Published online Jun 14, 2016. doi: 10.3748/wjg.v22.i22.5211
Peer-review started: December 22, 2015
First decision: January 13, 2016
Revised: March 1, 2016
Accepted: March 18, 2016
Article in press: March 1, 2016
Published online: June 14, 2016
AIM: To evaluate gut microbial dysbiosis in two visceral hypersensitive models in comparison with irritable bowel syndrome (IBS) patients and to explore the extent to which these models capture the dysbiosis of IBS patients.
METHODS: Visceral hypersensitivity was developed using the maternal separation (MS) rat model and post-inflammatory rat model. The visceral sensitivity of the model groups and control group was evaluated using the abdominal withdraw reflex score and electromyography in response to graded colorectal distention. The 16S ribosomal RNA gene from fecal samples was pyrosequenced and analyzed. The correlation between dysbiosis in the microbiota and visceral hypersensitivity was calculated. Positive findings were compared to sequencing data from a published human IBS cohort.
RESULTS: Dysbiosis triggered by neonatal maternal separation was lasting but not static. Both MS and post-inflammatory rat fecal microbiota deviated from that of the control rats to an extent that was larger than the co-housing effect. Two short chain fatty acid producing genera, Fusobacterium and Clostridium XI, were shared by the human IBS cohort and by the maternal separation rats and post-inflammatory rats, respectively, to different extents. Fusobacterium was significantly increased in the MS group, and its abundance positively correlated with the degree of visceral hypersensitivity. Porphyromonadaceae was a protective biomarker for both the rat control group and healthy human controls.
CONCLUSION: The dysbiosis MS rat model and the post-inflammatory rat model captured some of the dysbiosis features of IBS patients. Fusobacterium, Clostridium XI and Porphyromonadaceae were identified as targets for future mechanistic research.
Core tip: Dysbiosis of the gastrointestinal microbiota and hypersensitivity to colonic distension are critical features of irritable bowel syndrome (IBS). For animal models, the correlation between dysbiosis in the microbiota and visceral hypersensitivity remains unknown. This study identified common biomarkers between the animal models and IBS patients, which may be targets for future mechanistic research.
- Citation: Zhou XY, Li M, Li X, Long X, Zuo XL, Hou XH, Cong YZ, Li YQ. Visceral hypersensitive rats share common dysbiosis features with irritable bowel syndrome patients. World J Gastroenterol 2016; 22(22): 5211-5227
- URL: https://www.wjgnet.com/1007-9327/full/v22/i22/5211.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i22.5211
The human intestinal tract is home to trillions of bacteria that have co-evolved with their host over millennia[1]. Their combined genomes, called a metagenome, contain 150-fold more genes than do the human hosts, and they provide functions that humans otherwise do not have[2]. Complex interactions exist between the gut microbiota and the host[3]. Irritable bowel syndrome (IBS) is a common gastrointestinal disorder that is characterized by abdominal pain and alterations in bowel habits; statistically, IBS affects 7%-10% of people worldwide[4]. Accumulating evidence has indicated that the gut microbiota may participate in the pathogenesis of IBS[5]. Because collecting fecal samples both before and after a gastrointestinal infection from the same IBS patients is unfeasible for clinics, only gut dysbiosis in standing IBS patients has been evaluated to date[6,7]. However, how gut microbiota abnormalities arise and are maintained over time is unclear. These questions are critical for interventions targeting the microbiota, such as probiotic usage. In this work, we used visceral hypersensitive rat models to investigate the longitudinal changes of gut microbiota.
Currently, both post-infectious/inflammatory models and stress-related models have been frequently used to study the pathophysiology of IBS[8,9]. There are more than 12 major post-infectious/post-inflammatory models to mimic post-infectious IBS, which occurs after an initial episode of acute gastrointestinal infection. Chemicals such as trinitrobenzene sulfonic acid (TNBS)[10], mustard oil[11] and dextran sulfate sodium[12] were used to cause mucosal injury in the post-inflammatory models, and pathogens such as Trichinella spiralis[13] and Campylobacter[14] were used to infect the gut; both led to visceral hypersensitivity. Stress-related models[15] could also induce the modulation of visceral pain, and this may involve changes in the brain-gut axis[9]. However, one of the unsolved problems is the extent to which these models recapture the characteristics of gut dysbiosis in IBS patients. In this work, we used two visceral hypersensitive models, the TNBS post-inflammatory (pTNBS) model and the maternal separation (MS) model, to investigate: (1) whether and the extent to which these models reproduce the disturbance of gut microbiota in a similar way to that of the IBS patients; and (2) whether microbial dysbiosis, if it exists, is static or shifting in these visceral hypersensitive models. We also hoped to identify targets in the models’ gut microbial communities that are suitable for use in developing probiotics to specifically modulate the microbiota.
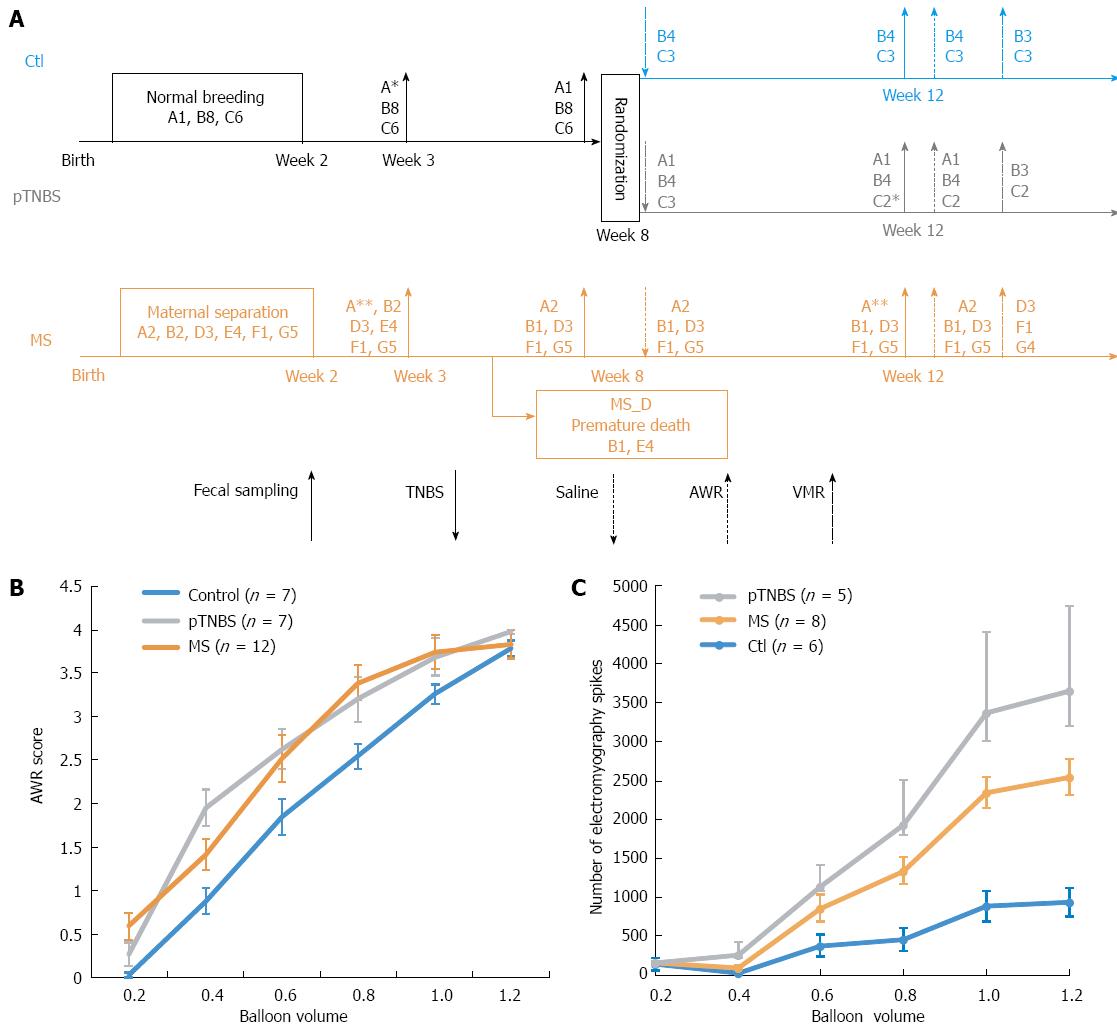
Sprague-Dawley rats were purchased from the animal center of Shandong University of Traditional Chinese Medicine. The rats were allowed to habituate for 7 d to the breeding facility prior to mating. They were kept under standardized specific pathogen-free conditions (21-22 °C, 12:12-h light-dark cycle) with access to pellet food and water ad libitum. All experiments were approved by the Ethical Committee and Institutional Animal Care and Use Committee of Qilu Hospital (KYLL-2013-005), and the methods were performed in strict accordance with the Animal Management Rules of the Chinese Ministry of Health. The overall design and co-housing relationship of involved rats are indicated in Figure 1A.
