Published online Mar 15, 2022. doi: 10.4251/wjgo.v14.i3.628
Peer-review started: March 10, 2021
First decision: June 4, 2021
Revised: June 4, 2021
Accepted: February 25, 2022
Article in press: February 25, 2022
Published online: March 15, 2022
Adenocarcinomas of the gastrointestinal tract (esophagus, stomach, and colon) represent a heterogeneous group of diseases with distinct etiology, clinical features, treatment approaches, and prognosis. Studies are ongoing to isolate molecular genetic subtypes, perform complete biological characterization of the tumor, determine prognostic groups, and find predictive markers to the effectiveness of therapy. Separate molecular genetic classifications were created for esophageal adenocarcinoma [The Cancer Genome Atlas (TCGA)], stomach cancer (TCGA, Asian Cancer Research Group), and colon cancer (Colorectal Cancer Subtyping Consortium). In 2018, isolation of TCGA molecular genetic subtypes for adenocarcinomas of the gastrointestinal tract (esophagus, stomach, and colon) highlighted the need for further studies and clinical validation of subtyping of gastrointestinal adenocarcinomas. However, this approach has limitations. The aim of our work was to critically analyze integration of molecular genetic subtyping of gastrointestinal adenocarcinomas in clinical practice.
Core Tip: Here we describe our opinion on molecular genetic subtyping of gastrointestinal adenocarcinomas (esophageal, gastric, and colon adenocarcinomas). The identification of combined molecular and genetic subtypes gave us insights to understanding gastrointestinal adenocarcinoma biology, determining aims for future clinical research, and helping to simplify the implementation of a unified system for subtyping gastrointestinal adenocarcinomas.
- Citation: Ignatova EO, Kozlov E, Ivanov M, Mileyko V, Menshikova S, Sun H, Fedyanin M, Tryakin A, Stilidi I. Clinical significance of molecular subtypes of gastrointestinal tract adenocarcinoma. World J Gastrointest Oncol 2022; 14(3): 628-645
- URL: https://www.wjgnet.com/1948-5204/full/v14/i3/628.htm
- DOI: https://dx.doi.org/10.4251/wjgo.v14.i3.628
Adenocarcinomas of the gastrointestinal tract represent a heterogeneous group of diseases. Their management depends on localization of the tumor, clinical and morphological characteristics of disease, and tumor biology.
To improve treatment outcomes we actively search for molecular biomarkers that can predict drug effectiveness and foretell the disease course. Molecular genetic typing of gastrointestinal adenocarcinomas is thought to be a promising approach.
Some data show that gastrointestinal tract adenocarcinomas represent several distinguished molecular subtypes that could be associated with different pathogenesis, prognosis, and treatment options. Separate molecular genetic classifications were created for colon cancer (Colorectal Cancer Subtyping Consortium) and gastric cancer (GC) [Asian Cancer Research Group (ACRG)]. In 2018, a pooled analysis of gastrointestinal adenocarcinomas of The Cancer Genome Atlas (TCGA) (esophageal cancer, GC, colon cancer, and rectal cancer) revealed five molecular genetic subtypes based on molecular studies with high-capacity methods.
To date, we have no doubt of the clinical significance of isolating molecular genetic subtypes of gastrointestinal tract adenocarcinomas. The main challenge is to adapt this classification for routine clinical use by defining “surrogate markers” of biological subtypes.
Esophageal cancer is one of the most aggressive cancers. According to GLOBOCAN, more than 600000 new cases of esophageal cancer were registered in 2020[1]. The main morphological form of esophageal cancer is squamous cell carcinoma (keratinizing or non-keratinizing) (95%). In 5% of cases there is adenocarcinoma of the esophagus (EA) and in rare cases, small cell carcinoma[2]. The distribution of histological tumor subtypes varies widely by country of residence, race, and gender. The pathogenesis of esophageal adenocarcinomas is known to be associated with gastroesophageal reflux disease, Barrett’s esophagus, obesity, and smoking[3]. Gastroesophageal reflux is one of the key DNA damaging factors. In a rat esophageal cancer model, reflux induction has been shown to increase mutation rates, mainly C/T and G/A transitions[4].
Drug treatment of esophageal cancer is determined by its histological type as well as the presence of molecular genetic markers such as microsatellite instability (MSI) status, PD-L1 expression, and HER2 expression (in adenocarcinoma). The range of therapeutic options is limited; the most effective drugs for both histological variants are cisplatin, fluoropyrimidines, and taxanes. Oxaliplatin, irinotecan, and trastuzumab (with overexpression/amplification of HER2) are also effective in adenocarcinomas[5]. For high levels of MSI (MSI-H) tumors, pembrolizumab may be prescribed as the second-line therapy. Pembrolizumab could be reasonable in combination with cisplatin and fluorouracil (KEYNOTE-590) as the first-line treatment option in patients with squamous cell carcinoma with a positive PD-L1 combined positive score (CPS) status ≥ 10. Nivolumab could be reasonable in folinic acid, fluorouracil, and oxaliplatin (FOLFOX) or oxaliplatin and capecitabine combinations (CheckMate 649) as the first-line treatment option in patients with esophageal adenocarcinoma with a positive PD-L1 CPS status of ≥ 5. However, the results of esophageal cancer treatment to date are still unsatisfactory with a 5-year overall survival rate of 20% and a median life expectancy of patients with metastatic cancer of less than a year[6]. Currently, we are actively searching to find new predictive biomarkers for treatment via molecular genetic analysis. It would allow us to identify individual subtypes of the disease and therefore a personalized treatment approach.
Dulak et al[7] were the first to publish work on advanced molecular genetic analysis of esophageal adenocarcinoma. They studied a spectrum of EA mutations with a pairwise analysis of tumor and normal controls in 149 EA patients via full-exome sequencing. However, 15 patients had full-genome sequencing analysis. They described a mutational signature having A>C transversions in AA dinucleotides[7].
In another study of Secrier et al[8], they analyzed results of full-genome sequencing of 129 EA samples. They showed that EA was usually prevalent with large rearrangements and extremely high heterogeneity of the tumor as well as a high frequency of mutations and coamplifications of tyrosine kinase receptors. The most common amplifications were ERBB2, EGFR, MET, and FGFRs. They managed to determine mutational signatures and stratify EA into 3 subtypes: the DNA damage repair induced (18%), C>A/T dominant (29%), and mutagenic (53%) subtype.
The DNA damage repair subgroup showed an increase in the frequency of violations of homologous recombination genes. Homologous recombination gene disorders might determine potential sensitivity to platinum-based chemotherapy (CT) and PARP inhibitors as well as sensitivity to radiation therapy[8].
The C>A/T dominant EA subtype was associated with aging, a lower level of duplications, and an increased frequency of interchromosomal translocations. In the C>A/T dominant subgroup there was a higher frequency of ERBB2/MET coamplifications. The use of tyrosine kinase receptor inhibitors[9] may be reasonable in this subtype.
The mutagenic subtype showed a high mutational load and load of neoantigens. These characteristics may mediate sensitivity to immunotherapy. Development of the mutagenic subtype is associated with gastroesophageal reflux. The mutagenic subtype demonstrated sensitivity to WEE1/CHK1 inhibitors[10]. The authors concluded that the use of mutational signatures and subtyping could help in the selection of promising therapeutic options and determine rationale for further research. Limitations for the use of a personalized approach to EA treatment are significant heterogeneity and a high number of coamplifications in tyrosine kinase receptor genes.
Comparative molecular genetic analysis of EA with GC and cancer of the esophageal-gastric junction is of particular clinical interest. In 2017, TCGA project presented a similar analysis. They analyzed 164 EA samples, 359 GC samples, and 36 adenocarcinomas of the esophageal-gastric junction. They found that EA results were consistent with chromosomally instable (CIN) gastric phenotype with its molecular and genetic characteristics and could probably be treated as a single nosology[10] .
EA is considered as the tumor with the most frequent changes in copy number variations[11,12]. The most significant amplifications for both the EA and CIN subtypes of GC are ERBB2, MYC, IKZF3, CDK12, VEGFA, CDK6, FGF3, and FGF4. However, a detailed examination revealed some differences in the molecular genetic characteristics and differences in tumor methylation in accordance with its location[12]. Thus, hypermethylation is more frequent in EA than in the CIN subtype (70% vs 30% respectively, P = 1.0 × 10-8). Moreover, the incidence of some genes change depending on the anatomical site. For example, SMARC4 mutations and RUNX1 tumor suppressor deletions are more common for EA, but APC mutations are rare compared to GC. The Wnt/β-catenin pathway seems to play a less important role in EA. In addition, MYC and VEGFA amplifications are more frequent for EA. Thus, there is a significant level of intratumor heterogeneity among EA with obvious properties of the CIN phenotype, like amplification of tyrosine kinase receptors.
In 2018, Guo et al[13] conducted a cumulative analysis of EA subtyping using the Gene Expression Omnibus and TCGA databases. When analyzing gene expression profiles of three independent cohorts, they found two molecular genetic subtypes of EA with distinct expression and somatic mutation profiles. They showed that the first subtype (I) shared common molecular expression profiles with GC, and the second one (II) was similar to squamous cell carcinoma. Specific somatic mutations of SMAD4, SOCS4, and SKAP2 were specific for the first subtype[14]. Only 3 patients in the study received CT: 2 patients with type II EA and 1 patient with type I EA. Two patients with subtype II had a complete response to treatment. One patient with subtype I progressed during treatment. Due to the extremely small sample size of patients and insufficient clinical information, it would be reasonable to continue analyzing prognostic and predictive significance of molecular subtype selection.
Molecular genetic analysis of Barrett’s esophageal samples is of particular clinical interest and may be useful for understanding the biology of EA. It is known that Barrett’s esophagus is a precursor of EA. Barrett’s esophagus was shown to be polyclonal and exhibit high mutational properties even in the absence of dysplasia. The genome of Barrett’s esophageal tissues is relatively more stable compared to invasive tumors[15]. About 32% of Barrett’s esophagus cases perform massive localized chromosomal translocations (chromotripsis), which can result in the activation of oncogenes and inactivation of tumor suppressors mediating rapid development of EA[16]. Genetic changes in EA are usually accompanied by significant epigenome changes. With the development of high-grade metaplasia and EA there is an additional increase in the methylation of CpG islands[16]. Thus, detection of epigenetic changes specific to EA and Barrett’s esophagus can help in the subtyping of patients.
In 2020, there was a study identifying subtypes of Barrett’s esophagus and EA based on DNA methylation profiles and integration of transcriptomic and genomic data[17]. They analyzed 150 samples of Barrett’s esophagus and 285 samples of EA. They identified four molecular genetic subtypes; each had distinctive biological properties. The first subtype exhibited hypermethylation of DNA (CpG island methylator phenotype-like), high mutational load with numerous mutations in cell cycle genes (CCND1, CCNE1, MYC, CDK6) and tyrosine kinase receptors (GATA4, ERBB2, KRAS). The second one had expression of gene patterns associated with metabolic processes and absence of methylation at specific sites of transcription factors. This subtype was common for Barrett’s esophagus (83%). The third subtype had no methylation with gene expression indicating infiltration by immune cells (cytotoxic cells, B cells, mast cells and neutrophils, and tumor-associated fibroblasts), and decreased expression of T helper cells. These patients had a poor prognosis compared to the other subtypes. The fourth subtype presented with hypomethylation of DNA and a high frequency of CCNE1 amplifications. Several preclinical studies demonstrated drug treatment effectiveness depending on the tumor subtype. For instance, irinotecan, a topoisomerase I inhibitor, is known to be effective in the treatment of tumors with a high level of methylation. Given the similarity of the molecular characteristics of CIN GC with EA, the EA I subtype (CpG island methylator phenotype-like) might be sensitive to inhibitors of DNA methyltransferase and topoisomerase I. It was also reported that CDK4/6 inhibitors were effective in EA of all subtypes with CDK2 inhibitors more effective in subtype 4 due to CCNE1 amplification. In addition, organelles with reduced levels of MGMT and CHFR expression were sensitive to temozolomide and taxane drugs[18-20].
