Published online Feb 15, 2003. doi: 10.3748/wjg.v9.i2.291
Revised: August 4, 2002
Accepted: September 16, 2002
Published online: February 15, 2003
AIM: To study hepatic virus C (HCV) RNA and HCV protein expression in peripheral blood mononuclear cells (PBMCs) of patients with HCV infection, and explore the relationship between the HCV RNA in the PBMCs and response to interferon (IFN) therapy.
METHODS: Type-specific primers were designed and RT-nested PCR was used to detect the plus- and minus- strands of HCV RNA in PBMCs of 54 patients with HCV infection; Indirect immunofluorescence assay was applied to identify HCVNS5 protein expression in PBMCs; 6 month-, 3 MU-IFN regiment was administrated to observe the responses to IFN in 35 chronic hepatitis C patients with different HCV RNA status in PBMCs.
RESULTS: HCV plus strand RNA was found in 10 of 19 (52.6%) acute hepatitis C patients and 22 of 35 (62.9%) chronic hepatitis C patients. HCV minus strand RNA was detected in 14 of 35 (40.0%) chronic hepatitis C patients, but only one patient (5.3%) with acute HCV infection was found to be minus HCV RNA positive. Though no HCV NS5 protein expression was found in the examined 10 cases of acute HCV infection, it was positive in 17 of 20 (85.0%) chronic hepatitis C patients by indirect immunofluoresence assay. There are significant differences of positive rate of the minus-strand and HCVNS5 protein between acute and chronic hepatitis C groups (u = 2.07, P < 0.05 and u = 4.43, P < 0.01 respectively). The patients with minus-strand HCV RNA showed a significantly lower 6-month sustained response (SR-6) to IFN compared to those without minus-strand HCVRNA in PBMCs (biologically 14.3% vs 42.8%, χ2 = 4.12, P < 0.05 and virologically 7.1% vs 23.9%, χ2 = 4.24, P < 0.05).
CONCLUSION: HCV is capable of infecting and replicating in PBMCs, and HCVNS5 protein was expressed in PBMCs. The patients with minus strand HCV RNA in PBMCs showed a significantly lower 6-month sustained response to IFN, suggesting that minus-strand HCV RNA in PBMCs may be one of the factors influencing response to IFN therapy.
- Citation: Gong GZ, Lai LY, Jiang YF, He Y, Su XS. HCV replication in PBMC and its influence on interferon therapy. World J Gastroenterol 2003; 9(2): 291-294
- URL: https://www.wjgnet.com/1007-9327/full/v9/i2/291.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i2.291
Hepatitis C virus (HCV), a single positive-strand RNA, belongs to the Flaviviridae family, and is the major cause of post-transfusion hepatitis. Infection with HCV usually results in chronic hepatitis, which may progress to cirrhosis and finally to hepatocellular carcinoma. The mechanisms responsible for the chronicity are unclear, one of which is supposed to be that HCV has the ability to escape the host immunity by mutations in genome. There are numerous genotypes of HCV worldwide, and genotype 1b is found to be responsible for the most cases of HCV infection in southern China[1]. IFN has been a widely accepted drug for the treatment of patients with HCV infection for more than 10 years, and now combination of IFN with ribavirin becomes the choice of therapy[3,4]. Reinfection of HCV after orthotopic liver transplantation has postulated that there exists extrahepatic sites suitable for HCV repliacation[5,6]. The possible extrahepatic cells for HCV replication may be PBMC, cells in pancreas, adrenal gland, bone marrow and spleen, even in the cerebrospinal fluid[2,7-9], among them, PBMCs have been the most controversial, in which the minus strand HCV RNA, a replicative intermediate of HCV, has been found. It still remains unclear whether HCV replication in PBMC is a factor influencing IFN therapy response. In this study, we not only detected the minus strand HCVRNA and HCVNS5 protein expression in PBMC of patients with hepatitis C, but also analyzed the relationship between minus strand HCVRNA in PBMC and IFN response.