The MS visceral hypersensitive models were developed as previously described[15]. Briefly, rat pups that were randomly assigned to the MS group were stressed by separating them from their mothers for 3 h daily between postnatal days 2-14. The control group (Ctl) received normal breeding during this session. All pups were weaned on postnatal day 22, and only the male pups were used for the following study. Because some MS pups naturally died before they aged, 5 male rats were randomly chosen from those that were sampled at week 3 but did not survive to week 8. We indicated this group as the MS early death (MS_D) group. By including the MS_D group, we could test whether the dysbiosis caused by MS stress was more severe in the early dying pups.
After the second fecal collection at week 8, half of the control group was randomly assigned to the post-TNBS inflammation group (pTNBS). The pTNBS group was fasted for 24 h with free access to tap water, and then 0.4 mL of 5% (v/v) TNBS (P2297, Sigma, Shanghai, diluted to 0.8 mL using 50% ethanol) was administered into the colorectum.
After the last fecal collection at week 12, visceral hypersensitivity was evaluated using both the abdominal withdraw reflex (AWR) score and electromyography in response to graded colorectal distention (CRD). Graded CRD was induced by rapidly injecting (0.2, 0.4, 0.6, 0.8, 1.0, and 1.2 mL) saline into a urinary catheter balloon placed in the colon over 1 s and maintaining the distention for 20 s. AWR score was recorded according to a previously described method[16]. To represent the overall visceral sensitivity, a visceral hypersensitive index (VHI) for each rat was calculated by summing the rank of the AWR score at 0.4, 0.6, 0.8 and 1.0 of the total balloon volume.
The visceromotor responses (VMRs) to CRD were quantified through electromyography of the rat obliqus external abdominis. Briefly, 5 d after embedding an electrode in the rat obliqus externus abdominis, the raw electromyography was recorded, rectified and quantified by counting the increased spike bursts during a 20 s window after graded CRD stimulation. The VMR index for each rat was calculated by summing the rank of electromyography spikes at 0.4, 0.6, 0.8 and 1.0 of the total balloon volume.
Fecal samples were collected 3, 8 and 12 wk after birth. The samples from the pTNBS group at weeks 3 and 8 were indicated as Ctl-pTNBS and were analyzed as the controls because their treatment was the same as that for the Ctl group. A chart illustrating the overall treatment, fecal collection and model evaluation time points is shown in Figure 1A.
The samples were snap-frozen in liquid nitrogen and stored at -80 °C. Genomic DNA was extracted with a TIANamp Stool DNA Kit according to the manufacturer’s instructions (Cat# DP328, Tiangen, Beijing). DNA purity and concentration were measured using a Nanodrop2000 (Thermo Fisher). The DNA samples were shipped to Majorbio (Shanghai), where the DNA integrity check, PCR amplification, DNA quantification, emPCR (using Roche GS FLX Titanium emPCR Kits) and pyrosequencing of the 16S rRNA gene V3 to V1 region (using Roche Genome Sequencer FLX+) were performed according to their optimized protocols. The sequencing results were archived in the Short Reads Achieve (number pending).
Raw sequencing data were prepared using Mothur v 1.33.0 according to their proposed 454 SOP (http://www.mothur.org/wiki/454_SOP)[17]. The raw sff files were decoded, denoised, trimmed and then aligned to Silva references (Release 102) using the default parameters. Chimeras were detected using the chimera.uchime command and were then removed. Distances between sequences were calculated with a cutoff value of 0.15. The sequences were clustered to the same operational taxonomic units (OTUs) if their distances were less than 0.03. The Shannon index and the inverse Simpson index (1/D) were calculated to indicate the diversity in each sample. Both indexes were calculated using Mothur, and the detailed formula can be accessed online (http://www.mothur.org/wiki/Shannon and http://www.mothur.org/wiki/Simpson). The OTU table was converted to biom files and the taxa abundance from domain to genus levels was generated using the summarize_taxa.py command in QIIME v1.8.0.
The richness of each taxonomy and the Shannon index between groups were compared using the Kruskal-Wallis test (KW) or a student’s t-test in SAS V.9.3 statistical software. The heatmap plot with dendrograms was drawn using the heatplot function in the made4 packages in R (version 3.1.1). For primary component analysis (PCA), the axis value of all 80 samples was calculated together using the prcomp function in the stats package in R, and then the samples were plotted by each time point (week 3, 8, and 12). Within each time point, samples were clustered based on the Euclidian distance using the vegdist and hclust in the vegan package. According to the cluster results, the PCA plot points were grouped and connected using the ordispider and ordiellipse functions, where the ellipse was estimated to cover 75% of the dots in this group. The distribution of each group in each cluster was checked using Fisher’s exact test in SAS. The community dissimilarity was tested by the weighted and unweighted UniFrac test using Mothur. The specific taxa that were differentially present in each group were identified using the LEfSe [linear discriminant analysis (LDA) coupled with effect size measurements] method with an LDA cut-off value of 2.0[18]. The Spearman correlation between the VHI and the taxonomy richness was calculated using the cor.test function in the stats package in R.
We downloaded the published 16S rRNA V4 region Miseq sequencing data by Jeffery et al[6]. This data set was analyzed by the same pipeline described above. We used LEfSe analysis on this dataset. Each positive finding from the rat experiment was checked against the human cohort. The relationship between human and rat biomarkers was indicated using a Venn plot.
The design and co-housing relationship of the rats involved in this study is shown in Figure 1A. Twenty-six of the 27 rats in this study (7 Ctl, 7 pTNBS, and 12 MS, see Figure 1B) were evaluated using AWR. The VHI score was calculated by summing the rank of the AWR score at 0.4, 0.6, 0.8 and 1.0 of the total balloon volume. A significant difference existed in the VHI among the three groups (χ2 = 9.98, df = 2, P = 0.0068, KW). The VHI difference in the pTNBS to Ctl comparison was 38.4 (95%CI: 15.7 to 61.0, P < 0.05), and the VHI difference in the MS to Ctl comparison was 32.9 (95%CI: 12.7 to 53.0, P < 0.05). There was no significant difference in the MS to pTNBS comparison, with a VHI difference of 5.48 (95%CI: -14.7 to 25.7, P > 0.05). Nineteen of the 27 rats were evaluated by VMRs (Figure 1C). The VMR index for each rat was calculated by summing the rank of electromyography spikes at 0.4, 0.6, 0.8 and 1.0 of the total balloon volume. The VMR index among groups was insignificant although the control group tended to be lower than the MS and pTNBS groups (43.5 vs 86.5 and 60, respectively, χ2= 2.235, df = 2, P = 0.3271, KW). Overall, these data indicate that both the MS and pTNBS groups developed visceral hypersensitivity at a comparable level.
A total of 489556 valid reads were assigned to 80 sequenced samples after barcode trimming. Sequence length varied between 230 and 327 bp per read. After removing chimeras and non-bacterial reads, 434594 reads remained. Each fecal sample included 3313 to 8161 reads. Based on a 97% species similarity, 2413 OTUs were identified from all of the fecal samples. Good’s coverage for each sample varied from 95.57% to 99.72%. The rarefaction curve reached a plateau for most samples, suggesting that the present study captured the dominant phylotypes.
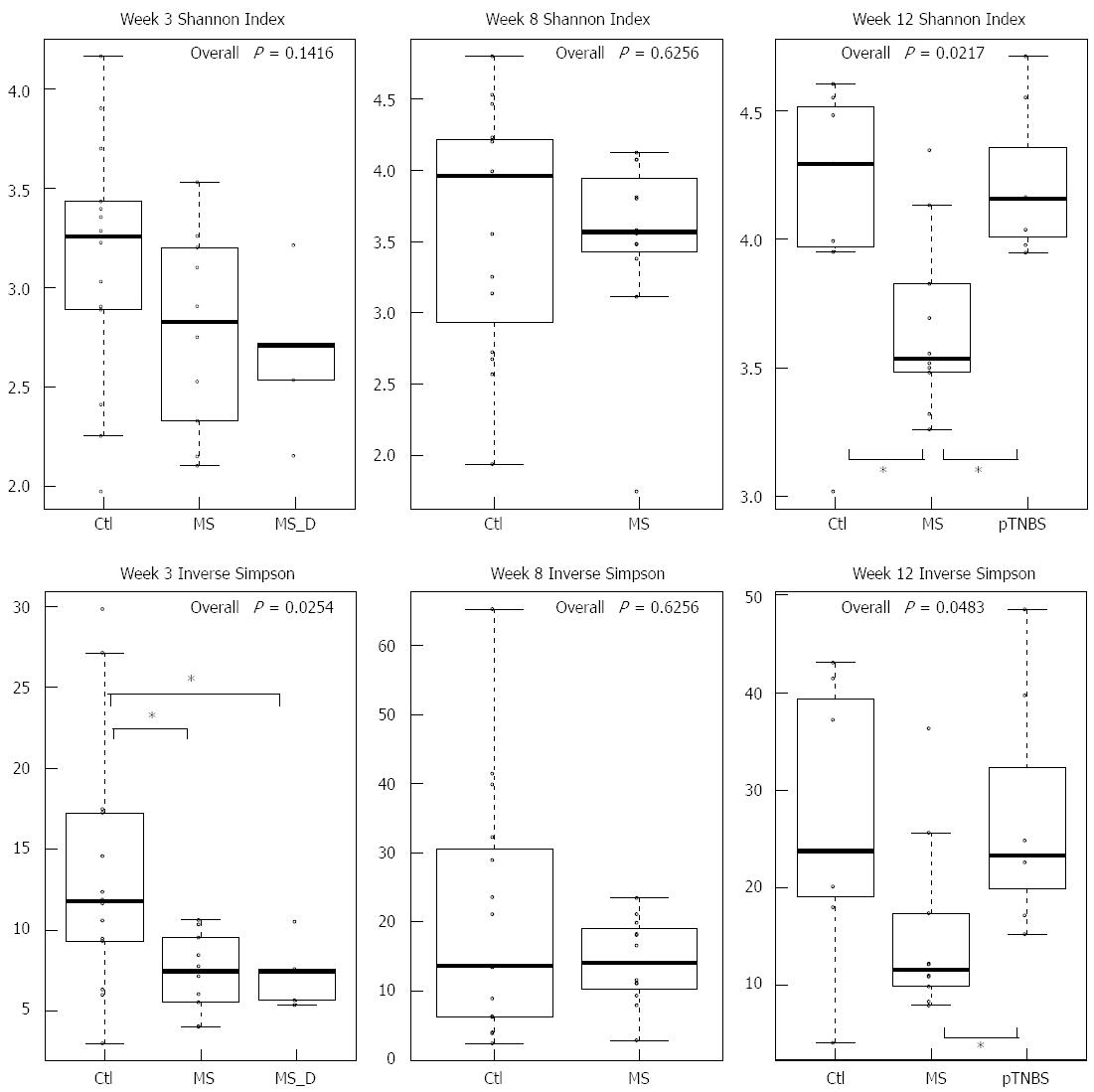
We first compared the microbial diversity among groups using the Shannon index and the inverse Simpson index (Figure 2). By week 3, the inverse Simpson index was significantly higher in the Ctl group (χ2 = 7.34, df = 2, P = 0.0254, KW). By week 8, the Shannon index and the inverse Simpson index were similar between the control group and the MS group. By week 12, the MS group has the lowest Shannon index (χ2 = 7.67, df = 2, P = 0.217, KW) and inverse Simpson index (χ2= 6.06, df = 2, P = 0.0483, KW) compared with other groups. The pTNBS group had roughly same diversity indexes compared to the Ctl group. These data indicate that the MS model, but not the pTNBS model, developed fecal microbiota with reduced diversity in a non-static manner.
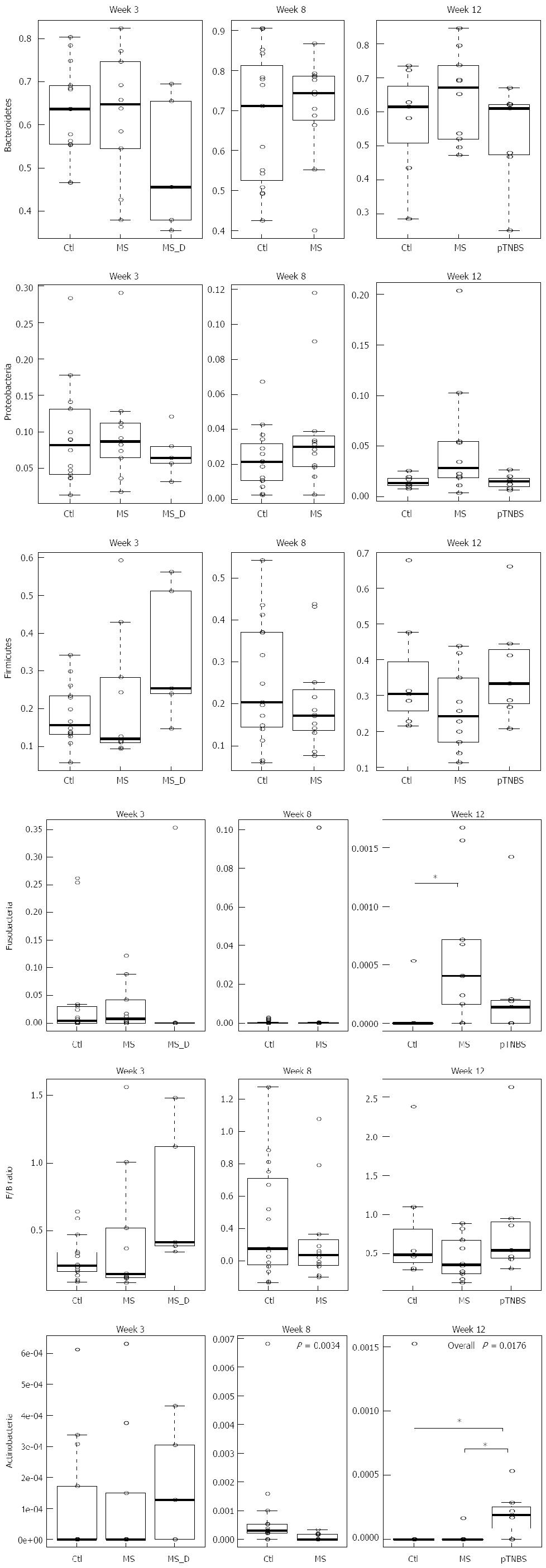
We then investigated whether differences in the phylum abundance exist at different time points (Figure 3). Bacteroidetes was the dominant phylum across all samples, and Firmicutes and Proteobacteria were the second and third most abundant phyla. No significance was reached for these three phyla at any of the 3 time points. The Firmicutes to Bacteroidetes (F/B) ratio was not significantly different (P > 0.05, KW).
Fusobacteria was abundant by week 3 (up to 0.25) and dropped to zero in most control rats. No difference in Fusobacteria existed by week 3 and week 8; however, by week 12, the MS group had significantly more Fusobacteria (χ2= 6.83, df = 2, P = 0.0328, KW, P < 0.05 in MS-Ctl comparison). The control group had significantly more Actinobacteria than the MS group at week 8 (P =0.0034, χ2 = 8.58, df =1, KW). However, by week 12, the pTNBS group had significantly more Actinobacteria than the Ctl and MS groups (χ2 = 8.07, df = 2, P = 0.0176, KW, P < 0.05 in the pTNBS-Ctl and pTNBS-MS comparison). These data suggest that the dysbiosis of the major phyla may be phase dependent and different among the visceral hypersensitive rat models.
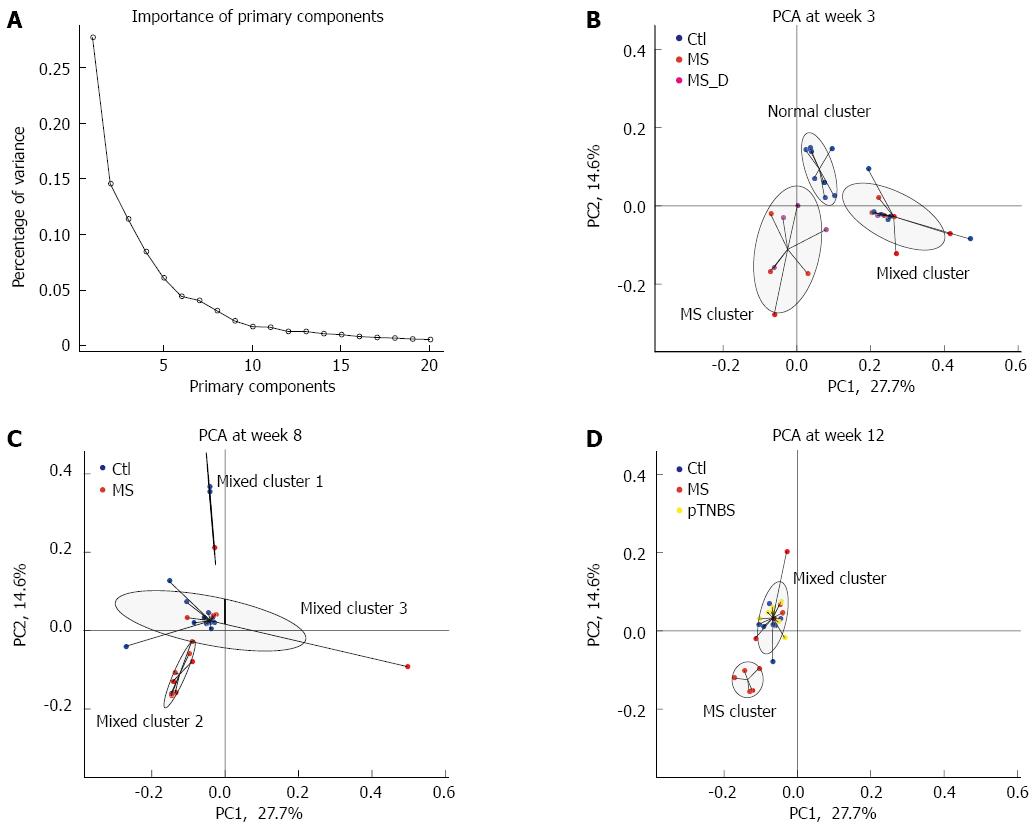
We based the cluster analysis and PCA on the OTU data from the 16S rRNA gene pyrosequencing. The primary components for all 80 samples were calculated from the relative abundance of the 2413 OTUs. The relative importance of the first 20 primary components is plotted in Figure 4A. Primary component 1 and primary component 2 explained 27.7% and 14.6% of the total variance, respectively (Figure 4A). The differences in the primary components between the time points and experimental groups are listed in Table 1. Primary component 1 mainly reflected the effect of time points (χ2 = 37.7, df = 2, P = 0.0000, KW). Primary components 2 and 4 reflected the effect of experimental groups on fecal microbiota composition. The other 3rd, 6th, and 9th components were different both among time points and among groups.
| Primary component | % of variance | Difference among time points (n = 80, df = 2) | Difference among groups (n = 80, df = 3) | ||
| χ2 | P-value | χ2 | P-value | ||
| 1 | 27.7% | 37.67931 | 0.000001 | 4.6999109 | 0.19514 |
| 2 | 14.6% | 1.352165 | 0.50861 | 20.4278998 | 0.000141 |
| 3 | 11.4% | 6.165561 | 0.045831 | 8.6830899 | 0.033821 |
| 4 | 8.5% | 0.719395 | 0.69789 | 21.7793862 | 0.000071 |
| 5 | 6.1% | 0.623716 | 0.73209 | 4.6400976 | 0.20013 |
| 6 | 4.5% | 8.388254 | 0.015081 | 9.3688439 | 0.024771 |
| 7 | 4.1% | 0.844768 | 0.65548 | 0.7871023 | 0.85255 |
| 8 | 3.2% | 0.412918 | 0.81346 | 6.6021743 | 0.08572 |
| 9 | 2.2% | 8.880049 | 0.011801 | 9.9866961 | 0.018681 |
| 10 | 1.7% | 1.762893 | 0.41418 | 2.6274074 | 0.45270 |
We then used cluster analysis to test whether the groups would fall into the same or different clusters. At each time point, the samples were fitted into the 3 top clusters based on the Euclidean distance. We designated the names of each cluster according to the samples it included. By week 3, the normal cluster included 8 Ctl, and the MS cluster included 4 MS and 4 MS_D samples. The mixed cluster included 6 Ctl, 6 MS and 1 MS_D samples (Figure 4B). The 3 groups’ distribution in the clusters was significantly different (P = 1.702 × 10-4, Fisher’s test). This result suggests that MS caused dysbiosis in rat models at early ages.
By week 8, the 27 fecal microbiota samples clustered into 3 mixed clusters (Figure 4C). The control group dominated mixed cluster 3 (12/16) while the MS group dominated mixed cluster 2 (7/8). The cluster distribution of the MS and control groups was significantly different (P = 0.0114, Fisher’s test). By week 12, the 24 fecal microbiota samples formed 3 clusters (Figure 4D). The MS cluster included 5 MS samples, and the mixed cluster included roughly the same number of samples from the Ctl (n = 6), MS (n = 5), and pTNBS (n = 7) groups. Another “orphan” cluster included only one control sample. The difference among groups was significant (P = 0.0150, Fisher’s test). These data suggest that the dysbiosis triggered by MS during childhood is still substantial in a fraction of adult rats. Four weeks after TNBS administration, the fecal microbiota of the post-inflammatory rat model was more similar to that of the control group as revealed by PCA and cluster analysis.
We tracked the longitudinal dysbiosis of 23 rats whose fecal samples were collected at all 3 time points. We analyzed whether the 10 MS rats clustered to the different or same clusters at week 3 and week 12. Seven out of 10 rats shifted to different clusters (mixed-to-MS or MS-to-mixed) from week 3 to week 12. The agreement Kappa value for cluster classification at week 3 and week 12 was -0.4000 (95%CI: -0.9566 to 0.1566). This result indicates that although MS stress generated an isolated dysbiosis cluster in a fraction of rats, each rat’s gut microbiota might shift between the less disturbed (mixed) cluster and the severely disturbed (MS) cluster.
We further tested the effect of modeling and co-housing on fecal microbiota using the weighted and unweighted UniFrac test (Table 2). A phylogenetic tree was built for all samples at each time point, and weighted and unweighted UniFrac scores were calculated to evaluate the community similarity. According to the unweighted UniFrac test, we found that by week 3, the fecal community in Ctl and MS groups was significantly different (P < 0.001). The co-housing effect caused community dissimilarity in the 2 houses of control rats (B8 vs C6, P < 0.001) but not in the 2 houses of MS rats (D3 vs G5, P = 0.507). By week 8, the co-housing effect was non-significant within the MS and Ctl groups, but the difference was still significant between these two groups. By week 12, a significant community difference existed among the Ctl, MS, and pTNBS groups; the co-housing effect was not obvious within any of the groups. The weighted UniFrac test was significant in all of the above comparisons. Overall, these data suggest that both the MS model and the pTNBS model developed dysbiosis of the fecal microbiota, and the differences were not caused by the co-housing relationship.
| Time point | Comparison | Number of samples to build phylogenetic tree | Weighted UniFrac test | Unweighted UniFrac test | ||
| Weighted UniFrac score | P-value | Unweighted UniFrac score | P-value | |||
| Week 3 | Ctl-MS | 29 | 0.863902 | < 0.0010 | 0.957176 | < 0.00101 |
| Ctl-MS_D | 29 | 0.926521 | < 0.0010 | 0.963861 | 0.01301 | |
| MS-MS_D | 29 | 0.833168 | < 0.0010 | 0.909242 | 0.2790 | |
| Ctl(B8)-Ctl(C6) | 29 | 1.000000 | < 0.0010 | 1.000000 | < 0.00101 | |
| MS(D3)-MS(G5) | 29 | 1.000000 | 0.0070 | 1.000000 | 0.5070 | |
| Week 8 | Ctl-MS | 27 | 0.719683 | < 0.0010 | 0.951668 | 0.01901 |
| Ctl(B8)-Ctl(C6) | 27 | 0.83063 | < 0.0010 | 0.955082 | 0.0870 | |
| MS(D3)-MS(G5) | 27 | 0.617715 | < 0.0010 | 0.874196 | 0.5220 | |
| Week 12 | Ctl-MS | 24 | 0.754046 | < 0.0010 | 0.944729 | 0.03901 |
| Ctl-pTNBS | 24 | 0.942882 | < 0.0010 | 0.978698 | 0.01601 | |
| MS-pTNBS | 24 | 0.828407 | < 0.0010 | 0.973938 | 0.01301 | |
| Ctl(B4)-Ctl(C3) | 24 | 0.928051 | < 0.0010 | 0.976209 | 0.3200 | |
| MS(D3)-MS(G5) | 24 | 0.771421 | < 0.0010 | 0.912219 | 0.4550 | |
| pTNBS(B4)-pTNBS(C2) | 24 | 1.000000 | < 0.0010 | 1.000000 | 0.6390 | |
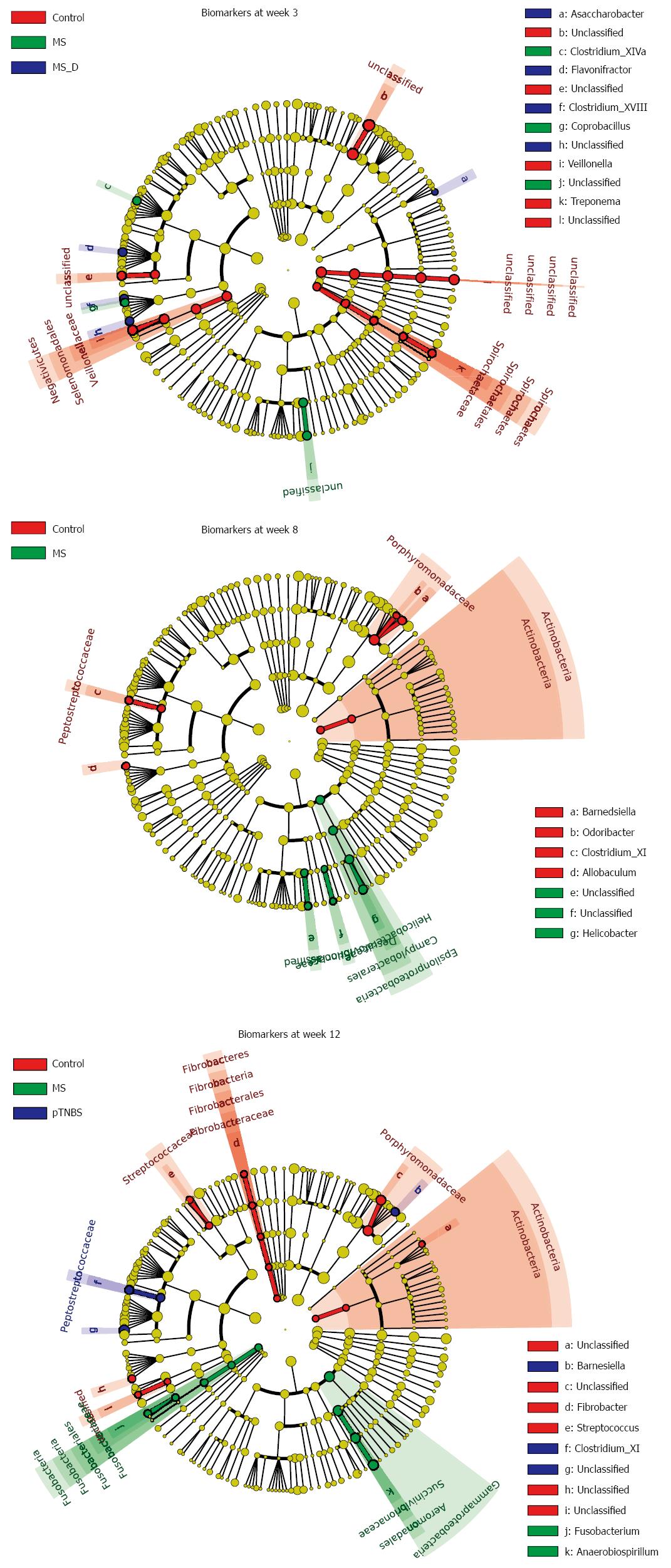
We performed linear discriminant analysis coupled with effect size measurement (LEfSe) analysis at different time points to screen biomarkers for each group (Figure 5, Figure S1-3). By week 3, 29 samples were analyzed using LEfSe. The control group was strongly associated with higher abundances of unclassified Bacteroidales, Veillonella, Treponema, and unclassified Clostridiales. The MS group was associated with higher abundances of unclassified Burkholderiales, Coprobacillus, and Clostridium_XIVa. By week 8, 27 samples were analyzed. The MS group was associated with higher abundances of Helicobacter, unclassified Burkholderiales, and unclassified Desulfovibrionaceae, which all belong to Proteobacteria. The control group was associated with higher abundances of Barnesiella, Actinobacteria, Clostridium_XI, Allobaculum, and Odoribacter.
To investigate whether the differentially abundant taxa correlated with the visceral hypersensitivity level, we listed the biomarkers in each group by week 12, and calculated their Spearman correlation to the VHI both within the respective group and across groups (Table 3). By week 12, Fusobacterium was associated with the MS group (LDA Score = 2.766). The Fusobacterium abundance was also significantly and positively correlated with the VHI across the all 24 samples by week 12 (r = 0.4564, P = 0.0250). Unclassified Erysipelotrichaceae was associated with the control group (LDA Score = 3.097) and significantly and negatively correlated with the VHI across groups (r = -0.4944, P = 0.0140). These data suggest that Fusobacterium may participate in the pathogenesis of visceral hypersensitivity and that Erysipelotrichaceae might protect against the hypersensitivity.
| Taxonomy | LDA score (Log10) | Group | Across groups (n = 24) | Within group | ||
| r_all_sample | p_all_sample | r_ingroup | p_ingroup | |||
| Actinobacteria | 2.338 | Controln = 7 | 0.1835 | 0.3906 | 0.2041 | 0.6606 |
| Actinobacteria|Actinobacteria | 2.338 | 0.1835 | 0.3906 | 0.2041 | 0.6606 | |
| Actinobacteria|Actinobacteria|Coriobacteriales|Coriobacteriaceae|Unclassified | 2.192 | 0.2639 | 0.2127 | 0.2041 | 0.6606 | |
| Bacteroidetes|Bacteroidia|Bacteroidales|Porphyromonadaceae | 4.610 | -0.1005 | 0.6405 | 0.1429 | 0.7825 | |
| Bacteroidetes|Bacteroidia|Bacteroidales|Porphyromonadaceae|Unclassified | 4.543 | -0.1344 | 0.5313 | 0.1429 | 0.7825 | |
| Fibrobacteres | 2.435 | -0.3601 | 0.0839 | -0.0741 | 0.8745 | |
| Fibrobacteres|Fibrobacteria | 2.435 | -0.3601 | 0.0839 | -0.0741 | 0.8745 | |
| Fibrobacteres|Fibrobacteria|Fibrobacterales | 2.435 | -0.3601 | 0.0839 | -0.0741 | 0.8745 | |
| Fibrobacteres|Fibrobacteria|Fibrobacterales|Fibrobacteraceae | 2.435 | -0.3601 | 0.0839 | -0.0741 | 0.8745 | |
| Fibrobacteres|Fibrobacteria|Fibrobacterales|Fibrobacteraceae|Fibrobacter | 2.435 | -0.3601 | 0.0839 | -0.0741 | 0.8745 | |
| Firmicutes|Bacilli|Lactobacillales|Streptococcaceae | 2.710 | 0.1215 | 0.5717 | 0.4447 | 0.3174 | |
| Firmicutes|Bacilli|Lactobacillales|Streptococcaceae|Streptococcus | 2.710 | 0.1215 | 0.5717 | 0.4447 | 0.3174 | |
| Firmicutes|Erysipelotrichia|Erysipelotrichales|Erysipelotrichaceae|Unclassified | 3.097 | -0.4944 | 0.01401 | -0.1429 | 0.7825 | |
| Firmicutes|Negativicutes|Selenomonadales|Unclassified | 2.396 | -0.2047 | 0.3374 | 0.7027 | 0.0782 | |
| Firmicutes|Negativicutes|Selenomonadales|Unclassified|Unclassified | 2.396 | -0.2047 | 0.3374 | 0.7027 | 0.0782 | |
| Fusobacteria | 2.766 | MSn = 10 | 0.4564 | 0.02501 | 0.2067 | 0.5667 |
| Fusobacteria|Fusobacteria | 2.766 | 0.4564 | 0.02501 | 0.2067 | 0.5667 | |
| Fusobacteria|Fusobacteria|Fusobacteriales | 2.766 | 0.4564 | 0.02501 | 0.2067 | 0.5667 | |
| Fusobacteria|Fusobacteria|Fusobacteriales|Fusobacteriaceae | 2.766 | 0.4564 | 0.02501 | 0.2067 | 0.5667 | |
| Fusobacteria|Fusobacteria|Fusobacteriales|Fusobacteriaceae|Fusobacterium | 2.766 | 0.4564 | 0.02501 | 0.2067 | 0.5667 | |
| Proteobacteria|Gammaproteobacteria | 4.622 | 0.1809 | 0.3976 | 0.0667 | 0.8648 | |
| Proteobacteria|Gammaproteobacteria|Aeromonadales | 4.622 | 0.1757 | 0.4115 | 0.0667 | 0.8648 | |
| Proteobacteria|Gammaproteobacteria|Aeromonadales|Succinivibrionaceae | 4.622 | 0.1757 | 0.4115 | 0.0667 | 0.8648 | |
| Proteobacteria|Gammaproteobacteria|Aeromonadales|Succinivibrionaceae|Anaerobiospirillum | 4.622 | 0.1757 | 0.4115 | 0.0667 | 0.8648 | |
| Bacteroidetes|Bacteroidia|Bacteroidales|Porphyromonadaceae|Barnesiella | 3.607 | pTNBSn = 7 | -0.1501 | 0.4840 | -0.0180 | 0.9694 |
| Firmicutes|Clostridia|Clostridiales|Peptostreptococcaceae | 3.776 | 0.1946 | 0.3623 | 0.0180 | 0.9694 | |
| Firmicutes|Clostridia|Clostridiales|Peptostreptococcaceae|Clostridium_XI | 3.776 | 0.1946 | 0.3623 | 0.0180 | 0.9694 | |
| Firmicutes|Clostridia|Clostridiales|Ruminococcaceae|Unclassified | 5.014 | -0.0322 | 0.8813 | -0.1982 | 0.6701 | |
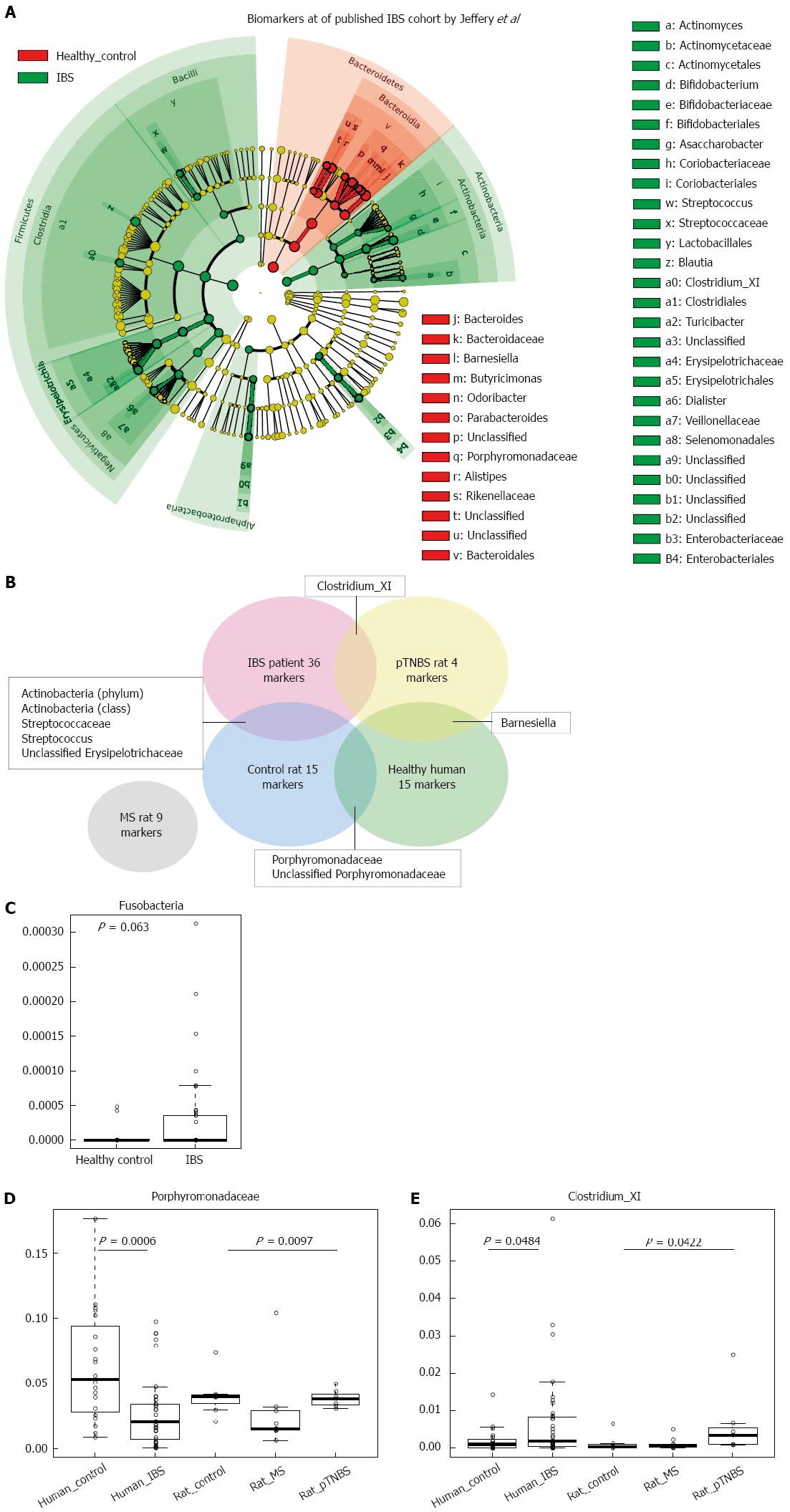
Next, we asked to what extent visceral hypersensitive rats’ dysbiosis resembles that of IBS patients. We downloaded the pyrosequencing data published by Jeffery et al[6], which included 37 IBS patients and 20 controls. We used the LEfSe method to analyze disease and healthy biomarkers in human fecal samples (Figure 6A); this data was further compared to the data from our animal models (Figure 6B). We identified 36 biomarkers of IBS patients and 15 biomarkers of human controls[6] (Figure 6A). The biomarkers of disease were largely different in human IBS and visceral hypersensitive rats, and only a few disease or control biomarkers were shared between the human study[6] and our rat model study. Fusobacteria marginally increased in IBS patients compared with human controls[6] (P = 0.063, KW, Figure 6C). The MS rat model did not share increased common biomarkers with human patients. Both control rats and human health controls have fecal microbial markers of Porphyromonadaceae and unclassified Porphyromonadaceae (Figure 6B). In the IBS cohort published by Jeffery et al[6], Porphyromonadaceae was significantly lower in the IBS groups (P = 0.0006, KW, Figure 6D), and the MS rats also had lower Porphyromonadaceae concentrations (P = 0.0097, KW, Figure 6D). The genus Clostridium_XI (belonging to family Peptostreptococcaceae, order Clostridiales) was a biomarker of both IBS patients and pTNBS rats (Figure 6E). Clostridium_XI accounted for up to 6% of the fecal microbiota in the IBS group and was significantly higher than the level in the healthy control group (P = 0.0484, KW). Additionally, Clostridium XI also colonized at a higher level in the pTNBS rats (P = 0.0422, KW).
In this study, we found that (1) both the MS and the pTNBS rat models developed dysbiosis of fecal microbiota; (2) the fecal microbiota of the MS model was characterized by a lower diversity and a higher level of Fusobacteria at week 3 and week 12. A fraction of the MS rats formed an isolated MS cluster that indicated clear-cut dysbiosis in comparison to the controls but the rats in this cluster tended to alternate; (3) the pTNBS model was characterized by higher Actinobacteria but did not develop any isolated clusters; (4) among the biomarkers observed by week 12, the Fusobacterium positively and unclassified Erysipelotrichaceae negatively correlated to visceral hypersensitivity; and (5) in comparison to a previously published fecal microbial profile in human IBS patients[6], Porphyromonadaceae was a protective biomarker for both healthy humans and rat controls; Clostridium_XI was a shared biomarker for human IBS patients and pTNBS rats.
Rodent models have been frequently used to study the pathogenesis and treatment of IBS, where the intestinal microbiota plays an important role. Before this study, dysbiosis in IBS rat models had not been specifically profiled, and whether any members in microbial community were correlated to visceral hypersensitivity was not known. We evaluated the fecal microbiota of MS and control rats at three time points and found that dysbiosis happened shortly after weaning (third week) and at the adult phase (12 wk). The pTNBS rat model did not develop an isolated microbial cluster but had biomarkers of Barnesiella, Clostridium_XI, and unclassified Ruminococcaceae. Through the unweighted UniFrac test, we found that the co-housing effect was no more significant at week 8 or week 12. Thus, both models developed significant dysbiosis of the fecal microbiota.
Approximately 10% of IBS patients believe that their symptoms began with an infectious illness, and prospective studies have shown that 3% to 36% of enteric infections lead to IBS symptoms[19]. Understanding underlying gut microbial dysbiosis associated with PI-IBS is critical for the prevention and management of this disease. Campylobacter jejuni, Campylobacter rodentium, and Salmonella enterica are available bacterial infectious murine models that mimic aspects of the pathogenesis of post-infectious IBS[8]. In this study, two rat models, MS and pTNBS, were not given any specific infector but were colonized more frequently by Fusobacterium and Clostridium XI, respectively. The latter includes the Clostridium difficile, Clostridium litorale, and Clostridium lituseburense. These two genera were also found to increase or tend to increase in the downloaded Miseq 16S rRNA gene sequencing data from Jeffery’s IBS cohort[6]. Thus, these two bacteria may be common dysbiosis features across human and rat models. In a chip-based study by Jalanka-Tuovinen et al[7], C. cellulosi and its relatives (members from Clostridium cluster IV) significantly decreased in IBS-D patients. Thus, whether Clostridium plays a mechanistic role in the pathogenesis of IBS warrants further study.
By week 12, Fusobacterium colonized in significantly greater abundance in MS rats, and its abundance was positively correlated with higher VHI scores. The Fusobacteria phylum also tends to be higher in published IBS cohorts[6]. Fusobacterium was invasive to the gut epithelial cells and has already been documented as being involved in the pathogenesis of colorectal adenoma[20-22] and inflammatory bowel disease[23,24]. This represents the first study documenting that Fusobacterium was involved in visceral hypersensitivity. Whether the increased colonization of Fusobacterium caused low grade inflammation and thus contributed to visceral hypersensitivity is not currently known. Moreover, both Fusobacterium and members in Clostridium are known short chain fatty acid (SCFA) producers[24-26]. The low fermentable oligo-, di-, and monosaccharides and polyol (FODMAP) diet has been shown to be an efficacious therapy for the reduction of IBS symptoms in randomized controlled trials[27-29]. Supplementing food containing FODMAP to IBS patients and healthy people would trigger gastrointestinal symptoms to a larger extent in the patient group[30]. However, the mechanism of treatment with a low FODMAP diet and why IBS patients are more sensitive to FODMAP remains unknown[31]. In this study, we identified two butyric producing taxa, Fusobacterium and Clostridium, which significantly correlate to visceral hypersensitivity or are disease biomarkers. Their colonization may render patients more ready to produce SCFA and gas in the presence of FODMAP. It was reported that the butyrate-producing Clostridium cluster XIVa significantly increased in IBS patients consuming a typical high FODMAP diet compared with those on a low FODMAP diet[32]. Farmer et al[33] showed that caecal intraluminal pH was significantly lower in IBS patients compared to controls. Thus, the detailed mechanistic role of Fusobacterium and Clostridium in IBS warrants further study.
Porphyromonadaceae is a family belonging to the order of Bacteroidales and the class Bacteroidetes. Among others, Barnesiella and Butyricimonas are genera under Porphyromonadaceae. Unfortunately, the biological function of Porphyromonadaceae has not been characterized in detail, and little attention has been paid to their role in gastrointestinal diseases. A previous study documented that Barnesiella was enriched in dextran sulfate sodium-induced colitis[34]. In our study, Barnesiella was also enriched 4 weeks after TNBS instillation. Whether Barnesiella promoted the inflammation or was passively enriched under inflammatory conditions was not determined in the current study. A recent review paper[35] summarized 29 relevant original research articles concerning microbiota analysis and IBS. Durbán’s pyrosequencing study[36] found that the family Porphyromonadaceae was increased in the fecal samples of IBS subjects. In our study, the Porphyromonadaceae was highest in the control group by week 12. The discrepancy may be explained by the different nature between human patients and rat models.
IBS is a human disease with multifactorial pathophysiology[37], and the prevalence of IBS is associated with social-economic factors[38]. To date no available model could ideally model the IBS pathogenesis. IBS is heterogeneous and thus unlikely to be modeled in any single model. Although common biomarkers were found between human IBS patients and rat models, the limitations of rat models should also be taken into consideration. The pTNBS model was triggered by a pro-inflammatory molecule (TNBS). Therefore, this model resembles the human inflammatory bowel disease to some extent and can only mimic the post-infectious IBS, which is associated only to a percentage of patients. Furthermore, the causal relationship between visceral hypersensitivity, dysbiosis, and the symptoms of IBS is not clear and remains to be untangled in the future.
In summary, both the MS and the post-inflammation rat models developed dysbiosis in the fecal microbiota, and the models captured parts of the dysbiosis features of human IBS patients. The potential pathogenic role of Fusobacterium and Clostridium XI, as well as the protective role of Porphyromonadaceae warrants further mechanistic study.
We acknowledge Dr. Ian Jeffery and Dr. Paul W. O’Toole for sharing their sequencing data from human IBS patients and healthy controls.
Previous studies have indicated that the gut microbiota participated in the pathogenesis of irritable bowel syndrome (IBS).
Dysbiosis of the gastrointestinal microbiota and hypersensitivity to colonic distension are critical features of IBS. For animal models, the correlation between dysbiosis in the microbiota and visceral hypersensitivity remains unknown.
Dysbiosis triggered by neonatal maternal separation (MS) was lasting but not static. Both MS and post-inflammatory rat fecal microbiota deviated from that of the control rats to an extent that was larger than the co-housing effect. Fusobacterium, Clostridium XI and Porphyromonadaceae were identified as targets for future mechanistic research.
This study indicated that the two animal models could capture part of the dysbiosis features of IBS. Further mechanistic study on the biomarkers’ role in the pathogenesis is warranted.
The manuscript is excellent and addresses adequately the relationship between dysbiosis and visceral hypersensitivity in experimental animals. The quality of the study design and experimental investigations are very high.
P- Reviewer: Guglielmetti S, Ierardi E, Soares RLS S- Editor: Ma YJ L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Burcelin R, Serino M, Chabo C, Garidou L, Pomié C, Courtney M, Amar J, Bouloumié A. Metagenome and metabolism: the tissue microbiota hypothesis. Diabetes Obes Metab. 2013;15 Suppl 3:61-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 94] [Cited by in F6Publishing: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 2. | Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59-65. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7052] [Cited by in F6Publishing: 7119] [Article Influence: 508.5] [Reference Citation Analysis (3)] |
| 3. | Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268-1273. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3003] [Cited by in F6Publishing: 2739] [Article Influence: 228.3] [Reference Citation Analysis (0)] |
| 4. | Spiegel BM. The burden of IBS: looking at metrics. Curr Gastroenterol Rep. 2009;11:265-269. [PubMed] [Cited in This Article: ] |
| 5. | Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159-176. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 594] [Cited by in F6Publishing: 601] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 6. | Jeffery IB, O’Toole PW, Öhman L, Claesson MJ, Deane J, Quigley EM, Simrén M. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut. 2012;61:997-1006. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 577] [Cited by in F6Publishing: 592] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 7. | Jalanka-Tuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63:1737-1745. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 226] [Cited by in F6Publishing: 234] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 8. | Qin HY, Wu JC, Tong XD, Sung JJ, Xu HX, Bian ZX. Systematic review of animal models of post-infectious/post-inflammatory irritable bowel syndrome. J Gastroenterol. 2011;46:164-174. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 9. | Larauche M, Mulak A, Taché Y. Stress-related alterations of visceral sensation: animal models for irritable bowel syndrome study. J Neurogastroenterol Motil. 2011;17:213-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 62] [Cited by in F6Publishing: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 10. | Zhou Q, Price DD, Caudle RM, Verne GN. Visceral and somatic hypersensitivity in TNBS-induced colitis in rats. Dig Dis Sci. 2008;53:429-435. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Laird JM, Martinez-Caro L, Garcia-Nicas E, Cervero F. A new model of visceral pain and referred hyperalgesia in the mouse. Pain. 2001;92:335-342. [PubMed] [Cited in This Article: ] |
| 12. | Larsson MH, Rapp L, Lindström E. Effect of DSS-induced colitis on visceral sensitivity to colorectal distension in mice. Neurogastroenterol Motil. 2006;18:144-152. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Yang X, Sheng L, Guan Y, Qian W, Hou X. Synaptic plasticity: the new explanation of visceral hypersensitivity in rats with Trichinella spiralis infection? Dig Dis Sci. 2009;54:937-946. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Spiller RC, Jenkins D, Thornley JP, Hebden JM, Wright T, Skinner M, Neal KR. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in post-dysenteric irritable bowel syndrome. Gut. 2000;47:804-811. [PubMed] [Cited in This Article: ] |
| 15. | Coutinho SV, Plotsky PM, Sablad M, Miller JC, Zhou H, Bayati AI, McRoberts JA, Mayer EA. Neonatal maternal separation alters stress-induced responses to viscerosomatic nociceptive stimuli in rat. Am J Physiol Gastrointest Liver Physiol. 2002;282:G307-G316. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 323] [Cited by in F6Publishing: 316] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 16. | Xu D, Wu X, Grabauskas G, Owyang C. Butyrate-induced colonic hypersensitivity is mediated by mitogen-activated protein kinase activation in rat dorsal root ganglia. Gut. 2013;62:1466-1474. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537-7541. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14372] [Cited by in F6Publishing: 12825] [Article Influence: 855.0] [Reference Citation Analysis (0)] |
| 18. | Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7042] [Cited by in F6Publishing: 8606] [Article Influence: 662.0] [Reference Citation Analysis (0)] |
| 19. | Spiller R, Garsed K. Postinfectious irritable bowel syndrome. Gastroenterology. 2009;136:1979-1988. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 489] [Cited by in F6Publishing: 524] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 20. | Allen-Vercoe E, Jobin C. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett. 2014;162:54-61. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 85] [Cited by in F6Publishing: 94] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 21. | McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 360] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 22. | Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207-215. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1659] [Cited by in F6Publishing: 1581] [Article Influence: 143.7] [Reference Citation Analysis (0)] |
| 23. | Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971-1978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 352] [Cited by in F6Publishing: 377] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 24. | Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79-83. [PubMed] [Cited in This Article: ] |
| 25. | Jiang L, Zhu L, Xu X, Li Y, Li S, Huang H. Genome Sequence of Clostridium tyrobutyricum ATCC 25755, a Butyric Acid-Overproducing Strain. Genome Announc. 2013;1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Kong Q, He GQ, Chen F, Ruan H. Studies on a kinetic model for butyric acid bioproduction by Clostridium butyricum. Lett Appl Microbiol. 2006;43:71-77. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 27. | Shepherd SJ, Halmos E, Glance S. The role of FODMAPs in irritable bowel syndrome. Curr Opin Clin Nutr Metab Care. 2014;17:605-609. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | de Roest RH, Dobbs BR, Chapman BA, Batman B, O’Brien LA, Leeper JA, Hebblethwaite CR, Gearry RB. The low FODMAP diet improves gastrointestinal symptoms in patients with irritable bowel syndrome: a prospective study. Int J Clin Pract. 2013;67:895-903. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 214] [Cited by in F6Publishing: 211] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 29. | Magge S, Lembo A. Low-FODMAP Diet for Treatment of Irritable Bowel Syndrome. Gastroenterol Hepatol (N Y). 2012;8:739-745. [PubMed] [Cited in This Article: ] |
| 30. | Ong DK, Mitchell SB, Barrett JS, Shepherd SJ, Irving PM, Biesiekierski JR, Smith S, Gibson PR, Muir JG. Manipulation of dietary short chain carbohydrates alters the pattern of gas production and genesis of symptoms in irritable bowel syndrome. J Gastroenterol Hepatol. 2010;25:1366-1373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 407] [Cited by in F6Publishing: 387] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 31. | Staudacher HM, Irving PM, Lomer MC, Whelan K. Mechanisms and efficacy of dietary FODMAP restriction in IBS. Nat Rev Gastroenterol Hepatol. 2014;11:256-266. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 165] [Cited by in F6Publishing: 154] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 32. | Halmos EP, Christophersen CT, Bird AR, Shepherd SJ, Gibson PR, Muir JG. Diets that differ in their FODMAP content alter the colonic luminal microenvironment. Gut. 2015;64:93-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 462] [Cited by in F6Publishing: 449] [Article Influence: 49.9] [Reference Citation Analysis (0)] |
| 33. | O’Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry. 2009;65:263-267. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 753] [Cited by in F6Publishing: 745] [Article Influence: 49.7] [Reference Citation Analysis (0)] |
| 34. | Tan Y, Zou KF, Qian W, Chen S, Hou XH. Expression and implication of toll-like receptors TLR2, TLR4 and TLR9 in colonic mucosa of patients with ulcerative colitis. J Huazhong Univ Sci Technolog Med Sci. 2014;34:785-790. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Taverniti V, Guglielmetti S. Methodological issues in the study of intestinal microbiota in irritable bowel syndrome. World J Gastroenterol. 2014;20:8821-8836. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 16] [Reference Citation Analysis (0)] |
| 36. | Durbán A, Abellán JJ, Jiménez-Hernández N, Salgado P, Ponce M, Ponce J, Garrigues V, Latorre A, Moya A. Structural alterations of faecal and mucosa-associated bacterial communities in irritable bowel syndrome. Environ Microbiol Rep. 2012;4:242-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in F6Publishing: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 37. | Soares RL. Irritable bowel syndrome: a clinical review. World J Gastroenterol. 2014;20:12144-12160. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 110] [Cited by in F6Publishing: 123] [Article Influence: 12.3] [Reference Citation Analysis (5)] |
| 38. | Soares RL, dos Santos JM, Rocha VR. Prevalence of irritable bowel syndrome in a Brazilian Amazon community. Neurogastroenterol Motil. 2005;17:883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |