Published online May 21, 2017. doi: 10.3748/wjg.v23.i19.3427
Peer-review started: December 14, 2016
First decision: December 29, 2016
Revised: January 13, 2017
Accepted: March 15, 2017
Article in press: March 15, 2017
Published online: May 21, 2017
To determine how a normal human colon cell line reacts to microbial challenge as a way to study oxidative stress-induced responses associated with inflammatory bowel disease.
Normal human colon epithelial cells (ATCC® CRL.1790™) were stimulated with either heat killed E. coli or heat killed murine cecal contents (HKC) and examined for several relevant biomarkers associated with inflammation and oxidative stress including cytokine production, mitochondrial autophagy and oxidant status. TNFα, IL-1β and IL-8 protein concentrations were measured within the supernatants. Fluorescent microscopy was performed to quantify the production of reactive oxygen species (ROS) using an oxidation responsive fluorogenic probe. Mitochondrial morphology and mitochondrial membrane potential was assessed by dual staining using COXIV antibody and a dye concentrating in active mitochondria. Mitochondrial ROS scavenger was used to determine the source of ROS in stimulated cells. Autophagy was detected by staining for the presence of autophagic vesicles. Positive controls for autophagy and ROS/RNS experiments were treated with rapamycin and chloroquine. Mitochondrial morphology, ROS production and autophagy microscopy experiments were analyzed using a custom acquisition and analysis microscopy software (ImageJ).
Exposing CRL.1790 cells to microbial challenge stimulated cells to produce several relevant biomarkers associated with inflammation and oxidative stress. Heat killed cecal contents treatment induced a 10-12 fold increase in IL-8 production by CRL.1790 cells compared to unstimulated controls at 6 and 12 h (P < 0.001). Heat killed E. coli stimulation resulted in a 4-5 fold increase in IL-8 compared to the unstimulated control cells at each time point (P < 0.001). Both heat killed E. coli and HKC stimulated robust ROS production at 6 (P < 0.001), and 12 h (P < 0.01). Mitochondrial morphologic abnormalities were detected at 6 and 12 h based on reduced mitochondrial circularity and decreased mitochondrial membrane potential, P < 0.01. Microbial stimulation also induced significant autophagy at 6 and 12 h, P < 0.01. Lastly, blocking mitochondrial ROS generation using mitochondrial specific ROS scavenger reversed microbial challenge induced mitochondrial morphologic abnormalities and autophagy.
The findings from this study suggest that CRL.1790 cells may be a useful alternative to other colon cancer cell lines in studying the mechanisms of oxidative stress events associated with intestinal inflammatory disorders.
Core tip: The normal human colon cell line, CRL.1970, can recapitulate oxidative stress-induced responses associated with inflammatory bowel disease following microbial challenge including enhanced production of reactive oxygen species (ROS), inflammatory cytokines, and enhanced mitochondrial autophagic responses. Scavenging mitochondrial ROS inhibited mitochondrial morphologic changes and autophagy suggesting that CRL.1790 cells can be used to study oxidative events associated with intestinal inflammatory disorders.
- Citation: Packiriswamy N, Coulson KF, Holcombe SJ, Sordillo LM. Oxidative stress-induced mitochondrial dysfunction in a normal colon epithelial cell line. World J Gastroenterol 2017; 23(19): 3427-3439
- URL: https://www.wjgnet.com/1007-9327/full/v23/i19/3427.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i19.3427
Oxidative stress-induced damage to intestinal epithelial cells is a key event in the initiation and progression of pathologies associated with multiple intestinal inflammatory disorders including ulcerative colitis, colon cancer and enteritis[1-3]. The intestinal epithelial layer is uniquely tasked with maintaining tolerance to commensal bacteria while recognizing and initiating immune responses to pathogenic infectious agents. Once tolerance is breached, immune and epithelial cells respond to commensal and pathogenic bacteria in an exaggerated manner and produce inflammatory mediators and reactive oxygen species (ROS) that can not only damage DNA, proteins and lipids[4,5], but also eventually lead to activation of apoptotic pathway that destroys the epithelial cell layer. Mitochondria are a primary target for ROS-induced damage to epithelial cells and are also the primary source of intracellular ROS produced by oxidative phosphorylation. Exposure of organelles to modest amounts of ROS will activate important cytoprotective processes such as the autophagy pathway, which is designed to maintain cellular homeostasis during times of stress by clearing or recycling damaged organelles. However, excessive ROS generation overwhelms the protective function of the autophagy pathway leading to activation of apoptotic cell death and eventually causing loss of mucosal barrier[6,7].
In vitro models studying oxidative stress response in intestinal epithelial cells are needed to understand the pathophysiology of oxidative stress in causing cellular damage. Currently, there are many colon cancer cell lines including HCT116, SW620, and Caco-2 that are used to assess the oxidative damage induced dysfunction of epithelial cells in conditions like microbial gastro-enteritis, ulcerative colitis, and Crohn’s disease[8,9]. Many of these cell lines tend to underestimate or overestimate the cellular oxidative responses because of their inherent resistance to oxidative stress, changes in endogenous antioxidant levels, altered expression or activation of detoxifying systems, and altered susceptibility of mitochondria and genetic components to ROS attack[10,11]. Additionally, these cancer cell lines likely respond differently to microbial stimuli compared to normal human intestinal epithelium. For example, intestinal neoplastic cells have abnormal chromosome numbers (chromosome number: Caco-2 -96, HCT116-45, sw620-50)[12-14] and react differently to various stimuli and stress factors compared to primary cells[15,16]. Proteomic studies comparing cancer cell lines with primary cells lines showed distinct alterations in metabolic pathways suggesting that neoplastic cell lines may not be the best choice for disease models[17]. Primary colon epithelial cells obtained from patient biopsy samples can be used to model oxidative stress during gastrointestinal disorders. However, limited cell recovery, a lack of reproducibility of experimental data, and procedural costs make the use of primary cell model impractical[18]. The CRL.1790 cells are an intestinal epithelial cell line isolated from normal human neonatal intestine and are successfully maintained under laboratory conditions[19,20]. The CRL.1790 cells have a normal diploid chromosome number, are easy to propagate at laboratory conditions and are cost effective. The current study proposes an in vitro cell culture model using the CRL.1790 normal human colon epithelial cells as an alternative to using other cancer cell lines to study oxidative stress responses to microbial exposure. Murine heat killed cecal contents (HKC) and heat killed E. coli were used to induce inflammation and associated oxidative stress. Inflammatory cytokine production, ROS generation, mitochondrial and autophagic responses were measured. Our results suggest that CRL.1790 cells may be used to model in vitro characteristics of epithelial cell mitochondrial dysfunction during inflammation-induced oxidative stress.
CCD 841 CoN (ATCC® CRL.1790™; Manassas, VA, United States) normal human colon epithelial cells were obtained from ATCC and maintained at 37 °C, 5% CO2 in MEM supplemented with 3% FBS, 2 mmol/L L-glutamine, penicillin-G (100 U/mL), and streptomycin (100 μg/mL). Colon cells ≤ 9 passages were grown as monolayers until confluent, harvested with trypsin-treatment at 37 °C for 5 min and plated for experiments. Media was replaced 24 h after plating and the cells were allowed to adhere for 48 h prior to experimental treatments.
Escherichia coli (ATCC® 25922™) was obtained from ATCC. E. coli was heat killed and used for experiments. Briefly, E. coli were grown in trypticase soy broth with gentle shaking to 37 °C to stationary phase. The bacteria were washed with PBS before cultures were adjusted to 1.0 × 105 cells per 1 μL. Bacterial cultures were then heat-killed at 80 °C for 30 min and penicillin- G (100 U/mL) and streptomycin (100 μg/mL) added prior to freezing and storage at -80 °C. To ensure complete killing of E. coli, aliquots were plated on trypticase soy agar and checked for growth. Murine HKC contents were prepared according to previously published methods[21]. Briefly, 25 mg of cecal contents were mixed with 1 mL sterile HBSS and filtered twice through nylon mesh to remove large particles. Filtered supernatants were heat-killed at 80 °C for 30 min then centrifuged at 150 × g, for 5 min to remove remaining large particulate matter. Penicillin-G (100 U/mL) and streptomycin (100 μg/mL) were added to supernatants and aliquots frozen and stored at -80 °C.
Epithelial cell monolayers were treated with heat killed E. coli (ATCC® 25922™) at multiplicity of infection (MOI) = 1 or with 200 μg of HKC contents per 2.0 × 105 cells[21] which served as a positive control. To study mitochondrial dysfunction, a positive control was created by adding carbonyl cyanide 3-cholorphenylhydrazone (CCCP) (Sigma-Aldrich, St. Louis, MO, United States) to a final concentration of 10 μmol/L to control wells for 90 min at 37 °C, 5% CO2 to induce mitochondrial fission and abrogate outer membrane potential[22,23]. Positive controls for autophagy-induction were treated with 500 nmol/L rapamycin and 10 μmol/L chloroquine for 16 h at 37 °C, 5% CO2[24].
ROS generation from CRL.1790 cells was measured by loading Carboxy-H2DCFDA dye before exposure to microbial ligands as described earlier[25]. Carboxy-H2DCFDA is non-fluorescent but in the presence of ROS, this reagent is oxidized, and becomes green fluorescent, which is then detected using fluorescence plate reader. Briefly, CRL.1790 cells were cultured in 96 well tissue culture treated plate at a concentration of 5 × 105 cells/well for 18 h. Supernatants were removed and incubated with HBSS containing 5 μmol/L of the Carboxy-H2DCFDA dye for 30 min. Cells were then washed to remove excess dye and fresh media containing serum was added. Additionally, the cells were treated with heat-killed E. coli, HKC or H2O2 (100 μmol/L for 1 h) for specified time points. ROS generation was then detected by measuring fluorescence at 490/520 (Ex/Em) wavelengths using Tecan Infinite M200 Plate reader. Background fluorescence from the cells was subtracted from the fluorescence values obtained after loading the cells with carboxy-H2DCFDA dye. Data are represented as fluorescence intensity.
Additionally, ROS generation was measured microscopically using cell permeable CellROX® Deep Red dye (Life Technologies Corp., Grand Island, NY, United States). Briefly, CRL.1790 cells were grown as monolayers to confluence, harvested, and seeded onto sterile cover slips within 6-well dishes at 4 × 105 cells per well. Cells were allowed to adhere for 48 h at 37 °C before performing treatments. Cells were then treated with microbial ligands for specific time points. 5 μmol/L of CellROX® Deep Red dye was added to each well and cells were incubated at 37 °C, 5% CO2 for 30 min. Media containing CellROX stain was removed and monolayers were washed with 1 × PBS and fixed with 3.7% paraformaldehyde for 15 min at 37 °C. Coverslips were processed for microscopy as described below.
For microscopy experiments, CRL.1790 cells were grown as monolayers to confluence, harvested, and seeded onto sterile cover slips within 6-well dishes at 4 × 105 cells per well. Cells were allowed to adhere for 48 h at 37 °C before performing treatments as described above. For mitochondria experiments, 500 nmol/L MitoTracker® (Life Technologies Corp., Grand Island, NY, United States) was added to each well and incubated for 30 min at 37 °C. MitoTracker stain is preferentially absorbed by mitochondria with intact outer membrane potential and reflects viable mitochondria. Diminished staining by mitochondria reflects disrupted membrane potential of non-viable mitochondria. Excess MitoTracker stain was removed after the 30 min incubation, chased with 1 × PBS for 15 min at 37 °C and fixed with 3.7% paraformaldehyde at 37 °C for 15 min. Following fixation, cells were permeabilized for 10 min with 0.1% Triton X-100 and blocked for 1 h with PBS containing 3% bovine serum albumin (BSA). Mouse monoclonal anti-COXIV (cytochrome c oxidase IV) (1:1000) antibody (Abcam, Cambridge, MA, United States), rabbit polyclonal anti-DRP1(1:100) (Santa Cruz Biotechnology, TX, United States), anti-MFn2 (1;100) (Santa Cruz Biotechnology, TX, United States) were used to stain mitochondria and incubated at room temperature for 1 h. Anti-mouse Alexa Fluor® 555 and anti- rabbit Alexa Fluor® 488 (1:500 dilution) were used as secondary antibodies to detect COXIV, DRP1 and MFN2 (Life Technologies Corp. Grand Island, NY, United States).
For experiments using MitoTempo as mitochondrial ROS scavenger, a final concentration of 25 nmol/L MitoTempo (Sigma Aldrich, St. Louis, MO, United States) was added to the cell culture 12 h prior to stimulation with HKC or E. coli. All cell nuclei were stained with DAPI (4’,6-Diamidino-2-Phenylindole, Dihydrochloride) for 15 min at room temperature. Autophagy experiments utilized the Cyto-ID® Autophagy Detection Kit (Enzo Life Sciences, Farmingdale, NY, United States) to detect autophagic vesicles and cells were processed according to manufacturer specifications. Briefly, cells were washed with assay buffer and incubated with a dual detection reagent containing Cyto-ID Green Detection Reagent and Hoechst stain for 30 min at 37 °C. After incubation, cells were washed with PBS and fixed with 4% paraformaldehyde. Cells were imaged immediately. Coverslips with cells for all experiments were mounted with Prolong® Gold anti fade mounting agent (Life Technologies Corp, Grand Island, NY, United States). Fluorescent images were taken at 60x (oil) magnification with a Zeiss Axiovert 200M and black and white AxioCam MRm (Zeiss) camera. All treatments were performed in duplicate and experiments were repeated a minimum of 3 times.
Cytokines including TNFα, IL-1β and IL-8 were measured in the supernatants from CRL.1790 cells treated with HKC- and heat-killed E. coli. Briefly, CRL.1790 cells were grown as monolayer to confluence, harvested, and seeded in 6-well dishes at 4 × 105 cells per well. Cells were treated with HKC contents or heat-killed E. coli for 6 or 12 h and supernatants were collected. TNFα, IL-1β and IL-8 kits were obtained from eBiosciences (San Diego, CA, United States) and samples were analyzed per manufacturer’s instructions. All values were represented as pg/mL of media. All treatments were performed in duplicate and experiments were repeated a minimum of three times.
For each coverslip, 10 fields were captured and analyzed resulting in 20 fields per treatment for each experiment. CellROX and Mitotracker and autophagy microscopy experiments were analyzed using ImageJ 1.46/Java 8 software (National Institute of Health, Bethesda, MD, United States) as described previously[26]. Briefly, individual cells in each image were selected and analyzed using the measurement command. Area, integrated density and mean gray value were collected. Additional measurements were made of areas without fluorescence adjacent to cells as background. Corrected total cell fluorescence (CTCF) was calculated using the equation: CTCF = integrated density - (area of selected cell × mean fluorescence of background). Mitochondrial morphology was analyzed from MitoTracker images using ImageJ, Mito-Morphology Plugin as described previously[27]. Measurements of mitochondrial area, perimeter, circularity, minor and major axis as well as total mitochondrial counts were collected for each imaged field. Mitochondrial morphology was characterized by average circularity, area/perimeter ratio as a measure of interconnectivity and inverse circularity reported as a measure of elongation[27-29]. Statistical significance of cytokine measurements, ROS, and morphology measurements was examined by ANOVA and Tukey’s HSD post hoc comparison (P = 0.05) using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, United States). All results were expressed as mean ± SE.
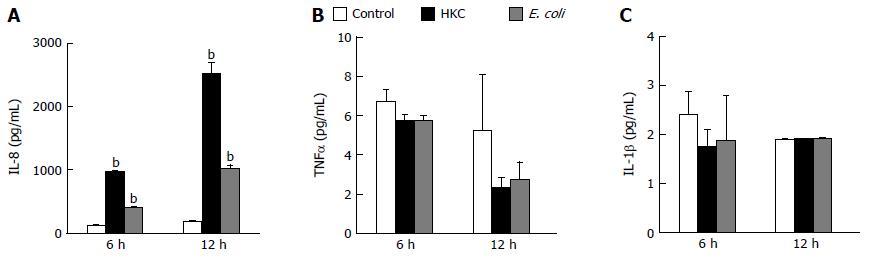
Excessive microbial stimulation of colonic epithelial cells is a key event in the progression of many intestinal disorders. Heat killed cecal contents obtained from wild type mice to mimic the population of intraluminal antigens and heat killed E. coli (ATCC 25922) were chosen to induce inflammatory responses in CRL.1790 cells. The cells were treated with HKC contents or heat killed E. coli and supernatants were analyzed for the production of inflammatory cytokines TNFα, IL-1β and IL-8 using ELISA. Of the 3 cytokines measured, only IL-8 production was significantly increased in HKC and E. coli treatment groups compared to untreated controls. Treatment with HKC contents induced 10-12 fold increase in IL-8 production by CRL.1790 cells compared to unstimulated controls at 6 and 12 h. Heat killed E. coli stimulation resulted in a 4-5 fold increase in IL-8 compared to the unstimulated control cells at each time point. (Figure 1A). No significant effects of either HKC or killed E. coli on TNFα or IL-1β production were detected at 6 or 12 h (Figure 1B and C).
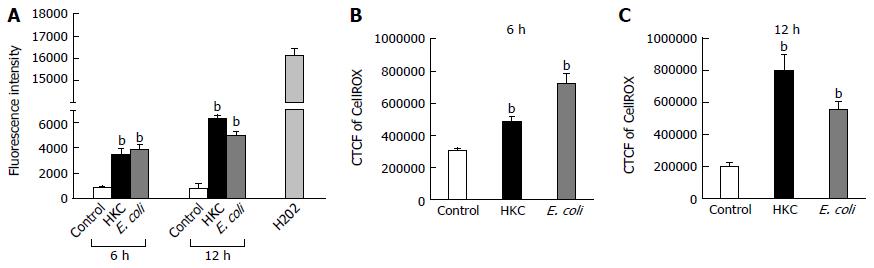
ROS are generated during mitochondrial oxidative metabolism as well as in response to bacterial invasion, xenobiotic metabolism and cytokine stimulation. ROS generation was detected microscopically using cell permeable CellROX deep red dye and also by measuring fluorescence emission using carboxy-H2DCFDA dye. CRL.1790 cells were grown on coverslips or 96 well plate and treated with HKC contents and heat-killed E. coli for 6 and 12 h. ROS generation was quantified microscopically by calculating corrected total cell fluorescence (CTCF) or by measuring fluorescence intensity using a fluorescence plate reader as described in the materials and methods. Both treatments induced a significantly higher production of ROS compared to untreated controls. At 6 h post-treatment, HKC contents and E. coli treatment induced 1.6 fold and 2 fold more ROS than untreated controls, respectively (Figure 2). By 12 h post-treatment, HKC and killed-E. coli produced 4 and 2.7 fold increases in ROS production, respectively, compared to untreated controls (Figure 2).
To test the impact of ROS generated in response to microbial stimulation, CRL.1790 cells treated with HKC contents and heat-killed E. coli were assessed microscopically for mitochondrial outer membrane potential and fission fusion dynamics, which are reflective of mitochondrial integrity.
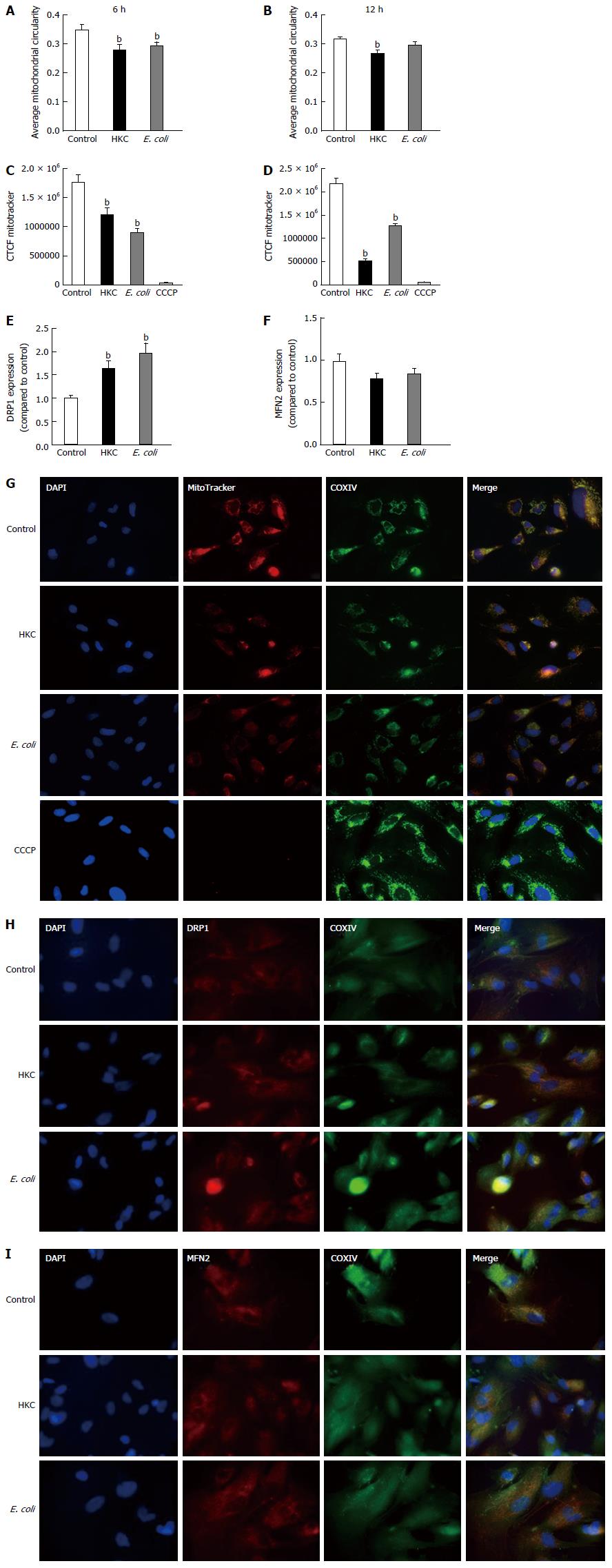
Mitochondrial fission-fusion dynamics can be measured by calculating and comparing average circularity of mitochondria[27]. Increased average circularity measurements denote more balanced fusion/fission dynamics and increased mitochondrial health. A decrease in average circularity measurements indicate accelerated fission suggestive of mitochondrial dysfunction and stress. Our results showed that average circularity was diminished in the cells stimulated with either HKC contents or E. coli (Figure 3A and B). At 6 h post stimulation a small but significant reduction in circularity was measured for both HKC contents and E. coli-treated cells. However, by 12 h, HKC stimulation alone showed a significant decrease in mitochondrial circularity. These data clearly demonstrate that CRL.1790 cells undergo alteration in mitochondrial fission/fusion dynamics during microbial stress potentially contributing to mitochondrial damage. Additionally, expression of Dynamin related protein 1 (DRP-1) (Marker for mitochondrial fission) and Mitofusin (MFN2) (Marker for mitochondrial fusion) were also measured after treating the cells with microbial ligands for 6 h. DRP1 expression was significantly increased in the HKC and E.coli treated groups suggesting increased mitochondrial fission. MFn2 expression was decreased in the HKC and E. coli treated groups, however the changes weren’t significant.
To assess mitochondrial integrity, the CRL.1790 cells were stained with MitoTracker after treatments and images were collected and analyzed using ImageJ software as described in the methods. MitoTracker stain is only absorbed by mitochondria with intact outer membrane potential (OMP). Mitochondria were double-labeled with anti-COXIV antibodies as a non-OMP dependent marker of mitochondria, confirming the presence of those organelles lacking fluorescent stain. A subset of cells were treated with CCCP, a potent mitochondrial oxidative phosphorylation uncoupler to inhibit mitochondrial activity and serve as negative control. Corrected total cell fluorescence (CTCF) was measured to assess OMP, which indicates intact mitochondrial integrity. Higher fluorescence intensity indicates functioning mitochondria and lower fluorescence reflects destabilization of the mitochondrial membranes. Our results show that, mitochondrial integrity was significantly decreased following both HKC and heat-killed E. coli treatments at 6 and 12 h (Figure 3C-E). These data demonstrate that CRL.1790 cells respond to microbial stress and undergo alteration mitochondrial function and integrity.
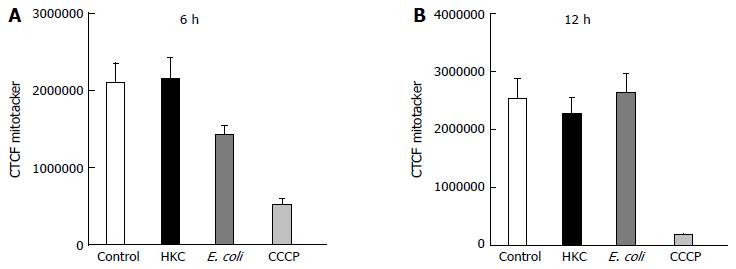
To test whether ROS produced during the microbial stress was responsible for the decrease in mitochondrial OMP, a mitochondrial-targeted oxidant scavenging molecule (MitoTempo) was used to inhibit ROS-induced mitochondrial damage. Briefly, MitoTempo was added to the cells for 12 h prior to treatments. Cells were stained with MitoTracker to assess the antioxidant impact on the OMP to evaluate mitochondrial integrity. Corrected total cell fluorescence was measured using ImageJ software as described in the methods. There was no significant difference in MitoTracker CTCF between HKC and untreated cells or heat-killed E. coli and untreated cells at 6 or 12 h of microbial stimulation. These data demonstrate that scavenging mitochondrial ROS prevented mitochondrial damage and dysfunction (Figure 4), suggesting that most of the ROS produced in the CRL.1790 cells following HKC and heat-killed E. coli treatment was of mitochondrial origin.
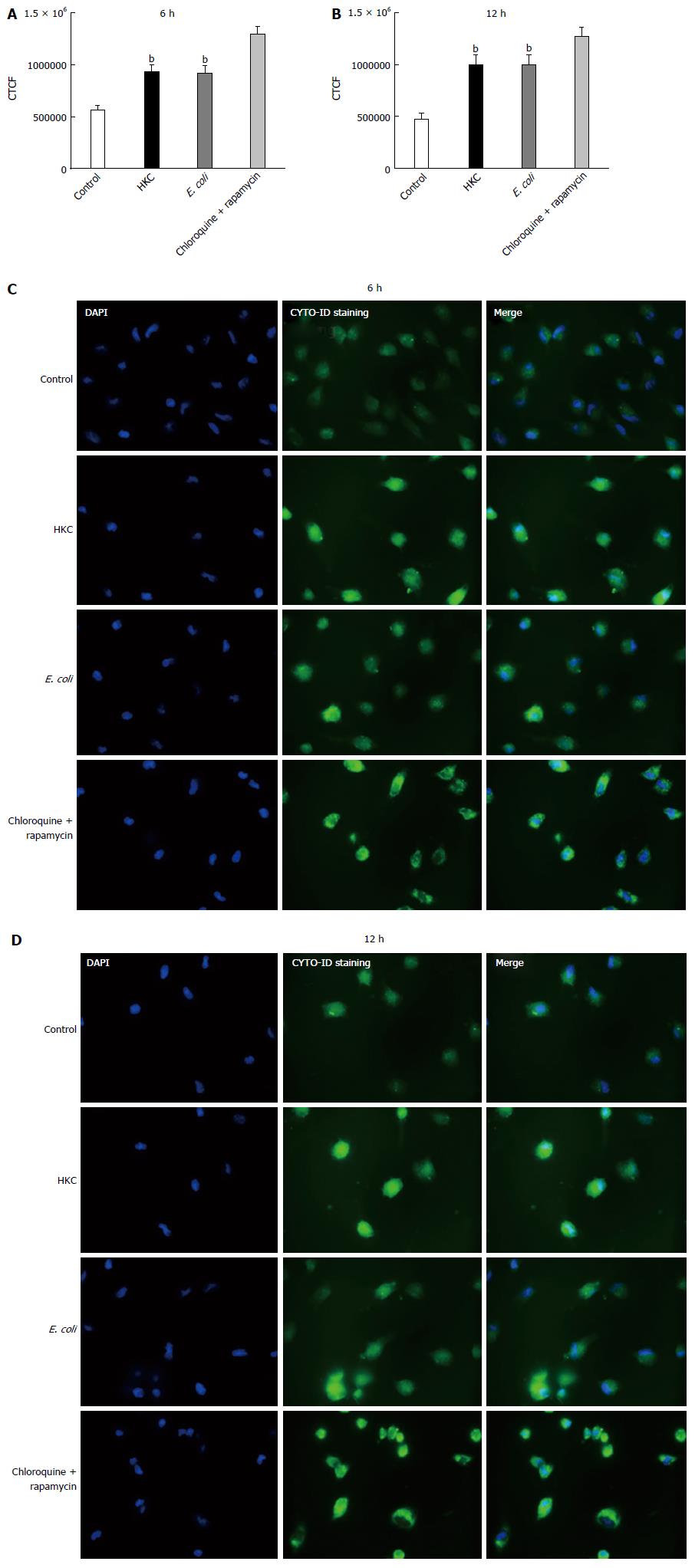
Excessive ROS production, damages organelles mainly targeting lipid and DNA molecules. Under normal conditions, the autophagy pathway promotes cell repair and survival. However, excessive production of ROS and the resultant oxidative stress can overwhelm the autophagy system and promote cell death. Thus, we evaluated autophagy activation in E. coli and HKC treated CRL.1790 cells. Staining of autophagic vacuoles using Cyto-ID kit was determined to measure autophagy activation through labeling of autophagic vacuoles. At 6 and 12 h post-stimulation both HKC and E. coli treatment induced a significant increase in autophagic vacuoles compared to untreated controls (Figure 5). These data suggest that CRL.1790 cells undergo sufficient damage to induce autophagy activation during microbial stress.
Studies have shown that cancer cell lines might be ineffective in recapitulating the oxidative stress response and mitochondrial dysfunction evident in inflammatory based intestinal diseases[10]. Studies comparing oxidative stress responses in normal and cancerous cell lines showed normal cell lines behaved differently when compared to cancerous cell lines like caco-2 cells, likely owing to their altered metabolic and genetic profile. Studies like these highlight the importance of using a cell line that is closely related to normal cells, but at a same time, are practical and efficient for in vitro studies. In this study, we tested the usability of CRL.1790 normal epithelial cells as a replacement for cancerous colon epithelial cell lines to study oxidative stress response and mitochondrial dysfunction induced by microbial ligands. The results of this study suggest that human colon epithelial CRL.1790 cells may be a good alternative for cancer cell lines used in studying cellular oxidative stress response and specifically mitochondrial dysfunction during microbial exposure. The CRL.1790 cells successfully recapitulated inflammatory response, induction of oxidative stress, mitochondrial dysfunction and autophagic responses which are key events leading to loss of epithelial cell barrier functions.
The CRL.1790 epithelial cells demonstrated pro-inflammatory responses following microbial challenge as evidenced by production of IL-8, which is a neutrophil attractant. Other cytokines specifically TNFα and IL1β were not significantly increased compared to unstimulated controls. This finding is in concurrence with other studies demonstrating that colon epithelial cells respond to TNFα[30,31] and IL1β[32] produced by immune cells but do not produce large quantities of these cytokines[33]. Similarly, in human patients suffering from Crohn’s disease and ulcerative colitis, only serum concentration of IL-8 is increased compared to control patients[34] suggesting IL-8 production can be served as a good biomarker to test colon epithelial cell responses.
During inflammatory responses, increased mitochondrial oxidative phosphorylation is required to meet increasing cellular demand and this may result in oxidative stress[29,35]. Excessive accumulation of ROS that leads to oxidative stress can elicit cellular damage through the oxidation of various macromolecules and thus alter their biological functions and potentiate cell death. For example, previous studies[36-38] highlighted the contribution of mitochondrial dysfunction to loss of epithelial cell integrity, leading to increased epithelial permeability promoting microbial translocation. One mechanism that promotes mitochondrial damage and dysfunction is excessive production of ROS[37,38]. Mitochondrial ROS generation is considered to be a continuous physiological process under aerobic conditions. During times of microbial stress, however, mitochondrial oxidative phosphorylation is increased leading to generation of additional ROS. Increased ROS production must be neutralized by anti-oxidant systems to prevent oxidative damage to mitochondria and other cellular organelles. CRL.1790 cells exhibited significant mitochondrial morphology changes including reduced circularity and diminished mitochondrial membrane integrity in response to microbial treatments. Previous studies[39] associated increased mitochondrial fission with decreased oxidative capacity, increased ROS generation and increased autophagy. In our current study, the damage to mitochondria also induced significant activation of autophagic responses demonstrating the ability of these cells to respond to oxidative stress and also potentially initiate recovery processes. We did not test activation of apoptotic pathway as a direct consequence of microbial stimulation in this study. However, mitochondrial stress and alterations in mitochondrial functions are observed in multitude of colon associated disease condition like ulcerative colitis, Crohn’s disease and colon cancer. Our results concur with other studies showing mitochondrial dysfunction may be an early event leading to epithelial cell dysfunction observed in and other intestinal infections[40] and the CRL.1790 cells could provide a model system for studying this potential initiating phase of the disease.
Finally, to determine whether scavenging mitochondrial ROS can reverse mitochondrial dysfunction, MitoTempo, a mitochondrial specific ROS scavenger was added prior to treatments and mitochondrial damage was studied. Our results clearly show, by pre-treating CRL.1790 cells with MitoTempo decreased the mitochondrial damage and dysfunction, confirming the mitochondria was one of the major sources of ROS generation. Additionally our study also revealed, HKC contents are more potent in inducing inflammation (IL-8 production and ,ROS generation) compared to heat killed E. coli. The increased activity of HKC contents is likely due to multiple microbial sources (bacterial, protozoal, fungal and viral antigens) and possibly due to presence of other luminal antigens, advantageous for researchers trying to model colitis. The different potencies of the microbial challenges may also account for the differential timing of ROS responses following exposure to either E. coli or HKC.
In conclusion, our findings indicated that the normal cell line, CRL.1790, could be used in a convenient and reliable way to recapitulate both physiological and pathological mitochondrial function associated with intestinal inflammatory disorders. CRL.1790 respond to microbial stimulation by increasing ROS and induced autophagic responses that serve as a good model to study oxidative stress responses.
The authors wish to thank Jeff Gandy, Vengai Mavangira and Valerie Ryman from the College of Veterinary Medicine at Michigan State University for their valuable scientific comments and advice on this study.
Cellular oxidative stress is implicated in the multifactorial etiology of inflammatory bowel diseases and is a key initiating event propagating cellular damage. Cancerous colon cell lines are used to model inflammatory and oxidative stress responses in inflammatory bowel disease (IBD), but their inherent alterations in genome and metabolism may lead to confounding results when investigating underlying mechanisms of disease. There is a need to determine if normal human colon cell lines can react to microbial challenge in a way that can recapitulate oxidative stress-induced responses that are associated with intestinal inflammatory disorders.
Previous studies have already proven that oxidative stress and mitochondrial dysfunction are key events leading to intestinal inflammatory disorders in humans and animal models.
This is the first study to evaluate the use of normal colonic epithelial cell line as a model to recapitulate oxidative stress-induced mitochondrial dysfunction that is known to be critical in the development of inflammatory bowel diseases.
The normal cell line, CRL.1790, can respond to microbial stimulation by increasing reactive oxygen species (ROS) and inducing autophagic responses that serve as a good model to study oxidative stress responses associated with intestinal inflammatory disorders. The results from this study confirm that CRL.1790 can be used in a convenient and reliable way to investigate both physiological and pathological mitochondrial function in response to microbial challenge as a way to investigate the underlying mechanisms associated with intestinal inflammatory disorders.
Oxidative stress is an imbalance between the production of ROS and other free radicals that can damage tissues and anti-oxidant defenses of the body that are needed to counteract pro-oxidant damage.
The authors investigated how normal human colon cells can react to microbial challenge as a way to investigate the role of oxidative stress on mitochondrial dysfunction. They were able to show that microbial challenge of CRL.1790 could induce oxidative stress-induced responses associated with IBD and that scavenging ROS within the mitochondrial during microbial challenge could overcome mitochondrial dysfunction.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Han J S- Editor: Yu J L- Editor: A E- Editor: Zhang FF
| 1. | Raza Y, Khan A, Farooqui A, Mubarak M, Facista A, Akhtar SS, Khan S, Kazi JI, Bernstein C, Kazmi SU. Oxidative DNA damage as a potential early biomarker of Helicobacter pylori associated carcinogenesis. Pathol Oncol Res. 2014;20:839-846. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 2. | Scarpa M, Cardin R, Bortolami M, Kotsafti A, Scarpa MC, Pozza A, Maran G, Picciocchi M, Ruffolo C, D’Incà R. Mucosal immune environment in colonic carcinogenesis: CD80 expression is associated to oxidative DNA damage and TLR4-NFκB signalling. Eur J Cancer. 2013;49:254-263. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 3. | Bhattacharyya A, Chattopadhyay R, Mitra S, Crowe SE. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev. 2014;94:329-354. [PubMed] [Cited in This Article: ] |
| 4. | Kruidenier L, Kuiper I, Lamers CB, Verspaget HW. Intestinal oxidative damage in inflammatory bowel disease: semi-quantification, localization, and association with mucosal antioxidants. J Pathol. 2003;201:28-36. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 262] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 5. | Kathiria AS, Butcher LD, Feagins LA, Souza RF, Boland CR, Theiss AL. Prohibitin 1 modulates mitochondrial stress-related autophagy in human colonic epithelial cells. PLoS One. 2012;7:e31231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 6. | Johansson ME, Gustafsson JK, Holmén-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjövall H. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281-291. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 582] [Cited by in F6Publishing: 642] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 7. | Wang Z, Li R, Tan J, Peng L, Wang P, Liu J, Xiong H, Jiang B, Chen Y. Syndecan-1 Acts in Synergy with Tight Junction Through Stat3 Signaling to Maintain Intestinal Mucosal Barrier and Prevent Bacterial Translocation. Inflamm Bowel Dis. 2015;21:1894-1907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 44] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 8. | Reddy KV, Naidu KA. Oleic acid, hydroxytyrosol and n-3 fatty acids collectively modulate colitis through reduction of oxidative stress and IL-8 synthesis; in vitro and in vivo studies. Int Immunopharmacol. 2016;35:29-42. [PubMed] [Cited in This Article: ] |
| 9. | Koh SJ, Kim JW, Kim BG, Lee KL, Chun J, Kim JS. Fexofenadine regulates nuclear factor-κB signaling and endoplasmic reticulum stress in intestinal epithelial cells and ameliorates acute and chronic colitis in mice. J Pharmacol Exp Ther. 2015;352:455-461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | Zhao D, Shah NP. Synergistic Application of Black Tea Extracts and Lactic Acid Bacteria in Protecting Human Colonocytes against Oxidative Damage. J Agric Food Chem. 2016;64:2238-2246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 11. | Halliwell B. Biochemistry of oxidative stress. Biochem Soc Trans. 2007;35:1147-1150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 889] [Cited by in F6Publishing: 838] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 12. | Jumarie C, Malo C. Caco-2 cells cultured in serum-free medium as a model for the study of enterocytic differentiation in vitro. J Cell Physiol. 1991;149:24-33. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 157] [Cited by in F6Publishing: 158] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981;41:1751-1756. [PubMed] [Cited in This Article: ] |
| 14. | Trainer DL, Kline T, McCabe FL, Faucette LF, Feild J, Chaikin M, Anzano M, Rieman D, Hoffstein S, Li DJ. Biological characterization and oncogene expression in human colorectal carcinoma cell lines. Int J Cancer. 1988;41:287-296. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in F6Publishing: 132] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 15. | Walczak K, Dąbrowski W, Langner E, Zgrajka W, Piłat J, Kocki T, Rzeski W, Turski WA. Kynurenic acid synthesis and kynurenine aminotransferases expression in colon derived normal and cancer cells. Scand J Gastroenterol. 2011;46:903-912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 16. | Kaur G, Dufour JM. Cell lines: Valuable tools or useless artifacts. Spermatogenesis. 2012;2:1-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 394] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 17. | Pan C, Kumar C, Bohl S, Klingmueller U, Mann M. Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol Cell Proteomics. 2009;8:443-450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 18. | Grossmann J, Maxson JM, Whitacre CM, Orosz DE, Berger NA, Fiocchi C, Levine AD. New isolation technique to study apoptosis in human intestinal epithelial cells. Am J Pathol. 1998;153:53-62. [PubMed] [Cited in This Article: ] |
| 19. | Siddiqui KM, Chopra DP. Primary and long term epithelial cell cultures from human fetal normal colonic mucosa. In Vitro. 1984;20:859-868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 56] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 20. | ATCC Product Sheet CCD 841 CoN (ATCC® CRL1790™). Available from: http://wwwatccorg/Products/All/CRL-1790aspx - generalinformation. [Cited in This Article: ] |
| 21. | Ocuin LM, Bamboat ZM, Balachandran VP, Cavnar MJ, Obaid H, Plitas G, DeMatteo RP. Neutrophil IL-10 suppresses peritoneal inflammatory monocytes during polymicrobial sepsis. J Leukoc Biol. 2011;89:423-432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 22. | MacVicar TD, Lane JD. Impaired OMA1-dependent cleavage of OPA1 and reduced DRP1 fission activity combine to prevent mitophagy in cells that are dependent on oxidative phosphorylation. J Cell Sci. 2014;127:2313-2325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 23. | Arasaki K, Shimizu H, Mogari H, Nishida N, Hirota N, Furuno A, Kudo Y, Baba M, Baba N, Cheng J. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev Cell. 2015;32:304-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 119] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 24. | Babcook MA, Shukla S, Fu P, Vazquez EJ, Puchowicz MA, Molter JP, Oak CZ, MacLennan GT, Flask CA, Lindner DJ. Synergistic simvastatin and metformin combination chemotherapy for osseous metastatic castration-resistant prostate cancer. Mol Cancer Ther. 2014;13:2288-2302. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Packiriswamy N, Steury M, McCabe IC, Fitzgerald SD, Parameswaran N. Bacterial Dose-Dependent Role of G Protein-Coupled Receptor Kinase 5 in Escherichia coli-Induced Pneumonia. Infect Immun. 2016;84:1633-1641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 26. | Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533-543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 572] [Cited by in F6Publishing: 614] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 27. | Dagda RK, Cherra SJ, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843-13855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 774] [Cited by in F6Publishing: 756] [Article Influence: 50.4] [Reference Citation Analysis (0)] |
| 28. | Burkewitz K, Morantte I, Weir HJ, Yeo R, Zhang Y, Huynh FK, Ilkayeva OR, Hirschey MD, Grant AR, Mair WB. Neuronal CRTC-1 governs systemic mitochondrial metabolism and lifespan via a catecholamine signal. Cell. 2015;160:842-855. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 29. | Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, Becker C, Altmueller J, Frommolt P, Sardiello M. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol. 2014;207:213-223. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 30. | Gitter AH, Bendfeldt K, Schulzke JD, Fromm M. Leaks in the epithelial barrier caused by spontaneous and TNF-alpha-induced single-cell apoptosis. FASEB J. 2000;14:1749-1753. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in F6Publishing: 204] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 31. | Mashukova A, Wald FA, Salas PJ. Tumor necrosis factor alpha and inflammation disrupt the polarity complex in intestinal epithelial cells by a posttranslational mechanism. Mol Cell Biol. 2011;31:756-765. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in F6Publishing: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 32. | Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14:329-342. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1545] [Cited by in F6Publishing: 1693] [Article Influence: 169.3] [Reference Citation Analysis (1)] |
| 33. | Daig R, Rogler G, Aschenbrenner E, Vogl D, Falk W, Gross V, Schölmerich J, Andus T. Human intestinal epithelial cells secrete interleukin-1 receptor antagonist and interleukin-8 but not interleukin-1 or interleukin-6. Gut. 2000;46:350-358. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 64] [Cited by in F6Publishing: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 34. | Korolkova OY, Myers JN, Pellom ST, Wang L, M’Koma AE. Characterization of Serum Cytokine Profile in Predominantly Colonic Inflammatory Bowel Disease to Delineate Ulcerative and Crohn’s Colitides. Clin Med Insights Gastroenterol. 2015;8:29-44. [PubMed] [Cited in This Article: ] |
| 35. | Haddad JJ. Antioxidant and prooxidant mechanisms in the regulation of redox(y)-sensitive transcription factors. Cell Signal. 2002;14:879-897. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 285] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 36. | Lewis K, Caldwell J, Phan V, Prescott D, Nazli A, Wang A, Soderhölm JD, Perdue MH, Sherman PM, McKay DM. Decreased epithelial barrier function evoked by exposure to metabolic stress and nonpathogenic E. coli is enhanced by TNF-alpha. Am J Physiol Gastrointest Liver Physiol. 2008;294:G669-G678. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 37. | Nazli A, Yang PC, Jury J, Howe K, Watson JL, Söderholm JD, Sherman PM, Perdue MH, McKay DM. Epithelia under metabolic stress perceive commensal bacteria as a threat. Am J Pathol. 2004;164:947-957. [PubMed] [Cited in This Article: ] |
| 38. | Song IS, Jeong YJ, Jeong SH, Heo HJ, Kim HK, Bae KB, Park YH, Kim SU, Kim JM, Kim N. FOXM1-Induced PRX3 Regulates Stemness and Survival of Colon Cancer Cells via Maintenance of Mitochondrial Function. Gastroenterology. 2015;149:1006-1016.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 79] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 39. | Picard M, Shirihai OS, Gentil BJ, Burelle Y. Mitochondrial morphology transitions and functions: implications for retrograde signaling? Am J Physiol Regul Integr Comp Physiol. 2013;304:R393-R406. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 177] [Cited by in F6Publishing: 208] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 40. | Novak EA, Mollen KP. Mitochondrial dysfunction in inflammatory bowel disease. Front Cell Dev Biol. 2015;3:62. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 124] [Cited by in F6Publishing: 143] [Article Influence: 15.9] [Reference Citation Analysis (0)] |