Blood samples were collected from 54 patients with hepatitis C virus infection from January of 1994 to January of 1998, all of them are positive for anti-HCV by ELISA (Sino-American Biotech. Company, China) and HCV RNA by RT-PCR (Sino-American Biotech. Company, China). PBMCs were separated from 10 mL of whole blood mixed with sodium citrate by density gradient centrifugation with ficoll-hypaque. The separated PBMCs were washed four times in 10 mL of RPMI-1640 and then frozen and stored at -70 °C until use.
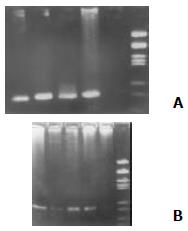
Total RNA of the PBMCs was extracted with an RNA isolation kit (Shanghai Huaxun Company, China) according to the manufacturer's instructions. Primers P1: 5'-CGCGCGACTAGGAAGACTTC-3'and P2: 5'-ATGTACCCCCATGAGGTCGGC-3' (as the external pair), and P3: 5'-AGGAAGACTTCCGAGCGCGGTC-3'and P4: 5'-GAGCCATCCTGCC CACCCCA-3' (as the internal pair) for RT-PCR were designed according to Okamoto et al[10]. 5 μL of PBMC RNA and 1 μL of P1 (for producing cDNA of minus HCVRNA) or 1 μL of P2 (for producing cDNA of plus HCVRNA) were added to the reverse transcription system (Promega, USA). The reverse transcription system includes 10 × buffer 2 μL, 25 mmol/L MgCl2 4 μL, RNasin 1 μL, AMVRT 15 U, 10 mmol/L dNTP 2 μL with a total volume of 20 μL by adding ddH2O. After incubation for 30 min at 42 °C, the synthesized HCV RNA cDNA was exposed at 95 °C for 30 min to destroy AMVRT. The first PCR was performed with the above synthesized HCVRNA cDNA as a template. The external primers (P1 and P2) were added into the PCR reaction mix. After pre-denaturing for 5 min, the reaction mix denatured at 94 °C for 60 s, annealed at 55 °C for 60 s and extended at 72 °C for 90 s for 35 cycles. The second PCR was performed same as in the first PCR except the production of the first PCR was used as the template and with the internal primers (P3 and P4). A total volume of 7 μL the second PCR product was loaded onto 2% agarose gel containing 0.5 ug/mL EB. After electrophoresis, the gel was placed under ultraviolet ray to analyze the results (Figure 1).
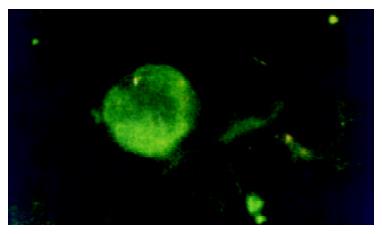
After being separated, PBMCs were suspended (1 × 107cell·mL-1) in RPMI-1640, dropped on to slides, and air dried. The slides were then fixed in acetone for 20 min at -20 °C, washed with PBS, and air-dried. Mouse anti-human HCV NS5 McAb (1:400, Virostat, U.S.A) was added onto the slides. After 30 min incubation at 37 °C, the slides were washed three times in PBS, and then isothiocyanate-conjugated rabbit anti-mouse IgG was added and incubated for 30 min. Slides were then washed and observed under microscope (Figure 2).
Three MU of Interferon α-2β (Tianjing Hualida, China) was administered intramuscularly three times a week for 6 mo. The efficacy of interferon therapy is defined biochemically as normalization of serum (adding full term of ALT here!!!) (ALT) and virologically as serum conversion of HCV RNA. End-treatment response (ETR) refers to the response to IFN when the treatment ends, and the sustained response (SR) refers to the response after the ending of treatment, i.e., SR-6 means response at 6 mo after the ending of treatment.
HCV plus-strand RNA was found in 32 of 54 (59.26%) patients with HCV infection, among them 10 of 19 (52.63%) with acute hepatitis C and 22 of 35 (62.85%) with chronic hepatitis C were positive respectively, and there is no statistically significant difference between the two groups. HCV minus-strand RNA was detected positive in 14 out of the 35 patients with chronic hepatitis C (14 of 35, 40%) and 1of 19 patients with acute hepatitis C and a significant difference was found between these two groups (u = 2.07, P < 0.05). Regarding to HCVNS5 expression in PBMCs of patients with hepatitis C, 17 out of 20 patients with chronic hepatitis C were found positive, but all the 10 patients with acute hepatitis C were found negative, and there is a remarkable statistically significant difference between the two groups (u = 4.43, P < 0.01. see Table 1).
Six-month regiment with 3MU of IFN-α 2b was completed in 35 patients with chronic hepatitis C, and the biochemical and viralogical ETR and SR-6 were evaluated. There is a tendency to have a lower response to IFN treatment in the patients with plus-strand HCVRNA positive in PBMCs, although no statistically significant difference was found when compared with the negative group. The patients with minus-strand HCV RNA in PBMCs showed a significantly lower SR-6 to IFN therapy than those without HCV RNA minus-strand, both biochemically (SR-6: 14.3% vs 42.8%, χ2 = 4.12, P < 0.05), and virologically (SR-6: 7.1% vs 23.9%, χ2 = 4.24, P < 0.05) (Table 2).
Extrahepatic HCV replication has long been a controversial topic since the finding of the high rate of re-infection of grafts after orthotopic liver transplantation in the patients with the end-stage HCV induced liver diseases. Weather PBMCs is suitable for HCV replication is still uncertain. The detection of the minus strand HCV RNA is thought to be reasonable for the discovery of HCV replication because the minus strand RNA is the replicative intermediate of HCV. In recent years, several reports on the detection of HCVRNA in PBMCs have been published[12-14]. Cribier et al[15] incubated PBMCs healthy donors with HCV positive sera, and detected HCV RNA plus-strand and minus-strand using RT-PCR and in situ hybridization. Our results showed that HCV RNA plus-strand were common in the PBMCs of patients, in both acute and chronic infection patients. This high rate of plus-strand HCV RNA is usually thought to be resulted from the contamination of plasma, therefore, minus-strand HCV RNA was explored in the PBMCs from hepatitis C patients, which indicates the replication of HCV in PBMCs. In acute HCV infection, HCV RNA minus-strand is rare in PBMCs, but in the chronic group, the minus-strand HCV RNA is not uncommon in the PBMCs (14 of 35, 40.0%), which is similar to what Chang et al[16] reported. The ratio of HCV RNA minus-strand detected in chronic hepatitis C is much higher than that in acute hepatitis C, suggesting that the replication of HCV in PBMCs may play an important role in the processes of chronicity, and the mechanism could be that HCV in PBMCs can escape from clearance resulting from host immununity, and make the infection of HCV persistent. On the other hand, the dysfunction of the HCV infected PBMCs leads to immune function decline or in disorder, and this becomes more difficult for the host to clear intrahepatic HCV, so that the injure of hepatocytes persists[17]. Although minus-strand HCV RNA is the replicative form and not found in patient's serum or plasma, indicating that is a more convincing parameter for HCV replication, some authors are still arguing that the minus-strand HCV RNA in the blood cells including PBMC may be artifacts from self-priming or mispriming during PCR reaction[18,19], or contamination or passive absorption by circulating virus[20,21]. To overcome that point, the expression of HCV related proteins in extrahepatic cells has become the key point for the identification of HCV replication. Sansonno et al[22], found HCV exists and replicates mainly in plasma of PBMCs, and the viral proteins, such as core protein, NS3 were found to be expressed in PBMCs. Chen et al[11], analyzed the relationship between HCV core expression in PBMCs and the diseased state of hepatitis C patients and found that the core protein was more intensely expressed in the nucleus of PBMCs from advanced chronic hepatitis C patients than that from the moderate patients. We further performed an indirect immunofluorescent assay for HCVNS5 protein and its expression was found mainly in cytoplasma of PBMCs from patients with chronic hepatitis C. Our results indicate that HCV not only replicates but also produces its related protein in PBMCs.
IFN is known to possess both immunomodulatory and antiviral activities. It is tempting to postulate that IFN therapy may enhance the host immune response to promote the clearance of HCV infection. IFN is currently the only approved efficient drug for hepatitis C infection, and combined with ribavirin, its antiviral activity will be increased[3,4,23]. Serum HCV load and the HCV subtypes have been considered as the major factors to influence the response to IFN therapy[28,29]. Others influencing factors include the increased amount of MxA mRNA, the higher complexity of HCV quasispecies and the frequency of mutations in NS5A region[30-33]. The extrahepatic HCV replication, especially in the PBMCs, acts as a predicator for the response to IFN therapy needs to be explored. Omata et al[24], reported a prospective IFN study, in which most of patients treated with IFN obtained normalization of serum aminotransferase, whereas only 3 cases from the control showed such change (P < 0.02); serum hepatitis C virus RNA became undetectable in 10 of 11 treated cases, but in only 1 of 12 patients of control group. IFN prevents the progression of acute hepatitis C to chronicity by eradicating HCV. The response of patients with chronic hepatitis C to IFN treatment was significantly lower than that of patients with acute hepatitis C. That the detection ratio of HC VRNA minus in PBMC of chronic hepatitis C is significantly higher than that of acute hepatitis C suggests that the replication of HCV in PBMCs is an important factor influencing the response to IFN treatment. LÖhr et al[25], reported that there was no relationship between HCVRNA minus-strand in PBMC and the response to IFN treatment. Others reported that the replication of HCVRNA in PBMC may be the source of relapse after IFN treatment in chronic hepatitis C[26,27]. The different HCV quasispecies in liver or PBMC may response to IFN differently and the quasispecies in PBMCs should be considered to predicator in response to IFN therapy[34-36]. Our results show the replication of HCV RNA in PBMC can influence the response to IFN. The patients with HCVRNA minus-strands in PBMC had a significantly lower 6-month sustained response to IFN, both biochemically and virologically, than those without minus-strand, suggesting HCV replication in PBMCs may be one reason for relapse after treatment with IFN.
Edited by Yuan HT
| 1. | Wang Y, Okamoto H, Mishiro S. HCV genotypes in China. Lancet. 1992;339:1168. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | Laskus T, Radkowski M, Bednarska A, Wilkinson J, Adair D, Nowicki M, Nikolopoulou GB, Vargas H, Rakela J. Detection and analysis of hepatitis C virus sequences in cerebrospinal fluid. J Virol. 2002;76:10064-10068. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 122] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485-1492. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2509] [Cited by in F6Publishing: 2416] [Article Influence: 92.9] [Reference Citation Analysis (0)] |
| 4. | Poynard T, Marcellin P, Lee SS, Niederau C, Minuk GS, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C. Randomised trial of interferon alpha2b plus ribavirin for 48 wk or for 24 wk versus interferon alpha2b plus placebo for 48 wk for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group (IHIT). Lancet. 1998;352:1426-1432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1667] [Cited by in F6Publishing: 1627] [Article Influence: 62.6] [Reference Citation Analysis (0)] |
| 5. | Féray C, Gigou M, Samuel D, Paradis V, Wilber J, David MF, Urdea M, Reynes M, Bréchot C, Bismuth H. The course of hepatitis C virus infection after liver transplantation. Hepatology. 1994;20:1137-1143. [PubMed] [Cited in This Article: ] |
| 6. | Radkowski M, Wang LF, Vargas HE, Rakela J, Laskus T. Detection of hepatitis C virus replication in peripheral blood mononuclear cells after orthotopic liver transplantation. Transplantation. 1998;66:664-666. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 7. | Schmidt WN, Wu P, Brashear D, Klinzman D, Phillips MJ, LaBrecque DR, Stapleton JT. Effect of interferon therapy on hepatitis C virus RNA in whole blood, plasma, and peripheral blood mononuclear cells. Hepatology. 1998;28:1110-1116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 8. | Laskus T, Radkowski M, Wang LF, Vargas H, Rakela J. Search for hepatitis C virus extrahepatic replication sites in patients with acquired immunodeficiency syndrome: specific detection of negative-strand viral RNA in various tissues. Hepatology. 1998;28:1398-1401. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 106] [Cited by in F6Publishing: 110] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 9. | Cheng JL, Liu BL, Zhang Y, Tong WB, Yan Z, Feng BF. Hepatitis C virus in human B lymphocytes transformed by Epstein-Barr virus in vitro by in situ reverse transcriptase-polymerase chain reaction. World J Gastroenterol. 2001;7:370-375. [PubMed] [Cited in This Article: ] |
| 10. | Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: Application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673-679. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 859] [Cited by in F6Publishing: 858] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 11. | Chen L, Chen P, Fan G, Li L, Liu C. [Localization of hepatitis C virus core protein in the nucleus of peripheral blood mononuclear cells of hepatitis C patients]. Zhonghua Shiyan He Linchuang Bingduxue Zazhi. 2002;16:37-39. [PubMed] [Cited in This Article: ] |
| 12. | Wang JT, Sheu JC, Lin JT, Wang TH, Chen DS. Detection of replicative form of hepatitis C virus RNA in peripheral blood mononuclear cells. J Infect Dis. 1992;166:1167-1169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 134] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 13. | Taliani G, Badolato C, Lecce R, Poliandri G, Bozza A, Duca F, Pasquazzi C, Clementi C, Furlan C, De Bac C. Hepatitis C virus RNA in peripheral blood mononuclear cells: relation with response to interferon treatment. J Med Virol. 1995;47:16-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Zignego AL, De Carli M, Monti M, Careccia G, La Villa G, Giannini C, D'Elios MM, Del Prete G, Gentilini P. Hepatitis C virus infection of mononuclear cells from peripheral blood and liver infiltrates in chronically infected patients. J Med Virol. 1995;47:58-64. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 66] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Cribier B, Schmitt C, Bingen A, Kirn A, Keller F. In vitro infection of peripheral blood mononuclear cells by hepatitis C virus. J Gen Virol. 1995;76:2485-2491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 84] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Chang TT, Young KC, Yang YJ, Lei HY, Wu HL. Hepatitis C virus RNA in peripheral blood mononuclear cells: comparing acute and chronic hepatitis C virus infection. Hepatology. 1996;23:977-981. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 49] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Müller HM, Pfaff E, Goeser T, Kallinowski B, Solbach C, Theilmann L. Peripheral blood leukocytes serve as a possible extrahepatic site for hepatitis C virus replication. J Gen Virol. 1993;74:669-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 167] [Cited by in F6Publishing: 170] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 18. | Mihm S, Hartmann H, Ramadori G. A reevaluation of the association of hepatitis C virus replicative intermediates with peripheral blood cells including granulocytes by a tagged reverse transcription/polymerase chain reaction technique. J Hepatol. 1996;24:491-497. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 19. | Lerat H, Berby F, Trabaud MA, Vidalin O, Major M, Trépo C, Inchauspé G. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845-851. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 205] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 20. | Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766-12771. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 702] [Cited by in F6Publishing: 688] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 21. | Meier V, Mihm S, Braun Wietzke P, Ramadori G. HCV-RNA positivity in peripheral blood mononuclear cells of patients with chronic HCV infection: does it really mean viral replication. World J Gastroenterol. 2001;7:228-234. [PubMed] [Cited in This Article: ] |
| 22. | Sansonno D, Iacobelli AR, Cornacchiulo V, Iodice G, Dammacco F. Detection of hepatitis C virus (HCV) proteins by immunofluorescence and HCV RNA genomic sequences by non-isotopic in situ hybridization in bone marrow and peripheral blood mononuclear cells of chronically HCV-infected patients. Clin Exp Immunol. 1996;103:414-421. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 80] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 23. | da Silva LC, Bassit L, Ono-Nita SK, Pinho JR, Nishiya A, Madruga CL, Carrilho FJ. High rate of sustained response to consensus interferon plus ribavirin in chronic hepatitis C patients resistant to alpha-interferon and ribavirin: A pilot study. J Gastroenterol. 2002;37:732-736. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 24. | Omata M, Yokosuka O, Takano S, Kato N, Hosoda K, Imazeki F, Tada M, Ito Y, Ohto M. Resolution of acute hepatitis C after therapy with natural beta interferon. Lancet. 1991;338:914-915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 144] [Cited by in F6Publishing: 135] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 25. | Löhr HF, Goergen B, Meyer zum Büschenfelde KH, Gerken G. HCV replication in mononuclear cells stimulates anti-HCV-secreting B cells and reflects nonresponsiveness to interferon-alpha. J Med Virol. 1995;46:314-320. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 41] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 26. | Saleh MG, Tibbs CJ, Koskinas J, Pereira LM, Bomford AB, Portmann BC, McFarlane IG, Williams R. Hepatic and extrahepatic hepatitis C virus replication in relation to response to interferon therapy. Hepatology. 1994;20:1399-1404. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 63] [Cited by in F6Publishing: 66] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | Manesis EK, Papaioannou C, Gioustozi A, Kafiri G, Koskinas J, Hadziyannis SJ. Biochemical and virological outcome of patients with chronic hepatitis C treated with interferon alfa-2b for 6 or 12 mo: A 4-year follow-up of 211 patients. Hepatology. 1997;26:734-739. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 27] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Torres B, Martín JL, Caballero A, Villalobos M, Olea N. HCV in serum, peripheral blood mononuclear cells and lymphocyte subpopulations in C-hepatitis patients. Hepatol Res. 2000;18:141-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Knolle PA, Kremp S, Höhler T, Krummenauer F, Schirmacher P, Gerken G. Viral and host factors in the prediction of response to interferon-alpha therapy in chronic hepatitis C after long-term follow-up. J Viral Hepat. 1998;5:399-406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 24] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 30. | Hino K, Yamaguchi Y, Fujiwara D, Katoh Y, Korenaga M, Okazaki M, Okuda M, Okita K. Hepatitis C virus quasispecies and response to interferon therapy in patients with chronic hepatitis C: A prospective study. J Viral Hepat. 2000;7:36-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 31. | Antonelli G, Simeoni E, Turriziani O, Tesoro R, Redaelli A, Roffi L, Antonelli L, Pistello M, Dianzani F. Correlation of interferon-induced expression of MxA mRNA in peripheral blood mononuclear cells with the response of patients with chronic active hepatitis C to IFN-alpha therapy. J Interferon Cytokine Res. 1999;19:243-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 56] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Schiappa DA, Mittal C, Brown JA, Mika BP. Relationship of hepatitis C genotype 1 NS5A sequence mutations to early phase viral kinetics and interferon effectiveness. J Infect Dis. 2002;185:868-877. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 33. | Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Natl Acad Sci USA. 2002;99:3081-3086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 169] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 34. | Jiang J, Zhang L, Gigou M. [Compartmental distribution of hepatitis C virus quasispecies in mononuclear cells and liver]. Zhonghua Yixue Zazhi. 1998;78:265-268. [PubMed] [Cited in This Article: ] |
| 35. | Maggi F, Fornai C, Morrica A, Vatteroni ML, Giorgi M, Marchi S, Ciccorossi P, Bendinelli M, Pistello M. Divergent evolution of hepatitis C virus in liver and peripheral blood mononuclear cells of infected patients. J Med Virol. 1999;57:57-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 36. | Kessler HH, Pierer K, Santner BI, Vellimedu SK, Stelzl E, Marth E, Fickert P, Stauber RE. Evaluation of molecular parameters for routine assessment of viremia in patients with chronic hepatitis C who are undergoing antiviral therapy. J Hum Virol. 1998;1:314-319. [PubMed] [Cited in This Article: ] |