Published online Aug 21, 2016. doi: 10.3748/wjg.v22.i31.7146
Peer-review started: February 12, 2016
First decision: March 21, 2016
Revised: April 15, 2016
Accepted: May 4, 2016
Article in press: May 4, 2016
Published online: August 21, 2016
AIM: To compare expression of nicotinic cholinergic receptors (CHRNs) in healthy and squamous cell carcinoma-affected esophagus and determine the prognostic value.
METHODS: We performed RT-qPCR to measure the expression of CHRNs in 44 esophageal samples from healthy individuals and in matched normal surrounding mucosa, and in tumors from 28 patients diagnosed with esophageal squamous cell carcinoma (ESCC). Next, we performed correlation analysis for the detected expression of these receptors with the habits and clinico-pathological characteristics of all study participants. In order to investigate the possible correlations between the expression of the different CHRN subunits in both healthy esophagus and tissues from ESCC patients, correlation matrices were generated. Subsequently, we evaluated whether the detected alterations in expression of the various CHRNs could precede histopathological modifications during the esophageal carcinogenic processes by using receiver operating characteristic curve analysis. Finally, we evaluated the impact of CHRNA5 and CHRNA7 expression on overall survival by using multivariate analysis.
RESULTS: CHRNA3, CHRNA5, CHRNA7 and CHRNB4, but not CHRNA1, CHRNA4, CHRNA9 or CHRNA10, were found to be expressed in normal (healthy) esophageal mucosa. In ESCC, CHRNA5 and CHRNA7 were overexpressed as compared with patient-matched surrounding non-tumor mucosa (ESCC-adjacent mucosa; P < 0.0001 and P = 0.0091, respectively). Positive correlations were observed between CHRNA3 and CHRNB4 expression in all samples analyzed. Additionally, CHRNB4 was found to be differentially expressed in the healthy esophagus and the normal-appearing ESCC-adjacent mucosa, allowing for distinguishment between these tissues with a sensitivity of 75.86% and a specificity of 78.95% (P = 0.0002). Finally, CHRNA5 expression was identified as an independent prognostic factor in ESCC; patients with high CHRNA5 expression showed an increased overall survival, in comparison with those with low expression. The corresponding age- and tumor stage-adjusted hazard ratio was 0.2684 (95%CI: 0.075-0.97, P = 0.0448).
CONCLUSION: Expression of CHRNs is homogeneous along healthy esophagus and deregulated in ESCC, suggesting a pathogenic role for these receptors in ESCC development and progression.
Core tip: Esophageal squamous cell carcinoma (ESCC) is the main histological subtype of esophageal cancer, and is associated with alcohol and tobacco consumption. Tobacco components, such as nicotine and nitrosamines, are high-affinity agonists of nicotinic cholinergic receptors (CHRNs), the activation of which triggers cellular signaling pathways important for cancer progression. However, data regarding differential expression and regulation of CHRNs in healthy esophageal mucosa and ESCC are limited. This study shows homogeneous expression of CHRNs along healthy esophagus and deregulation in ESCC, CHRNB4 overexpression preceding the first histopathological alterations during ESCC development, and CHRNA5 expression as an independent predictor of prognosis.
- Citation: Chianello Nicolau M, Pinto LFR, Nicolau-Neto P, de Pinho PRA, Rossini A, de Almeida Simão T, Soares Lima SC. Nicotinic cholinergic receptors in esophagus: Early alteration during carcinogenesis and prognostic value. World J Gastroenterol 2016; 22(31): 7146-7156
- URL: https://www.wjgnet.com/1007-9327/full/v22/i31/7146.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i31.7146
Worldwide, esophageal cancer (EC) is the 8th most frequent type of cancer and the 6th most common cause of cancer-related deaths[1], reflecting the high mortality rate associated with the disease that is a direct consequence of late diagnosis and poor treatment response[1-4]. Prognosis of EC patients is directly affected by tumor invasion and since dissemination occurs very early during the natural history of the disease due to the lack of serosa in the esophagus, identifying the mechanisms involved in this process is of major interest[5].
Esophageal squamous cell carcinoma (ESCC) is the main histological type of EC, accounting for about 90% of all EC cases globally[2]. The highest incidence rates of EC occur in developing countries, such as Brazil, where ESCC is also the most common histological subtype[2,6]. Several epidemiological studies have indicated that alcohol consumption and tobacco smoking are the major risk factors for ESCC development[7-9]. Studies from Western countries have shown that the concomitant use of these products multiplies the risk for disease development, with tobacco smoking identified as an important contributor to both tumor initiation and promotion and alcohol characterized as acting as a tumor promoter primarily[7,8,10].
Cigarette smoke contains potent carcinogens, such as polycyclic aromatic hydrocarbons and nitrosamines, which have been demonstrated extensively as associated with induction of different types of tumors[11]. These compounds are capable of inducing DNA adducts and mutations and, therefore, have been suggested to participate in the initiation of tobacco-related cancers[11]. However, tobacco-specific nitrosamines and nicotine itself may contribute to tumorigenesis through other additional mechanisms[12-16]. Activation of the nicotinic cholinergic receptors (CHRNs) by these compounds is known to trigger cellular signaling pathways that play key roles in cancer progression, including cell proliferation, angiogenesis, apoptosis inhibition and cell migration[16]. Therefore, researchers have put forth extensive efforts towards characterizing the role(s) of CHRNs in tobacco-related tumors. Most of the studies to date have focused on lung cancer[17-19], and the results have shown that lung tumors lack expression of CHRNA3, due to promoter hypermethylation, but have overexpression of CHRNA5 and CHRNA7[17,19]. It has also been shown that nicotine can modulate the expression of CHRNs in lung cells[20,21]. Finally, in addition to stimulation of cell proliferation and survival, the nicotine-induced activation of CHRNs was also shown to stimulate epithelial-mesenchymal transition and to promote invasion of lung cancer cell lines as well as of cells derived from breast and pancreatic tumors[21].
Based on these findings, it has been proposed that nicotine is likely to contribute to the progression of tobacco-related cancers through binding to CHRNs and the consequent activation of cellular pathways involved in tumorigenesis. Therefore, we hypothesized that alterations in CHRN expression may take part in and contribute to the pathogenesis of ESCC. Since there are no detailed data regarding CHRN expression in human esophagus, this study was designed to determine the expression profile of these receptors in healthy esophagus and ESCC to explore the role of these receptors in this type of cancer.
Forty-four fresh esophageal samples were obtained from healthy donors undergoing endoscopy at the Hospital Universitário Pedro Ernesto (HUPE-UERJ, Rio de Janeiro, Brazil) for use, with informed consent, in this study. None of these volunteer donors showed any observable alterations in the esophageal structure at the time of the biopsy and none had a history of cancer. Samples were collected from each third of the esophagus (upper, middle and lower). Furthermore, esophageal samples were obtained from 28 patients with a confirmed diagnosis of ESCC, but who had not undergone any treatment at the time of biopsy at the Instituto Nacional do Câncer (INCA; Rio de Janeiro, Brazil) for use, with informed consent, in this study; for all patients, tumor and matched histologically normal surrounding mucosa (taken 4 inches from the tumor border) were collected and stored at the National Tumor Bank of the INCA (BNT/INCA).
Clinical and demographic data were obtained from the hospitals’ medical records for all study participants. Healthy donors provided additional data by answering a standardized questionnaire. All participants signed an informed consent form prior to study enrollment. The project was approved by the Ethics Committees of all involved institutions.
Total RNA was extracted from the respective samples by using the TRIzol® reagent (Invitrogen, United States), following the manufacturer’s protocol. All RNA samples were quantified by spectrophotometry and purity was verified by calculating the absorbance ratio of 260 nm/280 nm and ensuring the ratio was ≥ 1.7.
A total of 500 ng RNA was reverse transcribed using SuperScript II® (Invitrogen) and following the manufacturer’s protocol. The Rotor-Gene Q system (Qiagen, Germany) and QuantiFast SYBR Green PCR Kit (Qiagen) were used for the qPCR, and each reaction was optimized for the particular pair of primers comprising exon-exon junctions to evaluate the mRNA expression of CHRN subunits (Table 1)[20,22]. GAPDH served as the reference gene. Each reaction contained 7.5 μL of QuantiFast SYBR Green Buffer × 2 (Qiagen), specific oligonucleotides at a final concentration of 0.5 μmol/L, 1 μL of cDNA (diluted × 10) and sterile deionized water to complete the final volume of 15 μL. The amplification reaction was performed as follows: 5 min of pre-denaturation at 95 °C, followed by 40 cycles of denaturation for 5 s at 95 °C and an annealing and extension step for 10 s at 60 °C. For CHRNA1 and CHRNA9, annealing steps of 10 s at 58.1 °C and of 5 s at 62 °C were added, respectively; finally, an extension step of 10 s at 60 °C was performed for both of these subunits. After the reaction, the expression of each CHRN subunit was normalized to the GAPDH expression, using the comparative Ct method[23].
| Gene | Oligonucleotide sequences (5'-3') | Positive control | Ref. |
| CHRNA1 | Forward: ACCAGGAGTCTAACAATGCG | Glioblastoma cell line, U251 | Designed by the authors |
| Reverse: ACAAGCATGAAGACTCCGAG | |||
| CHRNA3 | Forward: AACCTGTGGCTCAAGCAAATCT | Lung cancer cell line, A549 | [20] |
| Reverse: CATGAACTCTGCCCCACCAT | |||
| CHRNA4 | Forward: ACACAGACTTCTCGGTGAAG | ESCC cell line, TE1 | Designed by the authors |
| Reverse: CAGCAGGCAGACGATGATGA | |||
| CHRNA5 | Forward: AGATGGAACCCTGATGACTATGGT | Lung cancer cell line, A549 | [20] |
| Reverse: AAACGTCCATCTGCATTATCAAAC | |||
| CHRNA7 | Forward: GCTGCTCGTGGCTGAGATC | Lung cancer cell line, A549 | [20] |
| Reverse: TGGCGAAGTACTGGGCTATCA | |||
| CHRNA9 | Forward: GATGGCCTAGACTCCATCAG | Glioblastoma cell line, U251 | Designed by the authors |
| Reverse: CTGAAGATTCATCATCAGCCTTG | |||
| CHRNA10 | Forward: CTACTCCCTGCAGAGTGCCTG | Glioblastoma cell line, U251 | Designed by the authors |
| Reverse: TCTGGTCTGTGTCTGCCACAG | |||
| CHRNB4 | Forward: TCACAGCTCATCTCCATCAAGCT | Lung cancer cell line, A549 | [20] |
| Reverse: CCTGTTTCAGCCAGACATTGGT | |||
| GAPDH | Forward: CAACAGCCTCAAGATCATCAGCAA | - | [22] |
| Reverse: AGTGATGGCATGGACTGTGGTCAT |
All statistical analyses were performed using the GraphPad Prism 5 software (GraphPad Software, United States). Differences were considered statistically significant when P was < 0.05. When comparing two groups, the unpaired t test or Mann-Whitney test was used. For comparison of paired samples, the paired t test or Wilcoxon signed rank test was applied. For determining significant differences of mRNA expression (expressed as medians) among the different groups of samples, the one-way ANOVA or Kruskal Wallis test and Tukey’s post-test or Dunn’s post-test was used.
Correlations between the expression levels of the different genes were determined using Pearson’s or Spearman’s correlation tests. A receiver operating characteristic (ROC) curve was plotted to determine the value of gene expression as a marker to distinguish healthy esophagus from normal-appearing ESCC-adjacent mucosa.
For the estimate of univariate survival, a Kaplan-Meier survival curve was generated and the statistical significance between the two groups was calculated using the log-rank test. Variables that have been shown to influence ESCC outcome, such as age and tumor stage, were selected for the multivariate analysis. Finally, we applied Cox regression using the stepwise forward method. All survival analyses were performed using the software package “Survival” in the R statistical program[24]. The statistical methods of this study were reviewed by Dr. Mariana Boroni from INCA (Rio de Janeiro, Brazil).
Table 2 shows that most of the healthy individuals were women (70.5% of the group), never smokers (61.4%) and ever drinkers (54.5%), with a median age of 56.5 years (range, 18-85 years). By comparison, the ESCC patients were mostly male (64.3%), ever smokers (96.4%) and ever drinkers (92.9%), with a median age of 59.5 years (range, 46-79 years). Most of the ESCC patients died as a consequence of the disease (71.4%), with most of the tumors affecting the middle third of the esophagus (53.6%), being moderately differentiated (75.0%), and corresponding to stages 3 and 4 (53.6%).
| Evaluated criteria | Number of individuals (range or %) | |
| Healthy individuals,n = 44 | ESCC patients,n = 28 | |
| Age (yr) | 56.50 (18-85) | 59.50 (46-79) |
| Sex | ||
| Men | 13 (29.5) | 18 (64.3) |
| Women | 31 (70.5) | 10 (35.7) |
| Smoking | ||
| Never | 27 (61.4) | 1 (3.6) |
| Ever | 17 (38.6) | 27 (96.4) |
| Alcohol consumption | ||
| Never | 20 (45.5) | 2 (7.1) |
| Ever | 24 (54.5) | 26 (92.9) |
| Death | ||
| No | NA | 8 (28.6) |
| Yes | NA | 20 (71.4) |
| Tumor localization | ||
| Upper third | NA | 9 (32.1) |
| Middle third | NA | 15 (53.6) |
| Lower third | NA | 3 (10.7) |
| Histological grade, differentiation | ||
| Poor | NA | 6 (21.4) |
| Moderate | NA | 21 (75.0) |
| Well | NA | 1 (3.6) |
| Tumor stage | ||
| 1 + 2 | NA | 7 (25.0) |
| 3 + 4 | NA | 15 (53.6) |
| Not determined | NA | 6 (21.4) |
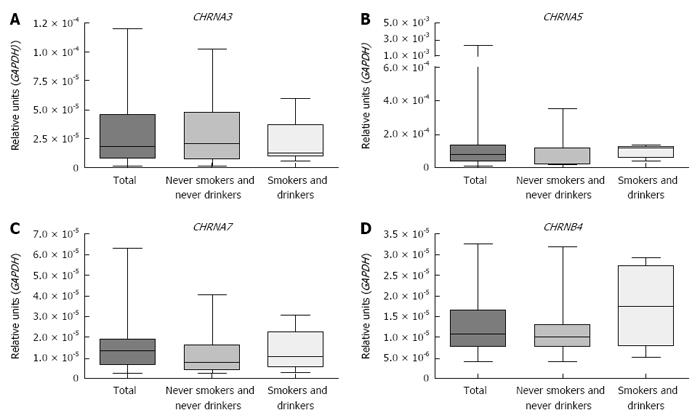
Figure 1 shows that CHRNA3, CHRNA5, CHRNA7 and CHRNB4 expression was detected in all healthy esophagus samples evaluated and that the expression levels did not differ significantly among the esophageal thirds nor according to the smoking and drinking status of the individuals (Kruskal Wallis test and Dunn’s post-test). There was no association found between the CHRNs expression and age, sex, smoking status or alcohol consumption (Table 3). Expression of CHRNA1, CHRNA4, CHRNA9 and CHRNA10 was undetectable in the healthy esophagus samples.
| Variable | CHRNA3 | CHRNA5 | CHRNA7 | CHRNB4 | ||||
| Expression, | P value | Expression, | P value | Expression, | P value | Expression, | P value | |
| median (min-max) | median (min-max) | median (min-max) | median (min-max) | |||||
| Esophageal thirds | ||||||||
| Upper | 1.563 × 10-5 | 0.769 | 8.431 × 10-5 | 0.945 | 1.023 × 10-5 | 0.789 | 9.027 × 10-6 | 0.153 |
| (2.735 × 10-6 - 7.183 × 10-5) | (1.277 × 10-5 - 2.157 × 10-3) | (1.033 × 10-6 - 5.616 × 10-5) | (2.534 × 10-6 - 2.828 × 10-5) | |||||
| Middle | 1.783 × 10-5 | 8.476 × 10-5 | 1.346 × 10-5 | 1.077 × 10-5 | ||||
| (1.512 × 10-6 - 1.196 × 10-4) | (1.286 × 10-5 - 2.388 × 10-3) | (2.588 × 10-6 - 6.319 × 10-5) | (3.913 × 10-6 - 3.259 × 10-5) | |||||
| Lower | 1.721 × 10-5 | 8.906 × 10-5 | 1.263 × 10-5 | 1.166 × 10-5 | ||||
| (2.914 × 10-6 - 6.796 × 10-5) | (1.442 × 10-5 - 1.990 × 10-3) | (1.678 × 10-6 - 5.636 × 10-5) | (3.621 × 10-6 - 4.453 × 10-5) | |||||
| Age, yr | 58 | 56.5 | 57 | 57 | ||||
| > median | 2.226 × 10-5 | 0.422 | 4.982 × 10-5 | 0.097 | 1.189 × 10-5 | 0.272 | 1.060 × 10-5 | 0.778 |
| (1.512 × 10-6 - 1.019 × 10-4) | (1.286 × 10-5 - 2.388 × 10-3) | (2.588 × 10-6 - 4.391 × 10-5) | (3.913 × 10-6 - 3.192 × 10-5) | |||||
| ≤ median | 1.274 × 10-5 | 9.054 × 10-5 | 1.449 × 10-5 | 1.094 × 10-5 | ||||
| (3.977 × 10-6 - 1.196 × 10-4) | (2.657 × 10-5 - 4.635 × 10-4) | (2.931 × 10-6 - 6.319 × 10-5) | (5.010 × 10-6 - 3.259 × 10-5) | |||||
| Sex | ||||||||
| Male | 1.344 × 10-5 | 0.253 | 1.104 × 10-4 | 0.243 | 1.316 × 10-5 | 0.807 | 1.772 × 10-5 | 0.372 |
| (3.977 × 10-6 - 5.947 × 10-5) | (2.657 × 10-5 - 3.351 × 10-4) | (2.931 × 10-6 - 4.060 × 10-5) | (5.559 × 10-6 - 2.917 × 10-5) | |||||
| Female | 1.812 × 10-5 | 7.842 × 10-5 | 1.375 × 10-5 | 1.034 × 10-5 | ||||
| (1.512 × 10-6 - 1.196 × 10-4) | (1.286 × 10-5 - 2.388 × 10-3) | (2.588 × 10-6 - 6.319 × 10-5) | (3.913 × 10-6 - 3.259 × 10-5) | |||||
| Smoking | ||||||||
| Never | 1.665 × 10-5 | 0.400 | 8.531 × 10-5 | 0.961 | 1.210 × 10-5 | 0.582 | 1.077 × 10-5 | 0.599 |
| (1.512 × 10-6 - 1.019 × 10-4) | (2.051 × 10-5 - 2.388 × 10-3) | (2.588 × 10-6 - 6.319 × 10-5) | (3.913 × 10-6 - 3.192 × 10-5) | |||||
| Ever | 1.948 × 10-5 | 8.405 × 10-5 | 1.375 × 10-5 | 1.078 × 10-5 | ||||
| (5.157 × 10-6 - 1.196 × 10-4) | (1.286 × 10-5 - 3.351 × 10-4) | (2.931 × 10-6 - 4.256 × 10-5) | (5.010 × 10-6 - 3.259 × 10-5) | |||||
| Alcohol consumption | ||||||||
| Never | 2.386 × 10-5 | 0.163 | 5.955 × 10-5 | 0.131 | 1.046 × 10-5 | 0.412 | 1.094 × 10-5 | 0.521 |
| (1.512 × 10-6 - 1.196 × 10-4) | (2.051 × 10-5 - 3.525 × 10-4) | (2.588 × 10-6 - 4.256 × 10-5) | (3.913 × 10-6 - 3.259 × 10-5) | |||||
| Ever | 1.274 × 10-5 | 1.018 × 10-4 | 1.375 × 10-5 | 1.007 × 10-5 | ||||
| (3.977 × 10-6 - 8.783 × 10-5) | (1.286 × 10-5 - 2.388 × 10-3) | (2.931 × 10-6 - 6.319 × 10-5) | (5.010 × 10-6 - 2.917 × 10-5) | |||||
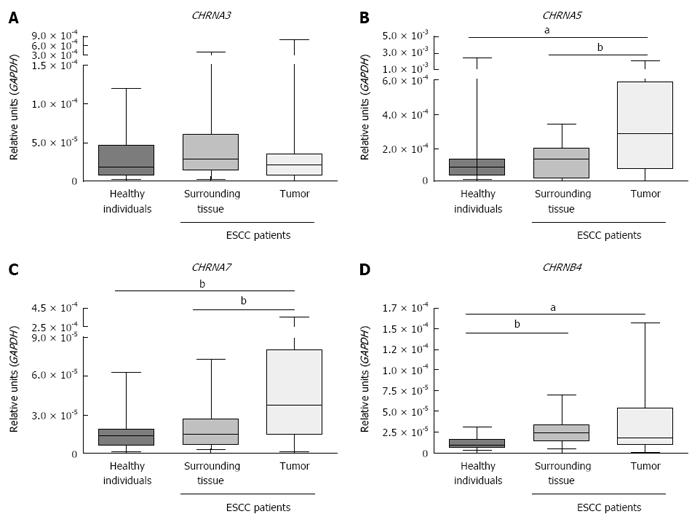
Figure 2 shows that CHRNA5 and CHRNA7 expression was higher in ESCC samples than in either the matched adjacent mucosa (P < 0.0001 and P = 0.0091, respectively; Wilcoxon signed rank test) or the esophageal mucosa samples from healthy individuals (P = 0.0157 and P = 0.0004, respectively; Mann-Whitney test). In addition, CHRNB4 expression was higher in both tumor samples and the matched surrounding mucosa than in the esophagus samples from healthy individuals (P = 0.0180 and P = 0.0005, respectively; Mann-Whitney test). The expression of CHRNA3 was similar in tumor, matched surrounding tissue (Wilcoxon signed rank test) and healthy esophageal mucosa (Mann-Whitney test).
Correlation matrices showed positive associations between the expression of CHRNA3 and CHRNB4 in the healthy esophagus samples (r = 0.47, P = 0.007, Spearman’s correlation test), in the normal surrounding mucosa samples from ESCC patients (r = 0.607, P = 0.006, Spearman’s correlation test) and in the tumor tissues from the ESCC patients (r = 0.544, P = 0.016, Spearman’s correlation test) (Table 4). Additionally, a positive correlation was shown to exist between the expression of CHRNB4 and CHRNA5 in the normal surrounding mucosa from ESCC patients (r = 0.556, P = 0.013; Spearman’s correlation test). Finally, the expression of CHRNA3 and CHRNA7 (r = 0.511, P = 0.026, Spearman’s correlation test) and of CHRNB4 and CHRNA7 (r = 0.561, P = 0.012, Spearman’s correlation test) were shown to be positively correlated in tumors.
| CHRNA3 | CHRNA5 | CHRNA7 | CHRNB4 | ||
| Healthy esophagus | CHRNA3 | P = 0.820 | P = 0.203 | P = 0.007a | |
| CHRNA5 | r = -0.040 | P = 0.727 | P = 0.672 | ||
| CHRNA7 | r = 0.220 | r = 0.060 | P = 0.193 | ||
| CHRNB4 | r = 0.470 | r = -0.079 | r = 0.240 | ||
| Normal surrounding mucosa | CHRNA3 | P = 0.486 | P = 0.753 | P = 0.006a | |
| CHRNA5 | r = 0.170 | P = 0.847 | P = 0.013a | ||
| CHRNA7 | r = -0.077 | r = -0.038 | P = 0.185 | ||
| CHRNB4 | r = 0.607 | r = 0.556 | r = -0.318 | ||
| Tumor Tissue | CHRNA3 | P = 0.450 | P = 0.026a | P = 0.016a | |
| CHRNA5 | r = 0.184 | P = 0.130 | P = 0.071 | ||
| CHRNA7 | r = 0.511 | r = 0.293 | P = 0.012a | ||
| CHRNB4 | r = 0.544 | r = 0.423 | r = 0.561 |
Evaluation of the potential association between the fold-change (ratio of mRNA expression between tumor and matched surrounding tissue) of the expression of the different subunits and the clinicopathological data indicated no statistically significant associations (Table 5).
| Clinicopathologicaldata | CHRNA3 | CHRNA5 | CHRNA7 | CHRNB4 | ||||
| FC, | P value | FC, | P value | FC, | P value | FC, | P value | |
| median | median | median | median | |||||
| (min-max) | (min-max) | (min-max) | (min-max) | |||||
| Age, yr | 60 | 59.5 | 59.5 | 60 | ||||
| (46-79) | (46-79) | (46-79) | (46-79) | |||||
| > median | 1.1 | 0.2775 | 2.166 | 0.1612 | 2.867 | 0.9451 | 1.984 | 0.1128 |
| (0.03136-14.69) | (0.3918-15.07) | (0.2630-20.53) | (0.1586-7.119) | |||||
| ≤ median | 0.5844 | 3.193 | 1.267 | 0.7518 | ||||
| (0.006324-10.62) | (0.8615-11.66) | (0.4813-23.10) | (0.1414-2.051) | |||||
| Sex | ||||||||
| Men | 0.9777 | 0.1053 | 3.032 | 0.1719 | 2.867 | 0.7192 | 1.685 | 0.1053 |
| (0.006324-14.69) | (0.3918-15.07) | (0.4813-23.10) | (0.2261-7.119) | |||||
| Women | 0.1153 | 2.102 | 1.657 | 0.3153 | ||||
| (0.03349-1.072 ) | (0.5510-3.593) | (0.2630-20.53) | (0.1414-2.809) | |||||
| Smoking | ||||||||
| Never | NA | NA | NA | NA | NA | NA | NA | NA |
| Ever | 0.6941 | 3.01 | 1.86 | 1.229 | ||||
| (0.006324-14.69) | (0.3918 - 15.07) | (0.2630-23.10) | (0.1414-7.119) | |||||
| Alcohol consumption | ||||||||
| Never | NA | NA | NA | NA | NA | NA | NA | NA |
| Ever | 0.5844 | 2.971 | 1.657 | 0.9571 | ||||
| (0.006324-14.69) | (0.3918-15.07) | (0.2630-23.10) | (0.1414-7.119) | |||||
| Tumor localization1 | ||||||||
| Upper third | 0.1153 | 0.4079 | 1.589 | 0.0737 | 0.8255 | 0.1525 | 0.3964 | 0.3510 |
| (0.03136-14.69) | (0.3918- 6.284) | (0.2630-7.917) | (0.1414-7.119) | |||||
| Middle third | 0.8265 | 3.01 | 2.567 | 1.5 | ||||
| (0.006324-10.62) | (0.8615-11.66) | (0.4813-23.10) | (0.2261-2.809) | |||||
| Lower third | 3.904 | 9.329 | 4.199 | 2.211 | ||||
| (0.07381-7.799) | (3.414-15.07) | (0.7220-13.36) | (1.613-2.552) | |||||
| Histological grade, differentiation | ||||||||
| Well | NA | 0.4902 | NA | 0.9767 | NA | 0.3661 | NA | 0.8610 |
| Moderate | 0.8039 | 3.031 | 1.454 | 0.9571 | ||||
| (0.03136-14.69) | (0.3918-15.07) | (0.2630-23.10) | (0.1414-7.119) | |||||
| Poor | 0.2908 | 2.6 | 2.514 | 1.5 | ||||
| (0.006324-1.626) | (1.826-4.993) | (1.057-18.57) | (0.5035-2.532) | |||||
| Tumor stage | ||||||||
| 1 + 2 | 0.5728 | 0.9599 | 2.505 | 0.778 | 1.454 | 0.7780 | 1.832 | 0.4457 |
| (0.03349-10.62) | (1-15.07) | (0.4813-20.53) | (0.1414-2.809) | |||||
| 3 + 4 | 0.8039 | 3.01 | 3.167 | 0.9571 | ||||
| (0.006324-7.799) | (0.4633-6.566) | (0.6082-23.10) | (0.2261-2.532) | |||||
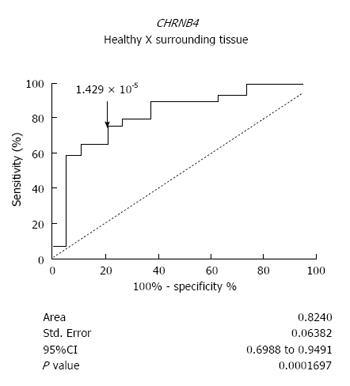
We next analyzed whether altered expression of CHRNs could precede histopathological modifications during esophageal carcinogenesis. Interestingly, the expression of CHRNB4 was able to distinguish the normal-appearing surrounding tissue of ESCC patients from the esophageal mucosa of healthy individuals. The sensitivity rate was 75.86% and the specificity rate was 78.95% (P = 0.0002, ROC curve) (Figure 3).
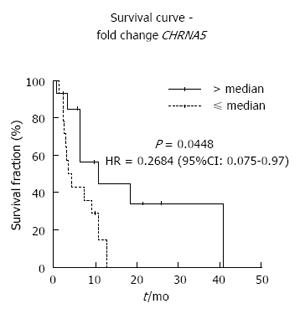
Finally, we evaluated the impact of CHRNA5 and CHRNA7 expression on overall survival. Multivariate analysis identified CHRNA5 expression as an independent prognostic factor of ESCC. ESCC patients with high CHRNA5 expression showed an increased overall survival, in comparison with the ESCC patients who had low expression (Figure 4). The corresponding age- and tumor stage-adjusted hazard ratio was 0.2684 (95%CI: 0.075-0.97, P = 0.045).
Expression of CHRNs in extra-neuronal tissues has been extensively reported[25]; however, to date, only one study has suggested the existence of a functional non-neuronal cholinergic system, present in the human esophageal epithelium. Nguyen and colleagues[26] showed that the human esophagus expresses the enzymes responsible for choline synthesis and degradation [i.e., choline acetyltransferase and acetylcholinesterase (AChE)], as well as four CHRN subunits (CHRNA3, CHRNA5, CHRNA7 and CHRNB2, which were evaluated in this study). The data in our current study agree with these previous findings; specifically, we were able to detect the mRNA of the same alpha subunits in the esophageal mucosa from healthy individuals. Furthermore, we also showed-for the first time-the presence of CHRNB4 in this epithelium; however, the mRNA of CHRNA1, CHRNA4, CHRNA9 and CHRNA10 were undetectable.
Following the confirmation of the expression of CHRNs in the human esophagus, we investigated their expression in ESCC samples and matched normal-appearing surrounding mucosa. Similar to the previous findings for lung cancer[27], we observed a statistically significant overexpression of CHRNA5 and CHRNA7 in tumors, as compared with expression in the matched adjacent tissue and in esophageal mucosa from healthy individuals. These findings suggest a role for these receptors in the pathogenesis of epithelial tumors, especially for tumors related to tobacco smoking. However, different from the previous findings reported for lung squamous cell carcinoma[27], the overexpression of these subunits in ESCC is probably not induced by tobacco components since there was no association found between the overexpression of these receptors and the smoking status of the ESCC patients in our study.
Whereas CHRNA5 and CHRNA7 overexpression seems to follow esophageal transformation, the induction of CHRNB4 expression seems to occur before the first histopathological alterations associated with development of ESCC. We showed that although there is no difference in the CHRNB4 expression of tumor samples and the matched surrounding mucosa samples of ESCC patients, both tissues present a significantly higher CHRNB4 expression in comparison with esophagus samples from healthy individuals. Therefore, it is tempting to speculate that the consumption of high doses of alcohol and/or tobacco, characteristic of ESCC patients, could influence the expression of this receptor, affecting both tumor and surrounding tissues. This speculation agrees with the hypothesis of field cancerization, which was proposed to explain the high propensity for development of multiple, independent tumors in the mucosal tissues of the head and neck as a consequence of risk factor exposure, a characteristic also observed in the esophagus[28]. In this context, the epithelium adjacent to the tumor may appear normal histologically but may already harbor molecular alterations, such as CHRNB4 overexpression. Following this hypothesis, we evaluated how efficiently CHRNB4 expression levels could distinguish the esophageal mucosa of healthy individuals from the surrounding normal-appearing epithelium of ESCC patients. Interestingly, CHRNB4 expression was able to discriminate the tissues with 75.86% sensitivity and 78.95% specificity, suggesting its potential utility as a predictive marker of field cancerization in the esophagus; further studies are necessary to verify this hypothesis, however.
The contribution of tobacco smoking to the development of several tumor types, such as lung, head and neck, and esophagus, is a consensus. More recently, the impacts of post-diagnosis exposure to tobacco components on treatment response and survival have emerged as a hot topic. In this context, patients with tobacco smoking-related lung cancer who continue smoking have a poorer prognosis[29,30]. Tobacco is, therefore, not only a source of carcinogens responsible for tumor initiation but also of tumor promoting agents, such as nicotine, which is able to induce cell proliferation, inhibit apoptosis and induce epithelial-mesenchymal transition, to name a few of its known effects[29]. However, studies that have evaluated the impact of the cholinergic system on prognosis of patients diagnosed with tobacco-related tumors are still scarce. Yoo and colleagues[31] reported that demethylation of CHRNB4, which is correlated with increased mRNA expression, confers a poorer prognosis in patients with non-small cell lung cancer. Castillo-González and colleagues[32] reported that low AChE activity is correlated with poor overall survival in patients with head and neck cancers. For ESCC, the current study is the first to report an impact of alterations of the cholinergic system on patient prognosis. Specifically, CHRNA5 expression was identified as an independent prognostic factor for ESCC, with patients who present a higher expression of this receptor showing a better overall survival. It is unclear how this overexpression could protect against tumor progression, but it has already been shown by others that under- or over-activation of the CHRNA5 promoter is protective against lung cancer development[33], suggesting a role of this receptor in tissue homeostasis.
The current study is also the first to investigate the expression profile of CHRNs in both healthy esophagus and ESCC (tumor and tumor-adjacent) tissues. Although the number of samples was limited, the results show homogeneous expression of CHRNA3, CHRNA5, CHRNA7 and CHRNB4 along the entire esophagus under normal, non-cancerous condition and suggest that nicotine and/or alcohol exposure are not capable of affecting the expression of these receptors in the healthy mucosa. Additionally, CHRNA1, CHRNA4, CHRNA9 and CHRNA10 were not detected in the esophageal epithelium, but this lack of expression should be validated by other techniques. A similar evaluation should also be carried out for the other subunits that were not assessed in the present study due to the lack of positive controls; these unexamined receptor genes include CHRNA2, CHRNA6, CHRNB2 and CHRNB3. Furthermore, this study also showed deregulation of CHRNA5 and CHRNA7 expression in ESCC, which may contribute to the esophageal carcinogenesis process. Finally, CHRNB4 overexpression was shown to be an early alteration of ESCC carcinogenesis and CHRNA5 expression as an independent prognostic factor. Such characterization provided evidence that the esophageal epithelium possesses a functional cholinergic system, which is deregulated in ESCC, but further analyses are now necessary to better comprehend which pathways could be affected by this deregulation and how this could contribute to the progression of esophageal cancer.
The authors would like to thank Dr. Mariana Boroni for revising all the statistical analysis performed in this study, and the Endoscopy Sections of the Hospital Universitário Pedro Ernesto and the Instituto Nacional de Câncer as well as the National Tumor Bank from INCA (BNT/INCA) for sample collection.
Esophageal squamous cell carcinoma (ESCC) is one of the most incident and lethal tumors worldwide. Although tobacco and alcohol are recognized as the main risk factors of the disease, the molecular mechanisms involved in its development remain unclear. Tobacco components are well recognized for their ability to induce mutations and to activate cellular pathways correlated with tumor progression. In this context, the nicotinic cholinergic receptors (CHRNs) may play a central role in ESCC, but data regarding their expression in the esophagus, both under normal and pathologic conditions, is still very limited.
So far, only one study in the publicly available literature has suggested the existence of a functional cholinergic system in normal esophageal epithelium and the expression of CHRNs have not been evaluated in ESCC tissues.
This is the first study to show that CHRNA3, CHRNA5, CHRNA7 and CHRNB4 are homogeneously expressed in the esophageal mucosa of healthy individuals. Moreover, the expression of these CHRNs does not seem to be modulated by tobacco and/or alcohol exposure. We also show, for the first time, overexpression of CHRNA7 and CHRNA5 in ESCC, with the latter showing an impact on prognosis. Finally, the findings from our study support the possibility that CHRNB4 overexpression is an early alteration during esophageal carcinogenesis, preceding the first histopathological alterations.
Identifying the molecular alterations that take place during esophageal carcinogenesis may help not only to elucidate which mechanisms contribute to ESCC development and progression but may also identify new biomarkers of diagnosis and prognosis. This knowledge is of utmost relevance for improving overall survival of ESCC patients.
Nicotinic CHRNs are recognized as important proteins that mediate chemical neurotransmission at neurons, ganglia, interneurons and the motor end plate. However, the ubiquitous expression of CHRNs in mammalian cells has suggested they may play an additional role in extra-neuronal tissues. In fact, different studies have shown their participation in maintaining communication and phenotypic functions of non-neuronal cells and the deregulation of these receptors has been observed in different tumor types. CHRN activation by tobacco components triggers different cellular pathways involved in survival and apoptosis blockade and may contribute to tumor progression by these mechanisms.
The study shows expression of CHRN subunits α3, α5, α7 and β4, but not α1, α4, α9 and α10, in normal esophageal mucosa. In ESCC, CHRNA5 and CHRNA7 subunits were found overexpressed when compared to matched surrounding mucosa. CHRNB4 was differentially expressed between healthy esophagus and normal-appearing ESCC adjacent mucosa. CHRNA5 expression is an independent prognostic factor in ESCC. Patients with high CHRNA5 expression showed an increased overall survival in comparison with those with low expression.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Bener A S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20108] [Cited by in F6Publishing: 19772] [Article Influence: 2196.9] [Reference Citation Analysis (17)] |
| 2. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13286] [Cited by in F6Publishing: 13414] [Article Influence: 706.0] [Reference Citation Analysis (1)] |
| 3. | Eslick GD. Epidemiology of esophageal cancer. Gastroenterol Clin North Am. 2009;38:17-25, vii. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in F6Publishing: 105] [Article Influence: 7.0] [Reference Citation Analysis (1)] |
| 4. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25175] [Article Influence: 1936.5] [Reference Citation Analysis (3)] |
| 5. | Napier KJ, Scheerer M, Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointest Oncol. 2014;6:112-120. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 499] [Cited by in F6Publishing: 539] [Article Influence: 53.9] [Reference Citation Analysis (3)] |
| 6. | Instituto Nacional de Câncer José Alencar Gomes da Silva. Estimativa 2016/2017: Incidência de Câncer no Brasil. Instituto Nacional de Câncer José Alencar Gomes da Silva, Coordenação de Prevenção e Vigilância. Rio de Janeiro: INCA 2016; [Internet]. Accessed on 2016 Jan 7 Available from: http://www.inca.gov.br/wcm/dncc/2015/index.asp. [Cited in This Article: ] |
| 7. | Castellsagué X, Muñoz N, De Stefani E, Victora CG, Castelletto R, Rolón PA, Quintana MJ. Independent and joint effects of tobacco smoking and alcohol drinking on the risk of esophageal cancer in men and women. Int J Cancer. 1999;82:657-664. [PubMed] [Cited in This Article: ] |
| 8. | Prabhu A, Obi KO, Rubenstein JH. The synergistic effects of alcohol and tobacco consumption on the risk of esophageal squamous cell carcinoma: a meta-analysis. Am J Gastroenterol. 2014;109:822-827. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 150] [Cited by in F6Publishing: 155] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 9. | Lin S, Wang X, Huang C, Liu X, Zhao J, Yu IT, Christiani DC. Consumption of salted meat and its interactions with alcohol drinking and tobacco smoking on esophageal squamous-cell carcinoma. Int J Cancer. 2015;137:582-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 45] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Tobacco smoking. IARC Monogr Eval Carcinog Risk Chem Hum. 1986;38:35-394. [PubMed] [Cited in This Article: ] |
| 11. | Hecht SS, Hoffmann D. Tobacco-specific nitrosamines, an important group of carcinogens in tobacco and tobacco smoke. Carcinogenesis. 1988;9:875-884. [PubMed] [Cited in This Article: ] |
| 12. | Schuller HM. Carbon dioxide potentiates the mitogenic effects of nicotine and its carcinogenic derivative, NNK, in normal and neoplastic neuroendocrine lung cells via stimulation of autocrine and protein kinase C-dependent mitogenic pathways. Neurotoxicology. 1994;15:877-886. [PubMed] [Cited in This Article: ] |
| 13. | Schuller HM. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat Rev Cancer. 2009;9:195-205. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 253] [Cited by in F6Publishing: 265] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 14. | Cattaneo MG, Codignola A, Vicentini LM, Clementi F, Sher E. Nicotine stimulates a serotonergic autocrine loop in human small-cell lung carcinoma. Cancer Res. 1993;53:5566-5568. [PubMed] [Cited in This Article: ] |
| 15. | Schaal C, Chellappan SP. Nicotine-mediated cell proliferation and tumor progression in smoking-related cancers. Mol Cancer Res. 2014;12:14-23. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 222] [Cited by in F6Publishing: 233] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 16. | Lee CH, Wu CH, Ho YS. From smoking to cancers: novel targets to neuronal nicotinic acetylcholine receptors. J Oncol. 2011;2011:693424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Paliwal A, Vaissière T, Krais A, Cuenin C, Cros MP, Zaridze D, Moukeria A, Boffetta P, Hainaut P, Brennan P. Aberrant DNA methylation links cancer susceptibility locus 15q25.1 to apoptotic regulation and lung cancer. Cancer Res. 2010;70:2779-2788. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633-637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 917] [Cited by in F6Publishing: 910] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 19. | Improgo MR, Schlichting NA, Cortes RY, Zhao-Shea R, Tapper AR, Gardner PD. ASCL1 regulates the expression of the CHRNA5/A3/B4 lung cancer susceptibility locus. Mol Cancer Res. 2010;8:194-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 20. | Lam DC, Girard L, Ramirez R, Chau WS, Suen WS, Sheridan S, Tin VP, Chung LP, Wong MP, Shay JW. Expression of nicotinic acetylcholine receptor subunit genes in non-small-cell lung cancer reveals differences between smokers and nonsmokers. Cancer Res. 2007;67:4638-4647. [PubMed] [Cited in This Article: ] |
| 21. | Dasgupta P, Rizwani W, Pillai S, Kinkade R, Kovacs M, Rastogi S, Banerjee S, Carless M, Kim E, Coppola D. Nicotine induces cell proliferation, invasion and epithelial-mesenchymal transition in a variety of human cancer cell lines. Int J Cancer. 2009;124:36-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 255] [Cited by in F6Publishing: 287] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 22. | de A Simão T, Souza-Santos PT, de Oliveira DS, Bernardo V, Lima SC, Rapozo DC, Kruel CD, Faria PA, Ribeiro Pinto LF, Albano RM. Quantitative evaluation of SPRR3 expression in esophageal squamous cell carcinoma by qPCR and its potential use as a biomarker. Exp Mol Pathol. 2011;91:584-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 23. | Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101-1108. [PubMed] [Cited in This Article: ] |
| 24. | Therneau T. A Package for Survival Analysis in S. version 2.38 [Internet]. (2015). Accessed on 2016 Jan 7. Available from: Available from: URL: http://CRAN.R-project.org/package=survival. [Cited in This Article: ] |
| 25. | Wessler I, Kirkpatrick CJ. Acetylcholine beyond neurons: the non-neuronal cholinergic system in humans. Br J Pharmacol. 2008;154:1558-1571. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 622] [Cited by in F6Publishing: 597] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 26. | Nguyen VT, Hall LL, Gallacher G, Ndoye A, Jolkovsky DL, Webber RJ, Buchli R, Grando SA. Choline acetyltransferase, acetylcholinesterase, and nicotinic acetylcholine receptors of human gingival and esophageal epithelia. J Dent Res. 2000;79:939-949. [PubMed] [Cited in This Article: ] |
| 27. | Brown KC, Perry HE, Lau JK, Jones DV, Pulliam JF, Thornhill BA, Crabtree CM, Luo H, Chen YC, Dasgupta P. Nicotine induces the up-regulation of the α7-nicotinic receptor (α7-nAChR) in human squamous cell lung cancer cells via the Sp1/GATA protein pathway. J Biol Chem. 2013;288:33049-33059. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 28. | Boyle P, Levin B. World cancer report 2008. Lyon, France: IARC press 2008; . [Cited in This Article: ] |
| 29. | Thunnissen FB. Acetylcholine receptor pathway and lung cancer. J Thorac Oncol. 2009;4:943-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 30. | Tanner NT, Kanodra NM, Gebregziabher M, Payne E, Halbert CH, Warren GW, Egede LE, Silvestri GA. The Association between Smoking Abstinence and Mortality in the National Lung Screening Trial. Am J Respir Crit Care Med. 2016;193:534-541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 133] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 31. | Yoo SS, Lee SM, Do SK, Lee WK, Kim DS, Park JY. Unmethylation of the CHRNB4 gene is an unfavorable prognostic factor in non-small cell lung cancer. Lung Cancer. 2014;86:85-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Castillo-González AC, Nieto-Cerón S, Pelegrín-Hernández JP, Montenegro MF, Noguera JA, López-Moreno MF, Rodríguez-López JN, Vidal CJ, Hellín-Meseguer D, Cabezas-Herrera J. Dysregulated cholinergic network as a novel biomarker of poor prognostic in patients with head and neck squamous cell carcinoma. BMC Cancer. 2015;15:385. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 33. | Zheng X, Duan W, Xu J, Nie C, Yang Z, Wang H, Wang W, Lu D. Functionally significant nicotine acetylcholine receptor subunit α5 promoter haplotypes are associated with susceptibility to lung cancer in Chinese. Cancer. 2011;117:4714-4723. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |