Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2373
Peer-review started: May 12, 2015
First decision: July 19, 2015
Revised: August 12, 2015
Accepted: November 30, 2015
Article in press: November 30, 2015
Published online: February 21, 2016
AIM: To investigate the role of single nucleotide polymorphisms (SNPs) in CD24 gene in susceptibility and overall survival of gastric cancer (GC).
METHODS: We genotyped 3 tagging SNPs of CD24-P-534 in the promoter region, P170 in the coding region of exon 2 and P1527 in the 3′ untranslated region - using polymerase chain reaction-restriction fragment length polymorphism in specimens from 679 histologically-confirmed GC cases, 111 gastric atrophy (GA) cases and 976 tumor-free controls. Serum immunoglobulin G antibodies to Helicobacter pylori (H. pylori) of all subjects were detected by enzyme-linked immunosorbent assay. CD24 expression was evaluated by immunohistochemistry in 131 GC specimens. Correlations between SNPs and risk of GC or GA were shown by P values and odd ratios (ORs) with 95% confidence intervals (95%CI) compared with the most common genotype of each SNP using the unconditional logistic regression model after adjusting for age, sex and H. pylori infection. Survival within each SNP group was plotted by Kaplan-Meier method and compared by log-rank test (recessive model). Hazard ratios with 95%CIs were computed by Cox regression model after adjusting for age, sex, histological type, tumor differentiation, clinical stage and post-operational chemotherapy.
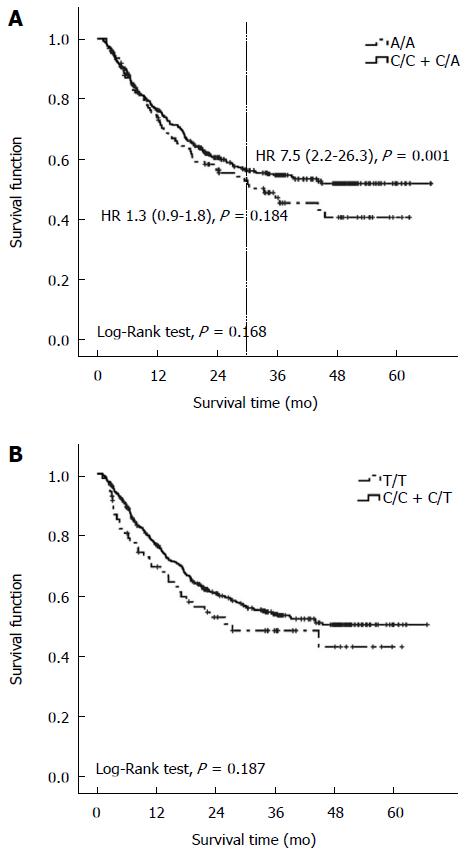
RESULTS: All of the three loci were in Hardy-Weinberg equilibrium in the control group. Median follow-up time for the 600 GC patients included in the survival analysis was 36.2 mo (range, 2.1-66.7 mo; 95%CI: 34.3-36.5 mo). Patients with the P-534 A/A genotype had significantly shorter survival (HR = 1.38, 95%CI: 1.01-1.88, P = 0.042) than did the C/C or C/A genotype carriers after adjusting for age, sex, histological type, tumor differentiation, clinical stage and post-operational chemotherapy. This trend was more evident in patients who lived longer than 2.5 years (HR = 7.55, 95%CI: 2.16-26.32, P = 0.001). The P170 T/T genotype was associated with a shorter lifespan than the non-T/T genotypes, but not significantly so. None of the three genetic variants was found to be associated with risk of GC (including tumor stage, grade and distant metastasis) or with risk of gastric atrophy. Furthermore, no difference of CD24 expression was found among the genotypes.
CONCLUSION: The P-534 site in CD24 gene affects the overall survival of gastric cancer and may serve as a prognostic marker for gastric cancer.
Core tip: We evaluated the role of three genetic variants of CD24 in gastric cancer (GC) risk and prognosis using 679 GC cases and 976 controls. We observed that GC cases with the A/A genotype of P-534 (which lies in the CD24 promoter) had a significantly shorter survival (HR = 1.38) especially among patients who lived longer than 2.5 years (HR = 7.55) after adjusting for age, sex, histological type, tumor differentiation, clinical stage and post-operational chemotherapy. Our study provides the first evidence that P-534 site in CD24 may serve as a prognostic marker for gastric cancer.
- Citation: Jia ZF, Wang LZ, Cao XY, Wang C, Cao DH, Wu X, You LL, Jin MS, Wang YP, Zhou BS, Jiang J. CD24 genetic variants contribute to overall survival in patients with gastric cancer. World J Gastroenterol 2016; 22(7): 2373-2382
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2373.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2373
Gastric cancer (GC) is one of the most common malignancies and the third cause of cancer-related death worldwide[1]. Helicobacter pylori (H. pylori) infection has been established to cause GC and was classified as carcinogenic to humans (Group 1) by IARC in 1994[2]. Although over half of the world’s population are estimated to be infected with H. pylori[3], relatively few develop GC, and gastric damages induced by H. pylori infection vary widely, which together imply a role for host genetic factors in response to chronic H. pylori infection and subsequent GC development.
CD24 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface glycoprotein with functions in signal transduction and cell adhesion. It is expressed in a large variety of human malignancies. Over-expression of CD24 results in a more aggressive malignant phenotype with greater proliferative and cell migration capabilities, whereas its down-expression shows a less malignant phenotype[4-6]. CD24 is reportedly associated with tumor growth, invasion, metastasis, recurrence and treatment response in various cancers, including breast cancer[7,8], prostate cancer[9], colorectal cancer[10,11], hepatocellular carcinoma[12], esophageal squamous cell carcinoma[13] and GC[14,15]. CD24 is also a potential marker of cancer stem cells, which possess capabilities for tumorigenesis, self-renewal and producing differentiated progeny[16-18].