Thus, a number of studies by large research groups are devoted to the EA subtyping. Their work aims to isolate molecular genetic subtypes, perform complete biological characterization of the tumor, determine prognostic groups, and find predictive markers to the effectiveness of therapy. The obtained data indicate the need for further research and require additional clinical validation for successful clinical use.
GC has an unfavorable prognosis with a median life expectancy in metastatic patients under a year. Standard molecular genetic diagnosis in metastatic GC includes expression and amplification of HER2/neu, MSI, and PD-L1 expression (CPS). If positive HER2/neu expression status is detected or HER2/neu is amplified, trastuzumab is added for first-line CT. A high level of MSI or positive expression of PD-L1 (CPS > 10) allows the use of pembrolizumab immunotherapy in the second-line setting. Positive expression of PD-L1 (CPS > 1) allows the use of pembrolizumab in subsequent lines of treatment. In addition, in 2020 a randomized phase 3 trial, CheckMate 649, showed a statistically significant (P < 0.001) increase in overall survival for combination of nivolumab with oxaliplatin and capecitabine or FOLFOX regimens for the first-line treatment of GC with PD-L1 CPS expression ≥ 5[21].
For the histological classification of GC, we use the World Health Organization classification, which distinguishes some major subtypes like papillary, tubular, and mucinous types[22]. Quite often, histological Lauren classification is used due to its simplicity and easy use. There are several Lauren subtypes according to dominant morphological picture, like intestinal, diffuse, and mixed types of GC[23]. The intestinal type of GC is more common in patients with severe atrophic gastritis and is associated with intestinal metaplasia and presence of persistent H. pylori infection. This subtype is more common in men or the elderly and usually has visceral metastasis. Diffuse GC is associated with low differentiation, treatment resistance, and poor prognosis. The diffuse subtype is more common in young women and usually presents with peritoneal dissemination.
There are a series of studies devoted to a predictive role of the GC histological subtypes. In a multicenter study, Messager et al[24] retrospectively analyzed the effectiveness of perioperative CT based on platinum and fluorouracil agents on the survival of patients with signet ring cell carcinoma GC. Signet ring cell carcinoma is a diffuse GC subtype according to the Lauren classification. Among 3010 patients receiving treatment in 19 clinics in France from January 1997 to January 2010, 1050 (34.9%) had signet ring cell carcinoma GC; 18.5% (171) of patients received perioperative CT, and 81.5% (753) received surgical treatment alone. With a median follow-up of 31.5 mo, median survival was lower in the signet ring carcinoma group (12.8 vs 14.0 mo, P = 0.043). Multivariate analysis showed that signet ring carcinoma was an independent factor of poor prognosis [hazard ratio (HR) = 1.4; 95% confidence interval (CI): 1.1-1.9; P = 0.042]. This study was retrospective and does not have the necessary power to change clinical practice. In randomized trials of perioperative and adjuvant CT (ACTS1[25], CLASSIC[26], INTERGroup0116[27], FNLCC[28]) there was no preplanned analysis of a patient subgroup with signet ring carcinoma diffuse GC.
In the MAGIC study[29], they compared surgical treatment only with perioperative CT, which included three courses of epirubicin, cisplatin, and 5-fluorouracil (ECF) before the operation and three courses after the operation. The addition of CT led to a significant increase in the 5-year life expectancy in patients from 23% to 36% (HR 0.75, 95%CI: 0.60 0.93, P = 0.009). After a subgroup analysis there were no differences in the frequency of pathological complete response in the diffuse or intestinal subtype GC according to Lauren.
The FLOT-4 study[30] evaluated two different perioperative CT regimens, the ECF/ECX regimen (epirubicin, cisplatin, fluorouracil or capecitabine) and docetaxel, oxaliplatin, leucovorin, and fluorouracil. The docetaxel, oxaliplatin, leucovorin, and fluorouracil combination benefited over the ECF/ECX regimen, significantly improving the 5-year overall survival rate (48% and 57%, HR = 0.77, P = 0.012). Subgroup analysis demonstrated that docetaxel, oxaliplatin, leucovorin, and fluorouracil efficacy was higher than the ECF/ECX regimen regardless of histological subtype, even in the subgroup of signet ring carcinoma GC.
Survey of experts at the 4th International Conference St. Gallen, dedicated to the treatment of operable GC and esophageal cancer, showed that signet ring carcinoma of diffuse histological subtype was an independent factor of poor prognosis and should be considered as a stratification factor in future studies[31]. The Lauren histological classification has been widely used over the past five decades, but its clinical significance is limited because it does not reflect the full complexity and molecular heterogeneity of the disease. With the use of molecular platforms (NGS, DNA microarrays, RPPA), it became possible to classify GC into molecular subtypes. Various research groups worked to determine molecular subtypes of GC.
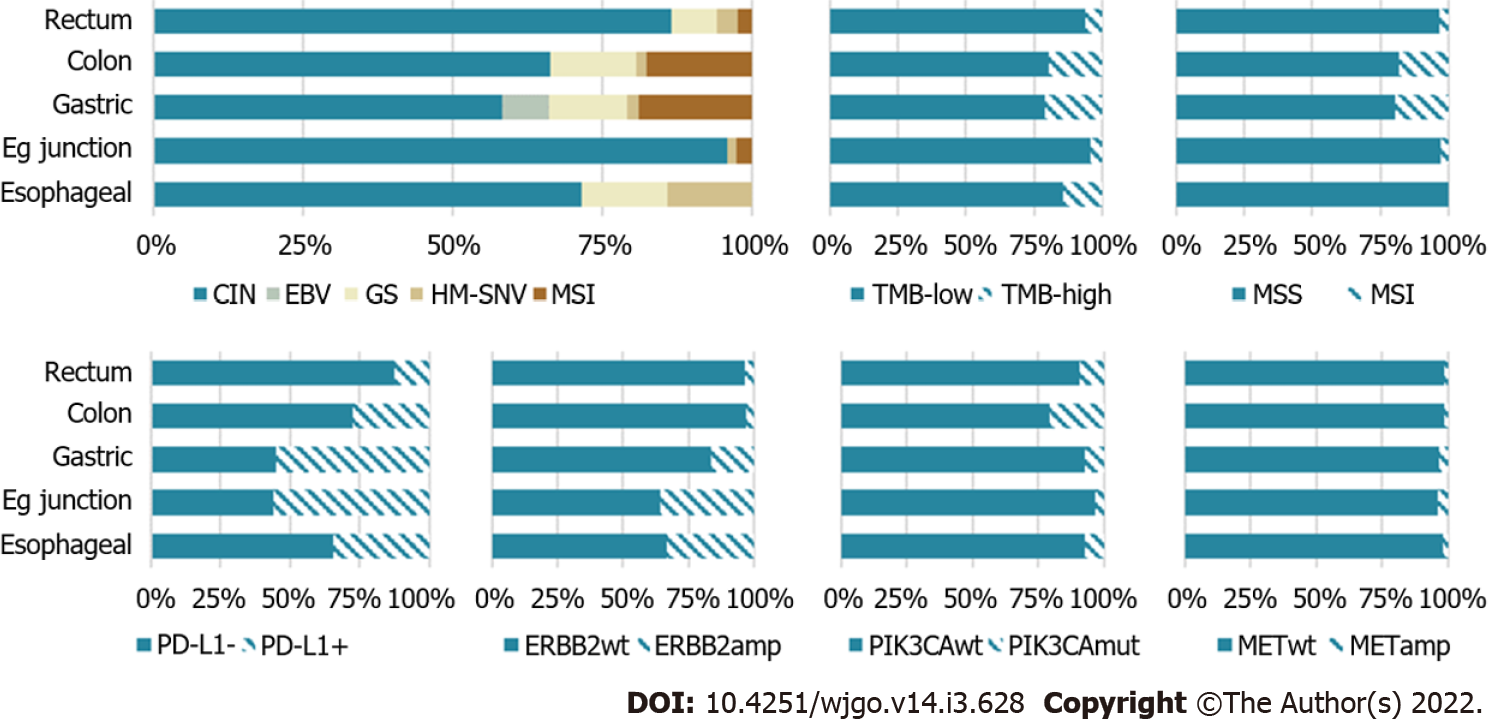
The most interesting work in subtyping of GC were the studies of Tan et al[32] in 2011 and 2013 (Figure 1)[33], TCGA project in 2014[12], and ACRG study in 2015 (Table 1).
| Subdividing and data level | Subtype | Prevalence | Defining characteristics |
| Esophagus subtypes | |||
| Liu et al[34] obtained subtypes based on SCNAs, WES, DNA methylation, mRNAseq, microRNAseq, RPPA (TCGA) | EA-CIN | 14.1 | EA similarity with CIN phenotype of GC. Methylation patterns and gene alterations differ in terms of localization |
| Guo et al[13] determined differences in expression profiles and somatic mutation profiles by using RNA-Seq and exome-Seq data | EA I | 40 | EA I shares the common expression profiles with GC |
| EA II | 60 | EA II was clustered with esophageal squamous cell carcinomas | |
| Jammula et al[17] divided OAC and Barrett’s esophagus by integration of WGS and RNA-seq data | Subtype I | 28.7 | SI: CIMP-like |
| Subtype II | 27.3 | SII: Expression of gene patterns associated with metabolic processes | |
| Subtype III | 22.7 | SIII: Immune cell infiltration | |
| Subtype IV | 21.1 | SIV: DNA hypomethylation; structural aberrations; CNA | |
| Secrier et al[8] received GC subtypes on the basis of mutation signatures obtained from WGS data | DDR-impaired | 15 | DDR: Enrichment for BRCA signature with prevalent defects in the homologous recombination pathway |
| C > A/T dominant | 32 | C > A/T: Aging imprint | |
| Mutagenic | 53 | Mutagenic: The highest mutational load and the highest load of neoantigens | |
| Gastric subtypes | |||
| Tan et al[32] obtained subtypes based on gene expression pattern (microarray) | G-INT | 58 | G-INT: Genes upregulated were related to carbohydrate and protein metabolism (FUT2) and cell adhesion (LGALS4; CDH17) |
| G-DIF | 42 | G-DIF: Cell proliferation (AURKB) and fatty acid metabolism (ELOVL5) functional annotations were enriched | |
| Lei et al[33] compared the patterns of gene expression samples of GC (mRNA, CNAs) | Proliferative | 45 | Proliferative: High levels of genomic instability; TP53 mutations and DNA hypomethylation |
| Metabolic | 23 | Metabolic: High expression of genes associated with metabolism | |
| Mesenchymal | 31 | Mesenchymal: Contain cells with features of cancer stem cells | |
| TCGA obtained subtypes based on SCNAs, WES, DNA methylation, mRNAseq, microRNAseq, RPPA[12] | EBV+ | 8.8 | EBV: Recurrent mutation of PIK3CA; intense hypermethylation; JAK2, CD274, PDCD1LG2 amplification |
| MSI | 21.7 | MSI: Increased frequency of mutations; aberrant epigenetic patterns | |
| CIN | 49.8 | CIN: The presence of multiple chromosomal rearrangements; localization mainly in the proximal gastric cancer and EGJ | |
| GS | 19.7 | GS: RHOA, CDH1 and ARID1A mutations; CLDN18-ARHGAP6 gene fusion | |
| Cristescu et al[55] received GC subtypes based on data of gene expression | MSI-high GC | 22.7 | MSI-high GC: Mutations in ARID1A, MTOR, KRAS, PIK3CA, ALK, and PTEN. Overexpression of PD-L1; T cell infiltrate |
| MSS/EMT GC | 15.3 | MSS/EMT GC: Loss of CDH1; Loss of cellular adhesion, angiogenesis, motility | |
| MSS/TP53- GC | 35.7 | MSS/TP53- GC: Highest prevalence of TP53 and RHOA mutations; APC, ARID1A, KRAS, PIK3CA, and SMAD4 enriched | |
| MSS/TP53+ GC | 26.3 | MSS/TP53+ GC: Frequent EBV infection; Frequent mutations in ARID1A, PIK3CA, SMAD4, APC | |
| Colon subtypes | |||
| Guinney et al[59] carried out combined molecular genetic analysis of 4151 colon tumor samples from 6 different scientific groups | CMS1 | 14 | CMS1: Hypermutated; microsatellite unstable; strong immune activation |
| CMS2 | 37 | CMS2: Epithelial, chromosomally unstable; marked WNT and MYC signaling activation | |
| CMS3 | 13 | CMS3: Epithelial; evident metabolic dysregulation | |
| CMS4 | 23 | CMS4: Prominent transforming growth factor β activation; stromal invasion and angiogenesis | |
| Liu et al[34] obtained subtypes based on SCNAs, WES, DNA methylation, mRNAseq, microRNAseq, RPPA (TCGA) | MSI | 17.5 | MSI: MSI tumors with MLH1 methylation were associated with BRAFV600E mutation |
| HM-SNV | 1.7 | HM-SNV: Hotspot mutations in polymerase E | |
| CIN | 66.6 | CIMP status is characteristic of CRC with associated mutations in KRAS and TGFβ pathways | |
| GS | 14 | GS: Lacking hypermutation and aneuploidy; enriched in DNA hypermethylation and mutations in KRAS, SOX9 and PCBP1 | |
In 2011, Tan et al[32] identified GC subtypes via analyzing gene expression with a panel of 37 GC cell lines. Gene expression analysis was performed using microarrays (HG-U133 Plus 2.0, Affymetrix). They identified the gene expression signature of 171 genes and identified two GC subtypes with distinct gene expression patterns, namely, the intestinal subtype (G-INT) and the diffuse subtype (G-DIF). These subtypes also were determined when analyzing primary gastric tumors in 270 patients in two independent groups. The G-INT subtype was found to be associated with activation of protein and carbohydrate metabolism (FUT2) and cell adhesion (LGALS4, CDH17) genes. The G-DIF subtype was associated with functional annotations of cell proliferation (AURKB) and fatty acid metabolism (ELOVL5). In addition, these subtypes had prognostic value. Patients with the G-DIF subtype had poor prognosis compared to patients with G-INT in several cohorts. In addition, 28 samples of cell lines (11 G-INT and 17 G-DIF) showed that G-INT cell lines were more sensitive to 5-fluorouracil and oxaliplatin, whereas G-DIF were more sensitive to cisplatin. Thus, the authors suggested that G-INT and G-DIF subtypes could be used to determine GC prognosis and individualize therapy[33].
Later in 2013, Lei et al[33] published a study with another attempt to identify molecular subtypes of GC. They analyzed 248 tumor samples from patients with GC. Three main subtypes of GC were identified: mesenchymal, proliferative, and metabolic. The mesenchymal subtype was named so because of the high activity of the epithelial-mesenchymal transition (EMT) process. EMT is a complex developmental program that allows malignant cells to suppress their epithelial properties, replacing them with mesenchymal ones. These changes allow cells to become mobile and be able to migrate from the primary focus. EMT is associated with metastasis. This subtype has high levels of CDH2 (N-cadherin) expression and low levels of CDH1 (E-cadherin) expression. In addition, cells of the mesenchymal subtype of GC proved to be particularly sensitive to PI3K-AKT-mTOR inhibitors in vitro. The proliferative subtype showed a high expression of genes associated with the cell cycle (E2F, MYC, RAS) as well as a high level of mutations in TP53 and copy number variation loci of CCNE1, MYC, and KRAS. Also, this type showed a decreased relapse-free survival compared to other types. The metabolic subtype showed high expression of genes associated with metabolism, which was also specific to healthy gastric mucosa. Therefore, this subtype is thought to be closer to healthy mucosa because of its molecular and genetic characteristics than the proliferative and mesenchymal types. Moreover, the metabolic subtype has higher sensitivity to 5-fluorouracil than the others[33].
In 2014, TCGA published the most promising and full-scale study, which also classified GC into molecular subtypes. They analyzed samples of 295 patients with GC who had not previously received CT and/or radiation therapy. They used six platforms for analysis: exome sequencing, comparative genomic hybridization, DNA methylation studies, matrix RNA and micro RNA sequencing, and proteomic analysis. They classified GC into four subtypes: Epstein-Barr virus (EBV)-positive tumors, tumors with MSI, tumors with a stable genome (GS), and tumors with CIN[12]. This work is important because it identified molecular and biological subtypes of GC, which determined further research for new therapeutic approaches of treatment[33].
EBV presents in 10% of GC cases worldwide. EBV-associated GC has specific molecular features. TCGA analysis showed that 80% of EBV-positive gastric tumors have PIK3CA mutations as well as amplifications of the JAK2, CD274 (PD-L1) and PDCD1LG2 (PD-L2) genes. Mutations in ARID1A (55%) and BCOR (23%) genes are also common, while TP53 defects are not specific for this subtype of GC[34].
PIK3CA mutations usually localize in hot spots: exon 9 (E542K and E545K) and exon 20 (H1047R)[35]. However, for EBV+ GC, the frequency of PIK3CA mutations in hot spots is only 28%, and mutations can be observed throughout the nucleotide sequence[39]. Genetic defects in PIK3CA may precede EBV infection, which then enhances the activation of the PI3K/Akt/mTOR pathway.
According to TCGA analysis, EBV-associated gastric tumors have a high frequency of DNA hypermethylation. In particular, hypermethylation of the CDKN2A gene promoter (p16INK4a) was observed in all the studied samples of EBV-positive gastric tumors. Epigenetic inactivation of this gene, along with such oncosuppressors as p14, APC, and TFAP2E, is specific for EBV-positive GC[36].
The EBV+ phenotype showed increased expression of PD-L1 and/or PD-L2 among another four molecular subtypes of GC[37,38]. The phase II study was the first to show a very high response rate to pembrolizumab therapy among patients with metastatic EBV-positive GC and MSI-GC (overall response rates of 100% and 85.7%, respectively). The authors concluded that EBV+ status and MSI-H serve as reliable predictors of response to immunotherapy, along with the high immunohistochemical expression of PD-L1 in the tumor. They proposed to introduce routine determination of EBV status into clinical practice in order to identify patients with gastric adenocarcinomas who could benefit from immunotherapy[39].
According to various data, from 5% to 37% of cases of GC have an MSI phenotype: 8%-20% in operable GC and 3%-5% in metastatic setting. There was a large meta-analysis of 48 studies with 18000 patients to study clinical and morphological characteristics of MSI-H GC[40]. MSI-H tumor cases included primarily women, the elderly, and the intestinal subtype. The tumor localizes in the body or in the proximal part of stomach. MSI-H tumors present with an absence of lymph node involvement. The meta-analysis showed that MSI-H GC has a favorable prognosis and better survival rates[41].
MSI-positive GC tumors showed hypermethylation of various genes, gene methylation, unpaired DNA base repair system, and a high level of expression of genes regulating mitotic activity[42]. With adjuvant or preoperative CT, the prognosis of patients with MSI-H GC is worse than in patients with microsatellite stability (MSS) tumors[41]. Kim et al[42] discovered this phenomenon in a retrospective study.
Other authors confirmed these results later. According to the results of a retrospective analysis of the randomized MAGIC and CLASSIC trials, the rationale for additional CT is questionable in cases of MSI-H GC. Only MSS patients benefited from systemic treatment, whereas MSI-H GC had a favorable prognosis with surgery alone and a poor prognosis with perioperative or adjuvant CT[43,44].
The MSI-GC-01 meta-analysis performed a pooled analysis of MAGIC and CLASSIC studies depending on MSI in the tumor. They showed that CT in patients with MSI-H did not improve the survival rates[45]. In the treatment of metastatic GC, MSI-H subtype allows immunotherapy in the second and subsequent lines of therapy according to the National Comprehensive Cancer Network recommendations[46].
The GS subtype of GC accounts for approximately 20% of the total number of GC cases analyzed by TCGA. In addition, TCGA showed that 73% of diffuse type GC cases can be classified as the GS subtype. No copy number variations were found in the GS subtype. However, the mutations of RHOA, CDH1, and ARID1A were detected as well as the chimeric gene CLDN18-ARHGAP6[47]. The Rho family of GTPases is known to regulate the dynamics of actomyosin as well as the processes of cell adhesion, proliferation, and survival. The RhoA signaling pathway is associated with invasion and metastasis. TCGA project identified 16 cases of GC with mutations in the RHOA gene, which were specific to the GS subtype. The inactivating ARID1A mutation is specific for both GS and EBV subtypes. This protein mediates regulation of cellular processes, such as DNA damage, differentiation, and development. Loss of ARID1A expression significantly correlates with tumor grade as well as with poor prognosis for patients with the GS subtype[48].
CIN subtype of GC is the most extensive group and accounts for up to 50% of stomach cancer. This subtype shows multiple chromosomal rearrangements, deletions, and translocations. The CIN subtype localizes mainly in the proximal stomach and gastroesophageal junction. It is more common in the intestinal histological type. Its distinctive feature is high frequency of TP53 mutations (in 70% of cases) and activation of tyrosine kinase receptors. Singapore researchers found amplification and coamplification of tyrosine kinase receptors in 40% of stomach cancers. To evaluate the effectiveness of targeted therapy, phase II and III studies were initiated and conducted. The EXPAND[49] and REAL-3[50] studies evaluated anti-EGFR therapy. The MET-gastric study[51] evaluated the effectiveness of MET inhibitor, and the SHINE study evaluated the effectiveness of an anti-FGFR2 agent[52]. However, all these targeted agents were not effective; all studies were negative. However, these studies did not stratify patients according to molecular changes, but they evaluated the entire patient population regardless of biomarker expression. To date, anti-Her2 therapy in patients with high HER2 expression and HER2 amplification is the only successful targeted therapy. The ToGa study proved trastuzumab in combination with CT as a first-line therapy for metastatic HER2-positive GC[53].
