Published online Aug 15, 1999. doi: 10.3748/wjg.v5.i4.289
Revised: July 3, 1999
Accepted: July 19, 1999
Published online: August 15, 1999
AIM: To investigate the characteristics of newly established f our hepatocellular carcinoma cell lines (SNU-739, SNU-761, SNU-878 and SNU-886) from Korean hepatocellular cancer patients.
METHODS: Morphologic and genetic studies were done.
RESULTS: All four lines grew as a monolayer with an adherent pat tern, and their doubling times ranged from 20 to 29 h. The viability rate was relatively high (88%-94%). Neither mycoplasmal nor bacterial contamination was present. The lines showed different patterns in fingerprinting analysis. The hepatitis B virus (HBV) DNA was integrated in the genomes of all four lines, and in all of them HBx, HBc and HBs transcripts were detected by reverse transcriptase-PCR methods. Among the three cell lines used as control (Hep 3B, SK Hep1 and Hep G2), only Hep 3B showed HBx expression, and this line was used as a HBV integrated control. The RNA of albumin was detected in three lines (SNU-761, SNU- 878 and SNU-886), that of transferrin in two lines (SNU-878, SNU-886), and that of IGF- II was detected in none of the cell lines.
CONCLUSION: These well characterized cell lines may be very useful for studying the biology of hepatocellular carcinoma in association with the hepatitis B virus.
- Citation: Lee JH, Ku JL, Park YJ, Lee KU, Kim WH, Park JG. Establishment and characterization of four human hepatocellular carcinoma cell lines containing hepatitis B virus DNA. World J Gastroenterol 1999; 5(4): 289-295
- URL: https://www.wjgnet.com/1007-9327/full/v5/i4/289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v5.i4.289
Hepatocellular carcinoma (HCC) is one of the most prevalent malignant diseases encountered in the world, killing up to 1 million people annually. Geographically, its prevalence varies greatly, with a very high incidence in sub Saharan Africa and south and northeast Asia, including Korea[1-3]. Although many risk factors for HCC such as aflatoxin, persistent hepatitis C viral infection and alcoholic cirrhosis have been reported, hepatitis B viral infection has been known to be the most important etiologic factor. Epidemiological and laboratory studies have confirmed a strong association between the hepatitis B virus (HBV) and HCC. Prospective studies have shown that in those infected with HBV, the risk o f HCC was several hundred times higher than in uninfected individuals[4,5].
Despite this well-known close epidemiological relationship between HBV and HCC, the direct evidence of a causal relationship is inconclusive; HBV does not contain real oncogenes, and the mechanism by which HBV induces hepatocellular carcinoma has not been clearly demonstrated. It has been noticed that HBx, a protein encoded by HBV, is a molecule critically involved in HBV-induced hepatocellular carcinogenesis, and the HBx protein has been shown to transactivate various viral and cellular genes, including growth related genes such as c-fos[6], c-myc[7] and IGF[8]. Thus, the HBx protein has been suspected to be a critical molecule in the HBV induced carcinogenesis of HCC, which was supported further by report that the HBV x gene product induces HCC in transgenic mice[9]. HBV, however, has a very narrow host range and restricted tissue tropism to hepatocytes, and it is therefore difficult to study HBV-related HCC. The role of HBV in hepatic carcinogenesis has mainly been studied either by transfecting cloned viral DNA into permanent cell lines or by using HBV transgenic mice[10]. Another useful approach may be to use established HCC cell lines that are integrated with HBV DNA. Although HBV integrated HCC cell line s are very useful in hepatocellular carcinogenesis research, such cell lines are not yet sufficient. Furthermore, the establishment of HCC cell lines has been more difficult since the usage of transcatheter embolization with lipiodol and chemotherapeutic agents in the treatment of HCC. The preoperative application of t his therapeutic modality induces extensive necrosis of tumor tissue and leaves only a small amount of viable tumor cells available for culture at the time of operation[11].
After reporting eight HCC cell lines[12], we established four more lines from Korean HCC patients infected with HBV (SNU-739, SNU-761, SNU-878, SNU-886). We analyzed phenotypes such as in vivo and in vitro (cell line) cell morphology and in vitro growth pattern, DNA finger printing, expression o f HBV viral transcripts, albumin transferrin and insulin like growth factor II. The relationship between HBx and p53 gene mutation in eight previously reported cell lines and these four newly reporting cell lines was already described[13].
Cell lines were established from pathologically proven hepatocellular carcinomas. Solid tumors were finely minced and disassociated into small aggregates by pipetting. Appropriate amounts of fine neoplastic tissue fragments were seeded into 25 cm2 flasks. Initial culture was performed in ACL-4 medium supplemented wit h 5% heat inactivated fetal bovine serum (AR5 medium). ACL-4 is a fully defined medium formulated for the selective growth of the human lung adenocarcinoma[14] and has proved to be useful in the establishment of colorectal cancer c ell lines and HCC cell lines[12,15]. The composition of ACL-4 includes RPMI 1640 as the basal medium, insulin (20 mg/L), transferrin (10 mg/L), sodium selenite (25 nM), hydrocortisone (50 nM), epidermal growth factor (1 μg/L), ethanolamine (10 μM), phosphoryle-thanolamine (10 μM), triiodothyronine (100 pM), bovine serum albumin (2 g/L), HEPES buffer (10 mM), glutamine (2 mM) and sodium pyruvate (0.5 mM). RPMI 1640, glutamine and sodium pyruvate were obtained from GIBCO/BRL (Grand Island, NY); epidermal growth factor was obtained from Collaborative Research (Waltham, MA); all other reagents were obtained from Sigma (St. Louis, MO). For the maintenance of the cultures, the medium was replaced with RPMI 1640 supplemented with 10% heat inactivated fetal bovine serum (R10 medium). Initial cell passages were performed when heavy tumor cell growth was observed, and sub sequent passages were performed every 1 or 2 weeks. Adherent cultures were passaged at sub-confluence after trypsinization. If stromal cell growth was noted during initial growth, differential trypsinization was used to obtain a pure tumor cell population, as previously described by Park et al[15]. Culture s were maintained in humidified incubators at 37 °C in an atmosphere of 5% CO2 and 95% air. Well characterized and widely used HCC cell lines (Hep 3B and SK-He p1) and one hepatoblastoma cell line (Hep G2) were obtained from American Type Culture Collection (ATCC, Rockville, MD) and used as controls in DNA profile and transcript analysis.
Surgically removed tissue samples were fixed in 10% neutral formalin for 24 h. After routine processing, they were embedded in paraffin for light microscopic examination. Four mm thick sections were stained with hematoxylin and eosin (HE), and tumor cells were classified using the WHO classifications[16]. For the morphological studies of the cell lines, cells grown in a 75 cm2 culture flask were observed daily under a phase-contrast microscope (Olympus, CK2). For light microscopic examination, cells grown on Flaskette Chamber Slides (Nunc Inc., Naperville, IL) were washed with phosphate buffered saline (PBS), fixed in 4% paraformaldehyde in 0.1 M sodium cacodylate for 30 min, and then stained with HE.
To determine the population doubling time, about 3 × 105 viable cells were seed ed into 15-20 identical 25 cm2 flasks. The number of cells was counted daily f or at least 14 d. Cultures were fed every 3 or 4 d and 24 h prior to counting. To determine cell viability, a dye-exclusion procedure using 0.4% trypan blue staining methods performed and the number of viable cells was counted under a microscope using a hemocytometer.
Mycoplasma contamination was tested by direct agar isolation and the Hoechst 33342 stain method (Microbiological Associates, Bethesda, MD) and rRNA based PCR method[17]. All lines were tested for bacterial contamination.
Total cellular RNA and DNA were obtained from washed cell pellets by homogenization in guanidine thiocyanate followed by centrifugation over a cesium chloride cushion. Subsequently, total genomic DNA was prepared by the proteinase K digestion and phenol chloroform extraction method[15].
For DNA profile analysis, three polymorphic DNA probes, pYNH24 on chromosome No. 2 (ATCC, Rockville, MD), ChdTC 15 on chromosome No. 12 and ChdTC 114 on chromosome No. 20 (from Dr. H. Mizusawa, JCRB, Tokyo, Japan) were used to detect variable number of tandem repeats (VNTRs). The probes were labeled with [α-32P]dCTP (3000 Ci/mmol) using a random primed DNA labeling kit (Boeh ringer Mannheim, Indianapolis, IN). Prior to separation by 0.7% agarose gel electrophoresis, 10 μg of each DNA sample was digested with 70 units of Hinf I restriction endonuclease, and Southern blots were then per formed[18].
For the identification of HBV DNA integration, 10 μg of each D NA sample was digested with 10 units of Hind III, separated by electrophoresis, and transferred to a nylon membrane as described above. A-32P labeled 3.2 kb-full length HBV DNA clone (pAM6) was used as a probe. Hybridization and autoradiography were done by the same methods described in Southern blot.
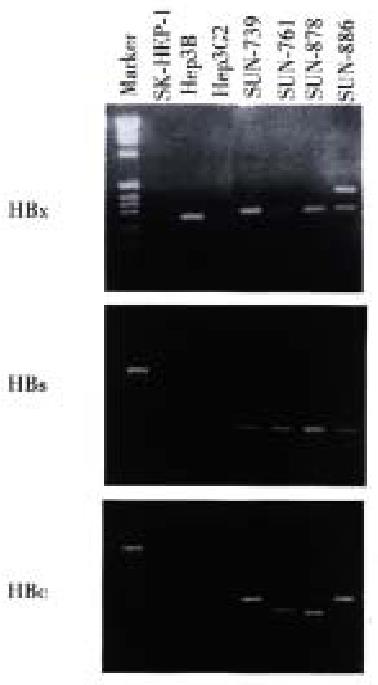
Total RNA was prepared from the cell lines and treated with DNase to exclude cellular DNA contamination. cDNA was made using MMLV reverse transcriptase (Boehringer Mannheim, Indianapolis, IN) and random hexamer. A reverse transcriptase reaction was performed at 42 °C for 1 h with 2 μg of template RNA. The PCR primer sequences were as follows: x gene (forward 5'-ACGGGGCGCACCTCTCTTTA-3' reverse 5'-TGCCTACAGCCTCCTAGTAC-3'), c gene (forward 5'-GCTTCTGTGGAGTTACTCTC-3', reverse 5'-GTGCGAATCCACACTCCAAA-3'), and s gene (forward 5'-TCTCAATTTTCTAGGGGGAG-3', reverse 5'-GCACTAGTAAACTGAGCCAG-3'). PCR reaction was performed with annealing temperature of 55 °C for 30 s. PCR product was resolved in 1% agarose gel and detected by EtBr staining. To confirm this result, we also performed Southern blot with HBx, HBs, and HBc probes labeled with [α-32P]dCTP (3000 Ci/mmol).
The four HCC cell lines designated SNU-739, SNU-761, SNU-878 and SNU-886 were established from Korean HCC patients, all of whom had previously been treated by transcatheter arterial embolization. Preoperative serologic testing had revealed the presence of hepatitis surface antigen in all patients from whom the specimens were obtained (Table 1).
All cell lines showed the monolayer growth pattern, and population doubling times ranged from 20 to 29 h. All lines were free from contamination with bacteria and mycoplasma.
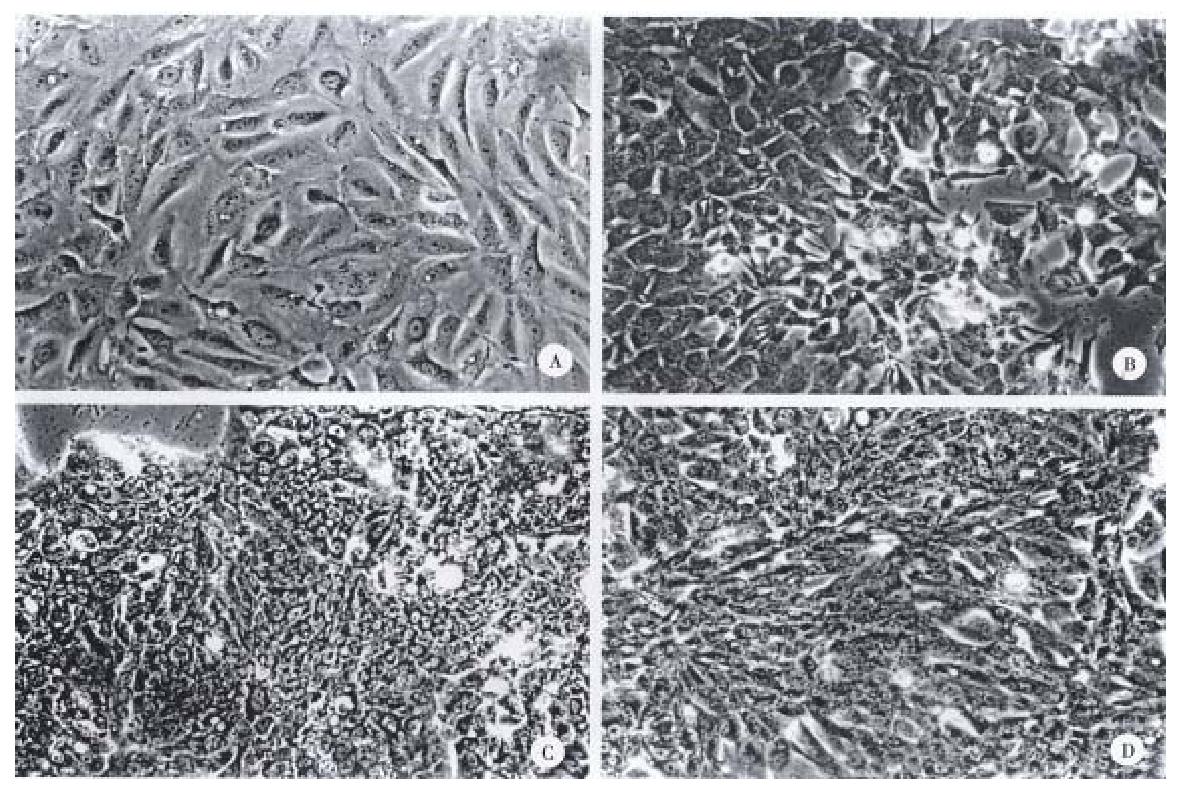
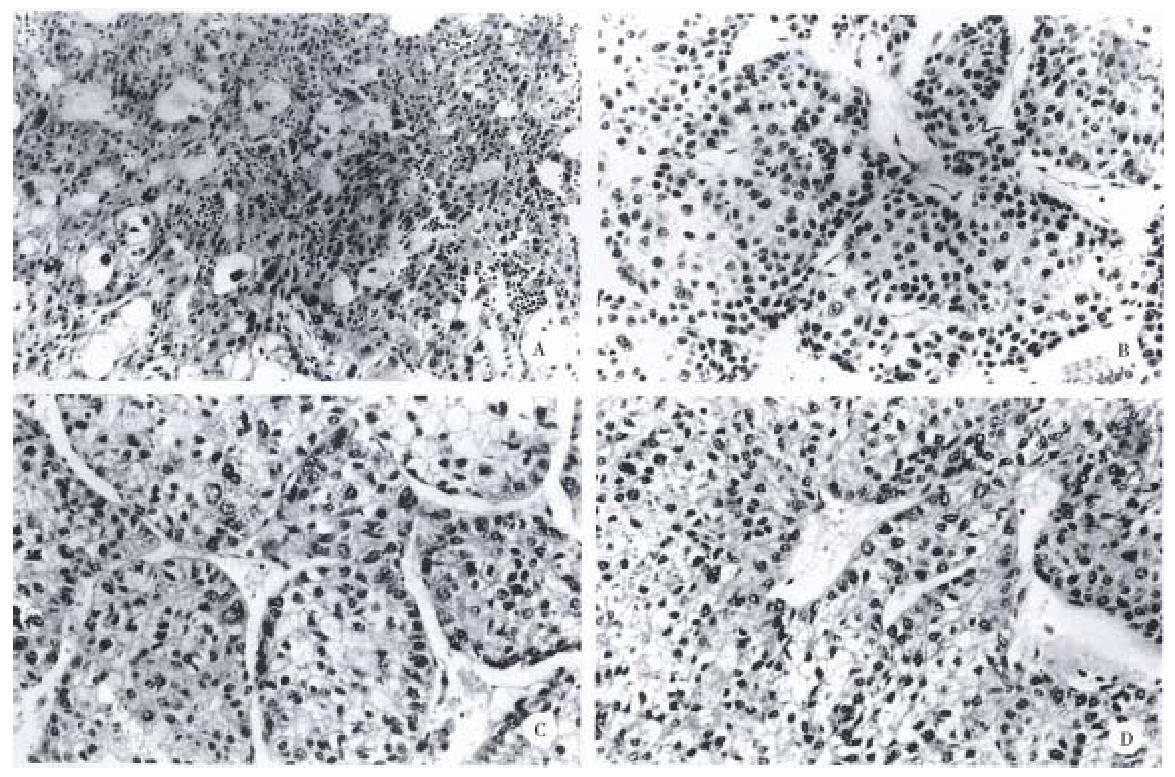
The gross and histologic features of the original tumors were categorized using the WHO classification. Grossly, the original tumors from which the cell lines were derived were either single nodular type or the single nodular with perinodal extension type (Table 2). Histologically, the original tumor of the SNU-739 cell line was the mixed microtrabecular and compact type; individual tumor cells were similar to normal hepatocytes (cirrhotomimetic type), but scattered multinuclear syncytial cells with clear cytoplasm were seen within the area of compact histology. Eosinophilic hyaline globules were occasionally seen, and scattered or aggregated neutrophils were encountered throughout the tumor (Figure 1A). Cultured cells grew as an adherent pattern; they were monotonous spindle shaped cells, containing single round nuclei and flattened cytoplasm. The nucleoli were inconspicuous (Figure 2A).
| Cell line | In vitro | In vivo | |||||
| Growth character | Cell morphology | Nuclear morphology | Histology | Cell type | Gross type | Grade | |
| SNU739 | Ada | Spindle | Single | Micro-trabecular compact | Cb/clean | SNc | II- IV |
| SNU761 | Ad | Polygonal | Single | Macro-trabecular | C | SN | III |
| SNU878 | Ad | Polygonal | Single multinuclear | Macro-trabecular | C | SEd | II- III |
| SNU886 | Ad | Polygonal-spindle | Single-double | Macro-trabecular | C | SN | II- IV |
The original tumor of cell line SNU-761 was the homogenous macrotrabecular type. The tumor cells were uniform cirrhotomimetic cells of grade III differentiation. Sinusoids were well developed and multinuclear giant cells were occasionally encountered (Figure 1B). The individual cultured cell was polygonal, with a single ovoid nucleus and abundant cytoplasm. The cells occasionally produced short cytoplasmic processes at the free lateral border. In large part, they formed an adherent monolayer, and tended to overlap (Figure 2B).
The original tumor of cell line SNU-878 was the macrotrabecular type with a minor area of compact histology. Its cells belong to the cirrhotomimetic group, and most were moderately differentiated (grade II or III). Clusters of cells containing clear cytoplasm were seen in the central part of tumor cell nests. The tumor cells occasionally contained Mallory body-like amorphous materials in their cytoplasm (Figure 1C). The cultured cells of SNU-878 grew as a monolayer of tightly packed colonies, consisting of polygonal cells of variable shapes in a mosaic pattern. Most tumor cells contained multiple nuclei, simulating syncytium, while a few were detached from the compact colony. Single or double conspicuous nucleoli were noted (Figure 2C).
The original tumor of cell line SNU-886 consisted of cirrhotomimetic cells in a macrotrabecular pattern. The nuclear shape of individual cells varied and large anaplastic cells were scattered throughout the tumors; the degree of cellular differentiation was categorized as grade II- IV (Figure 1D). The growing cells of cell line SNU-886 formed an adherent monolayer of mixed spindle shaped and polygonal cells. Most of the tumor cells contained one or two nuclei, with small multiple nucleoli (Figure 2D).
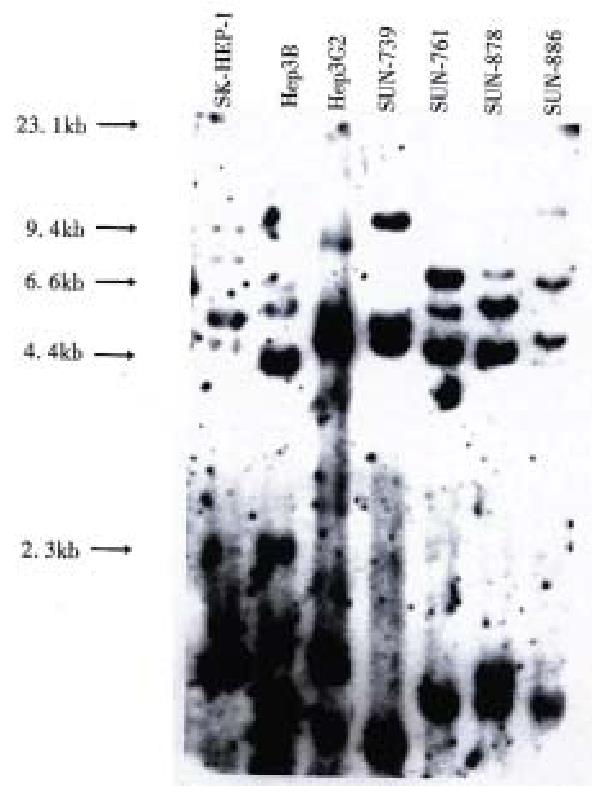
DNA profiles using restriction endonuclease Hinf I and polymorphic DNA probes pYNH24, ChdTC 15, and ChdTC 114 showed more than 12 allelic bands with the size range of 4.4 to 10 kb (Figure 3).
All four cell lines showed different band pattern, and this indicates that these cell lines were unique and unrelated. In addition, their DNA profiles were different from those of three previously reported ATCC cell lines, SK-Hep1, Hep3B, and Hep G2. These results exclude possible cross contamination between the cell lines.
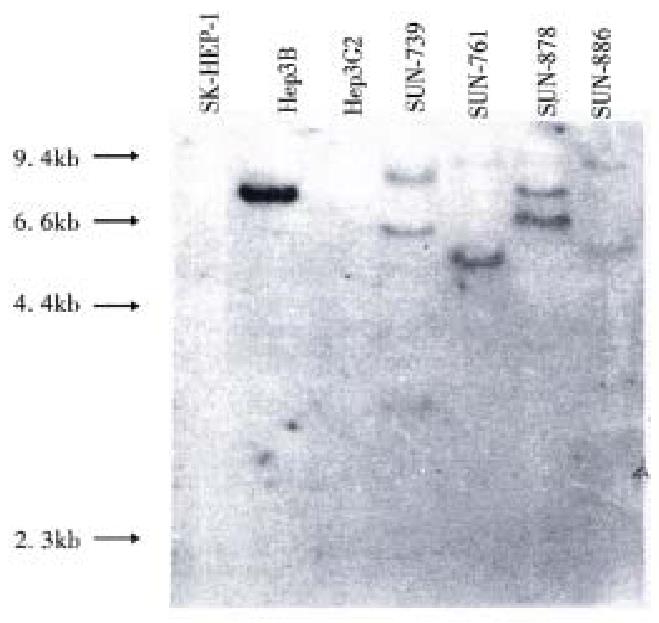
Hepatitis B virus DNA was detected in all cell lines by Southern blot hybridization with cellular DNA after digestion using Hind III. Relatively high molecular weight bands with enzyme digestion suggest integrated form of HBV DNA. Each cell line showed a different pattern of HBV integration (Figure 4).
Using the RT-PCR method, HBx RNA was detected in all four cell lines and the He p3B line. HBs and HBc RNA were detected in all four newly established cell lines (Figure 5). Among the three control cell lines, only Hep3B showed HBx RNA expression, and this line was known to be integrated with HBV DNA and express HBx RNA. SK-Hep1 and HepG2, reported to have no association with HBV, and did not show HBx RNA expression.
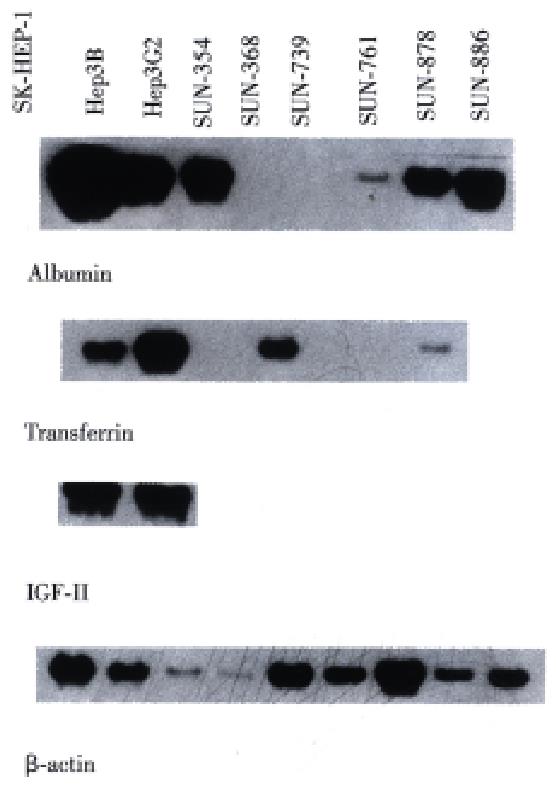
In Northern blot, the expression of RNA coding for albumin was detected in the SNU-761, SNU-878, and SNU-886 cell lines (Figure 6). RNA for transferrin was expressed in the SNU-878 and SNU-886 cell lines. RNA for IGF- II was expressed in none of these cell lines (Table 3).
| Cell lines | Viability (%) | Doubling time (hr) | HBV DNA integration | HBV RNA expression | Albumin | Transferrin | IGF- II | ||
| X | preS/S | C | |||||||
| SNU 739 | 94 | 20 | + | + | + | + | - | - | - |
| SNU 761 | 91 | 24 | + | + | + | + | + | - | - |
| SNU 878 | 88 | 25 | + | + | + | + | + | + | - |
| SNU 886 | 90 | 29 | + | + | + | + | + | + | - |
Cell lines established from human cancers provide very useful tools for studying the biology of cancer and the development and testing of new therapeutic approaches. In HCC, well-characterized HCC cell lines make possible various studies t hat cannot be performed with biopsy or postmortem tissues. The frequent integration of the HBV gene in hepatocellular carcinoma suggests that it play an essential role in carcinogenesis of hepatocellular carcinoma. Permanent cell lines derived from HCCs with the HBV gene integration offer good models not only for the investigation of HCC carcinogenesis but also for the elucidation of the regulatory mechanisms of HCC cell specific gene expression. In spite of their definite value as a resource for research in HCC, reports of established HCC cell lines have been decreasing since 1985, and only a few cases have been reported[11]. Furthermore, permanent HCC cell lines integrated with the HBV gene are rarer, and other than our previous report of eight lines, only a few cases have been re ported in the literature. Recent attempts to culture HCC cells as permanent line s have been rather disappointing. The use of transcatheter arterial embolization (TAE) with lipiodol or chemotherapeutic drugs before surgical resection is now common, and it is thus difficult to obtain HCC tissues suitable for isolation an d cultivation of intact viable cells. In our establishment of these herein reported cell lines, the success rate was only 9% (4 lines out of 44 trials), which is much lower than our previously reported 8 cell lines (success rate 29%, 8 lines out of 28 trials) due to extensive necrosis caused by the preoperative TAE. All four of these cell lines were in fact derived from HCC tissues of patients who underwent preoperative TAE. In these cases that the cell lines were successfully established, the necrotic portion was diverse, ranging from 30% to 70%. However, in cases that establishment of cell lines were failed, the necrotic portion was usually more extensive.
HBV is a 3.2 kb sized DNA virus with only four genes known to encode for the inner "c" and "e" antigen, "s" (surface) proteins, a DNA polymerase and the "x" protein. According to recent reports on the carcinogenetic mechanism of the HBS virus, the x gene of HBV (HBx), capable of gene transactivation, is considered to play an important role[19,20]. In order to investigate the transcriptional activity of HBV genes in our cell lines, HBx, preS/S and HBc RNA expression was examined using the RT-PCR methods. The HBV gene RNA transcript which encodes the x, preS/S and c was detected in these four lines and in Hep 3B; this is known to have one or two copies of HBV DNA integrated into the host genome and to express HBV viral transcripts of various size[21]. This RNA expression indicates that HBV genes integrated into the cell lines are transcriptionally active, though an exact quantitative assay was not possible.
Among our previously reported eight cell lines, x gene RNA was detected in only two cell lines (SNU-354, SNU-368) by Northern blot analysis[12]. W e re-examined the RNA transcript of x gene in those eight cell lines using the RT-PCR methods, and detected RNA expression in six of them (SNU-182, SNU- 354, SNU-368, SNU-387, SNU-449, SNU-475). This may indicate that for detecting a very tiny amount of viral transcript, RT-PCR is more sensitive than Northern blot; thus, the viral DNA actually transcribed should be determined by RT-PCR analysis.
These high expression rates of x gene transcription (10 of 12 lines) in HCC cell lines established from Korean HCC patients who were serologically proved t o be HBS antigen positive suggest that, in Korea, HBV plays an important role in the etiology of HCC. This is further supported by the prevalence of HBV infection in Korea, which affects as much as 8% of general population[22].
In this report, we have described the characteristics of four newly established HCC cell lines in which HBV DNA was integrated into the genomes. Considering that fresh specimens for the establishment of new cell lines are difficult to obtain, these four cell lines would be invaluable future resources.
This work was supported in part by a grant from the Korea Science and Engineering Foundation (KOSEF) through the Korean Cell-Line Bank and Cancer Research Center at Seoul National University, KOSEF-CRC-97-8
Edited by MA Jing-Yun
| 1. | World Health Organization. Causes of deaths by sex and age. In: World Health Statistics Annula. WHO, Geneva. : WHO 1993; . [Cited in This Article: ] |
| 2. | Omata M. Current perspectives on hepatocellular carcinoma in Oriental and African countries compared to developed western countries. Dig Dis. 1987;5:97-115. [PubMed] [Cited in This Article: ] |
| 3. | Simonetti RG, Cammà C, Fiorello F, Politi F, D'Amico G, Pagliaro L. Hepatocellular carcinoma. A worldwide problem and the major risk factors. Dig Dis Sci. 1991;36:962-972. [PubMed] [Cited in This Article: ] |
| 4. | Szmuness W. Hepatocellular carcinoma and the hepatitis B virus: evidence for a causal association. Prog Med Virol. 1978;24:40-69. [PubMed] [Cited in This Article: ] |
| 5. | Beasley RP. Hepatitis B virus as the etiologic agent in hepatocellular carcinoma epidemiologic considerations. Hepatology. 1982;2:21S-27S. [Cited in This Article: ] |
| 6. | Twu JS, Lai MY, Chen DS, Robinson WS. Activation of protooncogene c-jun by the X protein of hepatitis B virus. Virology. 1993;192:346-350. [PubMed] [Cited in This Article: ] |
| 7. | Balsano C, Avantaggiati ML, Natoli G. Transactivation of c-fos and c-myc protooncogenes by both full length and truncated versions of the HBV-X protein. Viral hepatitis and liver disease. Baltimore: Williams & Wilkins Co 1991; 572-578. [Cited in This Article: ] |
| 8. | Menzo S, Clementi M, Alfani E, Bagnarelli P, Iacovacci S, Manzin A, Dandri M, Natoli G, Levrero M, Carloni G. Trans-activation of epidermal growth factor receptor gene by the hepatitis B virus X-gene product. Virology. 1993;196:878-882. [PubMed] [Cited in This Article: ] |
| 9. | Kim CM, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317-320. [PubMed] [Cited in This Article: ] |
| 10. | Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145-1156. [PubMed] [Cited in This Article: ] |
| 11. | Miyazaki M, Namba M. Hepatocellular carcinomas. Atlas of human tumor cell lines. San Diego: Academic Press 1994; 185-212. [Cited in This Article: ] |
| 12. | Park JG, Lee JH, Kang MS, Park KJ, Jeon YM, Lee HJ, Kwon HS, Park HS, Yeo KS, Lee KU. Characterization of cell lines established from human hepatocellular carcinoma. Int J Cancer. 1995;62:276-282. [PubMed] [Cited in This Article: ] |
| 13. | Kang MS, Lee HJ, Lee JH, Ku JL, Lee KP, Kelley MJ, Won YJ, Kim ST, Park JG. Mutation of p53 gene in hepatocellular carcinoma cell lines with HBX DNA. Int J Cancer. 1996;67:898-902. [PubMed] [Cited in This Article: ] |
| 14. | Brower M, Carney DN, Oie HK, Gazdar AF, Minna JD. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 1986;46:798-806. [PubMed] [Cited in This Article: ] |
| 15. | Park JG, Oie HK, Sugarbaker PH, Henslee JG, Chen TR, Johnson BE, Gazdar A. Characteristics of cell lines established from human colorectal carcinoma. Cancer Res. 1987;47:6710-6718. [PubMed] [Cited in This Article: ] |
| 16. | Gibson JB, Sobin LH. Histologic typing of tumors of the liver, biliary tract and pancreas. Geneva: WHO 1978; 20. [Cited in This Article: ] |
| 17. | Deng S, Hiruki C, Robertson JA, Stemke GW. Detection by PCR and differentiation by restriction fragment length polymorphism of Acholeplasma, Spiroplasma, Mycoplasma, and Ureaplasma, based upon 16S rRNA genes. PCR Methods Appl. 1992;1:202-204. [PubMed] [Cited in This Article: ] |
| 18. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd Ed. Cold Spring Harbor: Cold Spring Harbor Laboratory Press 1989; . [Cited in This Article: ] |
| 19. | Shirakata Y, Kawada M, Fujiki Y, Sano H, Oda M, Yaginuma K, Kobayashi M, Koike K. The X gene of hepatitis B virus induced growth stimulation and tumorigenic transformation of mouse NIH3T3 cells. Jpn J Cancer Res. 1989;80:617-621. [PubMed] [Cited in This Article: ] |
| 20. | Balsano C, Billet O, Bennoun M, Cavard C, Zider A, Grimber G, Natoli G, Briand P, Levrero M. Hepatitis B virus X gene product acts as a transactivator in vivo. J Hepatol. 1994;21:103-109. [PubMed] [Cited in This Article: ] |
| 21. | Knowles BB, Howe CC, Aden DP. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980;209:497-499. [PubMed] [Cited in This Article: ] |
| 22. | Shin HR. HBV or HCV infection and risk of HCC in Korea. Proceedings of the Federation Meeting of Korean Basic Medical Scientist. Cold Spring Harbor: Cold Spring Harbor Laboratory Press 1993; 86. [Cited in This Article: ] |