In GC, CD24 can mediate carcinogenesis and promote GC progression[14,15]. CD24 expression gradually increases in the progression of normal gastric mucosa, non-atrophic chronic gastritis, chronic atrophic gastritis (CAG), CAG with intestinal metaplasia, dysplasia and finally, GC[14]. Mice with normal CD24 expression show more gastric inflammation, parietal cell atrophy and gland hyperplasia following Helicobacter felis infection compared with CD24-null mice which have the genetic background of inbred CD24-normal mice[19].
Given the important role of CD24 in GC, we tested whether single nucleotide polymorphisms (SNPs) in the CD24 gene are associated with genetic susceptibility to GC tumor progression and prognosis in a Chinese population.
From July 2008 to December 2012, patients with histologically diagnosed GC who underwent tumorectomies at the Department of Gastric and Colorectal Surgery of the First Hospital of Jilin University were invited to join this study. Patients with gastric atrophy (GA) and controls with no tumor history were recruited at the Physical Examination Center of the same hospital during the same period. A total of 1766 individuals, 679 GC cases, 111 GA cases and 976 controls signed the informed consent forms and agreed to participate in the study. The study protocol was reviewed and approved by the Ethics Committee of the First Hospital of Jilin University.
Gastric cancer cases were followed-up by telephone calls three months, six months, and one year after each patient’s tumorectomy and every one year thereafter until the end of the study or the death of the patients. Cases would not be included in the survival analysis if (1) they were lost to follow-up by the first telephone interview; or (2) they died of surgical complications in the perioperative period. Survival time was defined as the duration from the date of surgery to the date of death if the patients died, or to the date of the last successful interview if the patients were lost to follow-up or alive until the end of the study. Survival time was right-censored except for patients who died of GC.
Treatment information after surgery was also collected during the follow-up period. Post-operational chemotherapy is defined as at least 3 cycles of chemotherapy received after surgery. One third of the GC patients received this type of therapy. The treatment was classified into three regimens: FOLFOX-4 (combination of 5-fluorouracil, leucovorin and oxaliplatin); XELOX (capecitabine and oxaliplatin) and “other” (such as capecitabine or 5-fluorouracil alone).
Blood samples were collected in EDTA tubes and stored at -80 °C until DNA extraction. Genomic DNA was isolated following the protocol provided by the manufacturer (Axygen Biosciences, United States).
The full length of the CD24 gene was first identified by our previous study and was mapped to Chromosome 6q21 by fluorescence in situ hybridization (submitted to NCBI database, accession number FJ226006)[20]. Although SNP data on CD24 was unavailable in HapMap project or dbVar database, we have identified three linkage disequilibrium (LD) blocks that cover the promoter region and exons 1-2 of the CD24 locus[20]. Haplotype tagging SNPs (tag SNPs) were identified from the three LD blocks with the pairwise r2≥ 0.9 and miner allele frequency > 0.05.
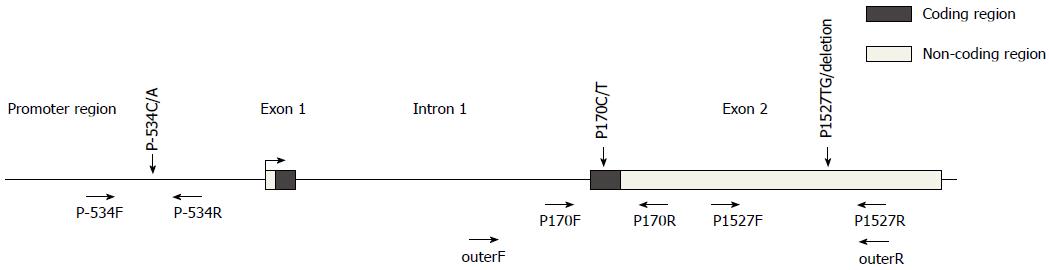
Three tagging SNPs, P-534C/A, P170C/T and P1527TG/del were genotyped to evaluate the association of CD24 and genetic susceptibility to GC. P-534C/A is located in the promoter region of CD24 and is 534bp away from the translation-starting site. P170C/T is located in the coding region of exon 2 and its C-to-T transition leads to an alanine to valine substitution at codon 57 of the CD24 protein. P1527TG/del, 1527bp down from the translation-starting site, is located in the 3’ untranslated region.
Genotypes of the selected sites were determined by polymerase chain reaction- restriction fragment length polymorphism (PCR-RFLP) as described by Li et al[21] and our previous study[20] (Primers used are listed in Table 1 and Figure 1 and were synthesized by Takara, Dalian, China). Briefly, the PCR products of P-534 were digested with endonuclease BsrFI overnight at 37 °C and the restriction site indicated the presence of the C allele (126 bp and 83 bp) (All restriction endonucleases were bought from New England Biolabs, United States). For P170 and P1527, a nested PCR in which one of the primers for the first PCR mapped to an intron was performed to increase specificity as the exon 2 of CD24 shows high homology with the intronless pseudogenes in Chromosomes 1, 15 and Y[22]. The second PCRs were amplified independently for P170 and P1527 using the 1000-fold-diluted products of the first PCR as templates (all reagents for PCR were from Tiangen, Beijing, China). These PCR products were then digested overnight with the restriction enzyme BstXI (for P170, 37 °C) and BsrI (for P1527, 65 °C). The T allele of P170 produced two fragments,275 bp and 129 bp; the TG allele of P1527 produced 645 and 202 bp fragments. All products were separated by electrophoresis on 1.5% agarose gels with ethidium bromide staining and scanned on gel imaging system (Gel DocTM XR+ system, Bio-Rad, United States). Fifty samples were randomly selected to be genotyped by direct sequencing to confirm the validity of PCR-RFLP; three of them with different genotypes were used as positive controls for each PCR-RFLP run. The overall concordance was 100%.
| Primer | Sequence | Length of product | Endonucleases | Bands |
| P-534F | 5’-AGAGATAACCCTGCCCGAG-3’ | 209 bp | BsrFI | C: 126 bp + 83 bp |
| P-534R | 5’-CCAAGTTTCCTTTGTTTCCC-3’ | |||
| outerF | 5’-CCACTTGGCATTTTTGAGGCATCT-3’ | 1882 bp | - | - |
| outerR | 5’-TGTGTCGAGGCAGTTGTAAAAG-3’ | |||
| P170F | 5’-CTAAAGAGAATGACCTTGGTGGGTTGAG-3’ | 404 bp | BstXI | T: 275 bp + 129 bp |
| P170R | 5’-GGATTGGGTTTAGAAGATGGGGAAA-3’ | |||
| P1527F | 5’-GCCAGGGCAATGATGAATGAG-3’ | 847 bp | BsrI | TG: 645 bp + 202 bp |
| P1527R | 5’-TGTGTCGAGGCAGTTGTAAAAGAT-3’ |
Serum immunoglobulin G (IgG) antibodies to H. pylori were detected by enzyme-linked immunosorbent assay (ELISA) using H. pylori-IgG ELISA kits (Biohit, Finland) according to the manufacturer’s protocol. Titers higher than the cut off value of 30 EIU were considered positive for H. pylori infection. The inter-day coefficient variations (CV) of the negative and the positive control samples were 4.5% and 1.4%, respectively.
Diagnosis of GA was described elsewhere[23]. Briefly, serum pepsinogen I (PGI) and II (PGII) were quantified by ELISA kits (Biohit, Finland). Individuals with PGI < 82.3 ng/mL and PGI/PGII < 6.05 were diagnosed as GA.
CD24 expression was assessed in tumor tissue of 131 gastric cancer patients by immunohistochemistry (IHC) method. The detailed procedure is described elsewhere[24]. Briefly, the 4-μm-wide tissue sections were deparaffinized and stained using a streptavidin-biotin immunoperoxidase technique. All slides were then incubated with anti-human CD24 polyclonal antibody (1:100 diluted, sc-7034, Santa Cruz, United States) and developed by 3, 3-diaminobenzidine (DAB). As negative controls, the slides were treated with the IgG isotypes in place of primary antibodies and all negative controls demonstrated negligible background staining. The stained slides were independently evaluated by two pathologists (MSJ and YPW) who were blinded to clinical data and outcomes. The HSCORE system was used to assess the staining results and was calculated by a following equation: HSCORE = ΣPi(i) (I = 0, 1, 2, 3, Pi = 0-100). The i means the intensity of staining (no staining: 0; weak staining: 1; moderate staining: 2; and strong staining: 3). Pi represents percentages (0-100) of stained cells with intensities. The HSCORE ranges from 0 to 300.
Continuous data were summarized as medians (25th to 75th percentiles) and compared by Mann-Whitney U test or Kruskal-Wallis test. Categorical variables were described as frequencies and percentages and compared using χ2-test. Correlations between SNPs and risk of GC or GA were demonstrated by P values and odd ratios (ORs) with 95% confidence intervals (95%CI) compared with the most common genotype of each SNP. The P values and ORs with 95%CIs were calculated using the unconditional logistic regression model after adjusting for age, sex and H. pylori infection. Survival functions of the GC patients within each SNP were plotted by Kaplan-Meier method and compared by log-rank test using the recessive model. Hazard ratios (HRs) with 95%CIs were used to quantify the influence of genotypes of each SNP on overall survival and were calculated with Cox regression model after adjusting for age, sex, histological type, tumor differentiation, clinical stage and post-operational chemotherapy. For haplotypes with frequencies > 1%, their associations with risk of GC or GA were assessed compared to the most common haplotype using the logistic regression model with the HAPSTAT software 3.0[25]. Unless otherwise stated, analyses were performed in SAS 9.1.3 software (SAS Institute Inc, United States). A two-tailed P value < 0.05 was considered to be statistically significant.
A total of 679 GC cases, 111 GA cases and 976 tumor-free controls were included in the study. The baseline characteristics of the subjects are summarized in Table 2. The GC group was oldest in the three group and the control group was youngest (median age: 61.0 years vs 50.0 years vs 48.5 years, P < 0.001 for all pairwise comparisons). And there were more males in the cancer group (71.7% vs 59.5% vs 59.2%, P < 0.001 for comparisons to the atrophy cases or the controls). In the GC group, 67.8% were positive for H. pylori infection, significantly higher than the control group (49.7%, P < 0.001) but non-significantly lower than the GA group (75.7%, P = 0.097). Therefore, comparisons of genotype distribution below were adjusted by age, sex and H. pylori infection.
| Cancer | Atrophy | Controls | P value | |
| n | 679 | 111 | 976 | |
| Gender | ||||
| Male | 487 (71.7) | 66 (59.5) | 578 (59.2) | < 0.0001 |
| Female | 192 (28.3) | 45 (40.5) | 398 (40.8) | |
| Age (yr) | 61.0 (54.0-70.0) | 50.0 (47.0-57.0) | 48.5 (44.0-55.0) | < 0.0001 |
| ≤ 55 | 204 (30.0) | 76 (68.5) | 756 (77.5) | < 0.0001 |
| 55-70 | 310 (45.7) | 28 (25.2) | 181 (18.5) | |
| > 70 | 165 (24.3) | 7 (6.3) | 39 (4.0) | |
| H. pylori | ||||
| Positive | 453 (67.8) | 84 (75.7) | 478 (49.7) | < 0.0001 |
| Negative | 215 (32.2) | 27 (24.3) | 483 (50.3) | |
| Differentiation | ||||
| Poor | 437 (68.4) | |||
| Moderate to well | 202 (31.6) | |||
| Pathologic type | ||||
| Tubular adenocarcinoma | 545 (82.2) | |||
| Signet ring cell | 50 (7.5) | |||
| Other | 68 (10.3) | |||
| TNM stage | ||||
| I | 97 (15.0) | |||
| II | 227 (35.1) | |||
| III | 224 (34.6) | |||
| IV | 99 (15.3) | |||
| Distant metastasis | ||||
| Positive | 573 (85.3) | |||
| Negative | 99 (14.7) | |||
| Post-operational Chemotherapy | ||||
| No | 456 (67.2) | |||
| FOLFOX-4 | 100 (14.7) | |||
| XELOX | 47 (6.9) | |||
| Other | 76 (11.2) | |||
The GC cases were mainly of tubular adenocarcinoma type (82.2%), with low-grade differentiation (68.4%), at TNM stage II (35.1%) or III (34.6%). One third of the cases received chemotherapy after operation (32.8%); 14.7% received FOLFOX-4 and 6.9% received XELOX.
All of the three SNPs were in Hardy-Weinberg equilibrium in the control group (P-534: P = 0.612; P170: P = 0.413; P1527: P = 0.423). Distributions of genotypes and alleles are listed in Table 3. Compared with the most common genotype of each SNP, no difference was observed for the distributions of the three loci between the GC and control groups after adjusting for age, sex and H. pylori infection. No allele or haplotype was associated with risk of GC. Similar negative results were obtained for GA risk (Table 3). Moreover, no associations were observed between SNPs and risk of H. pylori infection (data not shown).
| Controls | Cancer | P value | OR1 (95%CI) | Atrophy | P value | OR1 (95%CI) | |
| P-534 | |||||||
| C/C | 271 (27.8) | 181 (26.7) | - | Reference | 29 (26.1) | - | Reference |
| C/A | 488 (50.0) | 358 (52.7) | 0.099 | 1.1 (0.86-1.48) | 62 (55.9) | 0.204 | 1.2 (0.74-1.91) |
| A/A | 217 (22.2) | 140 (20.6) | 0.145 | 0.9 (0.61-1.20) | 20 (18.0) | 0.328 | 0.8 (0.46-1.54) |
| C | 1030 (52.8) | 720 (53.0) | 0.886 | - | 130 (54.1) | 0.716 | - |
| A | 922 (47.2) | 638 (47.0) | 1.0 (0.86-1.14) | 102 (45.9) | 0.9 (0.72-1.25) | ||
| P170 | |||||||
| C/C | 419 (42.9) | 295 (43.4) | - | Reference | 60 (54.1) | - | Reference |
| C/T | 439 (45.0) | 308 (45.4) | 0.277 | 1.1 (0.82-1.38) | 37 (33.3) | 0.082 | 0.6 (0.40-0.96) |
| T/T | 118 (12.1) | 76 (11.2) | 0.369 | 0.9 (0.60-1.29) | 14 (12.6) | 0.724 | 0.9 (0.47-1.65) |
| C | 1277 (65.4) | 898 (66.1) | 0.673 | - | 157 (70.7) | 0.114 | - |
| T | 675 (34.6) | 460 (33.9) | 1.0 (0.83-1.12) | 65 (29.3) | 0.8 (0.58-1.06) | ||
| P1527 | |||||||
| TG/TG | 826 (84.6) | 559 (82.3) | - | Reference | 95 (85.6) | - | Reference |
| TG/del | 142 (14.6) | 117 (17.2) | 0.069 | 1.3 (0.97-1.82) | 15 (13.5) | 0.940 | 0.9 (0.50-1.59) |
| del/del | 8 (0.8) | 3 (0.4) | 0.162 | 0.4 (0.09-1.77) | 1 (0.9) | 0.935 | 0.9 (0.10-7.30) |
| TG | 1794 (91.9) | 1235 (90.9) | 0.956 | - | 205 (92.3) | 0.821 | - |
| del | 158 (8.1) | 123 (9.1) | 1.1 (0.88-1.45) | 17 (7.7) | 0.9 (0.56-1.58) | ||
| Haplotype2 | |||||||
| ACTG | 45.6 | 47.0 | - | Reference | 45.4 | - | Reference |
| CTTG | 33.6 | 33.7 | 0.946 | 1.0 (0.85-1.16) | 28.7 | 0.407 | 0.9 (0.63-1.21) |
| CCTG | 11.4 | 10.2 | 0.382 | 0.9 (0.71-1.14) | 17.7 | 0.072 | 1.6 (0.99-2.37) |
| CCdel | 8.0 | 9.1 | 0.370 | 1.1 (0.87-1.46) | 7.7 | 0.919 | 1.0 (0.56-1.67) |
Genotypic distributions of SNPs were analyzed by clinicopathologic parameters such as histological type, tumor differentiation, TNM stage and distant metastasis in GC cases. However, no significant association was observed (Table 4).
| P-534 | P value | P170 | P value | P1527 | P value | ||||||
| C/C | C/A | A/A | C/C | C/T | T/T | TG/TG | TG/del | ||||
| n | 181 | 358 | 140 | 295 | 308 | 76 | 559 | 120 | |||
| Age | 60 (53-70) | 61 (54-71) | 61 (55-70) | 0.715 | 61 (53-71) | 61 (54-70) | 63 (56-70) | 0.658 | 61 (54-70) | 60 (51-71) | 0.307 |
| Sex | |||||||||||
| Male | 27.3 | 50.3 | 22.4 | 0.092 | 43.9 | 44.2 | 11.9 | 0.485 | 83.6 | 16.4 | 0.172 |
| Female | 25.0 | 58.9 | 16.1 | 42.2 | 48.4 | 9.4 | 79.2 | 20.8 | |||
| H. pylori | |||||||||||
| Positive | 26.3 | 53.4 | 20.3 | 0.905 | 42.6 | 46.1 | 11.3 | 0.769 | 82.3 | 17.7 | 0.996 |
| Negative | 27.0 | 51.6 | 21.4 | 45.6 | 43.7 | 10.7 | 82.3 | 17.7 | |||
| Differentiation | |||||||||||
| Poor | 24.7 | 55.8 | 19.5 | 0.070 | 43.5 | 47.4 | 9.2 | 0.055 | 81.5 | 18.5 | 0.500 |
| Moderate to well | 30.2 | 46.0 | 23.8 | 45.0 | 40.1 | 14.8 | 83.7 | 16.3 | |||
| TNM stage | |||||||||||
| I-II | 26.2 | 52.8 | 21.0 | 0.999 | 45.1 | 44.7 | 10.2 | 0.695 | 82.4 | 17.6 | 0.986 |
| III-IV | 26.3 | 52.6 | 21.1 | 42.7 | 45.2 | 12.1 | 82.4 | 17.6 | |||
| Pathologic type | |||||||||||
| Tubular adenocarcinoma | 26.4 | 52.3 | 21.3 | 0.754 | 45.0 | 43.9 | 11.2 | 0.173 | 82.8 | 17.2 | 0.751 |
| Signet ring cell | 26.0 | 60.0 | 14.0 | 34.0 | 60.0 | 6.0 | 80.0 | 20.0 | |||
| Other | 27.9 | 50.0 | 22.1 | 38.2 | 47.1 | 14.7 | 85.3 | 14.7 | |||
| Distant metastasis | |||||||||||
| Positive | 32.3 | 48.5 | 19.2 | 0.323 | 40.4 | 44.4 | 15.1 | 0.374 | 79.8 | 20.2 | 0.454 |
| Negative | 25.1 | 53.9 | 20.9 | 44.3 | 45.2 | 10.5 | 82.9 | 17.1 | |||
| CD24 staining | 60 (0-120) | 60 (0-120) | 100 (20-140) | 0.215 | 60 (0-120) | 60 (20-120) | 40 (0-100) | 0.482 | 60 (0-120) | 60 (40-100) | 0.922 |
Follow-up information was available for 610 of the 679 GC patients (89.8%). Ten patients died of postoperative complications within 30 d at the beginning of the study period (range, 0-29 d, median: 15.5 d) and these cases were excluded from analyses of effects of SNPs on survival. The median follow-up time for the remaining 600 GC patients was 36.2 mo (range, 2.1-66.7 mo; 95%CI: 34.3-36.5 mo). Two hundred and sixty patients (43.3%) died from GC during the follow-up, 272 patients (45.3%) lived and 68 (11.3%) died of other causes or were lost to follow up.
Survival curves were plotted and compared according to genotypes of each SNP using the recessive model. The patients who carried the A/A genotype of P-534 had shorter survival than those carrying C/C or C/A after adjusting for age, sex, histological type, tumor differentiation, clinical stage and post-operational chemotherapy (HR = 1.38, 95%CI: 1.01-1.88, P = 0.042; Table 5 and Figure 2A). This trend was more evident in patients who lived longer than 2.5 years (HR = 7.55, 95%CI: 2.16-26.32, P = 0.001). Similarly, the P170 T/T carriers tended to have shorter survival time than the C/C or C/T carriers, although not significantly so (Figure 2B). No meaningful correlation could be observed between the variation of “TG” deletion in P1527 and GC survival (P = 0.799).
| P value | HR | 95%CI | |
| P-534 | |||
| C/C + C/A | - | 1.00 | - |
| A/A | 0.0416 | 1.38 | 1.01-1.88 |
| Age | 0.1875 | 1.01 | 1.00-1.02 |
| Sex | |||
| Male | - | 1.00 | - |
| Female | 0.2315 | 0.83 | 0.61-1.13 |
| Differentiation | |||
| Moderate to well | - | 1.00 | - |
| Poor | 0.0089 | 1.50 | 1.11-2.04 |
| Pathologic type | |||
| Tubular adenocarcinoma | - | 1.00 | - |
| Signet ring cell | 0.4750 | 0.83 | 0.50-1.39 |
| Other | 0.9469 | 1.01 | 0.67-1.53 |
| TNM stage | |||
| I | - | 1.00 | - |
| II | 0.0003 | 5.50 | 2.18-13.86 |
| III | < 0.0001 | 22.29 | 9.05-54.89 |
| IV | < 0.0001 | 32.12 | 12.63-81.70 |
| Chemotherapy | |||
| No | - | 1.00 | - |
| FOLFOX-4 | 0.0195 | 0.64 | 0.45-0.93 |
| XELOX | 0.0021 | 0.43 | 0.25-0.74 |
| Other | 0.0004 | 0.45 | 0.29-0.70 |
Multivariate Cox regression analysis showed that three other factors-degree of differentiation, TNM stage and post-operational chemotherapy-were associated with the prognosis of GC. Patient whose GC had low-grade differentiation or advanced clinical stage or who did not received post-operational chemotherapy had shorter survival time (Table 5).
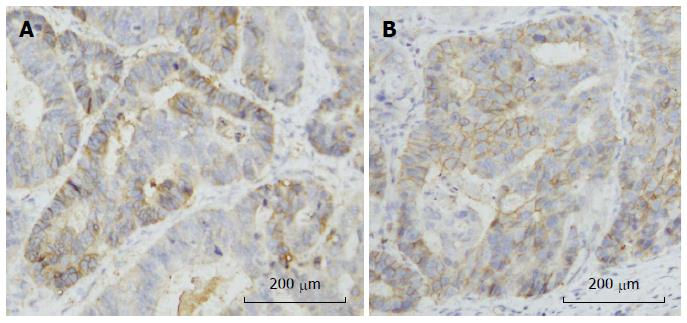
To assess whether SNPs were associated production of CD24 protein, CD24 expression was evaluated in cancerous tissue of 131 GC cases using IHC. Genotypic distributions of the three SNPs in selected cancer cases were similar to non-selected cases (data not shown). CD24 expression was seen mainly in membranes of tumor cells (Figure 3). However, CD24 expression did not observably differ among genotypes of each SNP (Figure 3 and Table 3).
In this study, we explored the association between variants of CD24 gene and GC. We found that patients who harbored the P-534 A/A genotype tended to have shorter survival than those who carry P-534 non-A/A genotypes.
This is the first study on the association between SNPs of CD24 gene and GC, as no CD24 SNPs were included in any genome-wide association studies of GC[26,27]. Distribution of the P-534 genotypes of CD24 differs slightly from that of Caucasian populations. The minor allele C of P-534 in Caucasian population (37.2%[20]) was the major allele (52.8%) in Han Chinese in our study. Distributions of P170 and P1527 were similar to those of other ethnicities[28,29].
Numerous studies have reported that SNPs of CD24 gene are correlated with risk of various autoimmune diseases, such as systemic lupus erythematosus (SLE)[28-31],multiple sclerosis[28,32-35] and inflammatory bowel disease[20,36]. Li et al[21] reported that P170 and P1527 of CD24 affected risk and progression of chronic hepatitis B infection and Sheng et al[37] showed that the T/T genotype of P170 correlated with a 2.96-fold increased of risk of hepatocellular carcinoma. In our study, however, we did not observe any influence of CD24 polymorphisms on risk of H. pylori infection (data not shown, but available on request), gastric atrophy, precancerous lesions of GC that are induced partly by H. pylori infection[38,39], or GC (Table 3).
Polymorphisms of CD24 have been related to prognosis in several cancers. In breast cancer, CD24 expression was associated with adverse prognosis[7,8] and CD24 P170 polymorphism could predict response to chemotherapy[40-42]. In esophageal cancer, P170 of CD24 was involved in regional lymph node metastasis[43]. In our study, we found that P-534 of CD24 affected long-term survival of GC, as P-534 A/A genotype carriers have a 7.5-fold increased mortality risk compared wirh non-A/A carriers among patients who lived longer than 2.5 years (Figure 2A). P170 T/T carriers also tended to have shorter survival than did non-T/T carriers, although not significantly so (Figure 2B).
The P-534, which is located in the promoter region of CD24, may influence transcriptional activity. Our previous work showed that a hypomorphic haplotype that contained the C allele of P-534 was associated with risk of multiple sclerosis and this haplotype was involved in higher transcriptional activity and increased expression of CD24 in peripheral blood lymphocytes[20]. The non-synonymous variant P170 may alter the quantity and quality of CD24. Zhou et al[32] found that the P170T/T genotype expressed more cell-surface CD24 than did the C/T or C/C genotypes using flow cytometry, which showed an increased risk and more rapid progression of multiple sclerosis. In our study, we evaluated CD24 expression using IHC and found that CD24 was expressed mainly in the membranes of GC cells. However, we did not observe any differences of CD24 expression among genotypes of P-534, P170 or P1527.
Three limitations should be noted in our study. The first one is that some baseline characteristics such as age are different among the three study groups, and we cannot rule out the possibility that individuals in the control group could develop GC when they are older. However, we control the potential influences to the greatest extent by adjusting for these factors in the multivariate analysis. The second is that the follow-up time of the GC cases seems insufficient because most of cases on the right side of the survival plots are censored. This may explain that although individuals carrying P-534 A/A genotype tended to have shorter observable survival time, P value from log-rank tests is not significant. However, when we adjust the potential influencing factors and divide the patients to subgroups who live shorter than 2.5 years and those who live longer than 2.5 years, the association of P-534 with long-term survival is statistically significant (HR = 7.5, 95%CI: 2.2-26.3). Nonetheless, longer follow-up time is needed to re-evaluate the role of CD24 SNPs in prognosis of GC. The last one is that we semi-quantified CD24 expression in tissue using IHC and the influence of SNPs on CD24 might be offset, as protein production can be regulated by factors known and unknown e.g., regulations of transcription, post-transcription and translation. Therefore, more rigorous design should be applied in our future study.
In summary, we find that polymorphisms of the CD24 gene affect the overall survival of GC, as patients who bear the P-534 A/A genotype tend to have shorter survival than do patients with P-534 non-A/A genotypes. However, we do not observe any associations between CD24 SNPs and risk of H. pylori infection, GA or GC. More studies with larger samples and longer follow-up time are needed to clarify the role of CD24 in GC.
The authors would like to thank all of those who participated in this study, especially to Ying Song for her work on following-up of subjects. The authors would also like to thank the Biospecimen Bank of the First Hospital of Jilin University for their kindness in supplying the specimens to our research.
CD24 expression as a potential biomarker is associated with poor prognosis in patients with gastric cancer (GC) but its genetic basis still remains to be elucidated.
GC is one of the most common malignancies and the third cause of cancer-related death worldwide. CD24 mediates gastric carcinogenesis and promotes GC progression. Elucidation of the association between CD24 genetic variants and risk and prognosis of GC may provide novel biomarkers to discriminate individuals with higher risk of GC or to predict prognosis for GC cases.
This study for the first time evaluates the effect of CD24 genetic variations in GC carcinogenesis and prognosis. Its results indicate that CD24 variants may serve as a marker for GC prognosis, but not for carcinogenesis.
P-534 site of CD24 might be used as a prognostic predictive marker for GC.
CD24 is a glycosylphosphatidylinositol-anchored cell-surface glycoprotein with functions in signal transduction and cell adhesion. CD24 over-expression is associated with a more aggressive malignant phenotype of greater proliferative and migration capability; down-expression shows a less malignant phenotype.
This study investigated the role of CD24 genetic variants in susceptibility and overall survival of GC and shows that P-534 of CD24 may serve as an independent prognostic marker for GC.
P- Reviewer: Zhang J S- Editor: Yu J L- Editor: A E- Editor: Liu XM
| 1. | Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer 2013; Available from: http://globocan.iarc.fr, accessed on 15/01/2015. [Cited in This Article: ] |
| 2. | Schistosomes , liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7-14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1-241. [PubMed] [Cited in This Article: ] |
| 3. | Cao XY, Jia ZF, Jin MS, Cao DH, Kong F, Suo J, Jiang J. Serum pepsinogen II is a better diagnostic marker in gastric cancer. World J Gastroenterol. 2012;18:7357-7361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 27] [Cited by in F6Publishing: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 4. | Naumov I, Zilberberg A, Shapira S, Avivi D, Kazanov D, Rosin-Arbesfeld R, Arber N, Kraus S. CD24 knockout prevents colorectal cancer in chemically induced colon carcinogenesis and in APC(Min)/CD24 double knockout transgenic mice. Int J Cancer. 2014;135:1048-1059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 5. | Baumann P, Cremers N, Kroese F, Orend G, Chiquet-Ehrismann R, Uede T, Yagita H, Sleeman JP. CD24 expression causes the acquisition of multiple cellular properties associated with tumor growth and metastasis. Cancer Res. 2005;65:10783-10793. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 246] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 6. | Sagiv E, Starr A, Rozovski U, Khosravi R, Altevogt P, Wang T, Arber N. Targeting CD24 for treatment of colorectal and pancreatic cancer by monoclonal antibodies or small interfering RNA. Cancer Res. 2008;68:2803-2812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 7. | Horiguchi K, Toi M, Horiguchi S, Sugimoto M, Naito Y, Hayashi Y, Ueno T, Ohno S, Funata N, Kuroi K. Predictive value of CD24 and CD44 for neoadjuvant chemotherapy response and prognosis in primary breast cancer patients. J Med Dent Sci. 2010;57:165-175. [PubMed] [Cited in This Article: ] |
| 8. | Athanassiadou P, Grapsa D, Gonidi M, Athanassiadou AM, Tsipis A, Patsouris E. CD24 expression has a prognostic impact in breast carcinoma. Pathol Res Pract. 2009;205:524-533. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Wang L, Liu R, Ye P, Wong C, Chen GY, Zhou P, Sakabe K, Zheng X, Wu W, Zhang P. Intracellular CD24 disrupts the ARF-NPM interaction and enables mutational and viral oncogene-mediated p53 inactivation. Nat Commun. 2015;6:5909. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 10. | Sahlberg SH, Spiegelberg D, Glimelius B, Stenerlöw B, Nestor M. Evaluation of cancer stem cell markers CD133, CD44, CD24: association with AKT isoforms and radiation resistance in colon cancer cells. PLoS One. 2014;9:e94621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 146] [Cited by in F6Publishing: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 11. | Weichert W, Denkert C, Burkhardt M, Gansukh T, Bellach J, Altevogt P, Dietel M, Kristiansen G. Cytoplasmic CD24 expression in colorectal cancer independently correlates with shortened patient survival. Clin Cancer Res. 2005;11:6574-6581. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 12. | Yang XR, Xu Y, Yu B, Zhou J, Li JC, Qiu SJ, Shi YH, Wang XY, Dai Z, Shi GM. CD24 is a novel predictor for poor prognosis of hepatocellular carcinoma after surgery. Clin Cancer Res. 2009;15:5518-5527. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 98] [Cited by in F6Publishing: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 13. | Sano A, Kato H, Sakurai S, Sakai M, Tanaka N, Inose T, Saito K, Sohda M, Nakajima M, Nakajima T. CD24 expression is a novel prognostic factor in esophageal squamous cell carcinoma. Ann Surg Oncol. 2009;16:506-514. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 14. | Wang YC, Wang JL, Kong X, Sun TT, Chen HY, Hong J, Fang JY. CD24 mediates gastric carcinogenesis and promotes gastric cancer progression via STAT3 activation. Apoptosis. 2014;19:643-656. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 15. | Takahashi M, Nakajima M, Ogata H, Domeki Y, Ohtsuka K, Ihara K, Kurayama E, Yamaguchi S, Sasaki K, Miyachi K. CD24 expression is associated with progression of gastric cancer. Hepatogastroenterology. 2013;60:653-658. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 7] [Reference Citation Analysis (0)] |
| 16. | Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137:1679-1686. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 145] [Cited by in F6Publishing: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 17. | Ke J, Wu X, Wu X, He X, Lian L, Zou Y, He X, Wang H, Luo Y, Wang L. A subpopulation of CD24⁺ cells in colon cancer cell lines possess stem cell characteristics. Neoplasma. 2012;59:282-288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 18. | Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030-1037. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2377] [Cited by in F6Publishing: 2352] [Article Influence: 138.4] [Reference Citation Analysis (0)] |
| 19. | Duckworth CA, Clyde D, Pritchard DM. CD24 is expressed in gastric parietal cells and regulates apoptosis and the response to Helicobacter felis infection in the murine stomach. Am J Physiol Gastrointest Liver Physiol. 2012;303:G915-G926. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 20. | Lisiansky V, Kraus S, Naumov I, Kazanov D, Nabiochtchikov I, Toledano O, Leshno M, Avivi D, Dotan I, Arber N. Role of CD24 polymorphisms in the susceptibility to inflammatory bowel disease. Int J Biol Markers. 2014;29:e62-e68. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 21. | Li D, Zheng L, Jin L, Zhou Y, Li H, Fu J, Shi M, Du P, Wang L, Wu H. CD24 polymorphisms affect risk and progression of chronic hepatitis B virus infection. Hepatology. 2009;50:735-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 22. | Zhou X, Cao Y, Luo J, Zeng X. [Association between CD24 polymorphism and genetic susceptibility to breast cancer: a case-control study]. Zhongnan Daxue Xuebao Yixueban. 2013;38:1122-1129. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 3] [Reference Citation Analysis (0)] |
| 23. | Jiang J, Jia Z, Cao D, Jin MS, Kong F, Suo J, Cao X. Polymorphisms of the DNA methyltransferase 1 associated with reduced risks of Helicobacter pylori infection and increased risks of gastric atrophy. PLoS One. 2012;7:e46058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 24. | Cao X, Cao D, Jin M, Jia Z, Kong F, Ma H, Wang Y, Jiang J. CD44 but not CD24 expression is related to poor prognosis in non-cardia adenocarcinoma of the stomach. BMC Gastroenterol. 2014;14:157. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29:299-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in F6Publishing: 100] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Sakamoto H, Yoshimura K, Saeki N, Katai H, Shimoda T, Matsuno Y, Saito D, Sugimura H, Tanioka F, Kato S. Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet. 2008;40:730-740. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 305] [Cited by in F6Publishing: 319] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 27. | Shi Y, Hu Z, Wu C, Dai J, Li H, Dong J, Wang M, Miao X, Zhou Y, Lu F. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215-1218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 199] [Cited by in F6Publishing: 219] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 28. | Wang L, Lin S, Rammohan KW, Liu Z, Liu JQ, Liu RH, Guinther N, Lima J, Zhou Q, Wang T. A dinucleotide deletion in CD24 confers protection against autoimmune diseases. PLoS Genet. 2007;3:e49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 66] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 29. | Sánchez E, Abelson AK, Sabio JM, González-Gay MA, Ortego-Centeno N, Jiménez-Alonso J, de Ramón E, Sánchez-Román J, López-Nevot MA, Gunnarsson I. Association of a CD24 gene polymorphism with susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2007;56:3080-3086. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Piotrowski P, Lianeri M, Wudarski M, Łacki JK, Jagodziński PP. CD24 Ala57Val gene polymorphism and the risk of systemic lupus erythematosus. Tissue Antigens. 2010;75:696-700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Dawidowicz K, Dieudé P, Avouac J, Wipff J, Hachulla E, Diot E, Tiev K, Cracowski JL, Mouthon L, Amoura Z. Association study of B-cell marker gene polymorphisms in European Caucasian patients with systemic sclerosis. Clin Exp Rheumatol. 2011;29:839-842. [PubMed] [Cited in This Article: ] |
| 32. | Zhou Q, Rammohan K, Lin S, Robinson N, Li O, Liu X, Bai XF, Yin L, Scarberry B, Du P. CD24 is a genetic modifier for risk and progression of multiple sclerosis. Proc Natl Acad Sci USA. 2003;100:15041-15046. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 85] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 33. | Goris A, Maranian M, Walton A, Yeo TW, Ban M, Gray J, Dubois B, Compston A, Sawcer S. CD24 Ala/Val polymorphism and multiple sclerosis. J Neuroimmunol. 2006;175:200-202. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Otaegui D, Sáenz A, Camaño P, Blázquez L, Goicoechea M, Ruíz-Martínez J, Olaskoaga J, Emparanza JA, López de Munain A. CD24 V/V is an allele associated with the risk of developing multiple sclerosis in the Spanish population. Mult Scler. 2006;12:511-514. [PubMed] [Cited in This Article: ] |
| 35. | Ronaghi M, Vallian S, Etemadifar M. CD24 gene polymorphism is associated with the disease progression and susceptibility to multiple sclerosis in the Iranian population. Psychiatry Res. 2009;170:271-272. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 36. | Diaz-Gallo LM, Medrano LM, Gómez-García M, Cardeña C, Rodrigo L, Mendoza JL, Taxonera C, Nieto A, Alcain G, Cueto I. Analysis of the influence of two CD24 genetic variants in Crohn’s disease and ulcerative colitis. Hum Immunol. 2011;72:969-972. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 37. | Sheng L, Shui Y. Clusters of differentiation 24 polymorphism and hepatocellular carcinoma. Hepatology. 2011;54:2273; author reply 2273-2274. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 38. | Matysiak-Budnik T, Mégraud F. Helicobacter pylori infection and gastric cancer. Eur J Cancer. 2006;42:708-716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 89] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 39. | Kabir S. Effect of Helicobacter pylori eradication on incidence of gastric cancer in human and animal models: underlying biochemical and molecular events. Helicobacter. 2009;14:159-171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 40. | Marmé F, Werft W, Walter A, Keller S, Wang X, Benner A, Burwinkel B, Sinn P, Hug S, Sohn C. CD24 Ala57Val polymorphism predicts pathologic complete response to sequential anthracycline- and taxane-based neoadjuvant chemotherapy for primary breast cancer. Breast Cancer Res Treat. 2012;132:819-831. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Zhou X. CD24 polymorphisms cannot predict pathologic complete response to anthracycline- and taxane-based neoadjuvant chemotherapy in breast cancer. Clin Breast Cancer. 2014;14:e33-e40. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 42. | Buck K, Hug S, Seibold P, Ferschke I, Altevogt P, Sohn C, Schneeweiss A, Burwinkel B, Jäger D, Flesch-Janys D. CD24 polymorphisms in breast cancer: impact on prognosis and risk. Breast Cancer Res Treat. 2013;137:927-937. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 43. | Sadot E, Kraus S, Stein M, Naboishchikov I, Toledano O, Kazanov D, Arber N, Kashtan H. CD24 gene polymorphism--a novel prognostic factor in esophageal cancer. Int J Biol Markers. 2014;29:e49-e54. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |