Published online May 28, 2017. doi: 10.3748/wjg.v23.i20.3643
Peer-review started: February 10, 2017
First decision: March 3, 2017
Revised: March 29, 2017
Accepted: May 4, 2017
Article in press: May 4, 2017
Published online: May 28, 2017
To identify metabolic signatures in urine samples from healthy and inflammatory bowel disease (IBD) children.
We applied liquid chromatography and gas chromatography coupled to targeted mass spectrometry (MS)-based metabolite profiling to identify and quantify bile acids and host-gut microbial metabolites in urine samples collected from 21 pediatric IBD patients monitored three times over one year (baseline, 6 and 12 mo), and 27 age- and gender-matched healthy children.
urinary metabolic profiles of IBD children differ significantly from healthy controls. Such metabolic differences encompass central energy metabolism, amino acids, bile acids and gut microbial metabolites. In particular, levels of pyroglutamic acid, glutamic acid, glycine and cysteine, were significantly higher in IBD children in the course of the study. This suggests that glutathione cannot be optimally synthesized and replenished. Whilst alterations of the enterohepatic circulation of bile acids in pediatric IBD patients is known, we show here that non-invasive urinary bile acid profiling can assess those altered hepatic and intestinal barrier dysfunctions.
The present study shows how non-invasive sampling of urine followed by targeted MS-based metabonomic analysis can elucidate and monitor the metabolic status of children with different GI health/disease status.
Core tip: Despite the limited number of subjects, the longitudinal experimental design with a healthy reference group reveals insights into childhood metabolic status in relation to growth and disease. Metabolite profiling of urine samples collected non-invasively enables identification and monitoring of biochemical signatures linked to metabolic and nutritional requirements in pediatric populations with inflammatory bowel disease (IBD). In the present study, a distinct biochemical process related to glutathione, glycine and bile acid metabolism distinguished children with IBD from healthy matched controls.
- Citation: Martin FP, Su MM, Xie GX, Guiraud SP, Kussmann M, Godin JP, Jia W, Nydegger A. Urinary metabolic insights into host-gut microbial interactions in healthy and IBD children. World J Gastroenterol 2017; 23(20): 3643-3654
- URL: https://www.wjgnet.com/1007-9327/full/v23/i20/3643.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i20.3643
The use of systems biology-oriented technologies (e.g., metabonomics, proteomics, genomics and microbiomics) redefines disease understanding and phenotyping of clinical characteristics in medical disorders such as in gastrointestinal deregulations[1,2]. This is particularly relevant for inflammatory bowel disease (IBD), where a systems-level approach of the pathogenesis can inform novel, integrated multiple pathway- therapies and contribute to the development of personalized disease management[2].
The prevalence of IBD has markedly increased over the years, with 25% of the population developing the disease during childhood and adolescence[2,3]. In addition to the clinical and immunological features of the disease, pediatric patients suffer from additional growth failure and delayed puberty[3-5]. During this critical period, IBD children can suffer from malnutrition, which is related to reduced energy intake, malabsorption and thus loss of nutrients, and inflammation[3,6-9]. These factors mainly contribute to delayed growth and physiological maturation. Moreover, there is a great lack of knowledge on how to address metabolic and nutritional requirements in these children and adolescents[5]. The use of omics-technologies for holistic molecular phenotyping over time can generate the data for a systems-level view of metabolic processes associated with growth and development of children in relation to the changing physiology in infancy and childhood[5].
Amongst the omic technologies, metabonomics enables the study of physiological regulatory processes through the simultaneous analysis of a large range of biochemical species[2,10-13]. The technology is based on either targeted or untargeted profiling of small biomolecular compounds in tissues and biological fluids, such as blood, urine or saliva. Metabolites profiles can be generated through the use of proton nuclear magnetic resonance (1H NMR) spectroscopy or mass spectrometry (MS) combined eventually with chromatographic separation. Recently, we have applied 1H NMR based profiling to urine samples from IBD and healthy matched children and highlighted differences related to central energy metabolism, as well as amino acid and gut microbial metabolism in relation to growth and disease activity[14]. In particular, we showed how specific metabolite patterns in urine could help monitoring metabolic status in relation to disease state. In the present study, we continue our metabolic exploration by targeting host-gut microbial metabolites in urine samples from the same children, including the aromatic amino acid and bile acid co-metabolites - two well-established examples of transgenomic metabolic cross-talk. For this purpose, we applied liquid chromatography (LC) and gas chromatography (GC) coupled to MS-based metabolite profiling. We here report urinary metabolic phenotypes specific to IBD children compared to healthy matched controls. Advanced clinical and anthropometric phenotyping was also performed on these IBD children over the course of one year of standard-care disease management (0, 6 and 12 mo) to identify relationships between biochemical processes and anthropometric phenotypes.
This clinical study was approved by the Ethical Committee of the University of Lausanne, Switzerland (protocol 69/10), and conducted in the Pediatric Gastroenterology outpatient clinic of the University Hospital of Lausanne, Switzerland. Informed written consent was obtained from the patients and their parents.
Eligible patients were aged between 10 and 18 years old, with a diagnosis of Crohn’s disease (CD) or ulcerative colitis (UC), confirmed according to international criteria[15]. IBD subjects were assessed at baseline (T0), after 6 (T6) and 12 mo (T12), respectively. All patients were in remission and underwent therapeutical management of the disease according to recommended drugs. To be noted that none was treated with enteral nutrition and no endoscopy was performed to assess mucosal inflammation. Control healthy subjects were recruited among the general pediatric population. They were matched for age, pubertal stage and gender to the IBD subjects. They had neither chronic inflammatory disease nor family history of inflammatory bowel. Anthropometric and clinical data and urine samples for metabolic analyses were collected at each time point.
Body weight, height and BMI were measured under standard clinical practice, as previously reported[14]. Height velocity was calculated as the amount of growth in centimeters divided by the time interval between measurements in years. All values were expressed in z-scores[16,17]. Pubertal stage was assessed according to Tanner score[18].
Bioimpedance analysis (BIA) was performed using Body Impedance Analyser Akern (Florence, Italy), as previously reported[14]. Fat free mass in kg (FFM) was then calculated using the software BodyGram Pro® supplied by the manufacturer [which uses weight, age, and an impedance index (height2/resistance)][19,20]. Percentage of FFM (%FFM) was calculated by dividing FFM with the body weight of the subject expressed in kg.
Disease activity was scored using the Pediatric Crohn’s Disease Activity Index (PCDAI)[21] for CD, a 100 point scale where a score > 30 indicates severe disease, and the Pediatric Ulcerative Colitis Activity Index (PUCAI)[22] for UC, a 85 points scale where a score > 35 indicates severe disease. Remission was defined as PCDAI or PUCAI score lower than 10. No endoscopic control was performed since all patients were in remission.
Inflammatory markers [Erythrocyte Sedimentation Rate (ESR), C-Reactive Protein (CRP)], urea, and growth factors [Insulin-like Growth Factor 1 and Insulin-like Growth Factor-Binding Protein 3, expressed in z-scores] were obtained after a fasting period of at least 6 h. Fecal calprotectin was measured and a cut-off value of 275 μg/g was set to determine possible relapse of disease[23].
All subjects underwent a 24-h food recall with the help of a questionnaire showing pictures of different sizes of plates for the different foods with the same examiner (dietician TC). Qualitative and quantitative analyses were made using the software Prodi 5.8 Expert. Daily intake was expressed as kcal per day.
Resting energy expenditure (REE, kcal) was measured using Quark RMR (Cosmed, pulmonary function equipment, Delta Medical, Italy), as previously reported[14].