To date, we actively validate clinical significance of TCGA classification in GC. Based on TCGA data, Sohn et al[54] developed the first prognostic model establishing statistically significant correlations between certain molecular subtypes of GC with patient survival rates and effectiveness of adjuvant CT.
EBV+ GC had the best prognosis in relation to both disease-free survival (P = 0.006) and overall survival (P = 0.004). The worst prognosis was associated with the GS subtype. The other two subtypes (MSI and CIN) had intermediate prognosis in relation to survival rates. They also confirmed that EBV+ GC is more common in men (79%) and at a younger age than the other subtypes (mean age 53, P = 0.01)[54].
Patients with the CIN subtype of GC had the greatest benefit from adjuvant CT showing a significant increase in disease-free survival (HR: 0.39; 95%CI: 0.16-0.94; P = 0.03). On the contrary, there was no statistically significant benefit from adjuvant CT (HR: 0.83; 95%CI: 0.36-1.89; P = 0.65) in patients with GS GC. It was not possible to assess the effectiveness of adjuvant CT in the EBV+ GC subgroup due to the absence of a control group. The authors also developed a single model for assessing risk of relapse after treatment (integrated risk assessment model), which is good predictor of disease-free survival (HR: 1.5; 95%CI: 1.2-1.9; P = 0.001).
Most samples (about 75%) in TGCA represent patients of the Western population, whose clinical course and biological characteristics differ from those of Eastern populations. Data from the ACRG[55] allowed further study of the clinical utility of TCGA classification. ACRG used gene expression data to characterize 300 postoperative GC samples from Korean patients. As in TCGA classification, they identified four subtypes: (1) MSS/EMT; (2) MSI; (3) MSS/p53+; and (4) MSS/p53-[55].
Thus, a lot of data from various researchers showed main four molecular genetic subtypes of stomach cancer. However, it is difficult to implement routine use of this classification system in routine use as multi-omics analysis would be required. Meanwhile, implementation in clinical practice may facilitate translational and retrospective studies, which would enhance understanding of clinical use of such classification systems. Therefore, the next step for the clinical implementation of this classification should be identification of surrogate biomarkers and their validation in further clinical trials.
In 2021, the standards of primary molecular genetic diagnostics for metastatic colon cancer included five biomarkers: mutational status of KRAS, NRAS, and BRAF genes, expression and amplification of HER2/neu, and MSI[56,57].
The first four are negative predictors to the effectiveness of anti-EGFR antibodies. BRAF gene (V600) mutation and expression of HER2/neu are predictors to the effectiveness of BRAF inhibitors (with anti-EGFR antibodies +/- MEK inhibitors) and anti-HER2 therapy, respectively. MSI is a predictor to the effectiveness of immunotherapy. At the same time, first-line management with targeted agents in wild-type RAS/BRAF tumors depends on clinical factors like localization of the primary tumor. Therefore, it is reasonable to suggest more complex molecular genetic differences between tumors[58,59]. Systematic work continues to determine patients who could benefit from certain targeted agents. Molecular classification of colon cancer implementation to the results of already conducted randomized trials is one option that should be considered first.
In 2015 six different scientific groups that previously proposed different genetic classifiers for colorectal cancer published results of a pooled molecular genetic analysis of 4151 colon tumors. Based on this work, they created a consensus on molecular genetic (expression) subtyping of colon cancer and identified five colon cancer subtypes (CMS1, CMS2, CMS3, CMS4, and unclassified subtype), which are characterized by certain clinical and molecular differences[59].
The immune subtype (CMS1) represents 14% of cases and predominantly describes tumors with a hypermutated phenotype, MSI, tumor infiltration lymphocyte expression, and activated immune cells. More often, such tumors localize in the cecum, colon ascendens, and hepatic flexure. Canonical (CMS2) represents 37% of samples and is characterized by the activity of WNT and MYC signaling pathways. The tumor mainly localizes in colon descendens, sigmoid colon, and rectum. The metabolic (CMS3) subtype represents 13% of cases and is characterized by alterations in the metabolic systems of the cell, KRAS gene mutations, low copy number of mutated genes, and it has a CpG island methylator phenotype. The primary tumor predominantly localizes in the sigmoid colon and rectum. Mesenchymal (CMS4) subtype represents 23% of tumors and is characterized by activation of transforming growth factor β (TGF-β), significant stromal response and angiogenesis. Localization of the primary tumor is similar to the specific CMS2 subtype. However, the researchers failed to classify almost every fifth sample.
Initially this classification was not meant to identify differences in disease prognosis. However, when the researchers looked at the survival of patients with different subtypes, they found that resectable tumors with the mesenchymal subtype had the worst prognosis, while the differences between immune, canonical, and metabolic subtypes were not detected [risks ratio (RR): 1.69, P < 0.001]. Beyond progression, the situation changed: patients with the immune subtype showed the lowest survival and canonical subtype had the best survival, while patients with the mesenchymal and metabolic subtypes had an intermediate prognosis (P < 0.001).
The next step was to use tumor subtypes as predictors to the effectiveness of targeted and CT agents. The FIRE3 study compared folinic acid, fluorouracil, and irinotecan (FOLFIRI) + cetuximab and FOLFIRI + bevacizumab in first-line treatment of patients with metastatic colon cancer with a wild-type KRAS gene. The 438 out of 514 patients were classified according to the CMS subtypes. The subtype incidence among tumors with wild-type KRAS gene was 14% for CMS1, 37% for CMS2, 15% for CMS3, and 34% for CMS4 subtype. Cetuximab was effective only in the CMS4 group. The distribution by subtype in tumors with wild-type RAS genes did not differ significantly and was 12%, 41%, 11%, and 34%, respectively.
When comparing the subtype distribution concerning primary tumor localization (right-sided or left-sided), the differences were 27% vs 11% for CMS1 subtype, 28% vs 45% for CMS2, 10% vs 12% for CMS3, and 35% vs 32% for CMS4 subtype. Prognostic differences between subtypes were consistent with the original work of Guinney et al[59]. In the group of wild-type RAS genes, only the mesenchymal (CMS4) subtype had a significant gain in overall survival in favor of cetuximab (RR = 0.57, 95%CI: 0.38-0.86, P = 0.008). Similar trends were in the metabolic (CMS3) subtype group (RR = 0.57, 95%CI: 0.27-1.23, P = 0.15). At the same time, the objective responses were the main endpoint in the FIRE3 study. Cetuximab numerically proved to be more effective in all subgroups. However, cetuximab statistically significantly benefited over the combination with bevacizumab in the CMS2 and CMS4 subtypes: 74% vs 42% (P = 0.043) and 76% vs 55% (P = 0.049), respectively[60].
The CALGB/SWOG 80405 study had a similar design, but it studied other CT regimens (73% of patients were treated with FOLFOX, 27% with FOLFIRI). There were no differences in overall survival between combinations of different targeted agents, even in the wild-type RAS genes group[61]. In contrast to the FIRE3 study, they managed to classify tumor subtypes only in half (n = 581) of the patients. In the entire group of patients, the subtypes were CMS1 in 17.90% of cases, CMS2 in 41.65% of cases, CMS3 in 11.70% of cases, and CMS4 in 28.74% of cases. In a comparative analysis of subtype distribution in regard to primary tumor localization (right-sided or left-sided) the differences were 37.34% vs 9.01% for CMS1 subtype, 23.42% vs 48.26% for CMS2, 11.39% vs 12.50% for CMS3, and 27.85% vs 30.24% for CMS4 subtype. That did not differ from the similar results in the FIRE3 study. Prognostic differences between subtypes also were consistent with the original work of Guinney et al[59]. However, the differences between the metabolic and mesenchymal subtypes were more significant in favor of the latter. In contrast to the results of the FIRE3 study, the CMS2 subtype had an improvement in overall survival, but not progression-free survival, for combinations with cetuximab (RR = 0.62, 95%CI: 0.45-0.86, P = 0.0046). The CMS1 subtype benefited from bevacizumab regimens in regards to overall survival (RR = 2.34, 95%CI: 1.48-3.7, P < 0.001) and progression-free survival (RR = 2.28, 95%CI: 1.47-3.55, P < 0.001). With the immune subtype, tumors with MSI-H had greater benefit from bevacizumab regimens (R = 0.42, P = 0.0091 for overall survival and RR = 0.46, P = 0.0109 for progression-free survival). The MSS CMS1 subtype had no difference between cetuximab and bevacizumab[62]. Researchers partially associate these findings with the tumor microenvironment and in particular the presence of tumor-associated macrophages and their M1/M2 polarization and possible angiogenic immunomodulatory effect of bevacizumab[62].
There are several reasons for the differences in the results of the discussed studies (FIRE3 and CALGB/SWOG 80405). For instance, there were differences in the chemotherapeutic component of the therapy regimen. In the FIRE3 trial all patients were treated with FOLFIRI, while in the CALGB/SWOG 80405 study 73% of patients were prescribed FOLFOX.
Previously, a randomized phase III study, NSABP C-07, examined the efficacy of adding oxaliplatin to leucovorin and fluorouracil in stage II-III colon cancer in an adjuvant setting. They also retrospectively subtyped 67.6% of tumors according to the CMS classification. The CMS2 (canonical) subtype showed a significant benefit from the oxaliplatin addition (RR = 0.61, 95%CI: 0.43-0.87, P = 0.006) but only in patients with enterocytic variant of expression data (CMS2-enterocyte: RR = 0.2, 95%CI: 0.07-0.59, P = 0.003)[63]. All other subgroups had no benefit from the addition of oxaliplatin to leucovorin and fluorouracil. These findings somewhat support the subtyping results of the CALGB/SWOG 80405 study (benefit from the addition of cetuximab in the canonical-CMS2 subtype). However, to ensure integrity of the study, they should have considered including a group of patients treated with cetuximab, leucovorin and fluorouracil (without oxaliplatin).
The ATITG MAX study compared the efficacy of first-line therapy with capecitabine, capecitabine with bevacizumab, and capecitabine with bevacizumab and mitomycin C. They also published results of subtyping according to the CMS classification. Subtype distribution among all patients did not differ significantly from other studies and represented 18% for CMS1, 47% for CMS2, 12% for CMS3, and 23% for CMS4 subtype. Prognostic significance of subtypes also were consistent with the original work of Guinney et al[59]. The researchers found that adding bevacizumab to CT significantly increased progression-free survival in CMS1 (RR = 0.83, 95%CI: 0.43-1.62), CMS2 (RR = 0.5, 95%CI: 0.33-0.76 ), and CMS3 (RR = 0.31, 95%CI: 0.13-0.75) but not in the CMS4 subtype (RR = 1.24, 95%CI: 0.68-2.25)[64].
Therefore, we could conclude that the FIRE3 results might indicate low efficacy of bevacizumab for the CMS4 subtype rather than high efficacy of cetuximab. Although angiogenesis prevails in the CMS4 subtype, it is likely primarily not due to the VEGF-mediated pathway but due to the TGF-β signaling pathway. This angiogenesis type is characterized with co-optation of vessels and vascular mimicry and usually prevails with the mesenchymal component[65,66,67], which explains the ineffectiveness of bevacizumab[68].
A phase III PETACC-8 study confirmed that addition of cetuximab to adjuvant FOLFOX regimen in stage III disease had no benefit. Subtyping with CMS classification revealed 17% of CMS1 tumors, 34% of CMS2 tumors, 4% of CMS3, and 45% CMS4 subtype. The study confirmed poor prognosis of the CMS4 subtype (RR = 1.7, P = 0.021) and revealed that addition of cetuximab worsened patient survival for the CMS1 subtype (P = 0.037)[74]. We could explain these results with fibroblast enrichment of the CMS1 subtype tumors, which decrease cetuximab activity due to the secretion of IL-16A and TGF-β[69,70]. These results might indicate low efficacy of cetuximab for the CMS1 subtype or bevacizumab high efficacy in the CALGB/SWOG 80405 study. On the other hand, the CMS1 subtype is usually observed in right-sided tumors, which usually have BRAF gene mutations[61]. The addition of anti-angiogenic drugs in the first-line or second-line therapy significantly increases the survival of patients with BRAF mutations[57,70].
However, subtype heterogeneity for each specific patient and evolution of gene expression upon progression on a particular regimen could result in prevalence of a certain subtype. Thus, we know that progression on oxaliplatin increases epithelial-mesenchymal transition gene expression, which is associated with fibroblast activity of the tumor. These changes determine resistance to anti-EGFR agents and might explain low efficacy of cetuximab in the second-line setting after progression on FOLFOX with bevacizumab[71].
Several groups studied subtype heterogeneity. Piskol et al[72] studied samples of 182 primary tumors and 130 metastases and found that the CMS2 and CMS4 subtypes are usually coincidental. However, the CMS1 subtype was somewhat more common for metastases (16.90% vs 9.34%), and the CMS3 subtype was less common for metastases (< 1% vs 11% in primary tumors). The CMS1 subtype was more typical for metastases in the liver and lungs and the CMS4 subtype for other localizations. This may indicate some tropism of subtypes to metastasize to certain organs. At the same time, the expression data for the CMS2 and CMS4 subtypes were rather consistent between primary tumor and metastases. To study concordance, 71 patient samples were taken. The concordance of the CMS subtypes between primary tumor and metastases was only 60%, primarily due to the transition of the CMS2 and CMS4 subtypes and changes in the expression of epithelial-mesenchymal transition genes. This indicates that results of subtype analysis of the primary tumor could be incorrect in metastatic disease. The researchers found drift and clone selection. In cases with discordance, it was more difficult to identify the subtype. Nevertheless, the authors concluded that specific features of a particular tumor like expression of tumor-specific genes did not change due to disease progression, but the microenvironment did. This might lead to the accuracy of CMS subtype classification[73]. However, Piskol et al[72] used a low-cost variant of genetic analysis with tumor material isolated from paraffin blocks, and the original study of Guinney et al[59] used fresh frozen samples, but their results were consistent with 85%.
Dunne et al[73] examined expression data at different sites of the primary tumor. In particular, when comparing the central part and invasive tumor front, concordance of CMS subtypes was only 38%. The concordance between the central part and lymph node metastases was 29%. Concordance between the invasive front and lymph node metastases was 21%. Concordance between all three zones was 17%. This is likely due to the CMS4 phenotype acquiring in the invasion front and metastasis area, whereas the CMS2 and CMS3 subtypes prevailed in the central zone of the tumor. As in previous work, only 46% of samples were unambiguously classified according to the CMS subtype. The authors also noted that surrounding tumor stroma significantly affects the expression data and therefore the assignment of the sample to the one subtype or another. This is especially true when studying transcription factors using a small sample of biopsy material[73].
In 2018, Laurent-Puig et al[74] presented more extensive data on tumor subtyping from the PETACC8 study patients discussed above. The authors noted that it was possible to identify the subtype unambiguously only in 42.6% of samples, while in 57.4% the tumor was treated as the mixed CMS subtype, combining at least up to 20% of two subgroups. The researchers managed to identify 16 variants of tumors with incidence from 2.1%-18.3% when they divided the mixed subgroups in accordance with combination of the largest and the smallest components. Interestingly, the pure metabolic (CMS3) subtype included only tumors with a mutation in the KRAS gene. Mixed tumors with the CMS4 component had the worst prognosis in terms of disease-free survival even taking into account clinical factors[74].
To study microenvironment influence on the therapy effectiveness and formation of certain transcriptome subtypes in colon cancer, Becht et al[75] created a CMP algorithm that allows determining tumor sample infiltration with various cells of the microenvironment (fibroblasts, macrophages, endotheliocytes, various subgroups of lymphocytes, etc.) via expression data. This approach was validated immunohistochemically by calculating the cellular composition of the sample. The authors compared the results with different CMS subtypes of colon cancer.
They found that the immune (CMS1) and mesenchymal (CMS4) subtypes were enriched with CD8 T lymphocytes and CD68 macrophages in contrast to the canonical (CMS2) and metabolic (CMS3) subtypes. The mesenchymal (CMS4) subtype proved to have a high density of tumor-associated fibroblasts, myeloid cells, and endothelial cells, which was confirmed with myeloid chemokine expression (CCL2), complement system components, proangiogenic factors (VEGFA, VEGFB, and PDGFC), and immunosuppression molecules (TGFβ1, TGFß3, and PDGFC).
The CMS1 subtype had a high expression of chemoattractants to T lymphocytes (CXCL9, CXCL10, and CXCL16) or molecules involved in tertiary lymphoid structure formation (CXCL13), increased expression of INFγ and IL-15, and high expression of genes encoding PD-1 ligands. Interestingly, the latter was also found in the mesenchymal subtype. The canonical (CMS2) subtype had low presentation of class 1 major histocompatibility complex proteins, and the tumor infiltration by lymphocytes was also low.
There is a strong positive correlation between the number of fibroblasts and myeloid and endothelial cells (in accordance with the CMP algorithm), but there is no such correlation between the fibroblasts and cytotoxic cells. The authors concluded that fibroblasts in the mesenchymal subtype promote angiogenesis, recruitment of proinflammatory cells, and the formation of the immunosuppressive phenotype[76].
Combining data on the tumor expression subtype with characteristics of the microenvironment allowed us to choose investigational therapeutic options. In particular, the combination of antiangiogenic drugs with immune checkpoint inhibitors or with inhibitors of proteins involved in the interaction of cells with extracellular matrix components could be beneficial in the mesenchymal subtype[77]. However, the combination of immune checkpoint inhibitors and bevacizumab failed in metastatic colon cancer without mismatch repair deficiency[78]. Similarly, bevacizumab had no benefit for the CMS4 subtype, although it expressed proangiogenic factors and endothelial cells via the CMS algorithm. We could possibly explain this with the fact that this subtype expresses TGF-β, which induces alternative pathways of angiogenesis. Also, tumors resistant to bevacizumab often have an increased expression of TGF-β[79]. We suggest the use of TGF-β inhibitors and immune checkpoint inhibitors in the CMS4 subtype.
The metabolic (CMS3) and canonical (CMS2) subtypes have low tumor lymphocyte infiltration and low expression of type I major histocompatibility complex components, which induces an unsatisfactory antigen-presenting function. Artificial saturation of tumors with lymphocytes in such situations could serve as a possible solution [for example, the use of bispecific antibodies to tumor antigens and lymphocytes (cibisatamab)][80]. Cibisatamab in combination with atezolizumab in a phase I study in patients with metastatic chemorefractory cancer and elevated CEA demonstrated 60% of metabolic responses with positron emission tomography/computed tomography[80]. However, due to heterogeneity and volatility of CEA expression in colon cancer cells, this approach might not prove to be beneficial[81].
Cells expressing CEA, as well as the canonical (CMS2) subtype, have increased activity of the WNT/β-catenin signaling pathway. This raises the rationale of studying inhibitors of this pathway in the corresponding subtype of colon cancer. Such inhibitors, BBI608 (napabucasin) and CGX1321 in combination with pembrolizumab, are thought to be therapy options. CGX1321 studies are still going, but napabucasin has already been studied in the phase II trial in accordance with the subtypes of colon cancer. The efficacy in patients with chemorefractory colon cancer and MSI-H was 50%, and in patients without mismatch repair deficiency it was 10%. In the CMS1 subtype, the objective response was in 1 out of 3 patients. In the CMS2 subtype, the objective response was in 0 out of 6 patients. In the CMS3 subtype, the objective response was in 1 (with polymerase E gene mutation) out of 4 patients. In the CMS4 subtype, the objective response was in 2 out of 6 patients. Therefore, the hypothesis that WNT inhibitors in combination with immune checkpoint inhibitors would be effective in the canonical and metabolic subtypes was not true. At the same time, the efficacy in the CMS4 subtype seemed to be encouraging.
However, the current state of colon cancer subtyping does not seem to be applicable in clinical practice. Nevertheless, CMS subtype classification could serve as a biological basis for searching for new targets. We believe that this approach for selecting targeted therapy should replace dividing patients by the primary tumor localization in the right or left half side of the colon. This seems to become a transitional stage towards real personalized therapy. In order to facilitate implementation of a classification system into clinical practice, further research should be focused on the development of feasible technology of subtyping based on conventional methods used in routine laboratory practice (immunohistochemistry or real time-PCR for example). Surrogate markers of each subtype should be described. Then, it would be possible to develop small panels, which would replace whole-exome or whole-transcriptome analysis.
In 2018, TCGA performed a comprehensive full-scale molecular genetic analysis of adenocarcinomas of the esophagus, stomach, and colon concerning their common endodermal origin. This analysis allowed subtyping of gastrointestinal adenocarcinomas and identified five distinct molecular subtypes: (1) EBV-associated GC (EBV+); (2) GC with MSI; (3) GC with CIN; (4) GS GC (GS); and (5) Hypermutated GC with single-nucleotide variants[34]. They initially determined EBV status in the tumor samples of gastrointestinal adenocarcinomas, then they divided EBV-negative gastrointestinal tumors into two groups according to the mutation load: adenocarcinomas with a high mutation load and gastrointestinal adenocarcinomas with a low mutation load. Adenocarcinomas with a high mutational load (hypermutated > 10 mutations per million nucleotides) were further classified into MSI and single nucleotide variant subtypes. They assigned hypermutated tumors with an indel mutation density > 1 mutation per million nucleotides and an indel/single nucleotide variant ratio > 1/150 to the MSI phenotype, while the remaining gastrointestinal adenocarcinomas were assigned to the single nucleotide variant subtype. Adenocarcinomas with low mutational density, in their turn, were divided into two groups depending on somatic copy number alteration presence or absence: tumors with CIN and a GS subtype. The identification of combined molecular and genetic subtypes gave us insights to understanding gastrointestinal adenocarcinomas biology, determining aims for future clinical research, and helping to simplify the implementation of a unified system for subtyping gastrointestinal adenocarcinomas.
However, to date, this approach has a number of limitations. One of these limitations is the significant heterogeneity between the primary tumor and distant metastases as well as evolution in the molecular and genetic properties of the tumor during treatment. One of the decisions may be the possibility of performing subtyping by liquid biopsy, though surrogate markers should be identified, which can be detected using such biological samples.
Full-scale molecular genetic analysis in most of the presented works used fresh frozen tumor tissue samples. The use of such tumor material and its storage is not routine in everyday clinical practice. Research is underway on the use of paraffinized tumor material for molecular typing, in particular for transcriptome analysis.
Another limitation for use of this classification in clinical practice is the significant volume of testing. Identification of surrogate biomarkers of molecular genetic subtypes or creation of small panels to determine these subtypes would accelerate clinical validation and its application in routine practice. Integration of subtyping into clinical studies as stratification factors is promising to assessing their clinical significance. Thus, we are still on the way to achieving successful application of the molecular genetic typing in routine clinical practice.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: Russia
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Gorgisen G, Turkey; Lee SW, South Korea; Yang Y, China S-Editor: Wu YXJ L-Editor: Filipodia P-Editor: Wu YXJ
| 1. | Ferlay J, Laversanne M, Ervik M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F (2020). Global Cancer Observatory: Cancer Tomorrow. Lyon, France: International Agency for Research on Cancer. Available from: https://gco.iarc.fr. [DOI] [Cited in This Article: ] |
| 2. | Gonzalez RS. WHO classification. PathologyOutlines.com website. . [DOI] [Cited in This Article: ] [Cited by in Crossref: 2213] [Cited by in F6Publishing: 1773] [Article Influence: 221.6] [Reference Citation Analysis (1)] |
| 3. | Abdel-Latif MM, Duggan S, Reynolds JV, Kelleher D. Inflammation and esophageal carcinogenesis. Curr Opin Pharmacol. 2009;9:396-404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 80] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 4. | Van Hoeck A, Tjoonk NH, van Boxtel R, Cuppen E. Portrait of a cancer: mutational signature analyses for cancer diagnostics. BMC Cancer. 2019;19:457. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 65] [Article Influence: 13.0] [Reference Citation Analysis (1)] |
| 5. | Smyth EC, Lagergren J, Fitzgerald RC, Lordick F, Shah MA, Lagergren P, Cunningham D. Oesophageal cancer. Nat Rev Dis Primers. 2017;3:17048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 587] [Cited by in F6Publishing: 590] [Article Influence: 84.3] [Reference Citation Analysis (1)] |
| 6. | Kauppila JH, Mattsson F, Brusselaers N, Lagergren J. Prognosis of oesophageal adenocarcinoma and squamous cell carcinoma following surgery and no surgery in a nationwide Swedish cohort study. BMJ Open. 2018;8:e021495. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 66] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 7. | Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478-486. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 601] [Cited by in F6Publishing: 566] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 8. | Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O'Donovan M, Miremadi A, Yang TP, Bower L, Chettouh H, Crawte J, Galeano-Dalmau N, Grabowska A, Saunders J, Underwood T, Waddell N, Barbour AP, Nutzinger B, Achilleos A, Edwards PA, Lynch AG, Tavaré S, Fitzgerald RC; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) Consortium. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016;48:1131-1141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 252] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 9. | Stewart A, Thavasu P, de Bono JS, Banerji U. Titration of signalling output: insights into clinical combinations of MEK and AKT inhibitors. Ann Oncol. 2015;26:1504-1510. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Cancer Genome Atlas Research Network. Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Centre; Harvard Medical School; Institute for Systems Biology; KU Leuven; Mayo Clinic; Memorial Sloan Kettering Cancer Center; National Cancer Institute; Nationwide Children’s Hospital; Stanford University; University of Alabama; University of Michigan; University of North Carolina; University of Pittsburgh; University of Rochester; University of Southern California; University of Texas MD Anderson Cancer Center; University of Washington; Van Andel Research Institute; Vanderbilt University; Washington University; Genome Sequencing Center: Broad Institute; Washington University in St. Louis; Genome Characterization Centers: BC Cancer Agency; Broad Institute; Harvard Medical School; Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University; University of North Carolina; University of Southern California Epigenome Center; University of Texas MD Anderson Cancer Center; Van Andel Research Institute; Genome Data Analysis Centers: Broad Institute; Brown University:; Harvard Medical School; Institute for Systems Biology; Memorial Sloan Kettering Cancer Center; University of California Santa Cruz; University of Texas MD Anderson Cancer Center; Biospecimen Core Resource: International Genomics Consortium; Research Institute at Nationwide Children’s Hospital; Tissue Source Sites: Analytic Biologic Services; Asan Medical Center; Asterand Bioscience; Barretos Cancer Hospital; BioreclamationIVT; Botkin Municipal Clinic; Chonnam National University Medical School; Christiana Care Health System; Cureline; Duke University; Emory University; Erasmus University; Indiana University School of Medicine; Institute of Oncology of Moldova; International Genomics Consortium; Invidumed; Israelitisches Krankenhaus Hamburg; Keimyung University School of Medicine; Memorial Sloan Kettering Cancer Center; National Cancer Center Goyang; Ontario Tumour Bank; Peter MacCallum Cancer Centre; Pusan National University Medical School; Ribeirão Preto Medical School; St. Joseph’s Hospital &Medical Center; St. Petersburg Academic University; Tayside Tissue Bank; University of Dundee; University of Kansas Medical Center; University of Michigan; University of North Carolina at Chapel Hill; University of Pittsburgh School of Medicine; University of Texas MD Anderson Cancer Center; Disease Working Group: Duke University; Memorial Sloan Kettering Cancer Center; National Cancer Institute; University of Texas MD Anderson Cancer Center; Yonsei University College of Medicine; Data Coordination Center: CSRA Inc; Project Team: National Institutes of Health. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169-175. [DOI] [Cited in This Article: ] |
| 11. | Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899-905. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2617] [Cited by in F6Publishing: 2804] [Article Influence: 200.3] [Reference Citation Analysis (0)] |
| 12. | Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202-209. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4230] [Cited by in F6Publishing: 4299] [Article Influence: 429.9] [Reference Citation Analysis (2)] |
| 13. | Guo X, Tang Y, Zhu W. Distinct esophageal adenocarcinoma molecular subtype has subtype-specific gene expression and mutation patterns. BMC Genomics. 2018;19:769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Ross-Innes CS, Becq J, Warren A, Cheetham RK, Northen H, O'Donovan M, Malhotra S, di Pietro M, Ivakhno S, He M, Weaver JMJ, Lynch AG, Kingsbury Z, Ross M, Humphray S, Bentley D, Fitzgerald RC. Whole-genome sequencing provides new insights into the clonal architecture of Barrett's esophagus and esophageal adenocarcinoma. Nat Genet. 2015;47:1038-1046. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 236] [Cited by in F6Publishing: 202] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 15. | Caspa Gokulan R, Garcia-Buitrago MT, Zaika AI. From genetics to signaling pathways: molecular pathogenesis of esophageal adenocarcinoma. Biochim Biophys Acta Rev Cancer. 2019;1872:37-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Islam F, Tang JC, Gopalan V, Lam AK. Epigenetics: DNA Methylation Analysis in Esophageal Adenocarcinoma. Methods Mol Biol. 2018;1756:247-256. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 4] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 17. | Jammula S, Katz-Summercorn AC, Li X, Linossi C, Smyth E, Killcoyne S, Biasci D, Subash VV, Abbas S, Blasko A, Devonshire G, Grantham A, Wronowski F, O'Donovan M, Grehan N, Eldridge MD, Tavaré S; Oesophageal Cancer Clinical and Molecular Stratification (OCCAMS) consortium, Fitzgerald RC. Identification of Subtypes of Barrett's Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data. Gastroenterology. 2020;158:1682-1697.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 55] [Cited by in F6Publishing: 49] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 18. | Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997-1003. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5037] [Cited by in F6Publishing: 4931] [Article Influence: 259.5] [Reference Citation Analysis (0)] |
| 19. | Li X, Francies HE, Secrier M, Perner J, Miremadi A, Galeano-Dalmau N, Barendt WJ, Letchford L, Leyden GM, Goffin EK, Barthorpe A, Lightfoot H, Chen E, Gilbert J, Noorani A, Devonshire G, Bower L, Grantham A, MacRae S, Grehan N, Wedge DC, Fitzgerald RC, Garnett MJ. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat Commun. 2018;9:2983. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 20. | Yun T, Liu Y, Gao D, Linghu E, Brock MV, Yin D, Zhan Q, Herman JG, Guo M. Methylation of CHFR sensitizes esophageal squamous cell cancer to docetaxel and paclitaxel. Genes Cancer. 2015;6:38-48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 21. | Moehler M, Shitara K, Garrido M, et al: Nivolumab plus chemotherapy vs chemo as first-line treatment for advanced gastric cancer/gastroesophageal junction cancer/esophageal adenocarcinoma: First results of the CheckMate 649 study. ESMO Virtual Congress 2020. Abstract LBA6_PR. Presented September 21, 2020. [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 137] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 22. | Lauwers GY, Carneiro F, Graham DY. Gastric carcinoma. In: Bowman FT, Carneiro F, Hruban RH, eds. Classification of Tumours of the Digestive System. Lyon:IARC; 2010. [Cited in This Article: ] |
| 23. | Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4011] [Cited by in F6Publishing: 4108] [Article Influence: 146.7] [Reference Citation Analysis (0)] |
| 24. | Messager M, Lefevre JH, Pichot-Delahaye V, Souadka A, Piessen G, Mariette C; FREGAT working group - FRENCH. The impact of perioperative chemotherapy on survival in patients with gastric signet ring cell adenocarcinoma: a multicenter comparative study. Ann Surg. 2011;254:684-93; discussion 693. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 140] [Cited by in F6Publishing: 154] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 25. | Sasako M, Sakuramoto S, Katai H, Kinoshita T, Furukawa H, Yamaguchi T, Nashimoto A, Fujii M, Nakajima T, Ohashi Y. Five-year outcomes of a randomized phase III trial comparing adjuvant chemotherapy with S-1 versus surgery alone in stage II or III gastric cancer. J Clin Oncol. 2011;29:4387-4393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 869] [Cited by in F6Publishing: 1004] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 26. | Noh SH, Park SR, Yang HK, Chung HC, Chung IJ, Kim SW, Kim HH, Choi JH, Kim HK, Yu W, Lee JI, Shin DB, Ji J, Chen JS, Lim Y, Ha S, Bang YJ; CLASSIC trial investigators. Adjuvant capecitabine plus oxaliplatin for gastric cancer after D2 gastrectomy (CLASSIC): 5-year follow-up of an open-label, randomised phase 3 trial. Lancet Oncol. 2014;15:1389-1396. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 690] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 27. | Kozak KR, Moody JS. The survival impact of the intergroup 0116 trial on patients with gastric cancer. Int J Radiat Oncol Biol Phys. 2008;72:517-521. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 28. | Ychou M, Boige V, Pignon JP, Conroy T, Bouché O, Lebreton G, Ducourtieux M, Bedenne L, Fabre JM, Saint-Aubert B, Genève J, Lasser P, Rougier P. Perioperative chemotherapy compared with surgery alone for resectable gastroesophageal adenocarcinoma: an FNCLCC and FFCD multicenter phase III trial. J Clin Oncol. 2011;29:1715-1721. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1216] [Cited by in F6Publishing: 1361] [Article Influence: 104.7] [Reference Citation Analysis (0)] |
| 29. | Cunningham D, Allum WH, Stenning SP, Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ, Falk SJ, Iveson TJ, Smith DB, Langley RE, Verma M, Weeden S, Chua YJ, MAGIC Trial Participants. Perioperative chemotherapy versus surgery alone for resectable gastroesophageal cancer. N Engl J Med. 2006;355:11-20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4120] [Cited by in F6Publishing: 4248] [Article Influence: 236.0] [Reference Citation Analysis (0)] |
| 30. | Al-Batran SE, Homann N, Pauligk C, Goetze TO, Meiler J, Kasper S, Kopp HG, Mayer F, Haag GM, Luley K, Lindig U, Schmiegel W, Pohl M, Stoehlmacher J, Folprecht G, Probst S, Prasnikar N, Fischbach W, Mahlberg R, Trojan J, Koenigsmann M, Martens UM, Thuss-Patience P, Egger M, Block A, Heinemann V, Illerhaus G, Moehler M, Schenk M, Kullmann F, Behringer DM, Heike M, Pink D, Teschendorf C, Löhr C, Bernhard H, Schuch G, Rethwisch V, von Weikersthal LF, Hartmann JT, Kneba M, Daum S, Schulmann K, Weniger J, Belle S, Gaiser T, Oduncu FS, Güntner M, Hozaeel W, Reichart A, Jäger E, Kraus T, Mönig S, Bechstein WO, Schuler M, Schmalenberg H, Hofheinz RD; FLOT4-AIO Investigators. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): a randomised, phase 2/3 trial. Lancet. 2019;393:1948-1957. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 974] [Cited by in F6Publishing: 1230] [Article Influence: 246.0] [Reference Citation Analysis (0)] |
| 31. | Lutz MP, Zalcberg JR, Ducreux M, Adenis A, Allum W, Aust D, Carneiro F, Grabsch HI, Laurent-Puig P, Lordick F, Möhler M, Mönig S, Obermannova R, Piessen G, Riddell A, Röcken C, Roviello F, Schneider PM, Seewald S, Smyth E, van Cutsem E, Verheij M, Wagner AD, Otto F. The 4th St. Gallen EORTC Gastrointestinal Cancer Conference: Controversial issues in the multimodal primary treatment of gastric, junctional and oesophageal adenocarcinoma. Eur J Cancer. 2019;112:1-8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Tan IB, Ivanova T, Lim KH, Ong CW, Deng N, Lee J, Tan SH, Wu J, Lee MH, Ooi CH, Rha SY, Wong WK, Boussioutas A, Yeoh KG, So J, Yong WP, Tsuburaya A, Grabsch H, Toh HC, Rozen S, Cheong JH, Noh SH, Wan WK, Ajani JA, Lee JS, Tellez MS, Tan P. Intrinsic subtypes of gastric cancer, based on gene expression pattern, predict survival and respond differently to chemotherapy. Gastroenterology. 2011;141:476-485, 485.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 266] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 33. | Lei Z, Tan IB, Das K, Deng N, Zouridis H, Pattison S, Chua C, Feng Z, Guan YK, Ooi CH, Ivanova T, Zhang S, Lee M, Wu J, Ngo A, Manesh S, Tan E, Teh BT, So JB, Goh LK, Boussioutas A, Lim TK, Flotow H, Tan P, Rozen SG. Identification of molecular subtypes of gastric cancer with different responses to PI3-kinase inhibitors and 5-fluorouracil. Gastroenterology. 2013;145:554-565. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in F6Publishing: 313] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 34. | Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, Kim J, Chatila W, Akbani R, Kanchi RS, Rabkin CS, Willis JE, Wang KK, McCall SJ, Mishra L, Ojesina AI, Bullman S, Pedamallu CS, Lazar AJ, Sakai R; Cancer Genome Atlas Research Network, Thorsson V, Bass AJ, Laird PW. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33:721-735.e8. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 337] [Cited by in F6Publishing: 318] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 35. | Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 2010;347:21-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 139] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 36. | Rennekamp AJ, Lieberman PM. Initiation of Epstein-Barr virus lytic replication requires transcription and the formation of a stable RNA-DNA hybrid molecule at OriLyt. J Virol. 2011;85:2837-2850. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 47] [Cited by in F6Publishing: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, Shen R, Taylor AM, Cherniack AD, Thorsson V, Akbani R, Bowlby R, Wong CK, Wiznerowicz M, Sanchez-Vega F, Robertson AG, Schneider BG, Lawrence MS, Noushmehr H, Malta TM; Cancer Genome Atlas Network, Stuart JM, Benz CC, Laird PW. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291-304.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1450] [Cited by in F6Publishing: 1308] [Article Influence: 218.0] [Reference Citation Analysis (0)] |
| 38. | Pereira MA, Ramos MFKP, Faraj SF, Dias AR, Yagi OK, Zilberstein B, Cecconello I, Alves VAF, de Mello ES, Ribeiro U Jr. Clinicopathological and prognostic features of Epstein-Barr virus infection, microsatellite instability, and PD-L1 expression in gastric cancer. J Surg Oncol. 2018;117:829-839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 42] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 39. | Kim ST, Cristescu R, Bass AJ, Kim KM, Odegaard JI, Kim K, Liu XQ, Sher X, Jung H, Lee M, Lee S, Park SH, Park JO, Park YS, Lim HY, Lee H, Choi M, Talasaz A, Kang PS, Cheng J, Loboda A, Lee J, Kang WK. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med. 2018;24:1449-1458. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 643] [Cited by in F6Publishing: 920] [Article Influence: 153.3] [Reference Citation Analysis (0)] |
| 40. | Polom K, Marano L, Marrelli D, De Luca R, Roviello G, Savelli V, Tan P, Roviello F. Meta-analysis of microsatellite instability in relation to clinicopathological characteristics and overall survival in gastric cancer. Br J Surg. 2018;105:159-167. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 164] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 41. | Sunakawa Y, Lenz HJ. Molecular classification of gastric adenocarcinoma: translating new insights from the cancer genome atlas research network. Curr Treat Options Oncol. 2015;16:17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 42. | Kim SY, Choi YY, An JY, Shin HB, Jo A, Choi H, Seo SH, Bang HJ, Cheong JH, Hyung WJ, Noh SH. The benefit of microsatellite instability is attenuated by chemotherapy in stage II and stage III gastric cancer: Results from a large cohort with subgroup analyses. Int J Cancer. 2015;137:819-825. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 43. | Smyth EC, Wotherspoon A, Peckitt C, Gonzalez D, Hulkki-Wilson S, Eltahir Z, Fassan M, Rugge M, Valeri N, Okines A, Hewish M, Allum W, Stenning S, Nankivell M, Langley R, Cunningham D. Mismatch Repair Deficiency, Microsatellite Instability, and Survival: An Exploratory Analysis of the Medical Research Council Adjuvant Gastric Infusional Chemotherapy (MAGIC) Trial. JAMA Oncol. 2017;3:1197-1203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 343] [Cited by in F6Publishing: 331] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 44. | Choi YY, Kim H, Shin SJ, Kim HY, Lee J, Yang HK, Kim WH, Kim YW, Kook MC, Park YK, Kim HH, Lee HS, Lee KH, Gu MJ, Choi SH, Hong S, Kim JW, Hyung WJ, Noh SH, Cheong JH. Microsatellite Instability and Programmed Cell Death-Ligand 1 Expression in Stage II/III Gastric Cancer: Post Hoc Analysis of the CLASSIC Randomized Controlled study. Ann Surg. 2019;270:309-316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 104] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 45. | Pietrantonio F, Raimondi A, Choi Y, Kang W, Langley RE, Kim Y, Kim K, Nankivell M, Perrone F, Kook M, Yeong J An, Grabsch H, Morano F, Cheong J, Noh S, Lee J, Cunningham D, Bartolomeo M, Fucà G, Smyth E. MSI-GC-01: Individual patient data (IPD) meta-analysis of microsatellite instability (MSI) and gastric cancer (GC) from four randomized clinical trials (RCTs). J Clini Oncol. 37:66. [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 46. | National Comprehensive Cancer Network. Gastric cancer (Version 1.2021). Available from: https://www.nccn.org/professionals/physician_gls/pdf/gastric_blocks.pdf. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 47. | Ling Y, Watanabe Y, Nagahashi M, Shimada Y, Ichikawa H, Wakai T, Okuda S. Genetic profiling for diffuse type and genomically stable subtypes in gastric cancer. Comput Struct Biotechnol J. 2020;18:3301-3308. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Garattini SK, Basile D, Cattaneo M, Fanotto V, Ongaro E, Bonotto M, Negri FV, Berenato R, Ermacora P, Cardellino GG, Giovannoni M, Pella N, Scartozzi M, Antonuzzo L, Silvestris N, Fasola G, Aprile G. Molecular classifications of gastric cancers: Novel insights and possible future applications. World J Gastrointest Oncol. 2017;9:194-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 43] [Cited by in F6Publishing: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 49. | Lordick F, Kang YK, Chung HC, Salman P, Oh SC, Bodoky G, Kurteva G, Volovat C, Moiseyenko VM, Gorbunova V, Park JO, Sawaki A, Celik I, Götte H, Melezínková H, Moehler M; Arbeitsgemeinschaft Internistische Onkologie and EXPAND Investigators. Capecitabine and cisplatin with or without cetuximab for patients with previously untreated advanced gastric cancer (EXPAND): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:490-499. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 589] [Cited by in F6Publishing: 640] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 50. | Waddell T, Chau I, Cunningham D, Gonzalez D, Okines AF, Okines C, Wotherspoon A, Saffery C, Middleton G, Wadsley J, Ferry D, Mansoor W, Crosby T, Coxon F, Smith D, Waters J, Iveson T, Falk S, Slater S, Peckitt C, Barbachano Y. Epirubicin, oxaliplatin, and capecitabine with or without panitumumab for patients with previously untreated advanced oesophagogastric cancer (REAL3): a randomised, open-label phase 3 trial. Lancet Oncol. 2013;14:481-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 504] [Cited by in F6Publishing: 554] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 51. | Shah MA, Bang YJ, Lordick F, Alsina M, Chen M, Hack SP, Bruey JM, Smith D, McCaffery I, Shames DS, Phan S, Cunningham D. Effect of Fluorouracil, Leucovorin, and Oxaliplatin With or Without Onartuzumab in HER2-Negative, MET-Positive Gastroesophageal Adenocarcinoma: The METGastric Randomized Clinical Trial. JAMA Oncol. 2017;3:620-627. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in F6Publishing: 218] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 52. | Van Cutsem E, Bang YJ, Mansoor W, Petty RD, Chao Y, Cunningham D, Ferry DR, Smith NR, Frewer P, Ratnayake J, Stockman PK, Kilgour E, Landers D. A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol. 2017;28:1316-1324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 132] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 53. | Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T, Aprile G, Kulikov E, Hill J, Lehle M, Rüschoff J, Kang YK; ToGA Trial Investigators. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687-697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4615] [Cited by in F6Publishing: 4839] [Article Influence: 345.6] [Reference Citation Analysis (1)] |
| 54. | Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, Lee KW, Kim EH, Yim SY, Lee SH, Cheong JH, Jeong W, Cho JY, Kim J, Chae J, Lee J, Kang WK, Kim S, Noh SH, Ajani JA, Lee JS. Clinical Significance of Four Molecular Subtypes of Gastric Cancer Identified by The Cancer Genome Atlas Project. Clin Cancer Res. 2017;23:4441-4449. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 211] [Cited by in F6Publishing: 298] [Article Influence: 42.6] [Reference Citation Analysis (0)] |
| 55. | Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, Gong L, Fu J, Jin JG, Choi MG, Sohn TS, Lee JH, Bae JM, Kim ST, Park SH, Sohn I, Jung SH, Tan P, Chen R, Hardwick J, Kang WK, Ayers M, Hongyue D, Reinhard C, Loboda A, Kim S, Aggarwal A. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-456. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1071] [Cited by in F6Publishing: 1332] [Article Influence: 148.0] [Reference Citation Analysis (0)] |
| 56. | Fedyanin MY, Gladkov OA, Gordeev SS, Rykov IV, Tryakin AA. Practical guidelines for drug treatment of colon and rectosigmoid junction cancer. Malignant tumors: Practical Recommendations of RUSSCO 3s2. 2020; 10. [DOI] [Cited in This Article: ] |
| 57. | National Comprehensive Cancer Network. Colon cancer (Version 2. 2021). Available from: https://www.nccn.org/professionals/physician_gls/pdf/gastric_blocks.pdf. [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 58. | Dienstmann R, Salazar R, Tabernero J. Molecular Subtypes and the Evolution of Treatment Decisions in Metastatic Colorectal Cancer. Am Soc Clin Oncol Educ Book. 2018;38:231-238. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 59. | Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350-1356. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3303] [Cited by in F6Publishing: 3040] [Article Influence: 337.8] [Reference Citation Analysis (0)] |
| 60. | Stintzing S, Wirapati P, Lenz HJ, Neureiter D, Fischer von Weikersthal L, Decker T, Kiani A, Kaiser F, Al-Batran S, Heintges T, Lerchenmüller C, Kahl C, Seipelt G, Kullmann F, Moehler M, Scheithauer W, Held S, Modest DP, Jung A, Kirchner T, Aderka D, Tejpar S, Heinemann V. Consensus molecular subgroups (CMS) of colorectal cancer (CRC) and first-line efficacy of FOLFIRI plus cetuximab or bevacizumab in the FIRE3 (AIO KRK-0306) trial. Ann Oncol. 2019;30:1796-1803. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 123] [Cited by in F6Publishing: 118] [Article Influence: 23.6] [Reference Citation Analysis (0)] |
| 61. | Lenz HJ, Ou FS, Venook AP, Hochster HS, Niedzwiecki D, Goldberg RM, Mayer RJ, Bertagnolli MM, Blanke CD, Zemla T, Qu X, Wirapati P, Tejpar S, Innocenti F, Kabbarah O. Impact of Consensus Molecular Subtype on Survival in Patients With Metastatic Colorectal Cancer: Results From CALGB/SWOG 80405 (Alliance). J Clin Oncol. 2019;37:1876-1885. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 154] [Cited by in F6Publishing: 149] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 62. | Peterson TE, Kirkpatrick ND, Huang Y, Farrar CT, Marijt KA, Kloepper J, Datta M, Amoozgar Z, Seano G, Jung K, Kamoun WS, Vardam T, Snuderl M, Goveia J, Chatterjee S, Batista A, Muzikansky A, Leow CC, Xu L, Batchelor TT, Duda DG, Fukumura D, Jain RK. Dual inhibition of Ang-2 and VEGF receptors normalizes tumor vasculature and prolongs survival in glioblastoma by altering macrophages. Proc Natl Acad Sci U S A. 2016;113:4470-4475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 222] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 63. | Song N, Pogue-Geile KL, Gavin PG, Yothers G, Kim SR, Johnson NL, Lipchik C, Allegra CJ, Petrelli NJ, O'Connell MJ, Wolmark N, Paik S. Clinical Outcome From Oxaliplatin Treatment in Stage II/III Colon Cancer According to Intrinsic Subtypes: Secondary Analysis of NSABP C-07/NRG Oncology Randomized Clinical Trial. JAMA Oncol. 2016;2:1162-1169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 122] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 64. | Mooi JK, Wirapati P, Asher R, Lee CK, Savas P, Price TJ, Townsend A, Hardingham J, Buchanan D, Williams D, Tejpar S, Mariadason JM, Tebbutt NC. The prognostic impact of consensus molecular subtypes (CMS) and its predictive effects for bevacizumab benefit in metastatic colorectal cancer: molecular analysis of the AGITG MAX clinical trial. Ann Oncol. 2018;29:2240-2246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 65. | Döme B, Hendrix MJ, Paku S, Tóvári J, Tímár J. Alternative vascularization mechanisms in cancer: Pathology and therapeutic implications. Am J Pathol. 2007;170:1-15. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in F6Publishing: 258] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 66. | Leenders WP, Küsters B, de Waal RM. Vessel co-option: how tumors obtain blood supply in the absence of sprouting angiogenesis. Endothelium. 2002;9:83-87. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 122] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 67. | Donnem T, Hu J, Ferguson M, Adighibe O, Snell C, Harris AL, Gatter KC, Pezzella F. Vessel co-option in primary human tumors and metastases: an obstacle to effective anti-angiogenic treatment? Cancer Med. 2013;2:427-436. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 186] [Cited by in F6Publishing: 194] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 68. | Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao ZH, Shi Y, Van den Eynden G, Daley F, Peckitt C, Tan X, Salman A, Lazaris A, Gazinska P, Berg TJ, Eltahir Z, Ritsma L, Van Rheenen J, Khashper A, Brown G, Nystrom H, Sund M, Van Laere S, Loyer E, Dirix L, Cunningham D, Metrakos P, Reynolds AR. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med. 2016;22:1294-1302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 254] [Cited by in F6Publishing: 281] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 69. | Colangelo T, Polcaro G, Muccillo L, D'Agostino G, Rosato V, Ziccardi P, Lupo A, Mazzoccoli G, Sabatino L, Colantuoni V. Friend or foe? Biochim Biophys Acta Rev Cancer. 2017;1867:1-18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 70. | Fedyanin MY, Polyanskaya EM, Elsnukaeva HM, Tryakin AA, Pokataev I. A., Ignatova E.O., Sergeev US, Bulanov AA, Tjulandin SA. Is it necessary to add anti-angiogenic therapy to chemotherapy in patients with metastatic colorectal cancer and BRAF mutation? Malignant tumours. 2020;10:36-44. [DOI] [Cited in This Article: ] |
| 71. | Bennouna J, Hiret S, Bertaut A, Bouché O, Deplanque G, Borel C, François E, Conroy T, Ghiringhelli F, des Guetz G, Seitz JF, Artru P, Hebbar M, Stanbury T, Denis MG, Adenis A, Borg C. Continuation of Bevacizumab vs Cetuximab Plus Chemotherapy After First Progression in KRAS Wild-Type Metastatic Colorectal Cancer: The UNICANCER PRODIGE18 Randomized Clinical Trial. JAMA Oncol. 2019;5:83-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 72. | Piskol R, Huw L, Sergin I, Kljin C, Modrusan Z, Kim D, Kljavin N, Tam R, Patel R, Burton J, Penuel E, Qu X, Koeppen H, Sumiyoshi T, de Sauvage F, Lackner MR, de Sousa E Melo F, Kabbarah O. A Clinically Applicable Gene-Expression Classifier Reveals Intrinsic and Extrinsic Contributions to Consensus Molecular Subtypes in Primary and Metastatic Colon Cancer. Clin Cancer Res. 2019;25:4431-4442. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 73. | Dunne PD, Alderdice M, O'Reilly PG, Roddy AC, McCorry AMB, Richman S, Maughan T, McDade SS, Johnston PG, Longley DB, Kay E, McArt DG, Lawler M. Cancer-cell intrinsic gene expression signatures overcome intratumoural heterogeneity bias in colorectal cancer patient classification. Nat Commun. 2017;8:15657. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 74. | Laurent-Puig P, Marisa L, Ayadi M, Blum Y, Balogoun R, Pilati C, Le Malico K, Lepage C, Emile JF, Salazar R, Aust D, Duval A, Selves J, Guenot D, Milano G, Seitz JF, Taieb J, Boige V, de Reynie`s A. Colon cancer molecular subtype intratumoral heterogeneity and its prognostic impact: An extensive molecular analysis of the PETACC-8. Ann Oncol. 2018;29:viii18. [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 75. | Becht E, de Reyniès A, Giraldo NA, Pilati C, Buttard B, Lacroix L, Selves J, Sautès-Fridman C, Laurent-Puig P, Fridman WH. Immune and Stromal Classification of Colorectal Cancer Is Associated with Molecular Subtypes and Relevant for Precision Immunotherapy. Clin Cancer Res. 2016;22:4057-4066. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 325] [Cited by in F6Publishing: 366] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 76. | Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6912] [Cited by in F6Publishing: 7713] [Article Influence: 482.1] [Reference Citation Analysis (0)] |
| 77. | Grothey A, Tabernero J, Arnold D. Fluoropyrimidine and bevacizumab plus or minus atezolizumab as first-line treatment for BRAF wild type metastatic colorectal cancer: Findings from the MODUL trial of biomarker-driven maintenance. ESMO 2018 Congress. Abstract LBA19. [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 64] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 78. | Itatani Y, Kawada K, Yamamoto T, Sakai Y. Resistance to Anti-Angiogenic Therapy in Cancer-Alterations to Anti-VEGF Pathway. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 193] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 79. | Melisi D, Hollebecque A, Oh DY, Calvo E, Varghese AM, Borazanci EH, Mercade TM, Simionato F, Park JO, Bendell JC, Faivre SJ, Zhao Y, Gueorguieva I, Man M, Estrem S, Benhadji KA, Lanasa M, Guba SC, Garcia-Carbonero R. A phase Ib dose-escalation and cohort-expansion study of safety and activity of the transforming growth factor (TGF) β receptor I kinase inhibitor galunisertib plus the anti-PD-L1 antibody durvalumab in metastatic pancreatic cancer. J Clin Oncol. 2019;37:4124. [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 80. |
Tabernero J, Melero I, Ros W, Argiles G, Marabelle A, Rodriguez-Ruiz ME, et al Phase Ia and Ib studies of the novel carcinoembryonic antigen (CEA) T-cell bispecific (CEA CD3 TCB) antibody as a single agent and in combination with atezolizumab: preliminary efficacy and safety in patients with metastatic colorectal cancer (mCRC).
|
| 81. | Gonzalez-Exposito R, Semiannikova M, Griffiths B, Khan K, Barber LJ, Woolston A, Spain G, von Loga K, Challoner B, Patel R, Ranes M, Swain A, Thomas J, Bryant A, Saffery C, Fotiadis N, Guettler S, Mansfield D, Melcher A, Powles T, Rao S, Watkins D, Chau I, Matthews N, Wallberg F, Starling N, Cunningham D, Gerlinger M. CEA expression heterogeneity and plasticity confer resistance to the CEA-targeting bispecific immunotherapy antibody cibisatamab (CEA-TCB) in patient-derived colorectal cancer organoids. J Immunother Cancer. 2019;7:101. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in F6Publishing: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |