecular profile is altered mainly following therapy and those alterations cannot be detected by analyzing the primary tumor material[87].
Nowadays, the presence of cell-free tumor derived nucleic acids (ctDNA/ctRNA) in cancer patients body fluids (plasma, serum, Broncho-alveolar, urine, stool, etc.) is well documented[84]. The term Liquid Biopsy has emerged indicating the use of these noninvasive materials for tumor characterization. The mutation status detected in a liquid biopsy reflects the status present in the patient’s tumor. Furthermore Liquid Biopsy analyses take into account intra-tumor or inter metastatic heterogeneity and could eventually detect more tumor alterations compared with the analysis of a specific area in a FFPE tissue[83,84]. A variety of sensitive methods can be used for the detection of ctDNA in plasma samples, including digital PCR, Real time PCR, Arms PCR and NGS[84].
The utility of liquid biopsy analysis has been proven in many studies that used ctDNA for the detection of tumor specific alterations in plasma with prognostic and/or predictive significance[87-98]. A liquid biopsy analysis can be performed before treatment as well as for patients monitoring during therapy. It is also very helpful in the detection of secondary mutations that arise due to targeted therapy. The detection of secondary mutations in plasma can modify the treatment strategy for those patients (Table 2).
| Gene | Mutation type | Tumor type | Possible clinical implications | Ref. |
| KRAS/NRAS | Point mutation/amplification | Colorectal/pancreatic cancer | Resistance to anti-EGFR therapy/sensitivity to MEK inhibitors | [88-90] |
| BRAF | Point mutation | Colorectal cancer | Resistance to anti-EGFR therapy/sensitivity to MEK inhibitors | [88,91] |
| MET | Amplification/alteration | Colorectal/esophageal cancer | Resistance to anti-EGFR therapy/sensitivity to MEK inhibitors | [92-94] |
| HER2 | Amplification | Colorectal/gastric cancer | Resistance to anti-EGFR therapy/sensitivity to Anti HER2 inhibitors | [95,96] |
| EGFR | Point mutation | Colorectal/pancreatic cancer | Panitumumab | [97,98] |
| PIK3CA | Point mutation | Colorectal/pancreatic cancer | mTOR inhibitor | [89] |
| Ckit | Point mutations | GISTS | Imatinib or dose escalation or alternative TKIs | [99,100] |
| PDGFRA | Point mutations | GISTS | Imatinib or dose escalation or alternative TKIs | [99,100] |
NGS is a general term referring to all post-Sanger sequencing technologies that are able to massively sequence millions of DNA segments[99,100]. The goal of these technologies is to increase sequencing capacity and speed at a lower cost. Furthermore, the sensitivity obtained is superior to that of the conventional sequencing technology, making possible the detection of mutations that are present at very low percentages in a background of normal DNA, which is very important for somatic mutation detection. Currently the most widely used platforms are those offered by Illumina, Inc. (United Sates); Thermo Fisher Scientific, Inc. (United Sates) and Roche Holding AG (Switzerland)[101-103]. The first NGS platform was created by Roche and used emulsion PCR (emPCR) to clonally amplify the fragments that are then sequenced via sequencing-by-synthesis (SBS) technology[101]. The Illumina platform is currently widely used in the NGS market and involves bridge amplification, of a solid surface-bound DNA, to clonally amplify the fragments that are then sequenced using SBS chemistry[102]. Unlike the previous two technologies, the Life technology platform uses Ion semiconductor sequencing, instead of fluorescence based sequencing, detecting the protons released as nucleotides are incorporated during synthesis[103,104].
The past years have seen an accelerating outbreak of publications in which NGS is applied for a variety of goals such as full-genome resequencing or more targeted mutation detection. Worldwide collaborative efforts, such as COSMIC database, International Genome Consortium (ICGC) and The Cancer Genome Atlas (TCGA) project, enabled to catalogue NGS data of thousands of cancer genomes across many disease types[105,106]. Targeted NGS, involving gene panels, is a quicker and cost effective alternative to whole genome sequencing or exome sequencing. Targeted NGS panels for somatic mutation detection include actionable cancer genes and allow the determination of the patient’s tumor molecular profile. The goal of their use is to increase the percentage of patients with detected actionable alterations and with copy number data, allowing them to be included in clinical trials[107-109].
Such panels are currently available or can be custom made. They exhibit high rates of sensitivity, specificity and repeatability; therefore they are optimal for diagnostic use. Benchtop NGS sequencers are now offered by both Illumina (MiSeq) and Thermo Fisher (PGM™ and Ion Proton™). The availability of the equipment required and the cost effectiveness of the analysis allows its implementation in local specialized laboratories[108,109]. However, the reliability of these tests should be reassured. Thus, NGS performing laboratories should have specialized personnel and equipment which will provide adequate data analysis management and interpretation with the aid of appropriate software and bioinformatics tools. Importantly, these tests should be operated under the guidelines of a quality assurance system[110,111].
Concerning the selection of the appropriate sequencing platform, it should be based on the individual laboratory’s needs. All NGS platforms have advantages and disadvantages and the choice of the platform used should be based on the application for which it is required. For example, the MiSeq platform (Illumina) has lower error rates especially in the homo-polymer regions compared to both Ion Proton and PGM (Life Technology). However it requires higher DNA concentration and quality, which is not always available when the starting material is FFPE tissue. On the other hand, the NGS platforms offered by Thermo Fisher provide a fast and cost-effective sequencing solution with good analytical performance. Additionally, they more compatible with low DNA concentrations and partially degraded poor quality DNA from FFPE samples[107-109,112]. Consequently, they provide an attractive option of clinical utility for the detection of cancer hotspot mutation analysis.
This review summarizes the use of biomarkers in the most common cancers of the GI tract. They are used for positive selection of patients who are likely to benefit from targeted therapy or for resistance prediction. Biomarker based targeted treatment is established in a subset of patients with gastrointestinal cancer. Meta-analysis studies have shown that biomarker based treatment is a promising approach and is associated with improved treatment outcome[113-115]. However, ongoing clinical trials, identification of novel biomarkers as well as further advances in high-throughput technologies will hopefully result in further development of therapeutic targets, treatment strategies and improved survival for these patients in the near future.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Greece
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Asic K, Zhao JB S- Editor: Qiu S L- Editor: A E- Editor: Wu HL
| 1. | Mendelsohn J. Personalizing oncology: perspectives and prospects. J Clin Oncol. 2013;31:1904-1911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 106] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 2. | Diaz Z, Aguilar-Mahecha A, Paquet ER, Basik M, Orain M, Camlioglu E, Constantin A, Benlimame N, Bachvarov D, Jannot G. Next-generation biobanking of metastases to enable multidimensional molecular profiling in personalized medicine. Mod Pathol. 2013;26:1413-1424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 3. | Kim TM, Lee SH, Chung YJ. Clinical applications of next-generation sequencing in colorectal cancers. World J Gastroenterol. 2013;19:6784-6793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 26] [Cited by in F6Publishing: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 4. | Pritzker KP. Predictive and prognostic cancer biomarkers revisited. Expert Rev Mol Diagn. 2015;15:971-974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Dienstmann R, Dong F, Borger D, Dias-Santagata D, Ellisen LW, Le LP, Iafrate AJ. Standardized decision support in next generation sequencing reports of somatic cancer variants. Mol Oncol. 2014;8:859-873. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 6. | Zhang Y. Epidemiology of esophageal cancer. World J Gastroenterol. 2013;19:5598-5606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 657] [Cited by in F6Publishing: 695] [Article Influence: 63.2] [Reference Citation Analysis (5)] |
| 7. | Findlay JM, Middleton MR, Tomlinson I. A systematic review and meta-analysis of somatic and germline DNA sequence biomarkers of esophageal cancer survival, therapy response and stage. Ann Oncol. 2015;26:624-644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 8. | Agrawal N, Jiao Y, Bettegowda C, Hutfless SM, Wang Y, David S, Cheng Y, Twaddell WS, Latt NL, Shin EJ. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 263] [Cited by in F6Publishing: 313] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 9. | Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat Genet. 2014;46:467-473. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 455] [Cited by in F6Publishing: 438] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 10. | Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 721] [Cited by in F6Publishing: 763] [Article Influence: 76.3] [Reference Citation Analysis (0)] |
| 11. | Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 566] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 12. | Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805-D811. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1815] [Cited by in F6Publishing: 1787] [Article Influence: 178.7] [Reference Citation Analysis (0)] |
| 13. | Wang K, Johnson A, Ali SM, Klempner SJ, Bekaii-Saab T, Vacirca JL, Khaira D, Yelensky R, Chmielecki J, Elvin JA. Comprehensive Genomic Profiling of Advanced Esophageal Squamous Cell Carcinomas and Esophageal Adenocarcinomas Reveals Similarities and Differences. Oncologist. 2015;20:1132-1139. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 14. | Fichter CD, Timme S, Braun JA, Gudernatsch V, Schöpflin A, Bogatyreva L, Geddert H, Faller G, Klimstra D, Tang L. EGFR, HER2 and HER3 dimerization patterns guide targeted inhibition in two histotypes of esophageal cancer. Int J Cancer. 2014;135:1517-1530. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Belkhiri A, El-Rifai W. Advances in targeted therapies and new promising targets in esophageal cancer. Oncotarget. 2015;6:1348-1358. [PubMed] [Cited in This Article: ] |
| 16. | Yamamoto H, Watanabe Y, Maehata T, Morita R, Yoshida Y, Oikawa R, Ishigooka S, Ozawa S, Matsuo Y, Hosoya K. An updated review of gastric cancer in the next-generation sequencing era: insights from bench to bedside and vice versa. World J Gastroenterol. 2014;20:3927-3937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 56] [Cited by in F6Publishing: 62] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 17. | Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149:1153-1162.e3. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 269] [Cited by in F6Publishing: 311] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 18. | Nadauld LD, Ford JM. Molecular profiling of gastric cancer: toward personalized cancer medicine. J Clin Oncol. 2013;31:838-839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Li X, Wu WK, Xing R, Wong SH, Liu Y, Fang X, Zhang Y, Wang M, Wang J, Li L. Distinct Subtypes of Gastric Cancer Defined by Molecular Characterization Include Novel Mutational Signatures with Prognostic Capability. Cancer Res. 2016;76:1724-1732. [PubMed] [Cited in This Article: ] |
| 20. | Liang H, Kim YH. Identifying molecular drivers of gastric cancer through next-generation sequencing. Cancer Lett. 2013;340:241-246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4230] [Cited by in F6Publishing: 4299] [Article Influence: 429.9] [Reference Citation Analysis (2)] |
| 22. | Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61:673-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 478] [Cited by in F6Publishing: 498] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 23. | Cui J, Yin Y, Ma Q, Wang G, Olman V, Zhang Y, Chou WC, Hong CS, Zhang C, Cao S. Comprehensive characterization of the genomic alterations in human gastric cancer. Int J Cancer. 2015;137:86-95. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 24. | Liu YJ, Shen D, Yin X, Gavine P, Zhang T, Su X, Zhan P, Xu Y, Lv J, Qian J. HER2, MET and FGFR2 oncogenic driver alterations define distinct molecular segments for targeted therapies in gastric carcinoma. Br J Cancer. 2014;110:1169-1178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 87] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 25. | O’Brien KM, Orlow I, Antonescu CR, Ballman K, McCall L, DeMatteo R, Engel LS. Gastrointestinal stromal tumors, somatic mutations and candidate genetic risk variants. PLoS One. 2013;8:e62119. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Zhao X, Yue C. Gastrointestinal stromal tumor. J Gastrointest Oncol. 2012;3:189-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 53] [Reference Citation Analysis (0)] |
| 27. | von Mehren M, Randall RL, Benjamin RS, Boles S, Bui MM, Casper ES, Conrad EU, DeLaney TF, Ganjoo KN, George S. Gastrointestinal stromal tumors, version 2.2014. J Natl Compr Canc Netw. 2014;12:853-862. [PubMed] [Cited in This Article: ] |
| 28. | Kee D, Zalcberg JR. Current and emerging strategies for the management of imatinib-refractory advanced gastrointestinal stromal tumors. Ther Adv Med Oncol. 2012;4:255-270. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 29. | Heinrich MC, Maki RG, Corless CL, Antonescu CR, Harlow A, Griffith D, Town A, McKinley A, Ou WB, Fletcher JA. Primary and secondary kinase genotypes correlate with the biological and clinical activity of sunitinib in imatinib-resistant gastrointestinal stromal tumor. J Clin Oncol. 2008;26:5352-5359. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 30. | Ferraro D, Zalcberg J. Regorafenib in gastrointestinal stromal tumors: clinical evidence and place in therapy. Ther Adv Med Oncol. 2014;6:222-228. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1215] [Cited by in F6Publishing: 1316] [Article Influence: 101.2] [Reference Citation Analysis (0)] |
| 32. | Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal Cancer: Epidemiology, Disease Mechanisms and Interventions to Reduce Onset and Mortality. Clin Colorectal Cancer. 2016;15:195-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 192] [Cited by in F6Publishing: 180] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 33. | Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262-271. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 34. | De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753-762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1491] [Cited by in F6Publishing: 1585] [Article Influence: 113.2] [Reference Citation Analysis (1)] |
| 35. | Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023-1034. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1610] [Cited by in F6Publishing: 1623] [Article Influence: 147.5] [Reference Citation Analysis (0)] |
| 36. | Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546-1558. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5242] [Cited by in F6Publishing: 5196] [Article Influence: 472.4] [Reference Citation Analysis (0)] |
| 37. | Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330-337. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5743] [Cited by in F6Publishing: 6144] [Article Influence: 512.0] [Reference Citation Analysis (0)] |
| 38. | Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in F6Publishing: 265] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 39. | Yu J, Wu WK, Li X, He J, Li XX, Ng SS, Yu C, Gao Z, Yang J, Li M. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64:636-645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 40. | Dumur CI. Available resources and challenges for the clinical annotation of somatic variations. Cancer Cytopathol. 2014;122:730-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Irahara N, Baba Y, Nosho K, Shima K, Yan L, Dias-Santagata D, Iafrate AJ, Fuchs CS, Haigis KM, Ogino S. NRAS mutations are rare in colorectal cancer. Diagn Mol Pathol. 2010;19:157-163. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 130] [Cited by in F6Publishing: 142] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 42. | Negru S, Papadopoulou E, Apessos A, Stanculeanu DL, Ciuleanu E, Volovat C, Croitoru A, Kakolyris S, Aravantinos G, Ziras N. KRAS, NRAS and BRAF mutations in Greek and Romanian patients with colorectal cancer: a cohort study. BMJ Open. 2014;4:e004652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 43. | Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705-5712. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1211] [Cited by in F6Publishing: 1218] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 44. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [PubMed] [Cited in This Article: ] |
| 45. | Kalady MF, Dejulius KL, Sanchez JA, Jarrar A, Liu X, Manilich E, Skacel M, Church JM. BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum. 2012;55:128-133. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 92] [Cited by in F6Publishing: 105] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 46. | Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28:1254-1261. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 492] [Cited by in F6Publishing: 513] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 47. | Van Cutsem E, Köhne CH, Láng I, Folprecht G, Nowacki MP, Cascinu S, Shchepotin I, Maurel J, Cunningham D, Tejpar S. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol. 2011;29:2011-2019. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1314] [Cited by in F6Publishing: 1452] [Article Influence: 111.7] [Reference Citation Analysis (0)] |
| 48. | Mao C, Liao RY, Qiu LX, Wang XW, Ding H, Chen Q. BRAF V600E mutation and resistance to anti-EGFR monoclonal antibodies in patients with metastatic colorectal cancer: a meta-analysis. Mol Biol Rep. 2011;38:2219-2223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 49. | Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8:627-644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1833] [Cited by in F6Publishing: 2007] [Article Influence: 60.8] [Reference Citation Analysis (0)] |
| 50. | Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 2012;18:2257-2268. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 214] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 51. | Huang L, Liu Z, Deng D, Tan A, Liao M, Mo Z, Yang X. Anti-epidermal growth factor receptor monoclonal antibody-based therapy for metastatic colorectal cancer: a meta-analysis of the effect of PIK3CA mutations in KRAS wild-type patients. Arch Med Sci. 2014;10:1-9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 52. | Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851-1857. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 560] [Cited by in F6Publishing: 577] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 53. | Bronte G, Silvestris N, Castiglia M, Galvano A, Passiglia F, Sortino G, Cicero G, Rolfo C, Peeters M, Bazan V. New findings on primary and acquired resistance to anti-EGFR therapy in metastatic colorectal cancer: do all roads lead to RAS? Oncotarget. 2015;6:24780-24796. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 62] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 54. | Therkildsen C, Bergmann TK, Henrichsen-Schnack T, Ladelund S, Nilbert M. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment in metastatic colorectal cancer: A systematic review and meta-analysis. Acta Oncol. 2014;53:852-864. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 275] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 55. | Moorcraft SY, Smyth EC, Cunningham D. The role of personalized medicine in metastatic colorectal cancer: an evolving landscape. Therap Adv Gastroenterol. 2013;6:381-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 56. | Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596-1606. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 651] [Cited by in F6Publishing: 626] [Article Influence: 52.2] [Reference Citation Analysis (0)] |
| 57. | Khan KH, Yap TA, Yan L, Cunningham D. Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J Cancer. 2013;32:253-265. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 151] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 58. | Wang XW, Zhang YJ. Targeting mTOR network in colorectal cancer therapy. World J Gastroenterol. 2014;20:4178-4188. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 75] [Cited by in F6Publishing: 74] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 59. | Berg M, Danielsen SA, Ahlquist T, Merok MA, Ågesen TH, Vatn MH, Mala T, Sjo OH, Bakka A, Moberg I. DNA sequence profiles of the colorectal cancer critical gene set KRAS-BRAF-PIK3CA-PTEN-TP53 related to age at disease onset. PLoS One. 2010;5:e13978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 60. | De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594-603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 417] [Cited by in F6Publishing: 443] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 61. | Dillon LM, Miller TW. Therapeutic targeting of cancers with loss of PTEN function. Curr Drug Targets. 2014;15:65-79. [PubMed] [Cited in This Article: ] |
| 62. | Liu J, Hu J, Cheng L, Ren W, Yang M, Liu B, Xie L, Qian X. Biomarkers predicting resistance to epidermal growth factor receptor-targeted therapy in metastatic colorectal cancer with wild-type KRAS. Onco Targets Ther. 2016;9:557-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 63. | Janku F, Hong DS, Fu S, Piha-Paul SA, Naing A, Falchook GS, Tsimberidou AM, Stepanek VM, Moulder SL, Lee JJ. Assessing PIK3CA and PTEN in early-phase trials with PI3K/AKT/mTOR inhibitors. Cell Rep. 2014;6:377-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 195] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 64. | Carpten JD, Faber AL, Horn C, Donoho GP, Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage S. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439-444. [PubMed] [Cited in This Article: ] |
| 65. | Davies BR, Guan N, Logie A, Crafter C, Hanson L, Jacobs V, James N, Dudley P, Jacques K, Ladd B. Tumors with AKT1E17K Mutations Are Rational Targets for Single Agent or Combination Therapy with AKT Inhibitors. Mol Cancer Ther. 2015;14:2441-2451. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 66. | Hechtman JF, Sadowska J, Huse JT, Borsu L, Yaeger R, Shia J, Vakiani E, Ladanyi M, Arcila ME. AKT1 E17K in Colorectal Carcinoma Is Associated with BRAF V600E but Not MSI-H Status: A Clinicopathologic Comparison to PIK3CA Helical and Kinase Domain Mutants. Mol Cancer Res. 2015;13:1003-1008. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 67. | Edmonston TB, Cuesta KH, Burkholder S, Barusevicius A, Rose D, Kovatich AJ, Boman B, Fry R, Fishel R, Palazzo JP. Colorectal carcinomas with high microsatellite instability: defining a distinct immunologic and molecular entity with respect to prognostic markers. Hum Pathol. 2000;31:1506-1514. [PubMed] [Cited in This Article: ] |
| 68. | Starostik P, Müller-Hermelink HK. Diagnosis of microsatellite instability-positive colorectal cancer. Expert Rev Mol Diagn. 2001;1:71-80. [PubMed] [Cited in This Article: ] |
| 69. | Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248-5257. [PubMed] [Cited in This Article: ] |
| 70. | Saridaki Z, Souglakos J, Georgoulias V. Prognostic and predictive significance of MSI in stages II/III colon cancer. World J Gastroenterol. 2014;20:6809-6814. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 60] [Cited by in F6Publishing: 56] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 71. | Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 2008;26:5783-5788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 584] [Cited by in F6Publishing: 617] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 72. | Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787-793. [PubMed] [Cited in This Article: ] |
| 73. | Parsons MT, Buchanan DD, Thompson B, Young JP, Spurdle AB. Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet. 2012;49:151-157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 193] [Cited by in F6Publishing: 206] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 74. | Jacob S, Aguado M, Fallik D, Praz F. The role of the DNA mismatch repair system in the cytotoxicity of the topoisomerase inhibitors camptothecin and etoposide to human colorectal cancer cells. Cancer Res. 2001;61:6555-6562. [PubMed] [Cited in This Article: ] |
| 75. | Kawakami H, Zaanan A, Sinicrope FA. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol. 2015;16:30. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 257] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 76. | Kawakami H, Zaanan A, Sinicrope FA. Implications of mismatch repair-deficient status on management of early stage colorectal cancer. J Gastrointest Oncol. 2015;6:676-684. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 33] [Reference Citation Analysis (0)] |
| 77. | Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509-2520. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6096] [Cited by in F6Publishing: 6542] [Article Influence: 726.9] [Reference Citation Analysis (0)] |
| 78. | Goel G. Evolving role of gene expression signatures as biomarkers in early-stage colon cancer. J Gastrointest Cancer. 2014;45:399-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 79. | O’Connell MJ, Lavery I, Yothers G, Paik S, Clark-Langone KM, Lopatin M, Watson D, Baehner FL, Shak S, Baker J. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937-3944. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 220] [Cited by in F6Publishing: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 80. | Gray RG, Quirke P, Handley K, Lopatin M, Magill L, Baehner FL, Beaumont C, Clark-Langone KM, Yoshizawa CN, Lee M. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611-4619. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 270] [Cited by in F6Publishing: 289] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 81. | Venook AP, Niedzwiecki D, Lopatin M, Ye X, Lee M, Friedman PN, Frankel W, Clark-Langone K, Millward C, Shak S. Biologic determinants of tumor recurrence in stage II colon cancer: validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31:1775-1781. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 125] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 82. | Yothers G, O’Connell MJ, Lee M, Lopatin M, Clark-Langone KM, Millward C, Paik S, Sharif S, Shak S, Wolmark N. Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31:4512-4519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 131] [Cited by in F6Publishing: 126] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 83. | Srivastava G, Renfro LA, Behrens RJ, Lopatin M, Chao C, Soori GS, Dakhil SR, Mowat RB, Kuebler JP, Kim G. Prospective multicenter study of the impact of oncotype DX colon cancer assay results on treatment recommendations in stage II colon cancer patients. Oncologist. 2014;19:492-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 84. | Francis G, Stein S. Circulating Cell-Free Tumour DNA in the Management of Cancer. Int J Mol Sci. 2015;16:14122-14142. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 85. | Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. 2013;10:472-484. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1114] [Cited by in F6Publishing: 1213] [Article Influence: 110.3] [Reference Citation Analysis (0)] |
| 86. | Bedard PL, Hansen AR, Ratain MJ, Siu LL. Tumour heterogeneity in the clinic. Nature. 2013;501:355-364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 807] [Cited by in F6Publishing: 808] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 87. | Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537-540. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1259] [Cited by in F6Publishing: 1292] [Article Influence: 107.7] [Reference Citation Analysis (0)] |
| 88. | Wong AL, Lim JS, Sinha A, Gopinathan A, Lim R, Tan CS, Soh T, Venkatesh S, Titin C, Sapari NS. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J Transl Med. 2015;13:57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 89. | Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015;16:937-948. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 242] [Cited by in F6Publishing: 244] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 90. | Misale S, Arena S, Lamba S, Siravegna G, Lallo A, Hobor S, Russo M, Buscarino M, Lazzari L, Sartore-Bianchi A. Blockade of EGFR and MEK intercepts heterogeneous mechanisms of acquired resistance to anti-EGFR therapies in colorectal cancer. Sci Transl Med. 2014;6:224ra26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 209] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 91. | Spindler KL, Pallisgaard N, Andersen RF, Jakobsen A. Changes in mutational status during third-line treatment for metastatic colorectal cancer--results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int J Cancer. 2014;135:2215-2222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 65] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 92. | Bardelli A, Corso S, Bertotti A, Hobor S, Valtorta E, Siravegna G, Sartore-Bianchi A, Scala E, Cassingena A, Zecchin D. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer Discov. 2013;3:658-673. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 465] [Cited by in F6Publishing: 513] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 93. | Kwak EL, Ahronian LG, Siravegna G, Mussolin B, Godfrey JT, Clark JW, Blaszkowsky LS, Ryan DP, Lennerz JK, Iafrate AJ. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET-Amplified Esophagogastric Cancer. Cancer Discov. 2015;5:1271-1281. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 94. | Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, Mussolin B, Kwak EL, Buscarino M, Lazzari L. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6:147-153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 275] [Cited by in F6Publishing: 301] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 95. | Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Komatsu S, Shiozaki A. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2016; Feb 13; Epub ahead of print. [PubMed] [Cited in This Article: ] |
| 96. | Takegawa N, Yonesaka K, Sakai K, Ueda H, Watanabe S, Nonagase Y, Okuno T, Takeda M, Maenishi O, Tsurutani J. HER2 genomic amplification in circulating tumor DNA from patients with cetuximab-resistant colorectal cancer. Oncotarget. 2016;7:3453-3460. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 97. | Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, Salido M, Gallen M, Marsters S, Tsai SP. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012;18:221-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 351] [Cited by in F6Publishing: 370] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 98. | Morelli MP, Overman MJ, Dasari A, Kazmi SM, Mazard T, Vilar E, Morris VK, Lee MS, Herron D, Eng C. Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol. 2015;26:731-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 188] [Article Influence: 20.9] [Reference Citation Analysis (1)] |
| 99. | Maier J, Lange T, Kerle I, Specht K, Bruegel M, Wickenhauser C, Jost P, Niederwieser D, Peschel C, Duyster J. Detection of mutant free circulating tumor DNA in the plasma of patients with gastrointestinal stromal tumor harboring activating mutations of CKIT or PDGFRA. Clin Cancer Res. 2013;19:4854-4867. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 100. | Kang G, Bae BN, Sohn BS, Pyo JS, Kang GH, Kim KM. Detection of KIT and PDGFRA mutations in the plasma of patients with gastrointestinal stromal tumor. Target Oncol. 2015;10:597-601. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 27] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 101. | Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin Biochem Rev. 2011;32:177-195. [PubMed] [Cited in This Article: ] |
| 102. | Tucker T, Marra M, Friedman JM. Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet. 2009;85:142-154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 246] [Cited by in F6Publishing: 184] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 103. | Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M. Comparison of next-generation sequencing systems. J Biomed Biotechnol. 2012;2012:251364. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 608] [Cited by in F6Publishing: 632] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 104. | Gagan J, Van Allen EM. Next-generation sequencing to guide cancer therapy. Genome Med. 2015;7:80. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 180] [Cited by in F6Publishing: 190] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 105. | Chin L, Hahn WC, Getz G, Meyerson M. Making sense of cancer genomic data. Genes Dev. 2011;25:534-555. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 241] [Cited by in F6Publishing: 213] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 106. | Wu TJ, Schriml LM, Chen QR, Colbert M, Crichton DJ, Finney R, Hu Y, Kibbe WA, Kincaid H, Meerzaman D. Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis. Database (Oxford). 2015;2015:bav032. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 107. | Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 999] [Cited by in F6Publishing: 1045] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 108. | Chen H, Luthra R, Goswami RS, Singh RR, Roy-Chowdhuri S. Analysis of Pre-Analytic Factors Affecting the Success of Clinical Next-Generation Sequencing of Solid Organ Malignancies. Cancers (Basel). 2015;7:1699-1715. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in F6Publishing: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 109. | Zhang L, Chen L, Sah S, Latham GJ, Patel R, Song Q, Koeppen H, Tam R, Schleifman E, Mashhedi H. Profiling cancer gene mutations in clinical formalin-fixed, paraffin-embedded colorectal tumor specimens using targeted next-generation sequencing. Oncologist. 2014;19:336-343. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 110. | Gargis AS, Kalman L, Berry MW, Bick DP, Dimmock DP, Hambuch T, Lu F, Lyon E, Voelkerding KV, Zehnbauer BA. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033-1036. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 381] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
| 111. | Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733-747. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 677] [Cited by in F6Publishing: 619] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 112. | de Leng WW, Gadellaa-van Hooijdonk CG, Barendregt-Smouter FA, Koudijs MJ, Nijman I, Hinrichs JW, Cuppen E, van Lieshout S, Loberg RD, de Jonge M. Targeted Next Generation Sequencing as a Reliable Diagnostic Assay for the Detection of Somatic Mutations in Tumours Using Minimal DNA Amounts from Formalin Fixed Paraffin Embedded Material. PLoS One. 2016;11:e0149405. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 113. | Jardim DL, Schwaederle M, Wei C, Lee JJ, Hong DS, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of a Biomarker-Based Strategy on Oncology Drug Development: A Meta-analysis of Clinical Trials Leading to FDA Approval. J Natl Cancer Inst. 2015;107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 118] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 114. | Schwaederle M, Zhao M, Lee JJ, Lazar V, Leyland-Jones B, Schilsky RL, Mendelsohn J, Kurzrock R. Association of Biomarker-Based Treatment Strategies With Response Rates and Progression-Free Survival in Refractory Malignant Neoplasms: A Meta-analysis. JAMA Oncol. 2016; Jun 6; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 208] [Cited by in F6Publishing: 232] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 115. | Schwaederle M, Zhao M, Lee JJ, Eggermont AM, Schilsky RL, Mendelsohn J, Lazar V, Kurzrock R. Impact of Precision Medicine in Diverse Cancers: A Meta-Analysis of Phase II Clinical Trials. J Clin Oncol. 2015;33:3817-3825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 288] [Cited by in F6Publishing: 311] [Article Influence: 34.6] [Reference Citation Analysis (0)] |