Morning spot urine samples were collected at baseline for all subjects, at the 6-mo and 12-mo visit for IBD patients only. Urine samples (1 mL) were collected by means of sterile plastic tubes, and stored at -80 °C, prior to analysis.
Chemicals: All of the 57 bile acid standards were obtained from Steraloids Inc. (Newport, RI) and TRC Chemicals (Toronto, ON, Canada), and 9 stable isotope- labeled standards were obtained from C/D/N Isotopes Inc. (Quebec, Canada) and Steraloids Inc. (Newport, RI).
Methanol (Optima LC-MS), acetonitrile (Optima LC-MS) and formic acid (Optima LC-MS) were purchased from Thermo-Fisher Scientific (FairLawn, NJ). Ultrapure water was produced by a Mill-Q Reference system equipped with a LC-MS Pak filter (Millipore, Billerica, MA).
Sample preparation: At the time of analysis, samples were thawed on ice-bath to diminish biochemical degradation. 100 μL of urine or standard solution of a bile acid-free matrix was directly lyophilized to dry powder using a freeze dryer. The residue was reconstituted in 50 μL of mobile phase B (acetonitrile/methanol = 95/5, v/v) and 25 μL of mobile phase A (water with formic acid, pH = 3.25), and centrifuged at 13500 g and 4 °C for 20 min. The supernatant was transferred to a 96-well plate for LC-MS analysis and the injection volume was 10 μL.
Data generation and analysis: An ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA) was used to quantitate bile acids in the human urine samples based on our previously published protocols[24,25]. The raw data was processed using the TargetLynx application manager (Waters Corp., Milford, MA) to obtain calibration equations and the measured concentration of each bile acid in the samples. The quality control samples were prepared with the addition of bile acid stock solutions to blank matrix at a final concentration of 10, 100, or 1000 nmol/L. The quality control samples (four QCs in a 96-well plate) were prepared along with the real samples, and injected at regular intervals to allow evaluating overall process variability and monitoring platform performance. Each kit plate is ideal for the analysis of 84 samples, 8 calibrators (CAL0 = blank matrix), and 4 QCs (QC1 = 10 nmol/L, QC2 and QC3 = 100 nmol/L, and QC4 = 1000 nmol/L). The noise baseline was established from reagent blank measurement and any metabolite with signal to noise ratio ≤ 3.0 is rejected from statistical analysis. The relative standard deviation (RSD) for the lower concentration metabolites in the reference standard mixture was less than 30% and the RSD for the higher concentration metabolites was better than 15% for each batch of sample analysis (100 injections for LC-MS analysis).
Chemicals: The derivatization reagents methyl and ethyl chloroformate, as well as HPLC grade solvents including methanol, ethanol, chloroform and pyridine were purchased from Sigma-Aldrich (St. Louis, MO, United States). Sodium hydroxide, sodium bicarbonate and anhydrous sodium sulfate were of analytical grade and obtained from JT Baker Co. (Phillipsburg, NJ). All standard compounds were commercially purchased from Sigma-Aldrich and Nu-Chek Prep (Elysian, MN, United States). Ultrapure water was produced by Milli-Q system (Millipore, Billerica, MA).
Extraction of metabolites from human urine: Urine samples were thawed on ice and prepared as follows: urine sample (100 μL) was added into a silanized glass vial and lyophilized directly under low temperature (-40 °C) by a Labconco freeze-dryer. The solids from urine samples after lyophilization process were sealed and stored at -80 °C for subsequent automated derivatization assay.
Automated chloroformate derivatization and GC/MS analysis: The sample derivatization protocols with MCF and ECF was based on the method described by Villas-Bôas et al[26] and our previously published procedures[27], with minor modifications. For routine large-scale sample analysis, sample derivatization and all liquid handling were performed by a commercially available robotic workstation (GERSTEL MPS Autosampler).
Gas chromatography/time-of-flight mass spectrometry analysis: Samples were randomly analyzed by GC/TOFMS (Agilent 6890N gas chromatography coupled with a LECO Pegasus HT time-of-flight mass spectrometer) using our optimized conditions in this study. One microliter of each derivatized sample was injected splitless into a DB-5 ms capillary column (30 m × 0.25 mm i.d., 0.25 μm film thickness; (5%-phenyl)-methylpolysiloxane bonded and cross-linked; Agilent JW Scientific, Folsom, CA) , with helium as the carrier gas at a constant flow rate of 1.0 mL/min. The solvent delay time was set to 2.5 min. The optimized temperature gradient was the following: 45 °C held for 1 min, then increased at a rate of 20 °C/min up to 260 °C and 40 °C/min to 320 °C/min, then held there for 2 min. The total time of analysis was 15.25 min. The temperature of injection, transfer interface and ion source were set to 270, 270 and 220 °C, respectively. Electron impact ionization (70 eV) at full scan mode (m/z 38-650) was used. The acquisition rate was 20 spectra/s.
Data processing: Non-processed MS files from GC/TOFMS analysis were exported in NetCDF format to ChromaTOF (v4.50, Leco Co., CA, United States) and subject to the following processing including baseline correction, smoothing, noise reduction, deconvolution, library searching, and area calculation. Compound identification was performed by comparing both MS and Kovats-RI with reference standards in author-constructed two alkyl chloroformates derivatives databases, with a similarity of more than 70%.
Chemometric analysis was performed on clinical and metabonomics data using the software package SIMCA-P+ (version 12.0, Umetrics AB, Umeå, Sweden). Principal component analysis (PCA) and a modification of Partial Least Squares Regression (PLSR) that removes all information orthogonal to the response variable during the fitting process were employed. This variant, Orthogonal Projection to Latent Structures (O-PLS)[28] provides sparser models (improving their interpretability) with the same degree of fit as PLSR. For group comparison, multivariate data analysis was performed using O-PLS-discriminant analysis (OPLS-DA). To highlight the weight of individual variables in the model, Variable Importance in the Projection (VIP) was used, with a value above 1 used as a threshold by convention. In addition, a Pearson correlation coefficient with a p-value significant at 95% confidence interval (e.g., with n = 46, r = 0.40). Metabolic pathway analysis was conducted by performing a metabolite set enrichment analysis, using the web-based MetaboAnalyst 3.0 tool[29], to the list of influential metabolites obtained through multivariate data analysis.
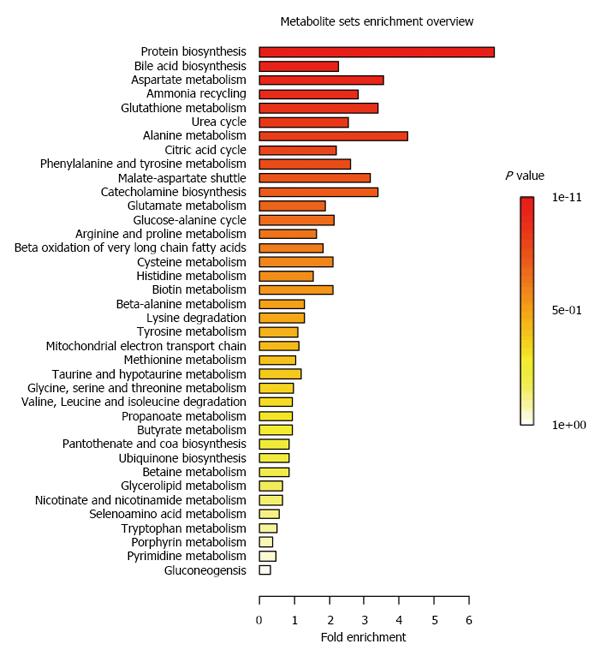
The patient information has been reported previously[14]. Briefly, the study population was composed of 21 pediatric IBD patients and 27 age and gender matched healthy children. IBD children were monitored over one year period, with a baseline visit, and follow-up visits after 6 and 12 mo. CD patients showed lower z-scores for body weight, height, BMI and resting energy expenditure at baseline and at follow-up visits[14]. Targeted MS metabolic profiling detected and quantified 69 metabolites by GC-MS and 30 bile acid species in urine. We report in Table 1 the urinary concentrations of all the metabolites at baseline for health and IBD children. Using this set of 99 metabolites, we have applied a Metabolite Set Enrichment Analysis[29] to summarize the main metabolic pathways captured by our systems approach. As reported in Figure 1, the analysis provides a comprehensive approach to generate insights in relation to protein biosynthesis, bile acid biosynthesis, aspartate, alanine, glutathione, urea, ammonia and citric acid (TCA) cycles.
| Metabolites | Healthy | IBD baseline | P value |
| 2-hydroxybutyric acid | 324.2 ± 228.5 | 401.9 ± 306.8 | 0.335 |
| 3-(3-hydroxyphenyl)-3-hydroxypropanoic acid | 85.1 ± 242.2 | 240.6 ± 488.6 | 0.168 |
| 3-aminoisobutanoic acid | 7278 ± 8267.9 | 12302.4 ± 15388.8 | 0.166 |
| 3-hydroxyisovaleric acid | 12675.2 ± 14366.2 | 13929.9 ± 6198.6 | 0.722 |
| 3-hydroxyphenylacetic acid | 6618.8 ± 7906.4 | 3579.7 ± 4269.6 | 0.134 |
| 3-methyl-2-oxovaleric acid | 12049.5 ± 5596.6 | 13764.2 ± 4303.2 | 0.269 |
| 4-hydroxybenzoic acid | 1659.4 ± 835.8 | 1660.6 ± 735.9 | 0.996 |
| 4-hydroxycinnamic acid | 1749 ± 2142.3 | 4655.3 ± 5127.8 | 0.013 |
| Adipic acid | 5757.5 ± 3488.3 | 6806.6 ± 6403.6 | 0.485 |
| Alpha-hydroxyisobutyric acid | 7514.4 ± 4562.7 | 10645.1 ± 5098.4 | 0.036 |
| Aminoadipic acid | 23462.2 ± 13757.6 | 24927 ± 13309.2 | 0.722 |
| Beta-alanine | 3554.8 ± 2690.1 | 3730.4 ± 2705.5 | 0.830 |
| Butyric acid | 594 ± 329.1 | 754.4 ± 551 | 0.230 |
| Caproic acid | 337.3 ± 393.0 | 423.4 ± 412.3 | 0.480 |
| Caprylic acid | 113.7 ± 233.6 | 114.2 ± 335.3 | 0.996 |
| Cis-aconitic acid | 126743 ± 94692.1 | 217610 ± 154442.2 | 0.019 |
| Citraconic acid | 1487.4 ± 719.3 | 1877.1 ± 529.1 | 0.051 |
| Citramalic acid | 13454 ± 11088.9 | 29200.5 ± 23835.6 | 0.005 |
| Citric acid | 1521627.7 ± 1117692.9 | 2010896.5 ± 1470323.3 | 0.211 |
| Dodecanoic acid | 76 ± 118.3 | 95.1 ± 230.1 | 0.718 |
| Dopamine | 25594.1 ± 15600.7 | 46132.8 ± 4070.5 | 0.000 |
| Ethylmethylacetic acid | 354.4 ± 161.6 | 300 ± 108.3 | 0.208 |
| Fumaric acid | 4447.5 ± 5258.8 | 8002.2 ± 5704.2 | 0.036 |
| Gamma-aminobutyric acid | 8011.9 ± 2433.5 | 9501.4 ± 2144.3 | 0.038 |
| Glutaric acid | 6350.9 ± 2956.9 | 8011.9 ± 3153 | 0.077 |
| Glyceric acid | 24294.2 ± 44555.4 | 34244.8 ± 54085.6 | 0.503 |
| Glycine | 90591.2 ± 73998.7 | 150330 ± 121868.4 | 0.048 |
| Glycolic acid | 85357.2 ± 57207.8 | 98641.7 ± 50868.8 | 0.424 |
| Hippuric acid | 579641.8 ± 409879.0 | 435904.9 ± 374381.5 | 0.234 |
| Homogentisic acid | 395731.6 ± 236443.5 | 706581.6 ± 61533.7 | 0.000 |
| Indoleacetic acid | 58737.6 ± 40343.9 | 107584.7 ± 107498.1 | 0.040 |
| Isocitric acid | 418285.5 ± 291898.4 | 767013.4 ± 476267.9 | 0.004 |
| Isovaleric acid | 96.7 ± 125.3 | 53.8 ± 103.4 | 0.228 |
| Itaconic acid | 7961.3 ± 6523.3 | 12949.6 ± 11408.9 | 0.071 |
| L-2-hydroxyglutaric acid | 14028.4 ± 7911.3 | 25645.4 ± 16050.4 | 0.003 |
| L-alanine | 26378.1 ± 18928.9 | 36445.6 ± 16618.3 | 0.070 |
| L-alpha-aminobutyric acid | 1777.6 ± 1093.0 | 1826.9 ± 1158.2 | 0.885 |
| L-asparagine | 16684.8 ± 15672.4 | 25642.5 ± 14904.9 | 0.059 |
| L-aspartic acid | 2801.2 ± 833.5 | 3708.1 ± 1242 | 0.005 |
| L-cysteine | 7833.5 ± 5012.7 | 12771 ± 4681.6 | 0.002 |
| L-glutamic acid | 18551.2 ± 13284.8 | 29044.7 ± 18203.4 | 0.030 |
| L-histidine | 300484.2 ± 313800.2 | 328182 ± 163648.8 | 0.726 |
| L-isoleucine | 1874.2 ± 954.0 | 3149.4 ± 1492.8 | 0.001 |
| L-leucine | 4028.1 ± 3120.4 | 5677.4 ± 4266.7 | 0.141 |
| L-lysine | 38409.7 ± 46071.6 | 46194.7 ± 36720.6 | 0.545 |
| L-methionine | 1236.4 ± 696.3 | 2195.1 ± 1206.4 | 0.002 |
| L-phenylalanine | 18545.8 ± 14394.6 | 26167 ± 13952.9 | 0.082 |
| L-proline | 1131.5 ± 512.2 | 2020.9 ± 1099.8 | 0.001 |
| L-tryptophan | 26500.2 ± 13737.3 | 33639.1 ± 12697.3 | 0.082 |
| L-tyrosine | 63971.6 ± 55716.2 | 84163.3 ± 63553.6 | 0.264 |
| L-valine | 4892.5 ± 2641.4 | 7343.2 ± 3296.4 | 0.008 |
| Malonic acid | 493 ± 486.4 | 663.3 ± 547.4 | 0.277 |
| Methylsuccinic acid | 1655.6 ± 861.4 | 2484.1 ± 1188.1 | 0.009 |
| N-acetyltryptophan | 82232.1 ± 58726.0 | 112567.4 ± 56382.5 | 0.088 |
| Nicotinic acid | 1293.3 ± 1311.8 | 2104.1 ± 1204 | 0.039 |
| Ornithine | 7948.8 ± 2592.9 | 11968.9 ± 4307.9 | 0.000 |
| Ortho-hydroxyphenylacetic acid | 1265.1 ± 770.7 | 1915.5 ± 1233.6 | 0.035 |
| Oxoglutaric acid | 23844.7 ± 20947.4 | 34260.9 ± 33676.2 | 0.209 |
| Phenol | 266.1 ± 168.1 | 282 ± 165.2 | 0.753 |
| Phenylacetic acid | 3055.4 ± 2506.3 | 5635.2 ± 5749.6 | 0.048 |
| Phenyllactic acid | 729.1 ± 1365.9 | 2726.7 ± 2958.9 | 0.004 |
| P-hydroxyphenylacetic acid | 8991.4 ± 5587.6 | 16438.3 ± 9973.0 | 0.003 |
| Pimelic acid | 2203 ± 1453.7 | 3343.5 ± 1728.2 | 0.021 |
| Pyroglutamic acid | 39396.7 ± 14482.8 | 52204.6 ± 19506.9 | 0.015 |
| Salicyluric acid | 3342.8 ± 1712.7 | 4135.9 ± 1498.5 | 0.112 |
| Suberic acid | 8538.8 ± 5594.7 | 12987.5 ± 5378.1 | 0.010 |
| Succinic acid | 23748.9 ± 18864.3 | 34383.7 ± 30189.9 | 0.154 |
| Tartaric acid | 5156.1 ± 9387.3 | 11376.5 ± 27499.5 | 0.291 |
| Vanillic acid | 2838.1 ± 5734.4 | 3091.8 ± 3028.7 | 0.861 |
| 12_ketoLCA | 11.5 ± 2.4 | 13.5 ± 4.5 | 0.072 |
| 3_DHCA | 13.4 ± 11.9 | 17 ± 15.7 | 0.396 |
| 7_DHCA | 145.5 ± 276.1 | 267.1 ± 439.7 | 0.267 |
| 7_ketoLCA | 10.9 ± 2.9 | 13.6 ± 7.2 | 0.093 |
| Beta-CA | 12.9 ± 10.4 | 26.1 ± 27.0 | 0.029 |
| Beta-UCA | 42.1 ± 93.8 | 114.8 ± 168.0 | 0.074 |
| CA | 182.3 ± 237.0 | 266 ± 322.5 | 0.327 |
| CDCA | 21.9 ± 9.7 | 24.9 ± 11.2 | 0.346 |
| CDCA_24GLN | 79.6 ± 49.1 | 168 ± 151.2 | 0.008 |
| CDCA_3GLN | 64.1 ± 45.2 | 149.1 ± 105.5 | 0.001 |
| DCA | 45.2 ± 28.7 | 40.2 ± 31.0 | 0.588 |
| GCA | 84.4 ± 43.2 | 151.8 ± 162.6 | 0.051 |
| GCDCA | 39.8 ± 20.6 | 51.3 ± 25.1 | 0.105 |
| GDCA | 34.5 ± 33.4 | 30.4 ± 15.9 | 0.627 |
| GHCA | 36.7 ± 17.6 | 49.4 ± 41.5 | 0.171 |
| GHDCA | 14.4 ± 2.6 | 12.3 ± 0.6 | 0.002 |
| GLCA | 8.4 ± 5.7 | 4.8 ± 4.2 | 0.026 |
| GUDCA | 15.5 ± 17.5 | 39.5 ± 47.9 | 0.025 |
| HCA | 10.3 ± 1.5 | 10.5 ± 1.5 | 0.768 |
| HDCA | 17.1 ± 35.3 | 15.2 ± 10.2 | 0.820 |
| LCA | 19.1 ± 6.6 | 22.4 ± 32.6 | 0.621 |
| LCA_S | 22.5 ± 11.9 | 22.6 ± 22.2 | 0.989 |
| NORCA | 154.3 ± 116.0 | 181.4 ± 133.4 | 0.476 |
| NORDCA | 9.2 ± 1.5 | 9.3 ± 0.4 | 0.679 |
| TCA | 24.6 ± 8.2 | 26 ± 11.9 | 0.652 |
| TCDCA | 23.4 ± 7.7 | 23.4 ± 5.6 | 0.99 |
| THCA | 13.7 ± 3.0 | 18.4 ± 11.4 | 0.052 |
| TUDCA | 8.2 ± 1.6 | 9 ± 1.2 | 0.099 |
| UCA | 108.2 ± 135.4 | 268.4 ± 310.0 | 0.024 |
| UDCA | 11.6 ± 1.4 | 13.1 ± 2.0 | 0.008 |
Due to sample dilution, the urine sample from hone healthy control was discarded from analysis by MS metabonomics. Initial inspection of the main source of variations between the samples was performed using principal component analysis (PCA). A model was generated using four principal components, explaining 30%, 8%, 6% and 4% of the total variance, respectively. The sources of variance captured by the third and fourth components highlighted a separation of the urine of healthy children from the IBD patients, irrespective of their time of collection.
Using OPLS-DA, we first assessed the occurrence of urinary signatures discriminating the urine samples from healthy subjects and IBD patients. A first OPLS-DA model generated on urine samples collected, with one predictive and one orthogonal component, described statistically robust metabolic differences at baseline. This is observed through the model parameters R2X = 0.37, R2Y = 0.72, Q2Y = 0.47, where R2X is the explained variance in the urine metabolic profile, R2Y the explained group variance and Q2Y an indicator of mode robustness. We further tested the reproducibility of the metabolic differences by comparing the urine collected from IBD patients after 6 and 12 mo of standard disease management with the reference samples from the healthy children. The models generated with one predictive and one orthogonal component were significant at 6 mo (R2X = 0.36, R2Y = 0.67, Q2Y = 0.41) and at 12 mo (R2X = 0.38, R2Y = 0.73, Q2Y = 0.47).
Examination of the three models resulted in the identification of consistent urinary metabolic differences between the group of healthy subjects and IBD patients. In Table 2, we report metabolites that were significantly different in at least two of the models with VIP > 1.0, and their corresponding Pearson correlation coefficient. These differences described variations in relation to glutathione metabolism (glycine, glutamate, pyroglutamate and cysteine), urea (fumarate, arginine, alanine, aspartate, and ornithine) and TCA (fumarate, isocitrate, 2-hydroxyglutaric acid, and methylsuccinic acid) cycles, catecholamines (dopamine and tyrosine), several amino acids (e.g., phenylalanine, tyrosine, proline, and glutamate), aromatic acid and bile acid bacterial co-metabolism.
| Metabolic pathways | Metabolites | Comparison healthy - IBD subjects at | |||||
| Baseline | 6 mo | 12 mo | |||||
| VIP | r | VIP | r | VIP | r | ||
| Tca cycle | Methylsuccinic acid | 1.3 | 0.4 | 1.2 | 0.2 | 1.0 | 0.1 |
| Fumaric acid | 1.2 | 0.3 | 1.0 | 0.3 | 1.1 | 0.2 | |
| 2-hydroxyglutaric acid | 1.4 | 0.5 | 1.3 | 0.4 | 1.3 | 0.3 | |
| Isocitric acid | 1.4 | 0.4 | 1.3 | 0.3 | 1.2 | 0.2 | |
| Glutathione metabolism | Cysteine | 1.5 | 0.5 | 1.2 | 0.3 | 1.4 | 0.4 |
| Glycine | 1.2 | 0.3 | 1.3 | 0.3 | 1.2 | 0.3 | |
| Glutamic acid | 1.2 | 0.3 | 1.3 | 0.3 | 1.2 | 0.3 | |
| Pyroglutamic acid | 1.3 | 0.4 | 1.3 | 0.3 | 1.0 | 0.1 | |
| Neurostransmitter metabolism | Gamma-Aminobutyric acid | 1.2 | 0.3 | 1.2 | 0.3 | 1.0 | 0.1 |
| Dopamine | 1.6 | 0.7 | 1.8 | 0.6 | 1.1 | 0.4 | |
| Homogentisic acid | 1.6 | 0.7 | 1.8 | 0.6 | 1.1 | 0.4 | |
| Bacterial metabolism | 3-hydroxyphenylacetic acid | 1.3 | -0.5 | 1.2 | -0.4 | 1.1 | -0.3 |
| Hippuric acid | 1.0 | -0.3 | 1.2 | -0.4 | 1.6 | -0.5 | |
| Indoleacetic acid | 1.3 | 0.4 | 1.3 | 0.4 | 1.0 | 0.1 | |
| 4-hydroxyphenylacetic acid | 1.3 | 0.5 | 1.1 | 0.3 | 1.0 | 0.2 | |
| Phenyllactic acid | 1.1 | 0.4 | 0.6 | 0.2 | 1.4 | 0.5 | |
| Other | Salicyluric acid | 1.2 | 0.4 | 0.9 | 0.2 | 1.0 | 0.4 |
| Dicarboxylic acid metabolism | Suberic acid | 1.3 | 0.5 | 1.5 | 0.5 | 1.1 | 0.3 |
| Bile acids | Beta-UCA | 1.5 | 0.6 | 1.0 | 0.4 | 0.7 | 0.2 |
| GUDCA | 1.3 | 0.5 | 1.5 | 0.5 | 1.9 | 0.7 | |
| THCA | 1.0 | 0.4 | 0.3 | 0.1 | 1.3 | 0.4 | |
| UDCA | 1.4 | 0.6 | 1.7 | 0.6 | 1.8 | 0.6 | |
| GHDCA | 1.3 | -0.5 | 1.3 | -0.5 | 1.0 | -0.3 | |
| Amino acid metabolism | Aspartic acid | 1.3 | 0.4 | 0.8 | 0.2 | 1.4 | 0.4 |
| Histidine | 1.1 | 0.3 | 1.1 | 0.2 | 0.9 | 0.1 | |
| Isoleucine | 1.5 | 0.4 | 1.6 | 0.4 | 1.3 | 0.2 | |
| Methionine | 1.5 | 0.5 | 1.6 | 0.5 | 1.4 | 0.4 | |
| Phenylalanine | 1.3 | 0.3 | 1.2 | 0.2 | 1.1 | 0.1 | |
| Proline | 1.4 | 0.5 | 1.4 | 0.4 | 1.2 | 0.3 | |
| Tryptophan | 1.2 | 0.3 | 1.2 | 0.2 | 1.1 | 0.2 | |
| Tyrosine | 1.1 | 0.2 | 1.0 | 0.1 | 1.0 | 0.1 | |
| Valine | 1.3 | 0.3 | 1.3 | 0.3 | 1.1 | 0.1 | |
| N-acetyltryptophan | 1.2 | 0.3 | 1.2 | 0.2 | 1.1 | 0.2 | |
| Ornithine | 1.4 | 0.5 | 1.7 | 0.5 | 1.7 | 0.5 | |
| Aminoadipic acid | 1.0 | 0.0 | 1.2 | 0.3 | 1.1 | 0.1 | |
Whilst most of the metabolites appeared to be excreted in higher concentrations in IBD patients, only 3 metabolites showed reduced concentrations, namely hippurate, 3-hydroxyphenylacetate and the bile acid glyco-hyodeoxycholic acid (GHDCA).
We further assessed if some urinary metabolites were specific to the few UC patients. OPLS-DA models were generated to compare the subjects at the different time points. The models generated with one predictive and two orthogonal components were significant at baseline (R2X = 0.42, R2Y = 0.94, Q2Y = 0.34), at 6 mo (R2X = 0.48, R2Y = 0.94, Q2Y = 0.55) and at 12 mo (R2X = 0.50, R2Y = 0.95, Q2Y = 0.55). Only three metabolites showed pattern differentiating UC from CD patients consistently over time, including higher urinary concentrations of deoxycholic acid (DCA), 7-ketolithocholic acid and beta-ursocholic acid (βUCA) in urine from UC patients (Table 3).
| Comparison UC - CD subjects at metabolites | Baseline | 6 mo | 12 mo | |||
| VIP | r | VIP | r | VIP | r | |
| DCA | 1.6 | 0.5 | 1.4 | 0.5 | 0.8 | 0.2 |
| 7-ketoLCA | 1.1 | 0.3 | 1.2 | 0.4 | 1.8 | 0.5 |
| Beta-UCA | 0.3 | 0.1 | 2.0 | 0.7 | 1.0 | 0.3 |
Finally, we explored urinary metabolic signatures linked to clinical and anthropometric endpoints in the IBD population. An O-PLS regression approach was applied using cross-validation by subjects, since clinical and metabonomics data were generated from each subject at 3 different time points. Some relationships between urinary metabolite profiles and anthropometric features were modeled, but not in relation to disease index (Q2Y < 0) or inflammatory status (CRP, calprotectin, Q2Y < 0). Statistically significant associations were modeled in relation to BMI (R2X = 0.40, R2Y = 0.82, Q2Y = 0.35), BMI-z scores (R2X = 0.40, R2Y = 0.83, Q2Y = 0.29), body weight (R2X = 0.43, R2Y = 0.74, Q2Y = 0.15), % of fat mass (R2X = 0.39, R2Y = 0.83, Q2Y = 0.36). A conservative approach was applied for selecting the most influential metabolites, namely using the VIP threshold > 1.0 and a r value significance at 95%CI (e.g., r > 0.265 or r < -0.265). A consolidated data overview is reported in Table 4 for the relevant anthropometric parameters and metabolites. Similar urinary metabolite patterns could be ascribed to the body composition parameters, including bile acids, amino acids and TCA cycle intermediates, as well as some dietary/gut microbial related metabolites (Table 4).
| Metabolic pathways | Metabolites | Anthropometric parameters | |||||||
| Fat mass % | BMI | BMI z-score | Body weight | ||||||
| r | VIP | r | VIP | r | VIP | r | VIP | ||
| TCA cycle | L-2-hydroxyglutaric acid | 0.41 | 1.57 | 0.22 | 1.23 | 0.29 | 1.36 | 0.14 | 1.07 |
| Oxoglutaric acid | 0.48 | 1.66 | 0.29 | 1.32 | 0.35 | 1.41 | 0.13 | 0.91 | |
| Succinic acid | 0.39 | 1.50 | 0.17 | 1.12 | 0.24 | 1.24 | 0.10 | 0.98 | |
| Bacterial metabolism | 3-hydroxyphenylacetic acid | 0.44 | 1.49 | 0.27 | 1.20 | 0.29 | 1.22 | 0.23 | 1.07 |
| Hippuric acid | 0.32 | 1.27 | 0.27 | 1.27 | 0.28 | 1.26 | 0.19 | 1.03 | |
| Phenol | 0.44 | 1.42 | 0.42 | 1.57 | 0.48 | 1.68 | 0.26 | 1.10 | |
| Bile acids | 12_ketoLCA | 0.34 | 1.13 | 0.41 | 1.53 | 0.40 | 1.40 | 0.39 | 1.48 |
| DCA | 0.48 | 1.62 | 0.48 | 1.82 | 0.50 | 1.76 | 0.52 | 2.02 | |
| GDCA | 0.48 | 1.58 | 0.49 | 1.85 | 0.47 | 1.66 | 0.53 | 2.01 | |
| GHDCA | 0.38 | 1.28 | 0.49 | 1.81 | 0.42 | 1.47 | 0.45 | 1.69 | |
| GLCA | 0.19 | 0.87 | 0.32 | 1.28 | 0.30 | 1.17 | 0.43 | 1.69 | |
| LCA | 0.35 | 1.16 | 0.37 | 1.40 | 0.34 | 1.20 | 0.41 | 1.57 | |
| NORDCA | 0.37 | 1.25 | 0.45 | 1.65 | 0.44 | 1.54 | 0.57 | 2.13 | |
| TUDCA | -0.12 | 0.81 | -0.32 | 1.35 | -0.31 | 1.27 | -0.43 | 1.65 | |
| UCA | -0.30 | 0.96 | -0.38 | 1.39 | -0.33 | 1.16 | -0.35 | 1.42 | |
| UDCA | -0.11 | 0.70 | -0.42 | 1.60 | -0.38 | 1.39 | -0.44 | 1.65 | |
| Amino acid metabolism | Aminoadipic acid | 0.36 | 1.41 | 0.22 | 1.20 | 0.25 | 1.23 | 0.17 | 1.06 |
| L-Asparagine | 0.26 | 0.89 | 0.34 | 1.33 | 0.36 | 1.30 | 0.27 | 1.21 | |
| L-Histidine | 0.30 | 1.33 | 0.33 | 1.48 | 0.34 | 1.47 | 0.31 | 1.42 | |
| L-Methionine | 0.31 | 1.39 | 0.16 | 1.16 | 0.24 | 1.29 | 0.09 | 1.03 | |
| L-Proline | 0.30 | 1.35 | 0.15 | 1.13 | 0.23 | 1.27 | 0.10 | 1.06 | |
| L-Tryptophan | 0.32 | 1.39 | 0.29 | 1.41 | 0.34 | 1.50 | 0.22 | 1.21 | |
| L-Tyrosine | 0.27 | 1.29 | 0.23 | 1.26 | 0.26 | 1.32 | 0.15 | 1.07 | |
| N-acetyltryptophan | 0.32 | 1.42 | 0.28 | 1.41 | 0.33 | 1.50 | 0.22 | 1.23 | |
Our MS based metabonomics approach allowed to characterize further metabolite differences between IBD and healthy children, by providing complementary metabolite readouts into the central energy metabolic pathways, bile acids, glutathione and glycine metabolism, which are discussed here after.
IBD is often associated with some variations in the enterohepatic circulation of bile acids[2,30,31]. Recently, Duboc et al[30] extensively discussed the connections between gut dysbiosis and bile acid metabolism in IBD, and sub-sequent implication for the management of inflammatory conditions. For instance, the alterations in gastrointestinal transit time link to altered bacterial metabolism of primary and secondary bile acids[32]. This is reflected through higher concentrations of conjugated bile acids in the stools from active IBD patients, and concomitant decreased concentrations of secondary bile acids[30]. Bile acid malabsorption has also been reported in children with CD[33]. In our study, we report higher urinary excretion of taurohyocholic acid (THCA, a product of TCDCA metabolism by CYP3A4), ursodeoxycholic acid (UDCA, a product of CDCA by the gut microbiota and the liver), tauroursodeoxycholic acid (TUDCA, a taurine conjugate of UCDA), and βUCA. This signature most likely reflects the alteration of the bile acid metabolism in the digestive tract, as well as modulation of the metabolism of the secondary bile acids by the liver. Previous reports showed an alteration in most circulating bile acids in CD and UC patients[31]. In particular, UC patients were reported with higher amount of LCA and DCA, two secondary bile acids, which may relate to their higher urinary excretion in the urine as noted here. We also observed a strong association between BMI z-scores and urinary levels in several bile acids, a trend that was previously reported as well in plasma in other cohort studies[34]. Interestingly, the secondary bile acids UDCA and TUDCA are associated with the lower BMI z-score, whilst bacterial metabolites hippurate and 3-hydroxyphenylacetate are positively associated with the higher ones. Considering IBD children have overall higher urinary UDCA and TUDCA, lower urinary hippurate and 3-hydroxyphenylacetate, and lower BMI z-scores, the signature may be reflecting a higher degree of gastrointestinal deregulations with IBD.
Pediatric IBD patients also showed a higher urinary loss in dopamine, gamma-aminobutyric acid (GABA) and homogentisic acid. This pattern may be reflecting a disbalance in serotonin - dopamine metabolism, which is being studied in patient with CD[35]. Recent evidences tend to describe excessive synthesis with associated increased tissue levels of serotonin, leading to a perturbation of the molecular homeostasis with dopamine and catecholamines. In particular, a genetic defect of the OCTN1 and OCTN2 of the colon has been identified in patients with CD, resulting in altered transportation of monoamines of the serotonin-dopamine system and their precursors[35]. The higher concentration in dopamine, closely related to higher levels of homogentisic acid (another product of tyrosine metabolism) may reflect this disbalance, yet serotonin metabolism could not be captured with our analytical approach in urine. Furthermore, the higher urinary loss of GABA may be reflecting the higher activity in the TCA cycle towards alpha-ketoglutarate (AKG) and glutamate, and altered colonic epithelial cell metabolism of serotonin-dopamine[36].
Another feature identified in the studied population is the occurrence of higher urinary levels of several metabolites involved in the glutathione metabolism, including pyroglutamic acid (PGA), glutamic acid, glycine and cysteine, which were significantly higher in IBD children in the course of the study. Since PGA is linked to glutathione turnover, the urinary pattern may reflect the higher requirement of the metabolism in glutathione to cope with oxidative stress and immune status[37,38]. In particular, higher urine PGA may indicate higher glutathione need concomitant to diminished glutathione utilization and/or deficient glutathione resynthesis. The higher urinary loss of cysteine, glycine and glutamic acid, the three main precursors for glutathione synthesis provides additional evidence that glutathione cannot optimally be resynthesized, and therefore is broken down and excreted in the urine via its constituent parts such as PGA. The observation of PGA may also point towards glycine deficiency[39], whilst the higher urinary excretion of many essential and non-essential amino acid mainly reflected higher protein breakdown related to higher metabolic requirements in IBD children. However, no statistically significant associations could be found between urinary metabolites and inflammatory endpoints (e.g., CRP, EST, calprotectin).
Moreover, it is well described, how before entering the urea cycle, glycine can be conjugated with benzoic acid to form hippurate, and be subsequently excreted in urine. An alternative pathway to excrete ammonia excess is through AKG to glutamic acid, glutamine and phenylacetylglutamine (PAG), which then exits the body via the urine. Glycine deficiencies would increase the demands on the urea cycle and the glutamate/AKG pathways. Moreover, the lower hippurate production may reflect this deficiency in glycine for specific metabolic processes. Previous studies reported that altered hippurate metabolic profile in IBD patients was not directly explain by variations in dietary intake or a deficit in the conjugation of benzoate to glycine in the kidney[40]. Moreover, our analysis describes a higher urinary excretion of salicyluric acid, a glycine conjugate of salicylic acid produced in the kidney via a pathway very similar to that of hippurate, suggesting selective requirement in glycine for metabolic functions. We previously discussed in the context of IBD[14], that carbamyl phosphate synthetase (CAD) - rate-limiting enzyme for the urea cycle - was identified as negative regulator of NOD2 and might be a pharmacologic target for CD therapies[41]. Our observations may provide additional evidence on the essential role of this pathway in case of higher metabolic requirements for glycine associated to malnutrition, higher requirement for glutathione, phase II metabolism and/or growth and development in pediatric populations.
We previously reported and discussed how the pediatric IBD patients show the typical phenotype associated with growth failure and body weight loss[6,14,42]. In particular, we reported that our CD pediatric patients showed lower z-scores for body weight, body height, BMI and resting energy expenditure compared to healthy controls, which linked to lower concentrations in IGF-1 and IGFBP-3[14]. Our MS metabolomics approaches provided more detailed insights into the changes in protein and energy metabolites. IBD children showed a significant increase in the urinary content of several closely related intermediates of the TCA cycle (fumarate, isocitrate, 2-hydroxyglutaric acid, methylsuccinic acid). Our previous NMR analysis identified higher urinary PAG in IBD patients, which indicated a different energy metabolic status[14]. PAG is a major nitrogenous metabolite for which synthesis depends on the availability of glutamine, mainly generated in the Krebs cycle from AKG. Therefore the increase in isocitrate (precursor of AKG) may be functionally related to higher metabolic turnover for AKG. Moreover, we noted an increased excretion of a 2-hydroxyglutaric acid (Table 2), normally in molecular equilibrium with AKG, which brings further evidence for a specific increased metabolic flux towards AKG within TCA cycle. Whilst succinate does not show a significantly increased urinary excretion (in agreement with our previous NMR observations[14]), closely related metabolites, namely methylsuccinate and fumarate, had higher urinary levels which may describe a selective oxidation of aspartate, tyrosine and phenylalanine.
Furthermore, the overall increased excretion in most amino acids and the sulfur containing amino acids cysteine and methionine (Table 2), indicates a different protein metabolism and handling of nitrogen in pediatric IBD patients when compared to healthy matched controls. Urinary metabolite patterns could be ascribed to body composition parameters, such a BMI, body weight and fat mass. Moreover, most of our IBD patients were in clinical remission, and whilst the patients tend to have lower z-scores for body weight, height and BMI at baseline, the higher the BMI z-scores during follow-up the better growth catch-up for the patient. We here observed an association between higher BMI z-score and higher urinary content in key intermediates of the TCA cycle, namely succinate and AKG, amino acids (asparagine, histidine, methionine, tyrosine, proline). This signature may be associated with overall changes in protein and energy requirements related to body weight gain. Our observations with BMI z-scores are similar to recent observations in blood with the Framingham study[43].
One main limitation of our study is the small number of subjects. However, the longitudinal design and the use of reference group of age- and gender match healthy children provide novel insights into childhood metabolic status in relation to growth and disease.
We demonstrate that a direct analysis of urine - a biological sample that can be collected non-invasively and in a repeatable fashion - using targeted MS based metabonomics might provide metabolic insights complementary to more invasive blood and stool metabolic analysis, whilst offering an opportunity to monitor the metabolic status in relation to disease state. Here, we identified a peculiar metabolite signature related to bile acids, central energy and glutathione biochemical pathways, which should be further followed up in future studies.
We would like to thank Tania Chatton, dietician, and Sylvie Poget, study nurse, for their substantial support during the study period, and Drs. Jessica Ezri and Ornella Cominetti for input during scientific discussions.
The prevalence of inflammatory bowel disease (IBD) has been increasing over the year, with 25% of the population developing the disease during childhood and adolescence. In the pediatric population, in addition to the clinical and immunological features of the disease, patients suffer of additional growth failure and delayed puberty.
There is a great lack of knowledge on how to address metabolic and nutritional requirements in children and adolescents. It is envisioned that the use of omics technologies can generate a system view of metabolic processes associated with the growth and the development of children in relation to the changing physiology. Metabonomics is a promising approach to explore physiological regulatory processes in human clinical research.
In the present study, they applied targeted mass spectrometry based metabolite profiling to explore bile acids and host-gut microbial metabolites in urine samples from IBD and healthy matched children. Metabolic differences encompass central energy, amino acids, and bile acid metabolism. The authors showed that non-invasive urine bile acid profiling reflect altered hepatic and intestinal barrier dysfunctions, as previously reported through blood or stool metabolic profiling. Moreover, levels of pyroglutamic acid, glutamic acid, glycine and cysteine were significantly higher in IBD children in the course of the study. This suggests that glutathione cannot be optimally synthesized and replenished, rather it is broken down to its constituent parts and excreted through the urine. In addition, a perturbation of the urinary levels of several glycine conjugated metabolites, may associate with differential metabolic requirements in children.
Non-invasive sampling of urine followed by targeted MS-based metabonomic analysis might elucidate and monitor the metabolic status of children in relation to disease state.
The excellent study gives a very good data set reflecting metabonomics in IBD children. Understanding the metabolic characteristics of pediatric patients with IBD compared to control is a key factor in diagnosis. The authors stuck to their specific hypothesis and eloquently presented their material. This opens the door for future trials looking at predictive modeling utilizing metabolomics and modifying various therapies to target patients with specific metabolic characteristics.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Switzerland
Peer-review report classification
Grade A (Excellent): A, A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Gassler N, Lilia ZM, Mattar MC S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | Baffy G. The impact of network medicine in gastroenterology and hepatology. Clin Gastroenterol Hepatol. 2013;11:1240-1244. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Moco S, Candela M, Chuang E, Draper C, Cominetti O, Montoliu I, Barron D, Kussmann M, Brigidi P, Gionchetti P. Systems biology approaches for inflammatory bowel disease: emphasis on gut microbial metabolism. Inflamm Bowel Dis. 2014;20:2104-2114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 3. | Ezri J, Marques-Vidal P, Nydegger A. Impact of disease and treatments on growth and puberty of pediatric patients with inflammatory bowel disease. Digestion. 2012;85:308-319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 4. | Sauer CG, Kugathasan S. Pediatric inflammatory bowel disease: highlighting pediatric differences in IBD. Gastroenterol Clin North Am. 2009;38:611-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 5. | Sperisen P, Cominetti O, Martin FP. Longitudinal omics modeling and integration in clinical metabonomics research: challenges in childhood metabolic health research. Front Mol Biosci. 2015;2:44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Shamir R, Phillip M, Levine A. Growth retardation in pediatric Crohn’s disease: pathogenesis and interventions. Inflamm Bowel Dis. 2007;13:620-628. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 7. | Day AS, Ledder O, Leach ST, Lemberg DA. Crohn’s and colitis in children and adolescents. World J Gastroenterol. 2012;18:5862-5869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 68] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 8. | Powell-Tuck J. Nutritional consequences of gastrointestinal disease. Baillieres Clin Gastroenterol. 1988;2:715-727. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 9. | Rigaud D, Angel LA, Cerf M, Carduner MJ, Melchior JC, Sautier C, René E, Apfelbaum M, Mignon M. Mechanisms of decreased food intake during weight loss in adult Crohn’s disease patients without obvious malabsorption. Am J Clin Nutr. 1994;60:775-781. [PubMed] [Cited in This Article: ] |
| 10. | De Preter V, Verbeke K. Metabolomics as a diagnostic tool in gastroenterology. World J Gastrointest Pharmacol Ther. 2013;4:97-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 55] [Cited by in F6Publishing: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 11. | Stephens NS, Siffledeen J, Su X, Murdoch TB, Fedorak RN, Slupsky CM. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J Crohns Colitis. 2013;7:e42-e48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 12. | Storr M, Vogel HJ, Schicho R. Metabolomics: is it useful for inflammatory bowel diseases? Curr Opin Gastroenterol. 2013;29:378-383. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Yau Y, Leong RW, Zeng M, Wasinger VC. Proteomics and metabolomics in inflammatory bowel disease. J Gastroenterol Hepatol. 2013;28:1076-1086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Martin FP, Ezri J, Cominetti O, Da Silva L, Kussmann M, Godin JP, Nydegger A. Urinary Metabolic Phenotyping Reveals Differences in the Metabolic Status of Healthy and Inflammatory Bowel Disease (IBD) Children in Relation to Growth and Disease Activity. Int J Mol Sci. 2016;17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Levine A, Koletzko S, Turner D, Escher JC, Cucchiara S, de Ridder L, Kolho KL, Veres G, Russell RK, Paerregaard A. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr. 2014;58:795-806. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 822] [Cited by in F6Publishing: 811] [Article Influence: 81.1] [Reference Citation Analysis (0)] |
| 16. | de Onis M, Onyango AW, Borghi E, Siyam A, Nishida C, Siekmann J. Development of a WHO growth reference for school-aged children and adolescents. Bull World Health Organ. 2007;85:660-667. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4196] [Cited by in F6Publishing: 4820] [Article Influence: 301.3] [Reference Citation Analysis (0)] |
| 17. | Lohman T, Roche A, Martorell R. Anthropometric standardisation reference manual. : Human Kinetics Pub 1988; . [Cited in This Article: ] |
| 18. | Tanner JM, Whitehouse RH. Clinical longitudinal standards for height, weight, height velocity, weight velocity, and stages of puberty. Arch Dis Child. 1976;51:170-179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2180] [Cited by in F6Publishing: 2162] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 19. | Pietrobelli A, Andreoli A, Cervelli V, Carbonelli MG, Peroni DG, De Lorenzo A. Predicting fat-free mass in children using bioimpedance analysis. Acta Diabetol. 2003;40 Suppl 1:S212-S215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Houtkooper LB, Lohman TG, Going SB, Hall MC. Validity of bioelectric impedance for body composition assessment in children. J Appl Physiol (1985). 1989;66:814-821. [PubMed] [Cited in This Article: ] |
| 21. | Hyams JS, Ferry GD, Mandel FS, Gryboski JD, Kibort PM, Kirschner BS, Griffiths AM, Katz AJ, Grand RJ, Boyle JT. Development and validation of a pediatric Crohn’s disease activity index. J Pediatr Gastroenterol Nutr. 1991;12:439-447. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 785] [Cited by in F6Publishing: 792] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 22. | Turner D, Otley AR, Mack D, Hyams J, de Bruijne J, Uusoue K, Walters TD, Zachos M, Mamula P, Beaton DE. Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology. 2007;133:423-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 694] [Cited by in F6Publishing: 716] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 23. | Diamanti A, Colistro F, Basso MS, Papadatou B, Francalanci P, Bracci F, Muraca M, Knafelz D, De Angelis P, Castro M. Clinical role of calprotectin assay in determining histological relapses in children affected by inflammatory bowel diseases. Inflamm Bowel Dis. 2008;14:1229-1235. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Xie G, Zhong W, Li H, Li Q, Qiu Y, Zheng X, Chen H, Zhao X, Zhang S, Zhou Z. Alteration of bile acid metabolism in the rat induced by chronic ethanol consumption. FASEB J. 2013;27:3583-3593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 139] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 25. | Ferslew BC, Xie G, Johnston CK, Su M, Stewart PW, Jia W, Brouwer KL, Barritt AS. Altered Bile Acid Metabolome in Patients with Nonalcoholic Steatohepatitis. Dig Dis Sci. 2015;60:3318-3328. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 197] [Cited by in F6Publishing: 228] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 26. | Villas-Bôas SG, Delicado DG, Akesson M, Nielsen J. Simultaneous analysis of amino and nonamino organic acids as methyl chloroformate derivatives using gas chromatography-mass spectrometry. Anal Biochem. 2003;322:134-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in F6Publishing: 154] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 27. | Qiu Y, Su M, Liu Y, Chen M, Gu J, Zhang J, Jia W. Application of ethyl chloroformate derivatization for gas chromatography-mass spectrometry based metabonomic profiling. Anal Chim Acta. 2007;583:277-283. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 123] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 28. | Trygg J, Wold S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integrated OSC filter. J Chemom. 2003;17:53-64. [DOI] [Cited in This Article: ] |
| 29. | Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251-W257. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2063] [Cited by in F6Publishing: 2043] [Article Influence: 227.0] [Reference Citation Analysis (0)] |
| 30. | Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531-539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 513] [Cited by in F6Publishing: 555] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 31. | Gnewuch C, Liebisch G, Langmann T, Dieplinger B, Mueller T, Haltmayer M, Dieplinger H, Zahn A, Stremmel W, Rogler G. Serum bile acid profiling reflects enterohepatic detoxification state and intestinal barrier function in inflammatory bowel disease. World J Gastroenterol. 2009;15:3134-3141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 53] [Cited by in F6Publishing: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 32. | Kruis W, Kalek HD, Stellaard F, Paumgartner G. Altered fecal bile acid pattern in patients with inflammatory bowel disease. Digestion. 1986;35:189-198. [PubMed] [Cited in This Article: ] |
| 33. | Gothe F, Beigel F, Rust C, Hajji M, Koletzko S, Freudenberg F. Bile acid malabsorption assessed by 7 alpha-hydroxy-4-cholesten-3-one in pediatric inflammatory bowel disease: correlation to clinical and laboratory findings. J Crohns Colitis. 2014;8:1072-1078. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Prinz P, Hofmann T, Ahnis A, Elbelt U, Goebel-Stengel M, Klapp BF, Rose M, Stengel A. Plasma bile acids show a positive correlation with body mass index and are negatively associated with cognitive restraint of eating in obese patients. Front Neurosci. 2015;9:199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 67] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | Stein A, Hinz M, Uncini T. Amino acid-responsive Crohn’s disease: a case study. Clin Exp Gastroenterol. 2010;3:171-177. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Sahoo S, Thiele I. Predicting the impact of diet and enzymopathies on human small intestinal epithelial cells. Hum Mol Genet. 2013;22:2705-2722. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 37. | Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489-492. [PubMed] [Cited in This Article: ] |
| 38. | Lord RS. Long-term patterns of urinary pyroglutamic acid in healthy humans. Physiol Rep. 2016;4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 39. | Metges CC, Yu YM, Cai W, Lu XM, Wong S, Regan MM, Ajami A, Young VR. Oxoproline kinetics and oxoproline urinary excretion during glycine- or sulfur amino acid-free diets in humans. Am J Physiol Endocrinol Metab. 2000;278:E868-E876. [PubMed] [Cited in This Article: ] |
| 40. | Williams HR, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJ, Teare JP. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol. 2009;104:1435-1444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 41. | Richmond AL, Kabi A, Homer CR, Marina-García N, Nickerson KP, Nesvizhskii AI, Sreekumar A, Chinnaiyan AM, Nuñez G, McDonald C. The nucleotide synthesis enzyme CAD inhibits NOD2 antibacterial function in human intestinal epithelial cells. Gastroenterology. 2012;142:1483-1492.e6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 42. | Heuschkel R, Salvestrini C, Beattie RM, Hildebrand H, Walters T, Griffiths A. Guidelines for the management of growth failure in childhood inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:839-849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 43. | Ho JE, Larson MG, Ghorbani A, Cheng S, Chen MH, Keyes M, Rhee EP, Clish CB, Vasan RS, Gerszten RE. Metabolomic Profiles of Body Mass Index in the Framingham Heart Study Reveal Distinct Cardiometabolic Phenotypes. PLoS One. 2016;11:e0148361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in F6Publishing: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |